Trehalose 6-phosphate binds to the KIN10 subunit of SnRK1 and inhibits it in a GRIK-dependent manner, positively regulating fatty acid synthesis by stabilizing WRINKLED1.
Abstract
WRINKLED1 (WRI1), the transcriptional activator of fatty acid synthesis, was recently identified as a target of KIN10, a catalytic α-subunit of the SUCROSE-NON-FERMENTING1-RELATED PROTEIN KINASE1 (SnRK1). We tested the hypothesis that trehalose 6-phosphate (T6P), a signal of cellular sucrose status, can regulate fatty acid synthesis by inhibiting SnRK1. Incubation of Brassica napus suspension cells in medium containing T6P, or overexpression of the Escherichia coli T6P synthase, OtsA, in Nicotiana benthamiana, significantly increased T6P levels, WRI1 levels, and fatty acid synthesis rates. T6P directly bound to purified recombinant KIN10 with an equilibrium dissociation constant (Kd) of 32 ± 6 μM based on microscale thermophoresis. GEMINIVIRUS REP-INTERACTING KINASE1 (GRIK1) bound to KIN10 (Kd 19 ± 3 μM) and activated it by phosphorylation. In the presence of T6P, the GRIK1-KIN10 association was weakened by more than 3-fold (Kd 68 ± 9.8 μM), which reduced both the phosphorylation of KIN10 and its activity. T6P-dependent inhibition of SnRK1 activity was reduced in extracts of individual Arabidopsis thaliana grik1 and grik2 mutants relative to the wild type, while SnRK1 activity in grik1 grik2 extracts was enhanced by T6P. These results indicate that the T6P sensitivity of SnRK1 in vivo is GRIK1/GRIK2 dependent. Based on our findings, we propose a mechanistic model that links sugar signaling and fatty acid homeostasis.
INTRODUCTION
Lipids are primary metabolites in cells, acting as structural components of cell membranes, energy-dense storage compounds, and cell signaling molecules. Fatty acids are major components of lipids, and triacylglycerols (TAGs) are storage lipids that accumulate mostly in oil bodies in plants (Li-Beisson et al., 2013). Imported sugar provides substrates including carbon skeletons in the form of acetyl-CoA, ATP, and reductants (NADPH) for de novo fatty acid synthesis in sink organs. Several lines of evidence have identified links between Suc levels and fatty acid synthesis. For example, wrinkled seed (rr), the classical Mendel pea (Pisum sativum) mutant in which a gene encoding a starch-branching enzyme is disrupted by a transposon-like insertion, showed both higher levels of Suc and increased lipid accumulation (Bhattacharyya et al., 1990). A 3-fold increase in Suc and a 30% increase in leaf TAG accumulation are observed in leaves of Arabidopsis thaliana in which starch synthesis is decreased by RNAi-mediated suppression of the small subunit of AGPase (ADG1) (Sanjaya et al., 2011). In addition, Arabidopsis exhibits a 4-fold TAG increase in roots when cultured on one-half-strength MS medium supplemented with 5% Suc, compared with medium without Suc (Kelly et al., 2013). To test the influence of endogenous sugar content on fatty acid and TAG accumulation, we generated a high-leaf-sugar mutant by simultaneously restricting sucrose phloem loading and blocking starch synthesis by crossing the suc2 (encoding a Suc/H+ symporter that loads Suc into phloem) mutant (Srivastava et al., 2008) and an adg1 mutant (Lin et al., 1988). The sugar content (combined Glc and Suc) in adg1 suc2 leaves was 80-fold higher than that of the wild type, and TAG accumulation increased more than 10-fold with respect to the wild type to 1% of dry weight, demonstrating a strong positive relationship between sugar accumulation and TAG accumulation (Zhai et al., 2017b).
That sugars play dual roles in providing carbon skeletons and sugar signaling is well established for the synthesis of starch (Nakamura et al., 1991; Harn et al., 2000; Wang et al., 2001; Nagata et al., 2012), fructans (Nagaraj et al., 2001; Noël et al., 2001), and anthocyanins (Tiessen et al., 2002). There is increasing evidence that sugar signaling is also important in regulating lipid synthesis. For example, glucose and fructose are necessary for seedlings ectopically overexpressing WRINKLED1 (WRI1) to accumulate TAG (Cernac and Benning, 2004). WRI1 is an APETALA2 (AP2) transcriptional factor that induces the expression of more than 20 genes involved in glycolysis and fatty acid synthesis (Cernac and Benning, 2004; Maeo et al., 2009; Ma et al., 2013). WRI1 expression is enhanced by Suc in Arabidopsis leaves (Masaki et al., 2005). Sanjaya et al. (2011) also observed that the expression of WRI1 in seedling of AGP-deficient lines is increased compared with the wild type. However, in the Arabidopsis adg1 suc2 mutant, WRI1 expression is not significantly different from the wild type, but the abundance of the WRI1 protein is increased due to its stabilization (Zhai et al., 2017b). Sugar-dependent regulation of WRI1 expression and WRI1 protein stability implies that sugar signaling is involved in regulating lipid (TAG) synthesis.
Previously, we reported that a catalytic α-subunit of SnRK1, KIN10, can directly phosphorylate WRI1 within its two AP2 DNA binding domains (Zhai et al., 2017a), close to the previously identified 14-3-3 binding sites (Ma et al., 2016), initiating its proteasomal degradation. This finding is consistent with observations from adg1 suc2 that high sugar levels posttranslationally stabilize WRI1. In the plant cell, SnRK1 is an important metabolic sensor that is activated when sugars are low, resulting in metabolic reprogramming that leads to a general inhibition of anabolism and stimulation of catabolism (Baena-González et al., 2007). The catalytic activity of the heterotrimeric (α/β/βγ) SnRK1 complex resides in its α-subunits, which in Arabidopsis are encoded by KIN10, KIN11, and SnRK1.3. These α-subunits are activated by phosphorylation of Thr-175 (or its equivalent) within their T-loops (Estruch et al., 1992; Hawley et al., 1996; Shen et al., 2009). The degree of T-loop phosphorylation reflects the metabolic status of the cell; under low-energy/sugar or stress conditions, it becomes phosphorylated and thereby activated. Conversely, under favorable, i.e., high-energy/sugar conditions, T-loop phosphorylation is reversed, leading to a decrease in KIN10 activity. In plants, GEMINIVIRUS REP-INTERACTING KINASE1 (GRIK1) and GRIK2 are responsible for phosphorylating Thr-175 in KIN10 (Shen et al., 2009; Glab et al., 2017), whereas protein phosphatases ABSCISIC ACID INSENSITIVE1 and PROTEIN PHOSPHATASE TYPE 2CA mediate its dephosphorylation (Rodrigues et al., 2013). It has also been reported that activated SnRK1 can phosphorylate and moderate the activity of GRIKs, providing a potential feedback control mechanism to regulate SnRK1 activation (Crozet et al., 2010).
Trehalose 6-phosphate (T6P) acts as a signal of sucrose availability connecting plant growth and development to its metabolic status (Schluepmann et al., 2003; Lunn et al., 2006; Yadav et al., 2014; Figueroa and Lunn, 2016). T6P is synthesized by the action of T6P synthase (TPS) on two activated forms of Glc, UDP-Glc and Glc 6-phosphate, both of which are central to plant metabolism (Cabib and Leloir, 1958). SnRK1 activity in crude extracts from developing Arabidopsis tissues is inhibited by T6P, and this inhibition depends on unknown protein factors principally expressed in young tissues (Zhang et al., 2009; Martínez-Barajas et al., 2011; Griffiths et al., 2016). T6P levels are quite variable with respect to organism, tissue types, and developmental states and can exhibit dynamic responses to environmental cues. Several reports from Arabidopsis and maize (Zea mays) have estimated T6P to be present in the low (4–47) micromolar range (Martins et al., 2013; Nuccio et al., 2015). However, as T6P is probably not distributed homogeneously in the tissues analyzed, these values should be regarded as minimum estimates of the in vivo concentrations of T6P.
Because WRI1 is a direct target of KIN10 and T6P is a potent inhibitor of SnRK1, we tested the hypothesis that T6P can regulate fatty acid synthesis via inhibition of SnRK1 and subsequent stabilization of WRI1. Data from experiments employing exogenous T6P treatment, or overexpression of TPS to elevate T6P levels, provided support for the hypothesis. Furthermore, we observed direct binding between T6P and purified recombinant KIN10 and quantitated their interaction. To do this, we used microscale thermophoresis (MST), a biophysical technique which provides precise equilibrium dissociation constants (Kd) for protein-protein and protein-ligand interactions (Wienken et al., 2010; Baaske et al., 2011). Using both MST and genetic approaches employing grik mutants, we demonstrated that T6P weakens the GRIK-KIN10 association, i.e., increases the equilibrium dissociation constant by more than 3-fold, identifying it as a mediator for T6P-dependent inhibition of SnRK1.
RESULTS
T6P Positively Regulates Fatty Acid Biosynthesis by Stabilizing WRI1 in Brassica napus Suspension Cells
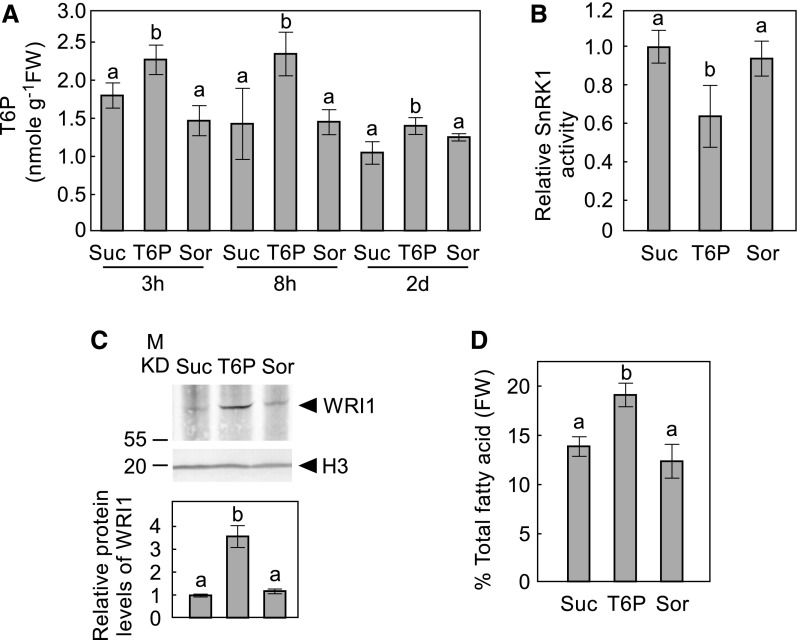
To test the regulatory effect of T6P on fatty acid biosynthesis, a microspore-derived cell suspension culture of B. napus cv Jet Neuf (subsequently referred to as B. napus) (Supplemental Figure 1) was used because it is a well-established model system for developing oilseeds (Shi et al., 2008; Andre et al., 2012). Because T6P does not efficiently penetrate the cell walls of intact plants (Griffiths et al., 2016), we exposed B. napus suspension cells to relatively high levels (100 µM) of T6P. This led to a rise in intracellular T6P levels after 3 h of T6P exposure, compared with cells incubated in the presence of the same concentration of sucrose or sorbitol (Figure 1A), indicating that under our experimental conditions B. napus suspension cells are able to take up exogenous T6P.
Figure 1.
The Effects of T6P on Microspore-Derived Cell Suspension Cultures of B. napus.
(A) T6P contents in B. napus cells cultured on plates supplemented with 100 µM T6P, Suc, or sorbitol (Sor) for 3 or 8 h, and 2 d.
(B) SnRK1 activity in the crude extracts of suspension cells treated for 2 d with sugars as indicated.
(C) WRI1 protein levels in (B) are shown by immunoblot with WRI1 specific antibody. Protein loading is shown by histone H3 (H3) from the same sample. A representative immunoblot is shown. The histogram under immunoblots shows relative WRI1 protein levels quantified with GelAnalyzer2010 and normalized against corresponding protein levels of histone H3.
(D) Total fatty acid at 2-d samples is shown for suspension cells treated with sugars as indicated.
In this figure, values represent mean ± sd of three independent biological replicates. Levels indicated with different letters above histogram bars are significantly different (Student’s t test for all pairs of sugars at the same time point, P < 0.05; Supplemental File 1).
A KIN10 kinase activity assay was performed using a sucrose phosphate synthase (SPS)-derived target peptide containing the KIN10 phosphorylation target sequence (Bhalerao et al., 1999). KIN10 activity was 40% lower in T6P-treated cells relative to sucrose-treated cells after 2 d of treatment (Figure 1B). We next investigated the effects of T6P treatment on WRI1. As shown in Figure 1C, WRI1 accumulated to ∼3-fold higher levels in T6P-treated cells relative to either sucrose- or sorbitol-treated controls after 2 d of treatment. Consistent with the observed increase in WRI1, total fatty acid levels increased by more than 30% in T6P-treated cells relative to sucrose- or sorbitol-treated controls after 2 d (Figure 1D).
It was previously reported that high concentrations of sucrose stabilize WRI1 and favor fatty acid synthesis (Zhai et al., 2017b). We found that incubation in medium containing 100 mM sucrose stabilized WRI1 and increased total fatty acid accumulation in B. napus suspension cells to a similar degree as incubation with 100 µM T6P (Supplemental Figure 2), consistent with previous reports that T6P is more potent than sucrose in inhibiting SnRK1 (Zhang et al., 2009).
Overexpression of a Bacterial T6P Synthase Increases de Novo Fatty Acid Biosynthesis in Nicotiana benthamiana Leaves
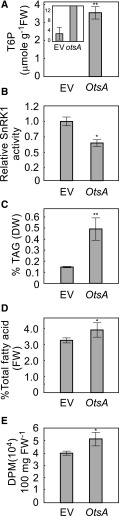
An alternative approach to increasing T6P content in plants is by heterologous expression of the Escherichia coli TPS, OtsA which converts UDP-Glc and Glc 6-phosphate to T6P (Schluepmann et al., 2003). To test whether T6P positively regulates fatty acid synthesis in plant vegetative tissue, OtsA was transiently expressed in leaves of N. benthamiana. After 3 d, T6P levels were elevated by 1000-fold relative to empty vector (EV) controls (Figure 2A). Consistent with the observation in T6P-treated B. napus cells, KIN10 activity in leaves expressing OtsA was significantly lower than EV controls (Figure 2B). OtsA-expressing leaves also accumulated significantly higher levels of TAG and total fatty acids than EV controls (Figures 2C and 2D). To test whether higher TAG accumulation in OtsA-expressing leaves resulted from de novo fatty acid biosynthesis, we performed [1-14C]acetate labeling studies in N. benthamiana leaves. The rate of fatty acid synthesis in leaves expressing OtsA was 29% higher than in leaves expressing EV (Figure 2E).
Figure 2.
Transient Expression of T6P Synthase OtsA Increases Biosynthesis of Both TAG and Total Fatty Acids in N. benthamiana Leaves.
(A) T6P contents in 5-week-old N. benthamiana leaves transformed with EV or expressing OtsA for 3 d. Values represent mean ± sd of four independent biological replicates.
(B) SnRK1 activity in crude extracts of infiltrated N. benthamiana leaves described in (A).
(C) and (D) TAG (C) and total fatty acids (D) were quantified in the samples described in (A).
(E) [1-14C]acetate incorporation into fatty acyl products by strips of N. benthamiana leaves transiently expressing genes as indicated. Values represent mean incorporation ± sd of five independent biological replicates after 30 min of labeling. Asterisks denote statistically significant differences from the EV control material (Student’s t test, *P < 0.05 and **P < 0.01; Supplemental File 1). DPM, disintegrations per minute.
T6P Specifically Binds to Arabidopsis KIN10 and Weakens Its Association with GRIK1
Next, we set out to determine how T6P inhibits SnRK1 to further understand the regulation of fatty acid synthesis by T6P. We tested the hypothesis that T6P directly binds to KIN10 and regulates its activity. As a first step, we used MST analysis to evaluate whether T6P, sucrose, sucrose-6ʹ-P, sorbitol, or trehalose could bind to purified recombinant KIN10. Only T6P showed evidence of binding to KIN10 when each of the sugars was presented at 50 µM (Table 1, Figure 3; Supplemental Figure 3). We also tested the possibility that T6P competes with ATP in binding to KIN10. The binding of T6P with KIN10 was compared for native KIN10 and a previously described KIN10 ATP binding mutant, K48A (Shen et al., 2009). As expected, KIN10 (K48A) did not bind ATP; however, it retained the ability to bind T6P, suggesting T6P likely binds at a site distinct from the ATP binding site on KIN10 (Table 1).
Table 1. T6P Specifically Binds KIN10.
| Ligand (50 µM) | Target | |
|---|---|---|
| KIN10 | KIN10 (K48A) | |
| ATP | + | – |
| T6P | + | + |
| Sucrose | – | n.d. |
| Trehalose | – | n.d. |
| Sorbitol | – | n.d. |
| Suc-6′-P | – | n.d. |
MST was performed to evaluate the interactions between target proteins [KIN10 and KIN10 (K48A)] and potential ligands. +, Interaction detected; −, lack of interaction; n.d., not determined.
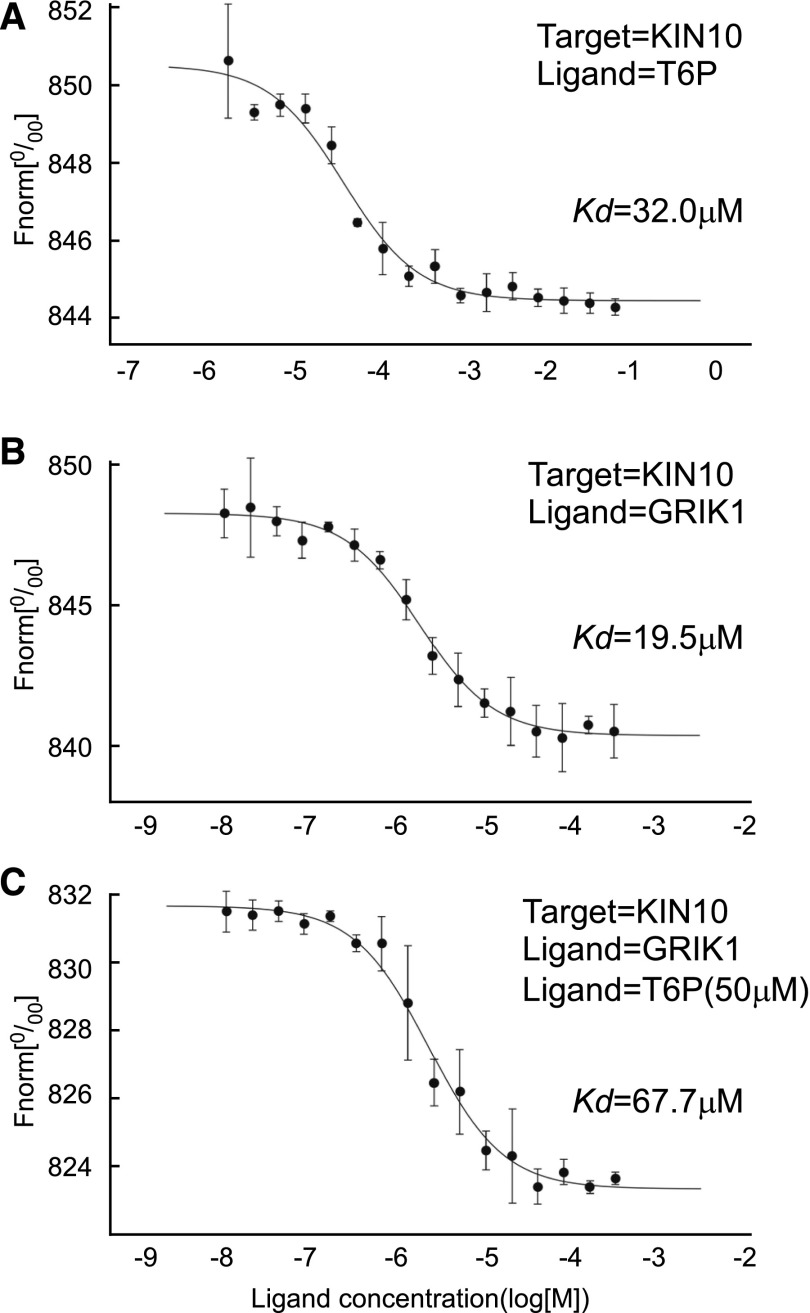
Figure 3.
T6P Weakens the Interaction between GRIK1 and KIN10.
Thermophoretic experiments were performed to determine the equilibrium dissociation constants between KIN10, GRIK1, and T6P (the means of three independent replicates are shown in Table 2). In the thermophoresis experiments, T6P was titrated against KIN10 (A), and GRIK1 was titrated against KIN10 in the absence of T6P (B) or in the presence of 50 µM T6P (C). Dissociation curves were fit to the data to calculate the Kd values shown. Data shown are the mean ± sd, n = 3 independent replicates.
A detailed MST study was performed to quantitate the binding of T6P to KIN10, in which serial dilutions of T6P were titrated against KIN10. The data were used to calculate an equilibrium dissociation constant (Kd) of 32.0 ± 5.6 µM (Figure 3, Table 2), demonstrating a direct interaction between T6P and KIN10 similar to that determined for ATP (Table 2). The observation that the Kd for ATP was unchanged in the presence of 50 µM T6P confirms that ATP and T6P bind at different sites of KIN10 (Table 2).
Table 2. T6P Binding to KIN10 Weakens the Interaction between GRIK1and KIN10.
| Target | Ligand(s) | Kd (µM) |
|---|---|---|
| KIN10 | ATP | 12.8 ± 1.1 |
| KIN10 | T6P | 32.0 ± 5.6 |
| GRIK1 | T6P | – |
| KIN10 | GRIK1 | 19.5 ± 2.6 |
| KIN10 | ATP+T6P (50 µM) | 12.9 ± 1.2 |
| KIN10 | GRIK1+T6P (50 µM) | 67.7 ± 9.8 |
MST was performed to determine the equilibrium dissociation constants (Kd) between KIN10, GRIK1, and T6P. Values represent mean ± sd, n = 3 independent replicates. Kd values for KIN10 and GRIK1 in the presence of 50 μM T6P were significantly different (P < 0.002 by Student’s t test of three independent replicates) from those in the absence of T6P. –, Lack of observed interaction between GRIK1 and T6P.
In plants, GRIK1 and GRIK2 can bind to KIN10 and phosphorylate it, resulting in its activation (Shen and Hanley-Bowdoin, 2006; Shen et al., 2009), so we tested whether T6P can also bind to purified GRIK1 (Supplemental Figure 4). MST analysis showed that T6P does not interact directly with GRIK1 (Table 2). To investigate whether the inhibition of SnRK1 activity by T6P is related to GRIK1 activation of KIN10, we tested whether the binding of T6P to KIN10 affected the equilibrium dissociation constant between KIN10 and GRIK1. In the absence of T6P, GRIK1 bound to KIN10 with a Kd of 19.6 ± 2.5 µM (Figure 3, Table 2), whereas in the presence of 50 µM T6P, the Kd between GRIK1 and KIN10 increased significantly (Student’s t test, P < 0.05) by 3.5-fold to 67.7 ± 9.8 µM (Figure 3, Table 2). These results demonstrate that binding of T6P to KIN10 significantly weakens the interaction between GRIK1 and KIN10.
T6P Decreases KIN10 Phosphorylation and Suppresses Its Activity in the Presence of GRIK1
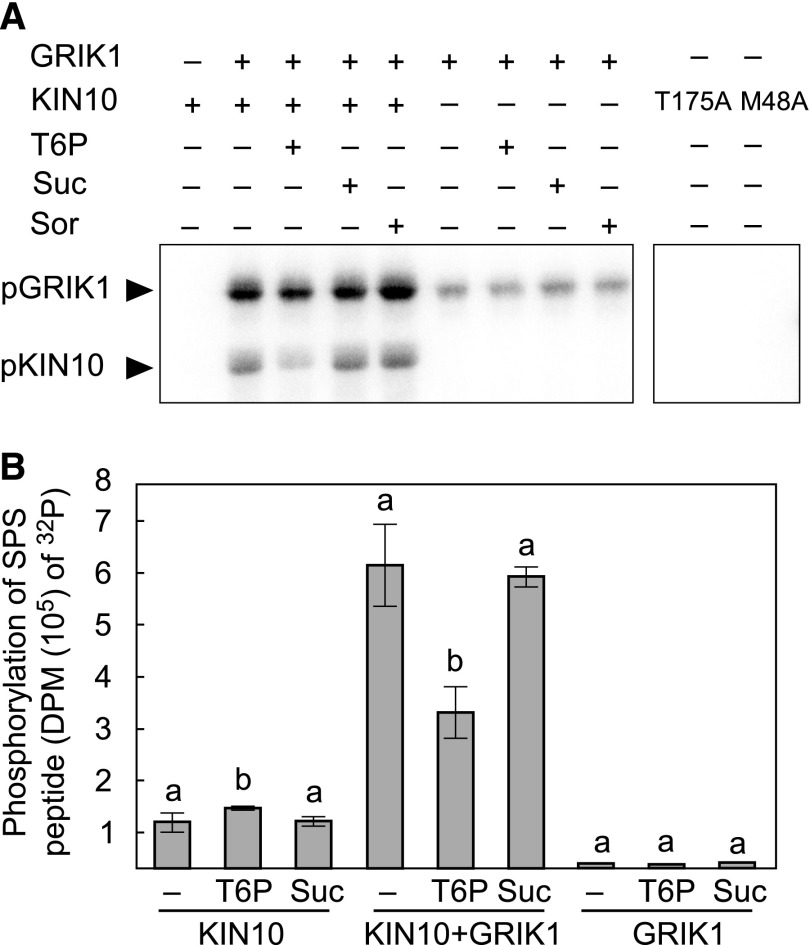
The results showing T6P weakens the interaction between GRIK1 and KIN10 prompted us to quantitate the effects of T6P on the GRIK1-dependent T-loop phosphorylation of KIN10, which in turn mediates its activation. 32P phosphorylation assays were conducted using purified preparations of GRIK1 and KIN10 in the presence or absence of various sugars (Figure 4). Under the assay conditions, phosphorylation of KIN10 was strictly dependent on GRIK1 (Figure 4A). The inclusion of T6P strongly suppressed GRIK1-dependent phosphorylation of KIN10, while sucrose or sorbitol had no discernable effect relative to the sugar-free control (Figure 4A). In the absence of KIN10, autophosphorylation of GRIK1 appears to be minimally influenced by the presence or absence of sugars (Figure 4A).
Figure 4.
T6P Suppresses KIN10 Activation by GRIK1 in Vitro.
(A) In vitro protein phosphorylation assays were performed using purified recombinant KIN10 and GRIK1. Reactions either contained (+) or did not (−) contain 1 µg of GRIK1, 1 µg of KIN10, or 1 mM T6P, Suc, or sorbitol (Sor), as indicated, in the presence of [γ-32P]ATP. After the reaction, proteins were separated using SDS-PAGE and transferred to PVDF membranes. 32P-labeled proteins were visualized by autoradiography. Inactive KIN10 kinase mutants, KIN10 (T175A) and KIN10 (M48A), were also included as negative controls. A representative autoradiograph is shown.
(B) KIN10 activity was determined by measuring the incorporation of 32P from [γ-32P]ATP into the SnRK1-target peptide of SPS. Kinase activity was quantified for 50 ng of KIN10 or for a mixture of 50 ng of KIN10 + 50 ng of GRIK1 or for 50 ng of GRIK1, in the absence (−) or presence of 1 mM T6P or Suc. Values represent mean ± sd of three independent replicates. Levels indicated with different letters above histogram bars are significantly different (Student’s t test for all pairs of the same proteins, P < 0.05; Supplemental File 1). DPM, disintegrations per minute.
As described above, GRIK-dependent phosphorylation of KIN10 results in its activation. We therefore evaluated KIN10 activity in the presence or absence of sugars using the well-established SPS peptide phosphorylation assay (Figure 4B, in its linear range; Supplemental Figure 5). GRIK1-dependent KIN10 phosphorylation of the SPS peptide was significantly suppressed (Student’s t test, P < 0.05) by ∼50% in the presence of T6P (Figure 4B). In the absence of GRIK1, KIN10 phosphorylation of SPS peptide was reduced by ∼5-fold and did not show the T6P-dependent inhibition observed for KIN10 phosphorylation in the presence of GRIK1. Indeed, we made an unexpected observation that KIN10 phosphorylation of the SPS peptide was stimulated ∼25% upon the inclusion of T6P (Figure 4B).
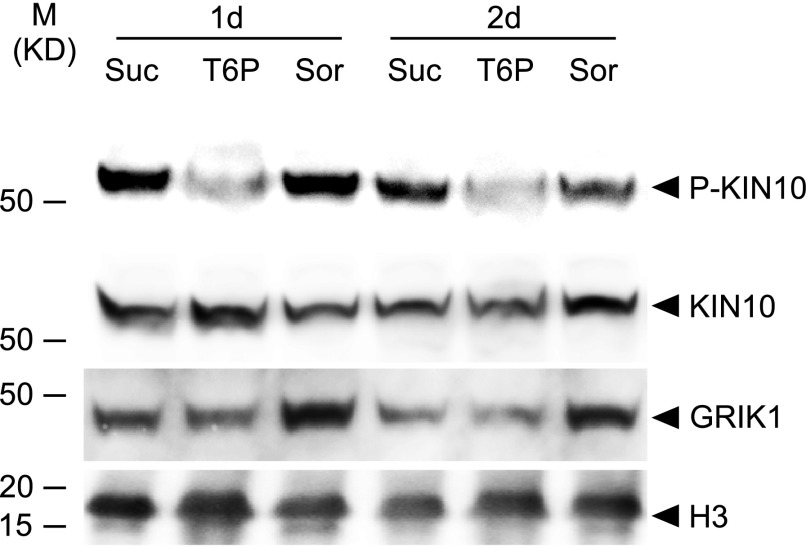
T6P Treatment Decreases KIN10 Phosphorylation in B. napus Suspension Cells
To test the effect of T6P on KIN10 in vivo, B. napus cells were incubated with 100 µM T6P, sucrose, or sorbitol for 1 or 2 d. Total proteins were extracted from cells and specific antibodies were used to detect T-loop phosphorylated KIN10 (P-KIN10) (Nukarinen et al., 2016), KIN10 (Rodrigues et al., 2013), and GRIK1 (Shen and Hanley-Bowdoin, 2006). As shown in Figure 5, phosphorylated KIN10 levels were substantially reduced relative to sorbitol-treated cells, while total KIN10 protein levels were quite similar across the treatments. This pattern is consistent with the decreases in GRIK1-dependent KIN10 phosphorylation in the presence of T6P observed for in vitro protein phosphorylation assays employing isolated purified enzymes described above (Figure 4).
Figure 5.
T6P Treatment Decreases KIN10 Phosphorylation in Microspore-Derived Cell Suspension Cultures of B. napus.
The levels of phosphorylated KIN10, P-KIN10, KIN10, and GRIK1 were quantified from B. napus cells cultured on plates supplemented with 100 µM T6P, Suc, or sorbitol (Sor) for 1 d (1d) or 2 d (2d) as indicated. A representative immunoblot for each protein is shown. Protein loading is shown by histone H3 from the same sample.
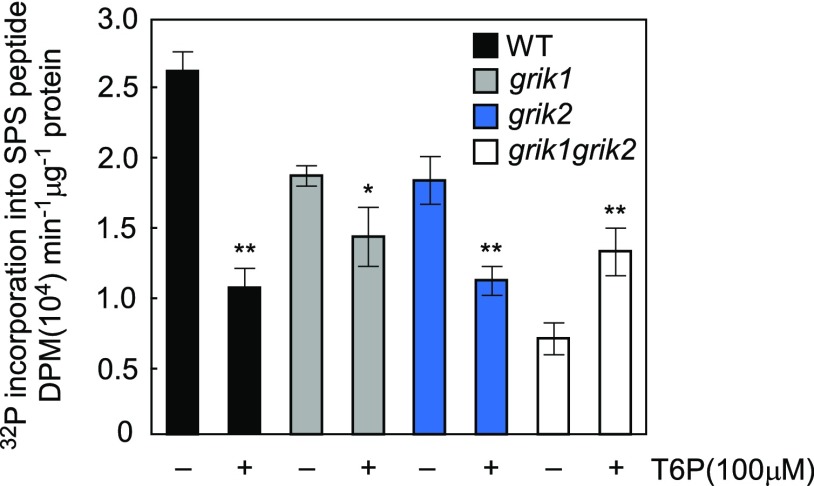
In Arabidopsis, T6P Inhibition of SnRK1 Is GRIK1 and GRIK2 Dependent
As described above, T6P inhibits GRIK1-dependent KIN10 activation in assays containing recombinant GRIK1and KIN10. To further explore these interactions, Arabidopsis grik1, grik2, and grik1 grik2 null mutants (Glab et al., 2017) were used to test whether T6P inhibition of SnRK1 is GRIK1- and GRIK2-dependent in planta (Supplemental Figure 6). SnRK1 activity in extracts of 12-d-old seedlings of grik1 grik2 was only 26% of the wild type (Figure 6), which is consistent with published findings that the phosphorylated form of KIN10 is undetectable in the grik1 grik2 double mutant (Glab et al., 2017). Inclusion of 100 µM T6P (i.e., ∼3-fold the Kd) significantly inhibited SnRK1 activity in crude extracts of the wild type, grik1, and grik2 by 60, 24, and 39%, respectively, but no inhibition was observed for extracts of the grik1 grik2 double mutant. Indeed, SnRK1 activity was significantly activated (by ∼2-fold) in the presence of 100 µM T6P, consistent with the in vitro assays described above employing purified KIN10 in the absence of GRIK1 (Figure 4B).
Figure 6.
Inhibition of SnRK1 by T6P Is GRIK Dependent.
SnRK1 activity is quantified by the incorporation of 32P from [γ-32P]ATP into the SPS peptide. Activity was measured in crude extracts from 12-d-old seedlings of Arabidopsis the wild type (WT), grik1, and grik2 single mutants and the grik1grik2 double mutant in the absence (−) or presence (+) of 100 μM T6P. Values represent mean ± sd of three independent biological replicates. Asterisks denote statistically significant differences from the respective control samples lacking T6P for each genotype (Student’s t test, *P < 0.05 and **P < 0.01; Supplemental File 1).
DISCUSSION
Accumulated evidence has established a role for sugar signaling in regulating lipid synthesis. The recent discovery that SnRK1, a key metabolic sensor in plants, phosphorylates WRI1 and promotes its proteasomal degradation connects plant metabolic status with fatty acid synthesis (Zhai et al., 2017a). In developing tissues, SnRK1 is reported to be inhibited by T6P (Zhang et al., 2009), a signal of cellular sucrose status (Lunn et al., 2006), by an unknown mechanism that involves undefined protein factor(s). In this work, we showed that T6P positively regulates fatty acid synthesis in B. napus cells and N. benthamiana leaves by incubation in medium supplemented with T6P and overexpression of a bacterial TPS, OtsA, respectively. Both treatments significantly elevated tissue T6P levels and stabilization of WRI1. These findings are consistent with previous observations that T6P inhibits SnRK1 and that SnRK1 in turn suppresses WRI1 via phosphorylation that predisposes the WRI1 protein to targeted for degradation (Zhai et al., 2017a).
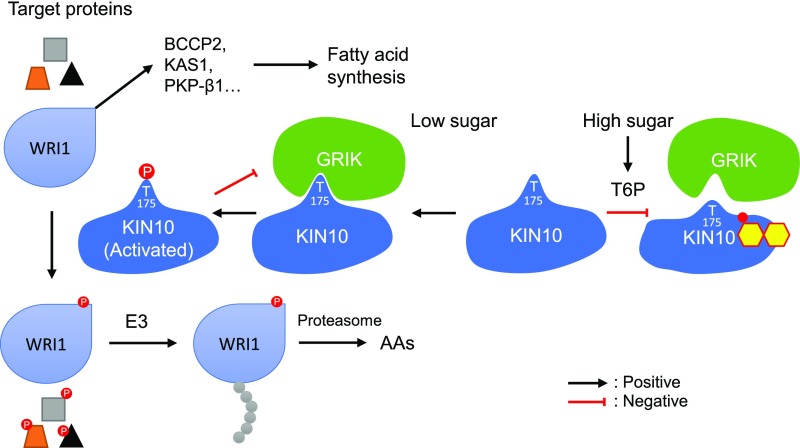
That T6P has been postulated to play a global role in sugar signaling and homeostasis (Lunn et al., 2006; Figueroa and Lunn, 2016) led us to ask how T6P was inhibiting SnRK1 in this study. In a surprise finding, we discovered that T6P binds directly to KIN10 and that this binding weakens the interaction between KIN10 and GRIK1. We therefore propose a model for T6P-regulated plant fatty acid synthesis in which GRIK is a factor that mediates T6P inhibition of SnRK1 (Figure 7). According to the model, in cells with low sucrose and T6P levels, GRIK associates with KIN10 and phosphorylates its activation loop. This enhances SnRK1 activity and the phosphorylation of its many targets including WRI1, which predisposes it to proteasomal degradation, thereby downregulating fatty acid synthesis and reducing cellular demand for energy and reductants. Alternatively, when sugars and T6P are abundant, T6P binds to KIN10, reducing its affinity for GRIK, lowering the likelihood of KIN10 activation loop phosphorylation. SnRK1 activity thus decreases and less WRI1 becomes phosphorylated and degraded, resulting in increased WRI1 accumulation, thereby upregulating fatty acid synthesis.
Figure 7.
T6P-Regulated Plant Fatty Acid Synthesis.
High cellular sugar (carbon) is associated with the accumulation of T6P, which binds to KIN10 (small molecule superimposed on KIN10), the catalytic subunit of SnRK1, weakening its affinity for GRIK. This results in decreased phosphorylation of the KIN10 activation loop, thereby reducing the proportion of activated KIN10. Conversely, when the cellular sucrose level is low, levels of T6P decrease and GRIK binds tightly to KIN10, phosphorylating its activation loop and increasing SnRK1 activity. Activated KIN10 phosphorylates and inactivates 3-HYDROXY-3-METHYLGLUTARYL COA REDUCTASE1, NITRATE REDUCTASE, SUCROSE PHOSPHATE SYNTHASE, and WRI1 to reduce energy-consuming pathways (anabolism) and at the same time, via phosphorylation of other target proteins including several transcription factors, promotes a variety of major catabolic pathways to generate energy. Activated KIN10 can also phosphorylate GRIK and reduce its activity.
Several lines of evidence support this model. (1) T6P feeding to B. napus cells or transient expression of OtsA in N. benthamiana leaves resulted in elevated T6P levels and significantly increased fatty acid content. (2) [1-14C]acetate labeling of N. benthamiana leaves expressing OtsA showed that increased fatty acid and TAG accumulation resulted from higher a higher rates of de novo fatty acid synthesis. (3) Increased WRI1 protein levels were observed in tissues with elevated levels of T6P. (4) T6P bound to KIN10, a catalytic subunit of SnRK1 with a Kd within the published physiological range. (5) There was a 3.5-fold reduction in the association between GRIK1 and KIN10 in the presence of physiologically relevant concentrations of T6P. (6) The incubation of B. napus cells in the presence of T6P led to a reduction of T-loop activation of KIN10 and of its activity. (7) The reduction in KIN10 activity in the presence of T6P was strictly GRIK1-dependent in reconstitution experiments employing bacterially expressed and purified KIN10 and GRIK1. (8) Extracts from young Arabidopsis leaves of the wild type and grik mutants exhibited GRIK-dependent T6P inhibition of SnRK1 activity, consistent with the purified GRIK1/KIN10 reconstitution experiments.
The observed reduction of binding affinity between GRIK1 and KIN10 in the presence of T6P can be attributed to T6P binding to KIN10 because T6P specifically binds to KIN10 but not GRIK1. We initially considered the possibility that T6P inhibited KIN10 activity by competing with its ATP binding site, but rejected the idea because T6P did not change the binding constant for ATP, consistent with previous reports based on assays in crude extracts (Zhang et al., 2009). In principle, the observed T6P-dependent increase in equilibrium dissociation constant for the KIN10 and GRIK1 association could result from a reduction in complex formation or an increase in its propensity to dissociate. How T6P interacts with KIN10 and how that in turn affects the interaction between KIN10 and GRIK1 is presently unknown. It is possible that T6P binds to a domain on KIN10 that overlaps with its GRIK1 interaction interface directly, or alternatively, T6P could bind to a remote site on KIN10 causing a conformational change that allosterically reduces the strength of the GRIK1-KIN10 interaction indirectly. Future structural studies will be required to distinguish between these possibilities.
The discovery that T6P binds to KIN10 initially led us to hypothesize the simplest scenario, i.e., that T6P binding directly inhibits its catalytic activity. To test this hypothesis, assays were performed to measure the phosphorylation activity of KIN10 in the presence or absence of T6P using the well-established SPS peptide substrate. These experiments employing bacterially expressed and purified KIN10 yielded the apparently paradoxical result that in the absence of GRIK1, T6P reproducibly activated, rather than repressed, KIN10 activity. Subsequent experiments with extracts of young leaves from the grik1 grik2 double mutant corroborated the activation of KIN10 by T6P in the absence of GRIK activity. Thus, KIN10 is subject to two apparently opposing effects with respect to T6P: In the presence of GRIK, T6P binding weakened the GRIK-KIN10 interaction decreasing T-loop phosphorylation and activation of KIN10, whereas in the absence of GRIK, T6P binding resulted in a small enhancement of KIN10 activity. Thus, under conditions favoring the accumulation of GRIK proteins, e.g., developing tissues, source tissues, or under some stress condition such as viral infection (Shen and Hanley-Bowdoin, 2006), the GRIK-dependent inhibitory effect would predominate, resulting in net inhibition of SnRK1 activity by T6P.
The findings presented here suggest a plant-specific mechanism in which SnRK1 senses cellular metabolic status changes through T6P. The sensitivity of SnRK1 to T6P and other sugar-phosphates (Glc6P and Glc1P; Nunes et al., 2013), and its insensitivity to AMP (Emanuelle et al., 2015), are consistent with SnRK1 acting as a general metabolic sensor, in contrast to its animal homolog, the AMP-activated protein kinase (AMPK), which is primarily a sensor of energy status. Showing how SnRK1 activity is influenced by sucrose status via T6P provides insights into the evolution of the plant-specific function of SnRK1 relative to its animal (AMPK) and yeast (SNF1) homologs. The details of WRI1 stabilization by T6P and its consequences for lipid synthesis underscore the complexity of SnRK1 regulation, which likely evolved due to its central role in integrating diverse cellular inputs and mediating cellular responses that contribute to metabolic and energy homeostasis (Sheen, 2014).
In summary, we recently identified SnRK1 as a regulator of WRI1 (Zhai et al., 2017a) and established a key role for sugar beyond simply supplying carbon skeletons for fatty acid synthesis by inhibiting SnRK1-dependent WRI1 phosphorylation, which predisposes it for degradation. This work adds a new layer of mechanistic detail to this sugar signaling pathway by identifying a role for T6P, a key sugar-signaling intermediate. T6P reduces SnRK1 activity by binding to KIN10 and weakening its interaction with GRIK1 (its activating kinase), thus reducing SnRK1 activity, WRI1 phosphorylation and degradation, thereby stimulating fatty acid synthesis.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia-0) grik1 (snak2), grik2 (snak1), and grik1 (+/−) grik2 (−/−) sesqui mutants were obtained from Nathalie Glab (Institute of Plant Sciences Paris-Saclay, France) (Glab et al., 2017). For experiments with the grik1 grik2 double mutants, double homozygous null individuals grik1(−/−) grik2(−/−) were selected from the progeny of the grik1(+/−) grik2(−/−) sesqui parental line as described in Supplemental Figure 6. Arabidopsis seeds were surface-sterilized and selected on agar plates containing one-half-strength Murashige and Skoog salts. After 1 week, seedlings were transplanted to moist soil (seed BM2 mix; Berger). All plants (Arabidopsis and Nicotiana benthamiana) were grown with a 16-h-light/8-h-dark photoperiod (combination of cool white fluorescent lamps and incandescent lamps, at a photosynthetic photon flux density of 250 μmol m−2 s−1) with a 23°/19°C day/night, 16-h/8-h temperature regime, and ∼75% relative humidity.
Genetic Constructs
The OtsA coding regions were amplified by PCR from Escherichia coli genomic DNA using primer pairs listed in Supplemental Table 1. The PCR products were then cloned into the Invitrogen Gateway pDONR/Zeo vector (Thermo Fisher Scientific) using the BP reaction and subcloned (LR reaction) into the plant Gateway binary vector pGWB414 (Nakagawa et al., 2007). Expression vectors for the recombinant His6-tagged kinase domain of Arabidopsis KIN10, KIN10 (K48A), and KIN10 (T175A) were obtained from Wei Shen (North Carolina State University) (Shen et al., 2009). The GRIK1 expression vector for producing recombinant His6-tagged GRIK1 protein was previously described (Zhai et al., 2017a).
Expression and Purification of Recombinant KIN10 and GRIK1
Recombinant KIN10 [including KIN10 (K48A) and KIN10 (T175A)] and GRIK1 proteins with N- terminal His6-tags were expressed in E. coli BL21(DE3). Protein purification was performed as described by Shen et al. (2009). Briefly, 1 liter of LB medium was inoculated with 10 mL of a saturated growth E. coli culture, incubated at 37°C with shaking at 225 RPM until its optical density at 600 nm reached 0.6, at which time the temperature of the culture was reduced by cooling on ice for 30 min before the addition of IPTG to a final concentration of 0.4 mM. The culture was incubated for a further 16 h at 16°C. Cells were collected by centrifugation at 6000g for 15 min at 4°C and resuspended in a lysis buffer containing 20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 10 mM imidazole, and 0.1% (v/v) Triton X-100. The cells were disrupted by triple passage through an EmulsiFlex-C3 (Avestin) at 1000 p.s.i. (6894 kPa). The cell lysate was clarified by centrifugation at 15,000g for 30 min at 4°C. The supernatant was applied to a 2 mL Ni-NTA resin column (Qiagen) equilibrated with lysis buffer supplemented. The column was washed twice with five column volumes of lysis buffer and once with wash buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 30 mM imidazole, and 0.1% [v/v] Triton X-100), and then bound proteins were eluted with 20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 10% (v/v) glycerol supplemented with 500 mM imidazole. The eluted protein samples were desalted using an Econo-Pac 10DG chromatography column (Bio-Rad) into 20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 50% glycerol, and 1 mM DTT. Protein preparations were separated into aliquots and stored at −20°C until use.
MST
Sucrose, sucrose-6ʹ-P, trehalose, sorbitol, and ATP were purchased from Sigma-Aldrich. T6P (purity ≥ 95%; catalog no. 136632-28-5) was purchased from Santa Cruz Biotechnology. Thermophoretic assays were conducted using a Monolith NT.115 apparatus (NanoTemperTechnologies). Target proteins (KIN10 and GRIK1) were fluorescently labeled according to the protocol for N-hydroxysuccinimide coupling of the dye NT647 (NanoTemper Technologies) to lysine residues. Target protein and dye were incubated on ice for 30 min. Free dye was separated from labeled protein by size-exclusion chromatography using MST buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 MgCl2, 1 mM DTT, and 0.05% [v/v] Tween 20). For binding tests, 100 nM target protein (fluorescently labeled) was incubated with 50 μM unlabeled ligand for 10 min prior to the measurement. To determine Kd, 100 nM target protein was incubated with a serial dilution of the ligand. Samples of ∼10 μL were loaded into capillaries and inserted into the MST instrument loading tray (Monolith NT.115). The thermophoresis experiments were performed using 40% MST power and 80% LED power at 25°C. T6P (50 µM) was included to test its effect on the associations of KIN10 with ATP or GRIK1.
Protein Phosphorylation Assay
The in vitro protein phosphorylation assay was performed according to Shen et al. (2009). Briefly, a 25-μL assay was performed with 1 µg of KIN10 and 1 µg of GRIK1 in reaction buffer: 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, and 92.5 kBq [γ-32P]ATP (Perkin-Elmer) at 30°C for 20 min. The reaction was terminated by addition of 5 μL of 6× SDS-PAGE loading buffer. A 10-μL aliquot of each assay was separated by SDS-PAGE and transferred to PVDF membrane. 32P-labeled proteins were visualized by autoradiography employing a phosphor screen in conjunction with a Typhoon FLA 7000 imager (GE Healthcare Life Sciences). For phosphorylation assays supplemented with sucrose, T6P, or sorbitol, 1 mM of each was added to the reaction mix containing KIN10 and incubated on ice for 5 min before GRIK1 was added to the reaction.
SnRK1 (KIN10) Activity Assay
Plant-soluble protein extraction and SnRK1 activity assays were performed according to the methods described (Ananieva et al., 2008; Zhang et al., 2009). Briefly, 50 mg (fresh weight) of suspension cells or plant tissues was extracted in 100 μL of ice-cold homogenization buffer: 100 mM Tricine-NaOH, pH 8.0, 25 mM NaF, 5 mM DTT, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM benzamidine, 1 mM PMSF, 1× protease inhibitor cocktail (P9599; Sigma-Aldrich), 1× phosphatase inhibitors (PhosStop; Roche Life Science), and 2% (v/v) (or w/v) insoluble polyvinylpolypyrrolidone (PVPP). The homogenate was centrifuged at 13,000g at 4°C for 15 min before spin-desalting of the supernatant (50 µL) (Illustra MicroSpin G-25 micro columns; GE Healthcare Life Science). Protein concentration was measured with Bradford reagent (B6916; Sigma-Aldrich) using BSA as standard. A 25-μL assay was assembled with 10 µg total protein per sample in kinase reaction buffer: 50 mM HEPES-NaOH, pH 7.5, 5 mM MgCl2, 200 µM SPS peptide (RDHMPRIRSEMQIWSED), 4 mM DTT, 0.5 µM okadaic acid, 0.2 mM ATP, and 12.2 kBq [γ-32P]ATP and incubated at 30°C for 5 min. The assay was stopped by blotting 10 μL per reaction on 4-cm2 squares of Whatman P81 Phosphocellulose paper, immersed in 1% (v/v) phosphoric acid, and then washed with four 800-mL volumes of 1% phosphoric acid, immersed in acetone, dried, and transferred to liquid scintillation vials. Radioactivity associated with phosphorylated SPS peptide was determined by liquid scintillation counting using a Tri-carb (Perkin-Elmer).
For the purified KIN10 activity assay, 0.05 µg KIN10 or 0.05 µg KIN10 and 0.05 µg GRIK1 were included in the kinase reaction buffer. For KIN10 kinase activity assays supplemented with sugars, 1 mM sucrose, T6P, or sorbitol was added to KIN10 and incubated on ice for 5 min before GRIK1was added to the reaction.
Cell Culture Growth
Brassica napus cv Jet Neuf suspension cell cultures were grown in NLN medium (containing 3% sucrose) as previously described (Shi et al., 2008). Prior to experiments suspension cell cultures were maintained in liquid media for 7 d with media being refreshed every 2 d. For sugar treatments, cell cultures were subsequently cultured on NLN solid media (containing 1% (w/v) sucrose and 8 g/L tissue culture grade agar) supplemented with 100 µM of different types of sugars: sucrose, sorbitol, or T6P. Cells were collected after 8 h, 1, 2, and 4 d by filtration, rinsed three times with water, flash frozen, and stored at 80°C until use.
T6P Quantification
Water-soluble metabolites were extracted from aliquots (10–20 mg) of frozen tissue powder using chloroform-methanol (Lunn et al., 2006) and evaporated to dryness using a centrifugal vacuum drier. The dried extract was dissolved in 350 μL purified water and filtered through MultiScreen PCR-96 Filter Plate membranes (Merck Millipore) to remove high molecular weight compounds. T6P, phosphorylated intermediates, and organic acids were measured by high-performance anion-exchange chromatography coupled to tandem mass spectrometry as described by Lunn et al. (2006), with modifications as described by Figueroa et al. (2016).
TAG and Total Fatty Acid Quantification
Total lipids (TAG plus polar lipids) were isolated from 100 mg of freshly harvested leaf tissue by the addition of 700 μL of methanol:chloroform:formic acid (2:1:0.1, by volume) by vigorous shaking for 30 min, after which 1 mL of 1 M KCl and 0.2 M H3PO4 was added. After mixing, the samples were centrifuged at 1500g (4°C) and total lipids were collected in the lower phase (chloroform). For TAG quantification, 60 μL of total lipid were separated by Silica Gel 60 (Merck Millipore) TLC developed with hexane:diethyl ether:acetic acid (70:30:1, by volume) and visualized by spraying with 0.05% (w/v) primuline (in 80% [v/v] acetone). TAG fractions identified under UV light were scraped from the plate and transmethylated to fatty acid methyl esters by incubation in 1 mL 12% (w/w) boron trichloride in methanol at 85°C for 40 min. For total fatty acid quantification, 10 μL of total lipids was directly transmethylated with boron trichloride-methanol as described above. For both assays, 5 µg heptadecanoic acid (C17:0) was added as internal standard prior to transmethylation. Fatty acid methyl esters were extracted into hexane and dried under a nitrogen stream before being dissolved in 100 μL hexane and analyzed by gas chromatography-mass spectrometry with an Agilent Technologies 7890A GC system equipped with an Agilent 60m DB23 capillary column (i.d. 250 μm) and a 5975C mass selective detector.
In Vivo [1-14C]Acetate Labeling
Labeling experiments were performed essentially as described by Koo et al. (2004). N. benthamiana leaves were incubated in 25 mM MES-NaOH, pH 5.7, buffer containing 0.01% (w/v) Tween 20 as wetting agent under illumination (180 µmol m−2 s−1) at 25°C. Labeling was initiated by the addition of 370 kBq of sodium [1-14C]acetate solution (2.15 GBq/mmol; American Radiolabeled Chemicals). Labeling was terminated by removal of the medium from the leaf, and the sample was washed three times with water. Total lipids were extracted and separated as described above. Radioactivity associated with total lipids was determined by liquid scintillation counting using a Tri-carb instrument (Perkin-Elmer).
Agroinfiltration of N. benthamiana
Transient gene expression in N. benthamiana by agroinfiltration was accomplished using a previously described procedure (Ohad and Yalovsky, 2010). Leaves were harvested 3 d after infiltration with different constructs and analyzed for T6P and lipid contents and for SnRK1 kinase activity and in vivo [1-14C]acetate labeling.
Antibodies and Immunoblotting
Anti-WRI1 polyclonal antibodies were described previously (Zhai et al., 2017a). Anti-KIN10 (catalog no. AS10919, lot no. 1610) and -histone H3 (catalog no. AS10710, lot no. 1411) polyclonal antibodies were purchased from Agrisera. Anti-phospho-AMPK α-1 (Thr-172) (catalog no. PA5-17831, lot no. TG2607252) was purchased from Thermo Fisher Scientific. Anti-GRIK1 (catalog no. PHY0844S, lot no. 4480A5) and GRIK2 (catalog no. PHY0845S, lot no. 5480A7) were purchased from PhytoAB. All primary antibodies were used at a 1:1000 dilution. Proteins were resolved by SDS-PAGE and transferred to PVDF membrane for immunoblot analysis. Immunoblots of targeted proteins were visualized using HRP-conjugated secondary antibodies (catalog no. AP187P; Millipore) with SuperSignal West Femto Maximum Sensitivity Substrate (catalog no. 34095; Thermo Fisher Scientific). Immunoblot signals were detected and digitalized with Image Quant LAS4000 and quantified with GelAnalyzer2010a.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource or UniProtKB under the following accession numbers: KIN10 (At3g01090), GRIK1 (At3g45240), GRIK2 (At5g60550), WRI1 (At3g54320), and OtsA (P31677).
Supplemental Data
Supplemental Figure 1. Microspore-derived cell suspension cultures of Brassica napus cv Jet Neuf in liquid culture and plate culture.
Supplemental Figure 2. Sucrose stabilizes WRI1 and increases fatty acid synthesis in Brassica napus suspension cells, but only if presented at a 1000-fold higher concentration than T6P.
Supplemental Figure 3. Microscale thermophoretic binding tests show a specific association between KIN10 and T6P at a concentration of 50 μM.
Supplemental Figure 4. Purified recombinant KIN10 and GRIK1 expressed from E. coli.
Supplemental Figure 5. Fit of DPM by the amount of purified KIN10 and GRIK1 in a 25-mL reaction.
Supplemental Figure 6. The phenotypes of grik mutants.
Supplemental Table 1. Oligonucleotide primer sequence pairs.
Supplemental File 1. Statistical analysis results for each figure.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-SC0012704, specifically through the Physical Biosciences Program of the Chemical Sciences, Geosciences, and Biosciences Division (Z.Z., J.K., H.L., and J.S.). T6P analysis was supported by the Max Planck Society (R.F. and J.E.L.).
AUTHOR CONTRIBUTIONS
J.S., Z.Z., and J.K. conceived the study. Z.Z., J.K., H.L., R.F., and J.E.L. performed experiments. Z.Z., J.K., R.F., J.E.L., and J.S. analyzed data. Z.Z., J.K., J.E.L., and J.S. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Ananieva E.A., Gillaspy G.E., Ely A., Burnette R.N., Erickson F.L. (2008). Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 148: 1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C., Haslam R.P., Shanklin J. (2012). Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc. Natl. Acad. Sci. USA 109: 10107–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baaske P., Wienken C., Willemsen M., Braun D., Duhr S. (2011). Protein-binding assays in biological liquids using microscale thermophoresis. J. Biomol. Tech. 22: S55. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Bhalerao R.P., Salchert K., Bakó L., Okrész L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96: 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M.K., Smith A.M., Ellis T.H., Hedley C., Martin C. (1990). The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60: 115–122. [DOI] [PubMed] [Google Scholar]
- Cabib E., Leloir L.F. (1958). The biosynthesis of trehalose phosphate. J. Biol. Chem. 231: 259–275. [PubMed] [Google Scholar]
- Cernac A., Benning C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40: 575–585. [DOI] [PubMed] [Google Scholar]
- Crozet P., Jammes F., Valot B., Ambard-Bretteville F., Nessler S., Hodges M., Vidal J., Thomas M. (2010). Cross-phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. J. Biol. Chem. 285: 12071–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelle S., et al. (2015). SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 82: 183–192. [DOI] [PubMed] [Google Scholar]
- Estruch F., Treitel M.A., Yang X., Carlson M. (1992). N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa C.M., et al. (2016). Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 85: 410–423. [DOI] [PubMed] [Google Scholar]
- Figueroa C.M., Lunn J.E. (2016). A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiol. 172: 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glab N., Oury C., Guérinier T., Domenichini S., Crozet P., Thomas M., Vidal J., Hodges M. (2017). The impact of Arabidopsis thaliana SNF1-related-kinase 1 (SnRK1)-activating kinase 1 (SnAK1) and SnAK2 on SnRK1 phosphorylation status: characterization of a SnAK double mutant. Plant J. 89: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Griffiths C.A., Sagar R., Geng Y., Primavesi L.F., Patel M.K., Passarelli M.K., Gilmore I.S., Steven R.T., Bunch J., Paul M.J., Davis B.G. (2016). Chemical intervention in plant sugar signalling increases yield and resilience. Nature 540: 574–578. [DOI] [PubMed] [Google Scholar]
- Harn C.H., Bae J.M., Lee S.S., Min S.R., Liu J.R. (2000). Presence of multiple cDNAs encoding an isoform of ADP-glucose pyrophosphorylase large subunit from sweet potato and characterization of expression levels. Plant Cell Physiol. 41: 1235–1242. [DOI] [PubMed] [Google Scholar]
- Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., Hardie D.G. (1996). Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271: 27879–27887. [DOI] [PubMed] [Google Scholar]
- Kelly A.A., van Erp H., Quettier A.-L., Shaw E., Menard G., Kurup S., Eastmond P.J. (2013). The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 162: 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J., Ohlrogge J.B., Pollard M. (2004). On the export of fatty acids from the chloroplast. J. Biol. Chem. 279: 16101–16110. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y., Shorrosh B., Beisson F., Andersson M.X., Arondel V., Bates P.D., Baud S., Bird D., DeBono A., Durrett T.P. (2013). Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi/10.1199/tab.e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-P., Caspar T., Somerville C., Preiss J. (1988). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol. 86: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J.E., Feil R., Hendriks J.H., Gibon Y., Morcuende R., Osuna D., Scheible W.-R., Carillo P., Hajirezaei M.-R., Stitt M. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Kong Q., Arondel V., Kilaru A., Bates P.D., Thrower N.A., Benning C., Ohlrogge J.B. (2013). Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS One 8: e68887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Kong Q., Mantyla J.J., Yang Y., Ohlrogge J.B., Benning C. (2016). 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 88: 228–235. [DOI] [PubMed] [Google Scholar]
- Maeo K., Tokuda T., Ayame A., Mitsui N., Kawai T., Tsukagoshi H., Ishiguro S., Nakamura K. (2009). An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 60: 476–487. [DOI] [PubMed] [Google Scholar]
- Martínez-Barajas E., Delatte T., Schluepmann H., de Jong G.J., Somsen G.W., Nunes C., Primavesi L.F., Coello P., Mitchell R.A., Paul M.J. (2011). Wheat grain development is characterized by remarkable trehalose 6-phosphate accumulation pregrain filling: tissue distribution and relationship to SNF1-related protein kinase1 activity. Plant Physiol. 156: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M.C.M., et al. (2013). Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 163: 1142–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T., Mitsui N., Tsukagoshi H., Nishii T., Morikami A., Nakamura K. (2005). ACTIVATOR of Spomin:LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 46: 547–556. [DOI] [PubMed] [Google Scholar]
- Nagaraj V.J., Riedl R., Boller T., Wiemken A., Meyer A.D. (2001). Light and sugar regulation of the barley sucrose: fructan 6-fructosyltransferase promoter. J. Plant Physiol. 158: 1601–1607. [Google Scholar]
- Nagata T., Hara H., Saitou K., Kobashi A., Kojima K., Yuasa T., Ueno O. (2012). Activation of ADP-glucose pyrophosphorylase gene promoters by a WRKY transcription factor, AtWRKY20, in Arabidopsis thaliana L. and sweet potato (Ipomoea batatas Lam.). Plant Prod. Sci. 15: 10–18. [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Ohto M.A., Yoshida N., Nakamura K. (1991). Sucrose-induced accumulation of β-amylase occurs concomitant with the accumulation of starch and sporamin in leaf-petiole cuttings of sweet potato. Plant Physiol. 96: 902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël G.M., Tognetti J.A., Pontis H.G. (2001). Protein kinase and phosphatase activities are involved in fructan synthesis initiation mediated by sugars. Planta 213: 640–646. [DOI] [PubMed] [Google Scholar]
- Nuccio M.L., Wu J., Mowers R., Zhou H.P., Meghji M., Primavesi L.F., Paul M.J., Chen X., Gao Y., Haque E., Basu S.S., Lagrimini L.M. (2015). Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 33: 862–869. [DOI] [PubMed] [Google Scholar]
- Nukarinen E., Nägele T., Pedrotti L., Wurzinger B., Mair A., Landgraf R., Börnke F., Hanson J., Teige M., Baena-Gonzalez E., Dröge-Laser W., Weckwerth W. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., Primavesi L.F., Patel M.K., Martinez-Barajas E., Powers S.J., Sagar R., Fevereiro P.S., Davis B.G., Paul M.J. (2013). Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol. Biochem. 63: 89–98. [DOI] [PubMed] [Google Scholar]
- Ohad N., Yalovsky S. (2010). Utilizing bimolecular fluorescence complementation (BiFC) to assay protein–protein interaction in plants. In Plant Developmental Biology. Methods in Molecular Biology (Methods and Protocols), Hennig L., ed Köhler C., Volume 655 (Totowa, NJ: Humana Press; ), pp. 347–358. [DOI] [PubMed] [Google Scholar]
- Rodrigues A., et al. (2013). ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25: 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya D., Durrett T.P., Weise S.E., Benning C. (2011). Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9: 874–883. [DOI] [PubMed] [Google Scholar]
- Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M. (2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6849–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2014). Master regulators in plant glucose signaling networks. J. Plant Biol. 57: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Hanley-Bowdoin L. (2006). Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142: 1642–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Reyes M.I., Hanley-Bowdoin L. (2009). Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 150: 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Xu G., Warrington T.B., Murdoch G.K., Kazala E.C., Snyder C.L., Weselake R.J. (2008). Microspore-derived cell suspension cultures of oilseed rape as a system for studying gene expression. Plant Cell Tissue Organ Cult. 92: 131–139. [Google Scholar]
- Srivastava A.C., Ganesan S., Ismail I.O., Ayre B.G. (2008). Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A., Hendriks J.H., Stitt M., Branscheid A., Gibon Y., Farré E.M., Geigenberger P. (2002). Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-J., Yeh K.-W., Tsai C.-Y. (2001). Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 161: 635–644. [Google Scholar]
- Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S. (2010). Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1: 100. [DOI] [PubMed] [Google Scholar]
- Yadav U.P., Ivakov A., Feil R., Duan G.Y., Walther D., Giavalisco P., Piques M., Carillo P., Hubberten H.-M., Stitt M., Lunn J.E. (2014). The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 65: 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Liu H., Shanklin J. (2017a). Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29: 871–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Liu H., Xu C., Shanklin J. (2017b). Sugar potentiation of fatty acid and triacylglycerol accumulation. Plant Physiol. 175: 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L.F., Jhurreea D., Andralojc P.J., Mitchell R.A., Powers S.J., Schluepmann H., Delatte T., Wingler A., Paul M.J. (2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149: 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]