Abstract
Human brain organoids, 3D self-assembled neural tissues derived from pluripotent stem cells, are important tools for studying human brain development and related disorders. Suspension cultures maintained by spinning bioreactors allow for the growth of large organoids despite the lack of vasculature, but commercially available spinning bioreactors are bulky in size and have low throughput. Here, we describe the procedures for building the miniaturized multiwell spinning bioreactor SpinΩ from 3D-printed parts and commercially available hardware. We also describe how to use SpinΩ to generate forebrain, midbrain and hypothalamus organoids from human induced pluripotent stem cells (hiPSCs). These organoids recapitulate key dynamic features of the developing human brain at the molecular, cellular and structural levels. The reduction in culture volume, increase in throughput and reproducibility achieved using our bioreactor and region-specific differentiation protocols enable quantitative modeling of brain disorders and compound testing. This protocol takes 14–84 d to complete (depending on the type of brain region–specific organoids and desired developmental stages), and organoids can be further maintained over 200 d. Competence with hiPSC culture is required for optimal results.
INTRODUCTION
Over the past 10 years, hiPSCs have emerged as an invaluable tool for modeling human diseases, especially those with complex genetic traits that are challenging to model in animals1,2. A new frontier of stem cell research is the generation of 3D tissue structures to model organogenesis and developmental disorders. Brain organoids are human pluripotent stem cell–derived 3D tissues that self-assemble into organized structures resembling the fetal human brain3–9. Unlike conventional 2D cell cultures, organoids resemble embryonic organs, not only at the cellular level, but also in architecture and developmental trajectory, therefore providing a unique opportunity to model human organogenesis. Here, we describe the methodology for the generation of three types of brain region–specific organoids from hiPSCs, using a custom-built multiwell spinning bioreactor8.
Development of the SpinΩ bioreactor
Suspension culture enabled by spinning bioreactors provides improved diffusion of oxygen and nutrients to support the growth and expansion of 3D tissue, despite the lack of vasculature10,11. Recently developed cerebral organoid methodology allows for the generation of large continuous ventricle-like structures resembling the structure of multiple regions of the fetal brain3. However, commercially available spin flasks are not designed for organoid culture, limiting the efficiency and throughput of organoid generation. First, these spin flasks are bulky and cannot operate independently without a magnetic stirrer plate, thereby limiting the number of flasks in a standard-sized incubator to ~10. Owing to space constraints, this makes it difficult for researchers to grow organoids from many different cell lines or explore the effects of several different conditions simultaneously. Second, it is expensive to maintain the organoid culture because each spin flask requires ~100 ml of culture medium and organoids are cultured for long periods of time, making it cost-prohibitive for many laboratories to supplement the media with growth factors and other essential compounds. Although the medium can be exchanged less frequently in spin flasks, the potency of growth factors may decay over time when the medium is not frequently refreshed. Disease modeling and compound testing require maintaining organoids under many different conditions in parallel, which is challenging to achieve using spin flasks. Therefore, we designed a spinning bioreactor that is customized for organoid culture via miniaturization and increased throughput. The device has a small footprint and an integrated spinning mechanism that does not require a large amount of dedicated incubator space. Organoids can be grown under many different conditions in parallel, with each condition requiring a minimal culture medium to reduce cost. In addition, the device is biocompatible and sterilizable for repeated use.
We designed the SpinΩ device on the basis of a standard 12-well tissue culture plate, with each well acting as a miniature spin flask to provide a suspension environment and improved oxygen delivery to the organoids. To generate enough suspension within a confined space, we could not directly adopt the geometry of the vertical dual fins used in spin flasks, but instead designed an original geometry with triple fins positioned at an angle to increase upward lift during rotation. We used computer-aided design (CAD) to custom-design various versions of each individual part and used 3D printing for quick prototyping to test different prototypes. Through trial and error, we eventually arrived at the current design, which reliably provides constant suspension and prevents aggregation of organoids. The power source driving all 12 wells to spin in synchrony originates from a single electric motor, and the wells are connected by a series of interconnecting gears. The body of the bioreactor is made with ULTEM, a heat-resistant plastic with no known biological side effects, to permit convenient sterilization by autoclaving. As such, SpinΩ offers markedly reduced medium consumption (3 ml in each well), a minimal footprint (standard 12-well plate) and substantially improved throughput (each unit = 12 independent conditions) as compared with commercially available spin flasks.
Development of methods for the generation of brain region–specific organoids
Reproducibility and consistency are critical to the success of organoid protocols in a quantitative model system for disease modeling and compound testing. Unlike directed 2D differentiation of target cell types, early organoid differentiation methodologies depended on the intrinsic signaling and self-assembly of stem cells10. However, although less extrinsic interference enables stem cells to follow their intrinsic program to grow and expand in a manner similar to in vivo development, tissue heterogeneity inevitably arises. This heterogeneity, along with batch and hiPSC line variability, makes it difficult to control experimental conditions. Inspired by organoid differentiation protocols based on the SFEBq method (serum-free floating culture of embryoid body (EB)-like aggregates with quick reaggregation), we developed protocols for the generation of brain region–specific organoids using EB cultures derived from hiPSCs5,12–15. To achieve a balance between intrinsic programming and stochasticity, we first use potent patterning cues to instruct hiPSC differentiation toward a homogeneous population of progenitors of specific brain regions (dorsal forebrain, midbrain and hypothalamus), and later switch to a medium that promotes tissue growth with fewer instructive signals. We start by detaching hiPSCs cultured on mouse embryonic fibroblast (MEF) feeder cells to form EBs, and culture the floating EBs with patterning factors toward the regional neural progenitor cell (NPC) identities. We later transfer the EBs to the SpinΩ bioreactor spinning culture, in which they grow and differentiate into organoids representative of specific brain regions. For forebrain organoids, we embed EBs in Matrigel, promoting formation and expansion of large neuroepithelial structures, which later become the ventricular zones (VZs) in organoids. Although feeder-independent hiPSCs are theoretically compatible with this method, we have comprehensively characterized the outcome of only this method using feeder-dependent hiPSC cultures.
SpinΩ allows researchers to test different differentiation protocols efficiently and cost-effectively. Guided by morphogen gradient models of the developing mouse brain and established 2D neural differentiation protocols for hiPSCs16–19, we tested multiple factor combinations, concentrations and durations of treatments while developing each of the three protocols described here. We characterized the cell types and structural organization of the resulting organoids by immunostaining and confocal microscopy at different stages. At the early stages (7–14 d), we aimed to achieve pure populations of progenitor cells with the corresponding brain region identities, whereas, at the later stages, we characterized the presence of more mature cell types representative of a specific brain region, such as TH+ dopaminergic neurons and various types of peptidergic neurons for midbrain and hypothalamus organoids, respectively. For the forebrain organoids, we focused on the formation of distinct laminated structures recapitulating the elaborate architecture of the embryonic human cerebral cortex. By characterizing the morphology and distribution of markers for NPCs, intermediate progenitor cells (IPCs) and neurons, we developed a protocol to generate forebrain organoids containing a well-defined VZ, subventricular zone (SVZ) and cortical plate (CP). Moreover, the dynamic formation and expansion of these layers resemble the progression of fetal development, despite the markedly miniaturized tissue size. In addition, we characterized the expression of molecular markers specific for human outer radial glia cells (oRGCs) and found that the forebrain organoids produce robust populations of oRGC-like cells, forming a distinct outer SVZ (oSVZ)8. These results are reproducible in many independent experiments and across multiple hiPSC cell lines and clones, including C1–1 (refs. 8,20–22), C1–2 (refs. 8,20,21,23–25), C2–1 (refs. 20,21), C3–1 (refs. 8,20,21), C3–2 (refs. 8,20,21), D3–1 (ref. 20), D3–2 (ref. 20), Y1–3 (refs. 21) and Y3–3 (ref. 21).
Comparison with alternative methods
We adopted the use of spinning bioreactors from the cerebral organoid method3, but adapted this protocol (which relied solely on intrinsic signaling and spontaneous differentiation of stem cells) by adding defined combinations of patterning factors to instruct hiPSCs to differentiate into progenitors for specific brain regions. The cerebral organoid method generates a mixture of tissues representing different brain regions, allowing comprehensive examination of brain development3, although the inherent tissue heterogeneity results in large outcome variability that limits its potential as a quantifiable model system3. Consistency and reproducibility are two main advantages of our methodology, and we have demonstrated that many parameters can be reliably quantified in these organoids8,23–25. Although each protocol produces organoids representative of a single brain region, SpinΩ could be used to generate various types of organoids, which can potentially be assembled to model interactions between brain regions26–28. Compared with prior organoid models for cerebral cortex development, our protocol for forebrain organoids produces more consistent results in terms of the structures generated. Forebrain organoids also contain more developed layers, including a well-defined oSVZ, to better recapitulate the architecture and cellular diversity of the developing human cortex4–6,29.
Applications of the protocol
Our protocols provide highly relevant model systems for studying human brain development. Specifically, the forebrain organoids recapitulate unique features of primate fetal cerebral cortex development, including the presence of oSVZ and human oRGC-like NPCs. As an in vitro model, the organoids can be easily manipulated genetically and pharmacologically for mechanistic studies that could shed light on basic questions regarding human brain development23–25. The use of hiPSCs has broad potential for disease modeling using patient-derived iPSC lines. Neurodevelopmental disorders with strong structural phenotypes, such as microcephaly, are particularly suitable for investigation using this forebrain organoid platform8,23,30. One example is recent modeling of Zika virus infection and its induced microcephaly-like phenotypes8,30,31. Neurodegenerative diseases and psychiatric disorders with genetic causes and early-onset symptoms may also be modeled, although more comprehensive examination is needed to identify relevant disease-like phenotypes. The increased throughput enabled by our SpinΩ bioreactor opens the door to large-scale compound testing and therapeutic development9,24. In addition, our pipeline of patterning EBs toward different lineages and growing them in suspension to form organoids can be used to develop protocols for other types of organoids by systematically testing different conditions using SpinΩ. The benefits of a 3D suspension environment and improved diffusion of oxygen and nutrients are not restricted to brain organoids, but could potentially be used to grow organoids for other organs, such as intestine, kidney and lung32–34, as well as for tumors such as glioblastoma.
Limitations of the protocol
Although SpinΩ provides a platform for growing organoids with improved efficiency and reduced cost, it is not currently available for mass production. 3D printing can be costly, and the assembly procedures are tedious. The dependency on 3D printing and individually purchased parts is sometimes accompanied by small irregular protrusions and crevices in the parts. These imperfections increase the risk of contamination in long-term cultures, so we recommend supplementing the medium with antibiotics and routinely sterilizing the bioreactor. The design of SpinΩ can theoretically be adapted to fit different dimensions, such as 24- and 48-well plates, for increased throughput. However, it is difficult to manufacture gears with exact dimensions at the necessary level of precision. Future collaborations with industry partners may improve the design and manufacture of SpinΩ.
The brain region–specific organoids that we developed recapitulate molecular, cellular and structural features of human brain development, but remain a simplified model as compared with in vivo conditions, as they contain only tissues from the neural lineage and lack overlying meninges and vasculature, which may also provide developmental cues. The lack of vasculature as a means to circulate essential factors also limits the size and viability of the organoids. Despite the improved diffusion in spinning cultures, as organoids expand in size, the interiors suffer from a lack of nutrients and oxygen, and eventually form a necrotic core. From our observations, the maximum distance from the surface of the organoid within which cell proliferation can take place is ~500 μm. Particularly for forebrain organoids, the NPCs are ‘buried’ in the interior by expanding neuronal layers and start to die after Day 100, although the neuronal layers will survive and continue to mature. As a result, forebrain organoids can mirror the developing architecture of human fetal cerebral cortex up to the end of the second trimester. Although incorporating functional vasculature could be difficult to achieve in vitro, alternative engineering approaches to improve organoid viability hold promise for the future.
Finally, although the protocols described here are highly reproducible, they still do not produce completely uniform organoids. The forebrain organoids contain a cluster of neural-tube-like structures, which later develop into individual protruding cortical units that we assume to be independent of each other. We have not yet found a way to precisely control the number of structures generated from each EB. In general, larger EBs tend to give rise to more neural tube structures, although the size of each individual structure is usually unaffected. Curiously, when we mechanically separate the neural tube clusters into individual neural tubes, they do not develop well and often form cysts. Therefore, we based most of our characterization on individual cortical units rather than whole organoids.
MATERIALS
REAGENTS
Cells
hiPSCs: We have successfully used feeder-dependent hiPSCs reprogrammed from newborn human foreskin fibroblasts (ATCC, cat. no. CRL-2522), as well as several other patient-derived iPSC lines20–22. ! CAUTION The use of human tissues and human stem cells must adhere to institutional and funding body regulations, as well as to relevant ethical guidelines. Patient consent must be obtained before acquiring and working with any patient-derived cells.
Mouse embryonic fibroblast feeder cells isolated from E13.5 CF1 mouse embryos (Charles River, strain code 023) that have been growth-arrested by γ-irradiation using a Cs-137 irradiator
Growth media components and supplements
DMEM (Corning, cat. no. 10–013)
DMEM/F-12, HEPES (Gibco, cat. no. 11330032)
Neurobasal medium (Gibco, cat. no. 21103049)
KnockOut Serum Replacement (KOSR; Gibco, cat. no. 10828028)
GlutaMAX supplement (Gibco, cat. no. 35050061)
MEM non-essential amino acids solution (MEM-NEAA; Gibco, cat. no. 11140050)
Penicillin–streptomycin (Pen/Strep; Gibco, cat. no. 15140122)
Amphotericin B (Thermo Fisher, cat. no. 15290026
2-Mercaptoethanol (Gibco, cat. no. 21985023)
N-2 supplement (Gibco, cat. no. 17502048)
B-27 supplement (Gibco, cat. no. 17504044)
Matrigel growth factor-reduced basement membrane matrix (Corning, cat. no. 354230)
Insulin solution (Sigma-Aldrich, cat. no. I9278–5ML)
FBS (Corning, cat. no. 35–010)
Small molecules and growth factors
A83–01 (Stem Cell Technologies, cat. no. 72022)
Dorsomorphin (Stem Cell Technologies, cat. no. 72102)
SB-431542 (Stem Cell Technologies, cat. no. 72232)
CHIR-99021 (Stem Cell Technologies, cat. no. 72052)
Purmorphamine (Stem Cell Technologies, cat. no. 72202)
LDN-193189 (Stem Cell Technologies, cat. no. 72146)
FGF-2 (Peprotech, cat. no. 100–18B)
FGF-8 (Peprotech, cat. no. 100–25A)
WNT-3A (R&D Systems, cat. no. 5036-WN-010)
SHH (Peprotech, cat. no. 100–45)
CNTF (Peprotech, cat. no. 450–13)
BDNF (Peprotech, cat. no. 450–02)
GDNF (Peprotech, cat. no. 450–10)
Ascorbic acid (Sigma-Aldrich, cat. no. 1043003)
cAMP (Sigma-Aldrich, cat. no. A9501)
Enzymes and other reagents
Collagenase, type IV (Thermo Fisher, cat. no. 17104019)
BSA (Sigma-Aldrich, cat. no. A9418)
Dulbecco’s PBS without calcium and magnesium (DPBS; Corning, cat. no. 21–031)
Donkey serum (Millipore, cat. no. S30)
0.1% (wt/vol) Gelatin in water (Stem Cell Technologies, cat. no. 7903)
Ethyl alcohol, 95% (vol/vol), 190 proof, pure (Pharmco-Aaper, cat. no. 111000190CSGL)
Sterile deionized water
DMSO (Sigma-Aldrich, cat. no. 472301)
1-Thioglycerol (Sigma-Aldrich, cat. no. M1753)
10% (vol/vol) Bleach
16% (vol/vol) Formaldehyde, methanol free (Polysciences, cat. no. 18814–10)
Sucrose (Sigma-Aldrich, cat. no. S5016)
Tissue-freezing medium (General Data, cat. no. TFM-5)
Triton X-100 (Sigma-Aldrich, cat. no. T9284)
Tween 20 (Sigma-Aldrich, cat. no. P7949)
Slide-mounting solution (Fisher Scientific, cat. no. 14-390-5)
EQUIPMENT
Building of SpinΩ
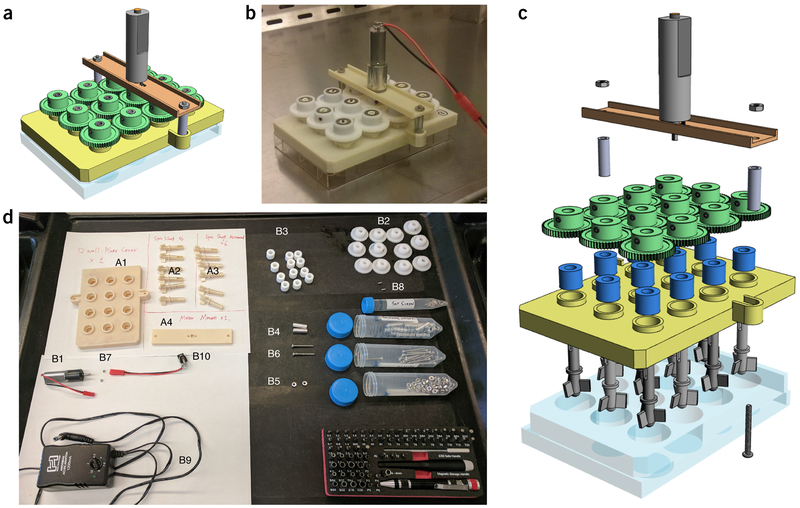
A full list of the equipment needed to build the SpinΩ (both 3D-printed parts and purchasable parts) is provided in Table 1 and Supplementary Data 1, and shown in Figure 1.
TABLE 1 |.
Parts needed to build the SpinΩ bioreactor.
| Part code | Part name | Number needed | Source |
|---|---|---|---|
| A1 | 12-well plate cover | 1 | 3D-printed |
| A2 | Spin shaft | 6 | 3D-printed |
| A3 | Mirror spin shaft | 6 | 3D-printed |
| A4 | Motor mount | 1 | 3D-printed |
| B1 | DC motor | 1 | SuperDroid Robots (cat. no. TD-060-090; IG16 6VDC 090 RPM gear motor) |
| B2 | Gear | 12 | SDP/SI (cat. no. A 1M 2MYZ05052; Module 0.5, 52 teeth, 20° pressure angle, acetal/no insert spur gear) |
| B3 | Bearing | 12 | McMaster-Carr (cat. no. 2685T11; metric PTFE sleeve bearing, for 6-mm shaft diameter, 12-mm o.d. and 10-mm length) |
| B4 | Spacer | 2 | McMaster-Carr (cat. no. 92320A327; stainless-steel unthreaded spacer, 1/4-inch o.d., 3/4-inch) |
| B5 | Nut | 2 | McMaster-Carr (cat. no. 91841A005; type 18–8 stainless-steel hex nut) |
| B6 | Long screw | 2 | McMaster-Carr (cat. no. 91772A118; 18–8 stainless-steel pan-head Phillips machine screw, 4–40 thread, 1–1/8-inch length) |
| B7 | Screw for motor | 2 | McMaster-Carr (cat. no. 91420A002; metric flat-head Phillips machine screw, zinc-plated steel, M2 size, 5-mm length, .4-mm pitch) |
| B8 | Set screw | 12 | McMaster-Carr (cat. no. 18–8 stainless-steel cup-point set screw, 4–48 thread, 3/16-inch long) |
| B9 | Charger | 1 | Amazon (Hosa Cable ACD477 Universal AC Power Supply. See Supplementary Table 1 for purchase URL) |
| B10 | Socket | 1 | Amazon (DC005 PCB Mount 3 Pins 2.1 mm × 5.5-mm DC female socket power jack, 10 pieces. See Supplementary Table 1 for purchase URL) |
CRITICAL For 3D-printing parts, CAD files can be found in Supplementary Data 1. For purchasable parts, see Supplementary Table 1 for URLs for online ordering.
Figure 1 |.
Building a SpinΩ bioreactor. (a,b) CAD drawing (a) and photo (b) of a fully assembled SpinΩ bioreactor. (c) Enlarged view of individual parts of a SpinΩ bioreactor. (d) Photo of all parts needed to build one SpinΩ bioreactor. Please refer to Table 1 for corresponding part codes. The 3D PDF files for a and c allow the user to view the model from any angle (Supplementary Data 2 and 3). To open the 3D PDF file in Adobe Reader, click ‘Options’ on the pop-up notification (yellow bar) and agree to trust this document always.
Generation of organoids
Eppendorf adjustable-volume pipette, P1000 (Fisher Scientific, cat. no. 13-690-032)
Eppendorf adjustable-volume pipette, P200 (Fisher Scientific, cat. no. 13-690-030)
Eppendorf adjustable-volume pipette, P20 (Fisher Scientific, cat. no. 13-690-027)
Pipette filler, portable (Fisher Scientific, cat. no. 13-675-42)
30-mm Tube rack (Fisher Scientific, cat. no. 14-809-44)
Microtube rack (Fisher Scientific, cat. no. 22-313630)
200-μl Pipette tips in racks (USA Scientific, cat. no. 1111–0800)
1,250-μl Pipette tips in racks (USA Scientific, cat. no. 1112–1820)• TC-treated six-well plates (Sigma-Aldrich, cat. no. CLS3506)
Non-treated 12-well plates (Sigma-Aldrich, cat. no. CLS3737)
Six-well clear flat-bottom ultra low attachment plates (Sigma-Aldrich, cat. no. CLS3471)
15-ml Conical centrifuge tubes (Thermo Fisher, cat. no. 14-959-70C)
50-ml Conical centrifuge tubes (Thermo Fisher, cat. no. 14-959-49A)
1.5-ml Microcentrifuge tubes (USA Scientific, cat. no. 1615–5500)
Disposable polystyrene serological pipettes, 1 ml (Fisher Scientific, cat. no. 13-675-15B)
Disposable polystyrene serological pipettes, 5 ml (Fisher Scientific, cat. no. 13-675-22)
Disposable polystyrene serological pipettes, 10 ml (Fisher Scientific, cat. no. 13-675-20)
Disposable polystyrene serological pipettes, 25 ml (Fisher Scientific, cat. no. 13-675-30)
Disposable filter units with 0.2-μm PES membrane, 500 ml (Thermo Fisher, cat. no. 566-0020)
Disposable filter units with 0.2-μm PES membrane, 1,000 ml (Thermo Fisher, cat. no. 567-0020)
Delicate task wipers (Fisher Scientific, cat. no. 34120)
Fine-tip marking pens (Fisher Scientific, cat. no. 13–379-4)
Biopsy cryomold (Tissue-Tek, cat. no. 4565)
Disposable microtome blades (Sakura Finetek USA, cat. no. 4689)
Microscope slides (Fisher Scientific, cat. no. 12-550-15)
Hydrophobic barrier PAP pen (Vector Laboratories, cat. no. H-4000)
Rectangle cover glasses (Fisher Scientific, cat. no. 12–545-F)
Slide-staining chamber (Electron Microscopy Sciences, cat. no. 62010–37)
Spray bottle containing 70% (vol/vol) ethanol
Weighing dishes (Sigma-Aldrich, cat. no. Z154873)
Scalpel blades (Sigma-Aldrich, cat. no. S2771)
Isopropanol (Sigma-Aldrich, cat. no. 437522)
Dry ice
Research cryostat (Leica, cat. no. CM3050 S)
Benchtop minicentrifuge (Benchmark Scientific, cat. no. C1012)
Benchtop centrifuge (Fisher Scientific, model no. AccuSpin 400)
Biological safety cabinet (Baker Company, Sterilgard Series)
CO2 incubator (Thermo Fisher, cat. no. 3307TS)
Inverted microscope (Zeiss, Axiovert 40 CFL model)
Confocal microscope (Zeiss, model no. LSM 800)
Software
Solid Works (Dassault Systèmes, http://www.solidworks.com/default.htm)
Insight (Stratasys, http://www.stratasys.com/3d-printers/fortus- 380mc-450mc)
Adobe Reader (Adobe, http://get.adobe.com/reader/)
REAGENT SETUP
Feeder-dependent hiPSC lines’
Culture human iPSCs on MEFs using standard procedures in a 5% CO2 incubator at 37 °C. Human iPSCs were passaged every week onto a new plate preseeded with γ-irradiated CF1 MEFs. The day before splitting or thawing hiPSCs, plate γ-irradiated MEFs on 0.1% gelatin-coated six-well plates at a density of roughly 2.0 × 105 cellsper well. Passage by incubating with 1 mg/ml collagenase IV in DMEM/F12 medium for 30–60 min until iPSC colonies have detached. Transfer the iPSC colonies to a 15-ml conical tube, and allow the colonies to sink. Wash with DMEM/F12 medium to remove collagenase and triturate with a 200-μl pipette tip to obtain smaller colonies before plating. Maintain iPSCs in standard hESC medium containing 20% (vol/vol) KnockOut Serum Replacement and 10 ng/ml FGF-2, as previously described21. Culture medium was changed each day. ▲ CRITICAL The quality of hiPSC culture is critical for the success of generating brain region–specific organoids. Good hiPSC colonies should form a uniform monolayer with clear boundaries and a homogeneous texture. We recommend that the hiPSCs used should be under 50 passages, as high passage number may substantially alter the results. Please refer to established protocols for additional details of hiPSC culture and quality control20,22,35. ▲ CRITICAL The size of hiPSC colonies will determine the size of the resulting EBs and eventually the size of the organoids. We recommend detaching the colonies when they are ~1.5 mm in diameter. Breaking the colony manually inevitably creates variations in colony size, but this variability is not substantially correlated with the structural organization and maturation of the organoids.
Reconstitution and storage of growth factors and other additives
Reconstitute growth factors (FGF2, FGF8, SHH, CNTF, BDNF and GDNF)in sterile DPBS + 0.1% (wt/vol) BSA to obtain 100 μg/ml stock solutions. Dissolve small molecules (A83–01, Dorsomorphin, SB-431542, CHIR-99021, Purmorphamine and LDN-193189) in sterile DMSO to obtain 5–10 mM stock solutions, depending on the solubility of individual compounds. Store aliquots at −80 °C for up to a year. ▲ CRITICAL The integrity of the growth factors and small molecules is essential to generating high-quality organoids. Minimize degradation of growth factors and small molecules by storingin a reliable −80 °C freezer and making small aliquots to avoid multiple freeze–thaw cycles. Small molecules should be dissolved in DMSO at a high starting concentration, as DMSO is toxic to cells.
Preparation of Matrigel aliquots
Matrigel should be stored at −20 °C upon receipt from the vendor. To prepare aliquots of Matrigel, thaw at 4 °C overnight and place the bottle on ice. Precool 1.5-ml microcentrifuge tubes and 1,250-μl pipette tips at 4 °C for 30 min. Inside a cell culture hood, transfer 500 μl of Matrigel to each microcentrifuge tube and immediately place the tube on ice. Store the aliquots at −20 °C for up to 1 year. Avoid repeated freeze–thaw cycles. ▲ CRITICAL This procedure should be done as quickly as possible because Matrigel solidifies at room temperature (23 °C). Precooling of tubes and pipette tips helps to minimize solidification during transfer. The quality of the Matrigel is important for ensuring the success of later steps.
Collagenase solution
Dissolve Collagenase, type IV, powder in DMEM/F12 to make a 10 mg/ml stock solution, and filter through a 0.2-μm filter. Add 1 ml of the stock solution to 15-ml conical tubes and store the aliquots at −20 °C for up to 1 year. Before use, thaw a tube of collagenase IV stock solution and dilute with 9 ml of sterile DMEM/F12 to obtain a 1 mg/ml working solution. Mix by pipetting before use. ▲ CRITICAL When kept at 4 °C, the effectiveness of collagenase decreases over time, resulting in longer incubation times for detaching iPSC colonies. Therefore, we recommend making a new working solution each time before use.
Culture medium
Prepare culture medium using a vacuum-driven disposable filter unit with a 0.2-μm PES membrane. A variety of different culture media are used for the generation of the three independent brain region–specific organoids, with the specific medium components summarized in Table 2. Prepare the base medium in a 500-ml or 1,000-ml volume and store at 4 °C for up to 1 month. The addition of antibiotics is optional for all culture media and does not alter the outcome noticeably. We recommend using antibiotics because the long-term culture of organoids in SpinΩ has potential risks of bacterial contamination.
TABLE 2 |.
Culture media.
| Stage | Fore brain | Mid brain | Hypothalamus |
|---|---|---|---|
| First | Forebrain first medium | Midbrain first medium | Hypothalamus first medium |
| DMEM/F12 | DMEM/F12 | DMEM/F12 | |
| 20% KOSR | 20% KOSR | 20% KOSR | |
| 1× GlutaMAX | 1× GlutaMAX | 1× GlutaMAX | |
| 1× MEM-NEAA | 1× MEM-NEAA | 1× MEM-NEAA | |
| 1× 2-Mercaptoethanol | 1× 2-Mercaptoethanol | 1× 2-Mercaptoethanol1-thioglycerol, 450 μM | |
| Pen/Strep | Pen/Strep | Pen/Strep | |
| *Dorsomorphin, 2 μM | *LDN-193189, 100 nM | *LDN-193189, 2.5 μM |
|
| *A-83, 2 μM | *SB-431542, 10 μM |
*SB-431542, 3 μM | |
| *SHH, 100 ng/ml |
|||
| *Purmorphamine, 2 μM |
|||
| *FGF8, 100 ng/ml |
|||
| Second | Forebrain second medium | Midbrain second medium | Hypothalamus second medium |
| DMEM/F12 | DMEM/F12 | DMEM/F12 | |
| 1× N2 supplement | 1× N2 supplement | 20% KOSR | |
| 1× GlutaMAX | 1× GlutaMAX | 1× GlutaMAX | |
| 1× MEM-NEAA | Pen/Strep | 1× MEM-NEAA | |
| Pen/Strep | *LDN-193189, 100 nM |
1× 2-Mercaptoethanol | |
| *CHIR-99021, 1 μM |
*CHIR-99021, 3 μM |
Pen/Strep | |
| *SB-431542, 1 μM |
*SHH, 100 ng/ml |
*WNT-3A, 10 ng/ml | |
| *Purmorphamine, 2 μM |
*SHH, 20 ng/ml | ||
| *FGF8, 100 ng/ml |
*Purmorphamine, 2 μM |
||
| Third | Forebrain third medium | Midbrain third medium | Hypothalamus third medium |
| DMEM/F12 | DMEM/F12 | 1:1 DMEM:F12/Neurobasal | |
| 1× N2 supplement | 1× N2 supplement | 1× N2 supplement | |
| 1× B27 supplement | 1× GlutaMAX | 1× B27 supplement | |
| 1× GlutaMAX | 1× MEM-NEAA | 1× GlutaMAX | |
| 1× MEM NEAA | Pen/Strep | 1× MEM-NEAA | |
| 1× 2-Mercaptoethanol | *LDN-193189, 100 nM | Pen/Strep 1× 2-Mercaptoethanol | |
| Pen/Strep | *CHIR-99021, 3 μM | *FGF-2, 10 ng/ml | |
| Insulin, 2.5 μg/ml |
*CNTF, 10 ng/ml | ||
| Fourth | Forebrain fourth medium | Midbrain fourth medium | Hypothalamus fourth medium |
| Neurobasal | Neurobasal | Neurobasal | |
| 1× B27 supplement | 1× B27 supplement | 1× N2 supplement | |
| 1× GlutaMAX | 1× GlutaMAX | 1× B27 supplement | |
| 1× MEM-NEAA | 1× MEM-NEAA | 1× GlutaMAX | |
| Pen/Strep | Pen/Strep | 1× MEM-NEAA | |
| Ascorbic acid, 0.2 mM | Ascorbic acid, 0.2 mM | Pen/Strep | |
| cAMP, 0.5 mM | cAMP, 0.5 mM | Ascorbic acid, 0.2 mM | |
| *BDNF, 20 ng/ml | *BDNF, 20 ng/ml | cAMP, 0.5 mM | |
| *GDNF, 20 ng/ml | *GDNF, 20 ng/ml | *BDNF, 20 ng/ml | |
| *GDNF, 20 ng/ml |
Components marked by an asterisk (*) should be added to 50 or 100 ml of the base medium before use, and should be stored at 4 °C for up to 1 week to ensure effectiveness of growth factors and small molecules.
EQUIPMENT SETUP
Building the SpinΩ bioreactor
The procedure for assembling the 12-well SpinΩ bioreactor is described and illustrated in detail in Box 1 and Supplementary Video 1. Table 1 provides a full list of the equipment needed (both the 3D-printed and purchasable parts; for URLs for online ordering, see Supplementary Table 1). The 3D-printed parts (A1–A4) are designed in SolidWorks (Dassault Systèmes) software, and the CAD files (Supplementary Data 1) are imported into Insight (Stratasys) software controlling a Fortus 400 mc large 3D printer. 3D-printing services are generally available at most university-associated machine shops. Many commercial vendors also offer 3D-printing services that can be ordered online by providing the appropriate CAD files. We have had success using 3D-printed parts from The Johns Hopkins University machine shop, as well as from online commercial vendors. Depending on the different hardware and software used, the specifications for 3D printing may vary. In our case, the following configurations are made:
Model material: ULTEM 9085
Model material color: Tran
Support material: ULTEM support
Invert build materials: No
Slice height: 0.0100
Model tip: T16
Support tip: T16
Part interior style: Solid-normal
Visible surface style: Normal
Support style: Basic
We chose ULTEM 9085 for its heat-resistant properties, which are compatible with sterilization by autoclaving. Other common 3D-printing materials, such as ABS and polycarbonate, tend to deform after autoclaving. ULTEM is not required if other sterilization methods are available, although we have not tested the biological compatibility of other plastic materials with organoid culture. The high melting temperature required for 3D printing using ULTEM also results in variability in product quality and dimensional precision. Please make sure that all parts are printed with smooth surfaces and precise dimensions before assembly.
Box 1 |. Procedure for SpinΩ assembly ● TIMING 3 h.
▲ CRITICAL Please refer to Table 1, Supplementary Video 1, Figure 1 and the 3D PDF files in Supplementary Data 2 and 3.
To open these files in Adobe Reader, click ‘Options’ on the pop-up notification (yellow bar) and agree to trust this document always.
▲ CRITICAL The gears (B2) do not come with holes for set screws (B8). A machine shop can help to drill holes on the side of the gears to fit the set screws.
Fully insert sleeve bearings (B3) into each of the 12 holes on the 12-well plate cover (A1). Depending on the 3D-printing precision, this could be a tight fit.
-
Insert the spin shaft (A2) and the mirror spin shaft (A3) through the sleeve bearings (B3) from the bottom of the plate cover (A1).
▲ CRITICAL STEP When the gears are connected and rotate in synchrony, neighboring gears in direct contact will rotate in the opposite directions (clockwise versus counterclockwise). Therefore, in order to create upward lift in all 12 wells as the spin shafts rotate, two versions with mirrored directionality are used. Make sure to alternate A2 and A3 for neighboring wells.A2 A3 A2 A3 A3 A2 A3 A2 A2 A3 A2 A3 Starting from the two holes in the middle, hold the A2 or A3 in place and place the gears (B2) on each shaft.
Align the directions of the screw hole with the groove on the spin shaft, and tighten the set screw (B8) to fix the gear to the spin shaft. Make sure the spin shaft rotates with the gear without strong friction.
Repeat Steps 3 and 4 for all 12 wells, and avoid misalignment of gears, which creates extra friction.
-
Check if every part is fixed firmly and aligned. Test the friction by rotating a gear by hand and see if it feels smooth.
? TROUBLESHOOTING
-
Observe the direction of the spin shafts and mark the correct direction for rotation to generate upward lift. When looking from the top, the A2 spin shaft should rotate clockwise, whereas the mirror A3 spin shaft should rotate counterclockwise.
▲ CRITICAL STEP The fins on the spin shaft are tilted to create an upward lift. When rotating in the opposite direction, a downward pressure is detrimental to organoid growth. Be sure to use the correct direction.
Wrap this assembly in aluminum foil for sterilization by autoclave using the standard solid autoclave cycle. To maintain sterility, store the assembly in wrapped aluminum foil in a clean lab drawer for up to 2 weeks.
Inside a sterile hood, place the assembly onto a standard 12-well plate to protect the interior from exposure to the outside. Carefully align the beveled corners on the SpinΩ with the beveled corners of the plate.
Solder wires to connect the two poles of the DC motor (B1) and socket (B10). Connect to the charger (B9) to test if the motor is functional.
Insert the DC motor into the center hole on the motor mount (A4) from the side with the flange.
Fix the motor to the mount by tightening the screws (B7).
Drop the motor assembly on top of the center gear, aligned with the two ‘ears’ on the plate cover, and insert the motor shaft into the top of the spin shaft. Align the flat side of the motor shaft with the side with the setscrew.
Tighten the set screw on the center gear so that the gear rotates with the motor. It is not necessary to screw it all the way in.
Fix the motor mount by assembling the supporting columns, consisting of B4, B5 and B6.
-
Connect the motor to the charger (B9) and observe if all gears are driven to rotate by the motor. If the rotation direction is opposite than previously marked in Step 7, reverse the polarity of the charger. The SpinΩ bioreactor is now ready to use.
? TROUBLESHOOTING
-
For routine sterilization between uses, detach the motor assembly by reversing steps 13–15 and autoclaving the bioreactor body. If contamination occurs within the SpinΩ, completely disassemble the bioreactor by reversing steps 1–15, and soak all nonelectronic parts in 10% bleach. Wash the parts extensively with sterile water to remove residual bleach and dry using sterile paper towels before repeating the assembly.
? TROUBLESHOOTING
If purchasable parts are out of stock from the indicated vendor, alternative parts with the same dimensions and specifications can be used. For example, the DC motor (B1) can be replaced with most standard 16-mm DC motors with similar rated speed from other vendors/manufacturers. The design of the motor mount (A4) can accommodate a range of dimensions of screw holes on the motor.
Preparing the SpinΩ bioreactor before use
Before each use, spray SpinΩ with 70% ethanol and place it inside the incubator at 37 °C and 5% CO2. Plug the DC charger into the wall or an extension board and move the power cable to reach inside the incubator. Tape the wires to hold them in place and check whether the incubator can be closed. The wire from the DC charger is thin and does not noticeably disrupt the seal of the incubator door. Plug the power into the socket on the motor and the motor should operate. Check once again if the spinning direction is correct. If the spinning direction is incorrect, reverse the two poles on the power cable (Supplementary Video 1). The bioreactor is now ready to use. When operating multiple units simultaneously, we recommend color-coding the wires and the DC charger with the corresponding units to avoid confusion.
PROCEDURE
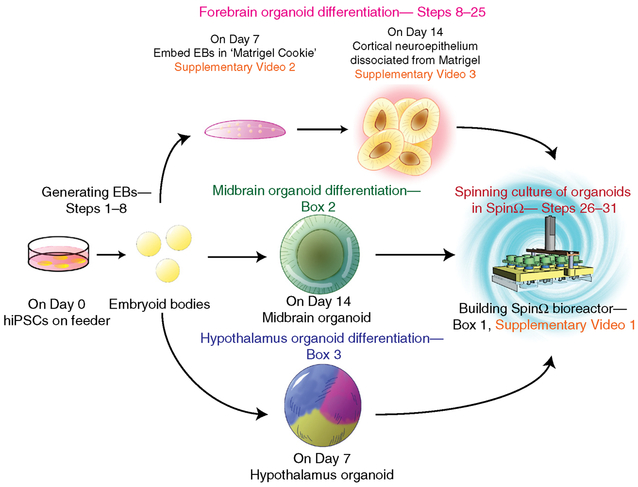
▲ CRITICAL Figure 2 summarizes the workflow for the generation of all three types of brain region–specific organoids. The process for the generation of EBs (Steps 1–7) is the same in all cases. From Step 8 onward, continue following the PROCEDURE to generate forebrain organoids, but refer to Box 2 for midbrain organoids and Box 3 for hypothalamus organoids. Refer to Table 2 for recipes for all media.
Figure 2 |.
Schematic diagram of brain region–specific organoid protocols. The protocols start with enzymatically detaching hiPSC colonies cultured on MEF at Day 0 to generate EBs (Steps 1–7 of the PROCEDURE). From here, the protocols diverge for the procedures to differentiate forebrain (Steps 9–32of the PROCEDURE), midbrain (Box 2) and hypothalamus (Box 3) organoids, respectively. The forebrain organoid protocol involves two unique procedures for embedding EBs in Matrigel at Day 7 and dissociating from Matrigel at Day 14, detailed in Steps 11–25. Finally, the procedures converge with culturing of organoids in the SpinΩ bioreactor, as described in Steps 26–30.
Box 2 |. Midbrain organoid differentiation ● TIMING 100 d, Days 1–100+.
After performing Steps 1–8 of the PROCEDURE on Day 0, change the medium to 3 ml of midbrain first medium on Day 1 (following the method described in the CRITICAL STEP note after Step 7 of the PROCEDURE). Remove dissociated single cells and fragmented colonies, which do not settle as quickly as healthy EBs.
Replace half the medium with fresh midbrain first medium daily.
On Days 5 and 6, change half the medium to midbrain second medium. This provides a smoother transition between the two mediums.
On Day 7, replace the medium with midbrain third medium. Change the medium every 2 d.
On Day 14, switch to midbrain fourth medium and follow Steps 26–31 of the PROCEDURE (growing organoids in the SpinΩ bioreactor). Change the medium every 2 d. The midbrain organoid can be maintained for up to 100 d.
Box 3 |. Hypothalamus organoid differentiation ● TIMING 100 d, Days 1–100+.
After performing Steps 1–8 of the PROCEDURE on Day 0, change the medium to 3 ml of hypothalamus first medium on Day 1 (following the method described in the CRITICAL STEP note after Step 7 of the PROCEDURE). Remove dissociated single cells and fragmented colonies, which do not settle as quickly as healthy EBs.
Replace half the media with fresh hypothalamus first medium daily.
On Days 4–7, change half the medium to hypothalamus second medium. This provides a smoother transition between the two mediums.
On Day 8, switch to hypothalamus third medium and follow Steps 26–31 of the PROCEDURE (growing organoids in the SpinΩ bioreactor). Change the medium every 2 d.
On Day 50, switch to hypothalamus fourth medium. Continue to change the medium every 2 d. The hypothalamus organoid can be maintained for up to 100 d.
Generation of EBs ● TIMING 1–2 h, Day 0
▲ CRITICAL These steps are performed for all three organoid types.
-
1
Day 0. Grow hiPSCs in six-well plates to colony size (a diameter of 1.5 mm). Remove all the hESC medium and replace with 1 ml of 1 mg/ml collagenase solution prewarmed in a 37 °C water bath. Place the cells back into the incubator at 37 °C and 5% CO2 for 45–60 min.
? TROUBLESHOOTING
-
2
Detached colonies should be floating without tapping after incubation. Add 1 ml of hESC medium to each well and use a 10-ml pipette to transfer the detached colonies to a 15-ml conical tube. Allow the colonies to settle to the bottom for ~2 min.
▲ CRITICAL STEP Pipette slowly and gently to minimize turbulence and prevent disruption of colonies.
-
3
Aspirate the supernatant from the top of the tube to just above the settled colonies and wash the colonies by gently adding 5 ml of hESC medium. Let the colonies settle again for ~2 min.
-
4
Aspirate the supernatant again and add 1 ml of the corresponding first medium (Table 2) prewarmed in a 37 °C water bath.
-
5
Add 3 ml of prewarmed first medium (Table 2) to each well of an ultra-low-attachment six-well plate.
-
6
Use a sterile razor blade or scissors to cut the opening of a 1,250-μl pipette tip to larger than 3 mm in diameter. Using the P1000 pipette with the cut tip, gently transfer detached colonies to an ultra-low-attachment six-well plate. When making multiple wells of EBs, distribute the colonies evenly in each well. Usually, hiPSC colonies in one well of a six-well plate will yield 30–50 EBs, which are placed into one well in this step.
▲ CRITICAL STEP The opening of the pipette tip must be cut large enough to prevent shearing of colonies through a tight opening. A 5-ml pipette can also be used, but we recommend using a P1000 pipette because it allows more precise manual control of the colonies placed in each well.
-
7
Place the cells back into the incubator at 37 °C and 5% CO2 for 24 h and avoid disturbance.
▲ CRITICAL STEP The method to change the medium for EBs is the same for all organoids. Gently swirl the plate to allow the EBs to gather at the center of the well. Tilt the plate by lifting the rear end of the plate 1–2 cm and let the EBs settle to the bottom of the front end for 3 min. You can place the rear end of the plate on another plate to tilt. Gently aspirate the medium from the top and always leave 0.5–1 ml of medium inside the well. Replace with fresh medium by pipetting slowly through the wall and avoid disturbing the EBs. Place the plate back into the incubator and gently shake the plate to distribute the EBs inside the well to prevent aggregation.
Forebrain organoid patterning ● TIMING 6d
▲ CRITICAL At this point, the procedures for organoids from different brain regions diverge. Continue to follow the PROCEDURE in order to generate forebrain organoids, but refer to Box 2 for midbrain organoids and Box 3 for hypothalamus organoids.
-
8
On Day 1, change the medium to 3 ml of forebrain first medium. While refreshing the medium, remove dissociated single cells and fragmented colonies, as they do not settle down as quickly as healthy EBs. Return the plate to the incubator at 37 °C and 5% CO2, and avoid disturbance.
? TROUBLESHOOTING
-
9
On Days 3 and 4, refresh the forebrain first medium.
-
10
On Days 5 and 6, use a 10-ml pipette to remove half the medium from each well (1.5 ml) and replace it with 1.5 ml of forebrain second medium. This provides a smoother transition between the two media.
? TROUBLESHOOTING
Embedding of EBs in Matrigel ● TIMING 1–2 h, Day 7
▲ CRITICAL Perform these steps only if generating forebrain organoids (not necessary for hypothalamus and midbrain organoids).
-
11
On Day 7, observe the EBs under an inverted microscope using phase contrast. Healthy EBs should look round and have a smooth surface. The edges of the EBs should look relatively bright (Supplementary Video 2).
▲ CRITICAL STEP The quality of the EBs is critical to a successful outcome. See Figure 3 for an example of high-quality EBs.
? TROUBLESHOOTING
-
12
Thaw a 500-μl aliquot of Matrigel on ice for ~2 h before use. Make sure that Matrigel is completely thawed and shows no signs of solidification. Always place Matrigel on ice when not using (for up to 4 h).
-
13
Cut the opening of a P200 pipette tip to ~1.5- to 2 mm in diameter and place aside for later use.
-
14
Using a 5-ml pipette, transfer the EBs to a 15-ml conical tube. Let the EBs settle and remove the supernatant from the top. Replace with 1 ml of forebrain second medium and let EBs settle to the bottom again.
▲ CRITICAL STEP EBs and small-sized organoids tend to stick to the walls of pipettes. To avoid sticking, prewet the pipette by pipetting the medium up and down once, and keep the pipette upright during transfer of EBs.
-
15
Set a P100 pipette to 67 μl and, using a cut pipette tip from Step 13, transfer ~20–30 EBs in medium to a microcentrifuge tube.
-
16
Using a cut tip, add 100 μl of Matrigel to the microcentrifuge tube and mix with the medium and EBs by pipetting up and down multiple times. This is to dilute the Matrigel with medium at 3:2 ratio, which yields optimal results for subsequent generation of neuroepithelial structures and later dissociation of the Matrigel.
-
17
Using a cut tip, pipette the Matrigel–EB mixture and spread it onto the center of an ultra-low-attachment six-well plate to make a Matrigel ‘cookie’ (imagine the EBs are the chocolate chips). The Matrigel ‘cookie’ should maintain a thickness of >1 mm to fully envelop the EBs in 3D. Distribute the EBs evenly as you pipette and avoid contacting the wall of the well. See Supplementary Video 2 for details.
▲ CRITICAL STEP The EBs should be spaced apart when embedding, as they will grow in the Matrigel and the EBs will fuse if they are too close to each other.
-
18
Incubate the Matrigel in a 37 °C incubator for 30 min to let it solidify.
-
19
Gently add 3 ml of forebrain second medium through the well wall in each well. Sometimes, the Matrigel ‘cookie’ will remain on the bottom and sometimes it will lift up and float. This does not affect the outcome of the resulting organoids. Place the plate back into the incubator.
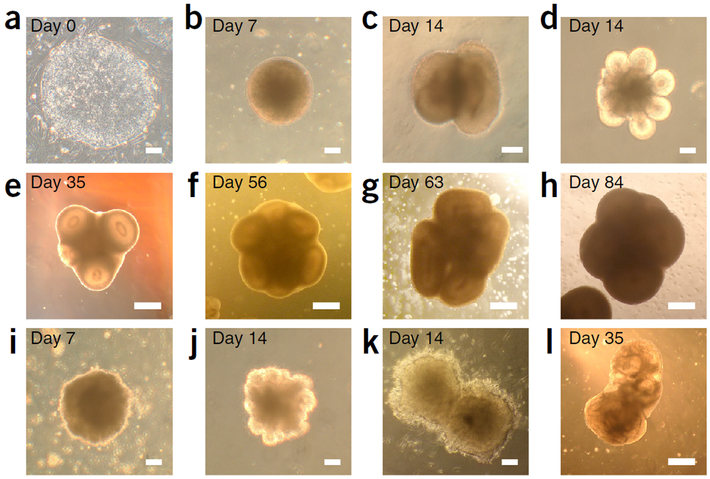
Figure 3 |.
Progression and quality control of forebrain organoid development. (a) Example of an optimal hiPSC colony cultured on MEF, displaying sharp boundaries and homogeneous texture, with a diameter of ~1.5 mm. (b) Example of an optimal EB at Day 7 before embedding into Matrigel, showing a round and smooth morphology and a translucent surface. (c,d) Optimal neuroepithelium formation at Day 14, with a smooth surface and no sign of neuronal differentiation. The number of structures within each cluster may vary, but this does not substantially affect the outcome. (e) Optimal organoid formation at Day 35. The VZ appears more optically translucent than the surrounding immature neuronal layer. Note that the apical surface and ventricular lumen are also clearly visible. (f,g) At Days 56 (f) and 63 (g), the SVZ and CP layers outside the VZ should have expanded, whereas the VZ still appears to be bright. (h) At Day 84, the outer layers should further thicken, and the organoid overall should appear darker, whereas the VZ remains vaguely visible. The forebrain organoid should always exhibit bulges, because individual cortical structures expand independently. (i) Example of a suboptimal EB at Day 7. Note the uneven surface withdead or unhealthy cells attached. (j) Example of suboptimal EB forming underdeveloped neuroepithelial buds at Day 14. (k) Example of a failureto form neuroepithelium at Day 14. Note that cells extending neuronal processes or migrating into Matrigel both indicate premature differentiation. (l) Example of cystic organoid formed from failed neuroepithelium at Day 35. Scale bars, 100 μm (a–d, i–k), 500 μm (e–h, l).
Induction of neuroepithelium inside the Matrigel ● TIMING 7 d, Day 7–14
▲ CRITICAL Perform these steps only if generating forebrain organoids (not necessary for hypothalamus and midbrain organoids).
-
20
Change the medium on Days 9, 11 and 13. Use a 10-ml pipette to remove 2–2.5 ml of medium and replace it with fresh forebrain second medium. Avoid pipetting the Matrigel. We recommend using a pipette instead of a vacuum for removing the medium because the vacuum could potentially aspirate the entire piece of Matrigel upon contact. Using a pipette is therefore much safer. Even if the Matrigel is accidentally pipetted up, just gently pipette it back into the plate.
? TROUBLESHOOTING
-
21
On Day 14, observe the EBs under an inverted microscope using phase contrast. Each EB should transform into a cluster of neuroepithelium buds, each resembling a neural tube. The neuroepithelium should look translucent, with a very smooth surface, and should lack cell processes extending into the Matrigel, which indicates direct neural differentiation. See Figure 3 for an example. The number of neural-tube-like structures from each EB is determined by EB size, whereas the size and thickness of the structures are not directly related to the EB size. Each neural tube will expand independently to form a cortical structure in the forebrain organoid.
▲ CRITICAL STEP The purity of the neuroepithelium is critical for subsequent mechanical dissociation of the organoids from the Matrigel. Pure neuroepithelium has a smooth surface and falls off the Matrigel easily upon pipetting. However, in the presence of neural processes from direct differentiation, these processes cling firmly onto the Matrigel, like roots of a tree, making it much more difficult to dissociate the Matrigel without damaging the organoids.
? TROUBLESHOOTING
Mechanical dissociation of organoids from Matrigel ● TIMING 1 h, Day 14
▲ CRITICAL Perform these steps only if generating forebrain organoids (not necessary for hypothalamus and midbrain organoids).
-
22
On Day 14, pipette up the entire Matrigel ‘cookie’ (together with the medium) using a 5-ml pipette. Point the tip of the pipette to the bottom front corner of the well and pipette down at an ~2 ml per s flow rate. Repeat the pipetting one more time (Supplementary Video 3).
▲ CRITICAL STEP The ‘Fast’ speed should be selected on the pipette aid we use (listed in Equipment). Effective dissociation requires enough shear force from the flow.
? TROUBLESHOOTING
-
23
Swirl the plate so that dissociated organoids gather in the center. Observe the organoids under the microscope to check that the majority are intact and have detached from the Matrigel. Pipette additional times after Step 22 if organoids are still attached to chunks of Matrigel. Small pieces of Matrigel may remain on the organoid surface but should quickly dissociate after 1 d in spinning culture.
▲ CRITICAL STEP Organoids consisting of pure neuroepithelium detach easily, and two rounds of pipetting are often sufficient. Additional pipetting risks breaking some organoids, but is sometimes required. Manual control of pipetting speed improves with experience. For inexperienced users, we recommend pipetting slowly for additional rounds, and frequently checking the condition of the organoids under the microscope.
-
24
(Optional). If you need to separate chunks of Matrigel and potential organoid debris, transfer the organoids and medium gently to a 15-ml conical tube using a 10-ml pipette. Let the organoids settle to the bottom for ~2 min and remove the supernatant. The spacing between organoids indicates chunks of Matrigel remaining. Then add 10 ml of forebrain third medium to the conical tubes from the bottom. The organoids should lift afloat and start to settle again. Observe the tube carefully as the organoids settle; intact organoids fully detached from Matrigel will reach the bottom faster than chunks of Matrigel and broken organoid debris. Aspirate the supernatant from the top immediately after the intact organoids reach the bottom to remove the remaining Matrigel and debris.
▲ CRITICAL STEP It is helpful to remove chunks of Matrigel before transfer to the SpinΩ because organoids could stick to the Matrigel during spinning and form aggregates.
Growth of brain region–specific organoids in the SpinΩ bioreactor ● TIMING from Day 14 up to Day 200+
▲ CRITICAL These steps are performed for all three organoid types.
-
25
Fill each well of a 12-well tissue culture plate with 3.5–4 ml of prewarmed corresponding medium (forebrain third medium, midbrain fourth medium or hypothalamus third medium).
-
26
Use a cut 1,250-μl pipette tip to transfer ~20 organoids to each well.
-
27
Replace the cap of the 12-well plate with the SpinΩ bioreactor by directly dropping it on top of the plate. Carefully align the beveled corners on the SpinΩ with the beveled corners of the plate.
-
28
Place the SpinΩ into the incubator and plug the power supply into the socket connecting the motor. Hold the SpinΩ horizontally and steadily to avoid medium spills.
-
29
As the shafts spin, observe the organoids from the side. The spinning speed can be adjusted by changing the voltage on the charger; we recommend setting it to the minimal speed required to lift the organoids in constant suspension. For a DC motor with a rated speed of 90 r.p.m. at 6 V, we usually set the power to 7.5 V, which yields ~100 r.p.m.
▲ CRITICAL STEP A high rotation speed may generate turbulence that is potentially damaging to the organoids. The DC motors are not perfectly reliable in their speed, so we recommend measuring the rotation speed by manually counting for initial installation. Small variations in spin speed have not been observed to alter the outcome for the three organoid types described here, but the spinning speed may need to be optimized for other organoid types with different tissue properties.
? TROUBLESHOOTING
-
30
Change the medium for organoids every 2 d. Inside the cell culture hood, lift the SpinΩ and place it on the base of an empty sterile 12-well plate before using its cap to cover the plate containing the organoids. The organoids can then be observed outside the hood under the microscope (Fig. 3). Tilt the plate to allow the organoids to sink to the bottom, aspirate ~3 ml of medium from the top and replace with 3 ml of fresh medium. Medium changes can be done less frequently if fewer organoids are cultured in each well.
▲ CRITICAL STEP As the organoids expand in size and reach a 1.5- to 2-mm diameter, culturing 20 organoids per well becomes too crowded. Divide the organoids into multiple wells by transferring them using a cut P1000 pipette. At a 2-mm diameter, we typically culture ten organoids per well, whereas at a ≥3-mm diameter, we culture five organoids per well.
? TROUBLESHOOTING
Reconstitution of extracellular matrix on forebrain organoids using Matrigel coating ● TIMING from Day 35 up to Day 200+
▲ CRITICAL Perform this step only if generating forebrain organoids (not necessary for hypothalamus and midbrain organoids).
-
31
Starting from Day 35, supplement Matrigel directly into forebrain third medium at a 1:100 dilution to create an extracellular matrix (ECM) coating on the organoid surface that substantially improves the alignment of radial fibers from radial glial cells. Thaw 500 μl of Matrigel on ice for 2 h and directly add it to 50 ml of cold forebrain third medium, followed by rigorous pipetting using a 10-ml pipette. From this step onward, use forebrain third medium supplemented with Matrigel for routine media changes.
▲ CRITICAL STEP To prevent the Matrigel from solidifying, medium supplemented with Matrigel should be kept at 4 °C for no longer than 1 week and should be kept at room temperature for no longer than 30 min before use. The medium should not be warmed to 37 °C.
Neuronal maturation of forebrain organoids ● TIMING from Day 70 up to Day 200+
▲ CRITICAL Perform this step only if generating forebrain organoids (not necessary for hypothalamus and midbrain organoids).
-
32
(Optional). To improve maturation during prolonged culture, replace forebrain third medium with forebrain fourth medium every 2 d from Day 70 onward. Follow the method described in Step 31 to continue supplementing Matrigel.
This step is optional because it does not noticeably alter the structural organization of forebrain organoids or the properties of neural progenitor cells. Supplementing media with BDNF and GDNF substantially improves the functional maturation of neurons in the forebrain organoids (as measured by electrophysiological assays).
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
TABLE 3 |.
Troubleshooting table.
| Steps | Problem | Possible reason | Solution |
|---|---|---|---|
| Box 1, step 6 | Strong friction | Spin shaft is not smooth The teeth of gears are not perfectly aligned | Eliminate small protrusions by sanding Redo 3D printing Take off the gears and repeat steps 3–5 |
| Box 1, step 16 | Large noise when operating | Misalignment of assembly | Check assembly and dimensional precision of 3D-printed parts |
| Motor dies | DC motor has a limited life span inside an incubator | This is normal after ~3 months of continuous operation. Monitor the motor daily for signs of failure, including slowing of rotational speed and jerky movements, and replace the motor periodically (every 1–2 months) to minimize the chance of motor failure | |
| Motor dies after less than a month | Humidity enters the motor | If motors other than the indicated one are used, make sure to use sealed motors and prevent liquid spills | |
| Strong friction | Check assembly and dimensional precision of 3D-printed parts | ||
| Set screw rust | Liquid spilled onto the screw | Avoid spills. Use medical-quality stainless-steel screws instead | |
| Box 1, step 17 | Contamination inside the bioreactor | Long-term culture increases the risk of contamination 3D- printing defects increase the risk of contamination | We recommend having extra SpinQ units for routine sterilization Avoid using parts with crevices and protrusions from 3D printing |
| 1 | Colonies do not detach after 90 min of incubation | Collagenase was not fresh hiPSCs show signs of differentiation | Prepare collagenase solution right before use Check the morphologies and pluripotency of hiPSC colonies |
| 8 | Large variation in EB sizes | hiPSC colony size has large variation | Break iPSC colonies into similar-sized clumps during passaging. Let the larger clumps sink to the bottom of a microcentrifuge tube and remove smaller clumps from the top |
| 10 | EBs stick to each other | EBs are not distributed with enough spacing inside the plate | Gently shake the plate when placing it into the incubator. Make sure that the incubator shelf is horizontal and that the EBs do not sink to one side. When sticking starts to occur, pipetting the EBs with the medium twice up and down using a 5-ml pipette usually separates them with no damage to the EBs |
| 11 | Suboptimal EB morphology | EBs were damaged during handling hiPSC culture quality was suboptimal | Make sure to transfer the colony and perform medium changes gently to avoid disturbances Check the morphology and pluripotency of the hiPSC colonies. Start from earlier passages of hiPSCs |
| 20 | EBs fuse with neighbors during expansion in Matrigel | Insufficient spacing between EBs | Do not put too many EBs into one Matrigel cookie. Mix the EBs inside the microcentrifuge tube extensively. If necessary, use a sterile P200 pipette tip to gently push the EBs to fine-tune the spacing |
| 21 | No neuroepithelium formation | EB quality was suboptimal Lot batch-to-lot batch variations in Matrigel | Check EB quality and iPSC quality Try another lot batch of Matrigel |
| 22 | Organoids are still attached to Matrigel after extensive pipetting | Neural processes from differentiated cells cling onto Matrigel | Need to obtain pure neuroepithelium formation with minimal differentiation. Check EB and iPSC quality |
| Incorrect dilution of Matrigel in Step 16 | Make sure to dilute Matrigel with medium 3:2 as more-concentrated Matrigel is difficult to detach | ||
| Lot batch-to-lot batch variations of Matrigel | Use another lot batch of Matrigel | ||
| Neuroepithelium was damaged | Damage was caused during pipetting | Control the speed of pipetting and avoid air bubbles. Frequently check under the microscope to determine if sufficient pipetting has been performed | |
| 29 | Organoids appear unhealthy and darkened after spinning | Spinning direction is opposite of intended spinning direction | Check the spinning direction. The opposite direction of spin pushes the organoid down and limits oxygen delivery. Refer to Box 1 and Supplementary Video 1 |
| Organoids form aggregates after spinning | Low spinning speed | At low spinning speeds, organoids may gather at the center of the well underneath the bottom of the spin shaft and form aggregates. Increase the speed until organoids are lifted in constant suspension | |
| Chunks of Matrigel remain in the medium | Perform the procedures in Step 24 to remove the remaining Matrigel | ||
| DC motor stopped for a prolonged time | Keep an eye on the bioreactor and replace broken or worn-out motors | ||
| 30 | Medium color turns noticeably yellow | Organoids inside the well are overcrowded | Divide organoids into multiple wells |
● TIMING
Steps 1–7, generation of EBs: 1–2 h, Day 0
Steps 8–10, forebrain organoid patterning: 6 d, Days 1–6
Steps 11–19, embedding of EBs in Matrigel: 1–2 h, Day 7
Steps 20 and 21, induction of neuroepithelium inside the Matrigel: 7 d, Days 7–14
Steps 22–24, mechanical dissociation of organoids from Matrigel: 1 h, Day 14
Steps 25–30, growth of brain region–specific organoids in the SpinΩ bioreactor: from Day 14 up to Day 200+
Step 31, reconstitution of ECM on forebrain organoids using Matrigel coating: from Day 35 up to Day 200+
Step 32, neuronal maturation of forebrain organoids: from Day 70 up to Day 200+
Box 1, procedure for SpinΩ assembly: 3 h
Box 2, midbrain organoid differentiation: 100 d, Days 1–100
Box 3, hypothalamus organoid differentiation: 100 d, Days 1–100
ANTICIPATED RESULTS
Evaluating the success of this protocol can be achieved by both bright-field microscopy and immunofluorescence. The forebrain protocol has three critical points for quality control. First, at Day 0, the hiPSC colony should have sharp boundaries surrounded by feeder cells, and the interior should exhibit a uniform monolayer with a homogeneous texture (Fig. 3a). Second, at Day 7, the EBs should be round and have smooth edges (Fig. 3b). The surface of the EBs should be more translucent than the interior. Sometimes two EBs may fuse together, but this should not affect the outcome of the organoid generated. EB quality typically varies from batch to batch and is due to suboptimal quality of hiPSC colonies or mechanical disturbance during EB formation. Third, at Day 14, the tissue should consist of almost pure populations of neuroepithelial cells organized into large neural-tube-like structures. The neuroepithelium appears translucent under phase contrast and has smooth edges interfacing with Matrigel (Fig. 3c,d). Unsuccessful tissue formation typically exhibits smaller, underdeveloped neuroepithelium (Fig. 3j) or premature neuronal differentiation, marked by neural processes extending into the Matrigel (Fig. 3k). These suboptimal tissues usually result in the formation of cystic organoids when grown for longer (Fig. 3l), and are thus not worth continuing to culture. Please refer to the troubleshooting advice in Table 3 if suboptimal outcomes recur. Such quality variation typically arises from suboptimal EB and hiPSC qualities. At this stage, each EB becomes a cluster of neural-tube-like structures, ranging from 2 to more than 10 structures per organoid (Fig. 3c,d). We have yet to establish a methodto precisely control the number of structures producedfrom each EB, but a general rule is that bigger EBs produce more structures within the cluster, whereas the thicknessof individual neuroepithelia is not substantially affected by this variation. Usually, smaller clusters with less than ten structures are favored because a larger cluster will develop into an oversized organoid, with a bigger necrotic core at later stages. From our observations, Matrigel has an important role in providing ECM scaffolding to maintain neuroepithelium expansion and suppress neuronal differentiation. After mechanically dissociating organoids from the Matrigel and transferring them to the SpinΩ at Day 14, neurogenesis is initiated. Removing Matrigel manually at a defined time point enables this protocol to synchronize the differentiation of neuroepithelium, subsequently leading to temporal consistency in the cortical tissues generated.
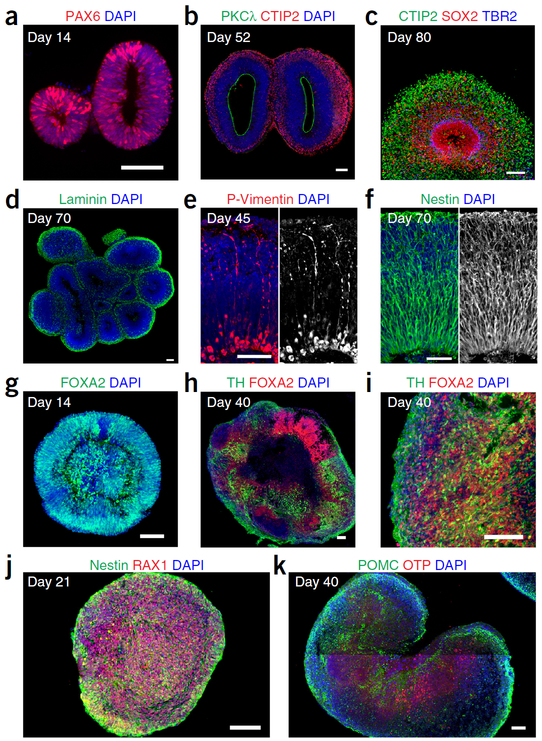
The organoids can be fixed and cryosectioned for immunostaining analysis by following standard procedures8,23. The specific antibodies and dilutions used are summarized in Table 4. At Day 14, almost all cells within the neural-tube-like structures should be positive for dorsal forebrain NPC marker Pax6 (Fig. 4a). Differences in the fluorescence intensity between Pax6+ nuclei are normal. Each neural-tube-like structure develops independently to form one cortical structure, producing a variety of cell types including NPCs, IPCs and neurons, which organize into distinct VZ, SVZ and CP layers. During organoid growth, large, continuous ventricular structures are clearly visible under both bright-field microscope (Fig. 3e) and with immunohistochemistry (Fig. 4b). Under phase contrast, the VZ is always more translucent than the surrounding neuronal layers (Fig. 3e–h), making it distinctive even at later stages (after Day 80). As each organoid contains multiple independent cortical structures, the overall geometry of a forebrain organoid is not spherical, but instead exhibits numerous large bulges (Fig. 3e–h). We most often use triple immunostaining of CTIP2, SOX2 and TBR2 to define the layers of forebrain organoids (Fig. 4c). The VZ contains a pure population of SOX2+ NPCs organized into signature ventricular morphology with an apical surface delineated by adherens junction marker PKCλ (Fig. 4b). Near the basal surface, pure CTIP2+ neuronal populations are deposited to form the CP. Between the VZ and CP, the region containing a mixed population of NPCs, neurons and TBR2+ IPCs form the SVZ.
TABLE 4 |.
Antibodies for organoid characterization.
| Cell/tissue type | Antibody | Species | Vendor | Cat. no. | Dilution |
|---|---|---|---|---|---|
| Dorsal forebrain progenitors | Pax6 | Mouse | BD | 561664 | 1:300 |
| Adherens junction | PKCλ | Mouse | BD | 610207 | 1:300 |
| Cortical neurons | CTIP2 | Rat | Abeam | abl8465 | 1:300 |
| NPC/RGC | SOX2 | Goat | Santa Cruz | sc-17320 | 1:300 |
| ECM | Laminin | Rabbit | Abeam | abll575 | 1:300 |
| Dividing RGCs | Phospho-Vimentin | Mouse | MBL | D076–3S | 1:1,000 |
| NPCs/RGCs | Nestin | Chicken | Aves | NES | 1:300 |
| Floor plate/midbrain progenitors | F0XA2 | Goat | R&D | AF2400 | 1:300 |
| Dopaminergic neurons | TH | Rabbit | Millipore | AB152 | 1:300 |
| Hypothalamus neurons | Rax1 | Rabbit | Takara Bio | M228 | 1:300 |
| Peptidergic neurons | POMC | Rabbit | Phoenix Pharm | H02930 | 1:250 |
| Hypothalamic neurons | OTP | Guinea pig | Takara Bio | M195 | 1:300 |
Figure 4 |.
Immunofluorescence characterization of brain region–specific organoids. (a) Forebrain organoid at Day 14, showing expression of dorsal forebrain NPC marker Pax6 (red). (b) A forebrain organoid with two cortical structures at Day 52. The ventricular surface is delineated by staining for adherens junction marker PKCλ (green). (c) Forebrain organoid at Day 84. The distribution of NPCs (SOX2, red), IPCs (TBR2, blue) and neurons (CTIP2, green) shows distinct VZ, SVZ and CP layers. (d) A forebrain organoid with multiple cortical structures coated by an ECM layer (laminin, green) from Matrigel treatment. (e,f) RGCs in forebrain organoids exhibit straight and radially oriented basal processes (P-Vimentin, red; Nestin, green) at Day45 (e) and Day 70 (f). (g) Midbrain organoid at Day 14, exhibiting radially organized neuroepithelium in the outer region positive for floor plate progenitor marker FOXA2 (green). (h, i) Midbrain organoid at Day 40, with clusters of FOXA2+ progenitor regions (red) and TH+ dopaminergic neurons (green). These panels show the same image, but i is a magnified view of a section of h. (j) Hypothalamus organoid at Day 21, consisting purely of RAX1+ progenitors (red). (k) Hypothalamus organoid at Day 40, containing clusters of OTP+ hypothalamic neurons (red) and POMC+ peptidergic neurons (green). Scale bars, 100 μm.
The addition of dissolved Matrigel in forebrain organoid medium (Step 32) creates and maintains a layer of ECM coating on the surface, visualized by laminin staining (Fig. 4d), similar to that which was previously reported5. This ECM layer provides anchoring points for the end-feet of basal processes from both ventricular RGCs and oRGCs to create tension, enabling these processes to straighten and orient radially (Fig. 4e,f). A recent publication also reported a similar approach to reconstitute the basement membrane36. When allowed to grow beyond Day 100, the forebrain organoid gradually stops expanding in size. As the VZ and SVZ containing proliferative cells are organized interior to the CP, the dramatic expansion of CP thickness prevents the interior from receiving sufficient nutrients and oxygen diffused through the surface. The neurons and astrocytes in the CP will remain healthy and mature over time when analyzed by electrophysiology; we have maintained them for up to 300 d. The organoids can also be dissociated for FACS analysis of cell cycle and marker expression25.
For the midbrain organoids, the EBs should appear round and smooth at Days 5–7, almost identical to EBs for forebrain protocol. On Day 14, the outer portion of the EB becomes a thick layer of radially oriented neuroepithelium tissue, curiously with the apical-like surface oriented toward the outside. This neuroepithelium structure contains a high percentage of FOXA2+ floor plate progenitors (Fig. 4g). After transfer to the SpinΩ, the midbrain organoids typically expand in size rapidly, forming clusters of FOXA2+ progenitor zones and TH+ dopaminergic neurons (Fig. 4h). Within the corresponding cluster, close to 90% of cells are FOXA2+, and TH staining reveals complex neuronal processes (Fig. 4i). The number of dopaminergic neurons within midbrain organoids substantially increases over time. Please refer to our previous publication for more comprehensive characterization8.
For the hypothalamus organoids, the EB expands in size to form organoids from day 7 to 21, but remains spherical, with less distinctive morphological features. At this stage, the majority of the cells should express the hypothalamic progenitor marker RAX1 (Fig. 4j). At Day 40, the hypothalamic neuron marker OTP and peptidergic neuron markers such as POMC appear as localized clusters similar to the nucleus organization found in vivo (Fig. 4k). Please refer to our previous publication for characterization of the diverse subtypes of peptidergic neurons generated8.
Supplementary Material
ACKNOWLEDGMENTS
We thank K.M. Christian for feedback on the manuscript, members of Ming and Song laboratories for discussions, and L. Liu, Y. Cai and D.G. Johnson for technical assistance. The research was supported by grants from the National Institutes of Health (R37NS047344, U19MH106434, P01NS097206 and R01AG057497 to H.S.; R21NS095348, R01MH105128, R35NS097370 and U19AI131130 to G.M.), the Simons Foundation (to H.S. and G.M.) and The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to G.M.).
Note:
Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Takahashi K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Wen Z, Christian KM, Song H & Ming GL Modeling psychiatric disorders with patient-derived iPSCs. Curr. Opin. Neurobiol 36, 118–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariani J et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadoshima T et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 110, 20284–20289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo J et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19, 248–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian X et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen HNN, Song H & Ming G Engineering human pluripotent stem cell-derived 3D brain tissue for drug discovery. J. Transl. Neurosci 1, 38–48 (2016). [Google Scholar]
- 10.Lancaster MA & Knoblich JA Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Yin X et al. Engineering stem cell organoids. Cell Stem Cell 18, 25–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danjo T et al. Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J. Neurosci 31, 1919–1933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K & Sasai Y Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537–550 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi H et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun 6, 8896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasai Y Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 12, 520–530 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Kriks S et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackshaw S et al. Molecular pathways controlling development of thalamus and hypothalamus: from neural specification to circuit formation. J. Neurosci 30, 14925–14930 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byerly MS & Blackshaw S Vertebrate retina and hypothalamus development. Wiley Interdiscip. Rev. Syst. Biol. Med 1, 380–389 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Chambers SM et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen Z et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon KJ et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang CH et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol. Psychiatry 16, 358–360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon KJ et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell 21, 349–358.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu M et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med 22, 1101–1107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon KJ et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagley JA, Reumann D, Bian S, Levi-Strauss J & Knoblich JA Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang Y et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383–398.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lui JH, Hansen DV & Kriegstein AR Development and evolution of the human neocortex. Cell 146, 18–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian X, Nguyen HN, Jacob F, Song H & Ming GL Using brain organoids to understand Zika virus-induced microcephaly. Development 144, 952–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ming GL, Tang H & Song H Advances in Zika virus research: stem cell models, challenges, and opportunities. Cell Stem Cell 19, 690–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T & Clevers H Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Chen YW et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juopperi TA et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington′s disease patient cells. Mol. Brain 5, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancaster MA et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol 35, 659–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.