The Ocd1 oxalyl-CoA decarboxylase catalyzes oxalyl-CoA into formal-CoA and CO2 for oxalate degradation and ocd1 mutations affect seed storage reserves, metabolome, and endosperm vitreousness.
Abstract
The organic acid oxalate occurs in microbes, animals, and plants; however, excessive oxalate accumulation in vivo is toxic to cell growth and decreases the nutritional quality of certain vegetables. However, the enzymes and functions required for oxalate degradation in plants remain largely unknown. Here, we report the cloning of a maize (Zea mays) opaque endosperm mutant that encodes oxalyl-CoA decarboxylase1 (EC4.1.1.8; OCD1). Ocd1 is generally expressed and is specifically induced by oxalate. The ocd1 mutant seeds contain a significantly higher level of oxalate than the wild type, indicating that the ocd1 mutants have a defect in oxalate catabolism. The maize classic mutant opaque7 (o7) was initially cloned for its high lysine trait, although the gene function was not understood until its homolog in Arabidopsis thaliana was found to encode an oxalyl-CoA synthetase (EC 6.2.1.8), which ligates oxalate and CoA to form oxalyl-CoA. Our enzymatic analysis showed that ZmOCD1 catalyzes oxalyl-CoA, the product of O7, into formyl-CoA and CO2 for degradation. Mutations in ocd1 caused dramatic alterations in the metabolome in the endosperm. Our findings demonstrate that ZmOCD1 acts downstream of O7 in oxalate degradation and affects endosperm development, the metabolome, and nutritional quality in maize seeds.
INTRODUCTION
Oxalate is the simplest dicarboxylic acid and is produced in most if not all organisms. The oxalate in plants mainly occurs as soluble free acid and insoluble metal oxalate crystals. In crop plants and many vegetables, it may account for as much as 3 to 10% of the dry weight and plays various functional roles in biological and metabolic processes such as metal tolerance, ion balance, and insect defense (Franceschi and Nakata, 2005). For example, micromolar concentrations of aluminum (Al) can inhibit root growth and decrease the acquisition of water and mineral nutrients. In response to Al stress, plants excrete oxalate into the environment surrounding their roots to prevent the external absorption of Al or sequester absorbed Al in the nontoxic form of Al-oxalate within live cells (Ma et al., 1997, 1998; Shen et al., 2002; Yang et al., 2008). Oxalate also participates in the detoxification of heavy metals such as lead, strontium, cadmium, and copper, via the formation of different oxalate crystals (Franceschi and Schueren, 1986; Mazen and El Maghraby, 1998; Yang et al., 2000; Choi et al., 2001).
Oxalate functions as an important counterion to inorganic cations, including sodium and potassium, thus contributing to ionic equilibrium (Osmond, 1963). Plant calcium (Ca) oxalate crystals have been known for a long time and many physiological and biochemical studies have demonstrated that the formation of these crystals plays a crucial role in the regulation of the Ca concentration in plant tissues and organs (Leeuwenhoek, 1975; Franceschi, 1989; Mazen and El Maghraby, 1998; Zindler-Frank et al., 2001; Volk et al., 2002; Prychid et al., 2008; Nakata, 2012; Dauer and Perakis, 2014; Islam et al., 2015). In addition to balancing the Ca content, the production of Ca oxalate crystals protects plants against chewing insects such as caterpillars, including the larvae of the beet armyworm, Spodoptera exigua (Korth et al., 2006; Park et al., 2009).
Although oxalate provides many benefits for plant adaption, excessive accumulation oxalate has negative effects. For example, the overexpression of a bacterial oxalic acid biosynthetic gene in Arabidopsis thaliana led to retarded growth and development as a result of higher oxalate production (Nakata, 2015). Certain pathogens secrete oxalate to enhance their pathogenicity in plants by stimulating stomatal opening and cell wall permeability, ultimately leading to programmed cell death (Bateman and Beer, 1965; Guimarães and Stotz, 2004; Kim et al., 2008). Oxalate in plant foods has an adverse impact on human health, as oxalate acts as an antinutrient that impairs the absorption of Ca and some other minerals (Weaver et al., 1987; Brzezicha-Cirocka et al., 2016). Indeed, compared with kale (Brassica oleracea), which has a low oxalate content, spinach (Spinacia oleracea) has a high level of oxalate and poor Ca bioavailability (Weaver et al., 1987; Heaney and Weaver, 1990). Moreover, ingestion of free oxalate or metal oxalate crystals may cause serious diseases. For example, oxalate can induce breast cancer and Ca oxalate crystals are associated with kidney stone composition and urinary metabolic disturbances (Taylor and Curhan, 2007; Gil et al., 2013; Lorenz et al., 2013; Kirejczyk et al., 2014; Castellaro et al., 2015). Therefore, the content of oxalate in food plants must be strictly regulated and a better understanding of the mechanisms underlying oxalate biosynthesis and degradation is of theoretical and practical importance for genetic improvement of crop plants.
To date, at least four potential pathways for oxalate biosynthesis have been reported in plants. The substrate glyoxylate can be oxidized by glycolate oxidase (Richardson and Tolbert, 1961; Yu et al., 2010) or dehydrogenated by lactate dehydrogenase to generate oxalate (Davies and Asker, 1983). In oxalate-secreting fungi, oxaloacetate acetylhydrolase catalyzes the hydrolytic cleavage of oxaloacetate to generate oxalate (Han et al., 2007). l-ascorbic acid was also reported to function as a substrate for oxalate synthesis (Horner et al., 2000), although the enzymes responsible for this pathway have not been identified.
A putative oxalate degradation pathway and its corresponding enzymes have been proposed in plants. The combined action of four enzymes converts one molecule of oxalate into two molecules of the final product CO2, with each step catalyzed by oxalyl-CoA synthetase, oxalyl-CoA decarboxylase, formyl-CoA hydrolase, and formate dehydrogenase (Foster et al., 2012, 2016; Foster and Nakata, 2014; Lou et al., 2016a). The gene encoding plant oxalyl-CoA synthetase was first cloned from a maize (Zea mays) classic endosperm mutant opaque7 (o7) (Miclaus et al., 2011; Wang et al., 2011). The zein storage proteins in the maize endosperm are devoid of the essential amino acid lysine, which results in poor-quality protein in the grain. The o7 seeds synthesize a reduced level of zeins (especially the 19-kD α-zeins), and as a consequence, they have elevated lysine contents. However, when o7 was cloned, the function of this gene had not been elucidated because of the lack of sufficient reference information in the public database; later, its homolog ACYL-ACTIVATING ENZYME3 (AtAAE3) in Arabidopsis was shown to encode an oxalyl-CoA synthetase, which catalyzes the first step of oxalate catabolism by ligating oxalate and CoA to generate oxalyl-CoA (Foster et al., 2012, 2016; Foster and Nakata, 2014; Lou et al., 2016a). Mutations in AtAAE3 result in increased oxalate accumulation, abnormal seeds, and reduced defense against fungal pathogens, suggesting that normal oxalate metabolism is crucial for plant growth and the immune system (Foster et al., 2012). Oxalyl-CoA synthetase was also found in Oxalobacter formigenes, a human gut bacterium, where it performs a similar function in oxalate metabolism and is associated with kidney stone formation (Stewart et al., 2004; Siener et al., 2013). In addition, oxalate oxidase in plants can oxidize oxalate into CO2 and H2O2 (Lane et al., 1993). The overexpression of the wheat (Triticum aestivum) oxalate oxidase gene in Brassica napus enhanced the resistance to exogenous oxalate (Dong et al., 2008).
In this study, we isolated a maize opaque mutant and cloned the causal gene. This gene (designated Ocd1) encodes an oxalyl-CoA decarboxylase functioning downstream of O7 (ZmAAE3) in oxalate degradation. The oxalate contents in o7 and ocd1 seeds are higher than those in the wild type. Reduced zein and starch synthesis and altered metabolic phenotypes have been characterized with ocd1. Pleiotropic effects on plant development caused by defects in oxalate degradation are also discussed.
RESULTS
Characterization of a Previously Undescribed Opaque Endosperm Mutant in Maize
Fifteen UFMu06493 seeds (with naturally induced mutations by the Mu transposable elements in the W22 background; https://www.maizegdb.org/uniformmu) were obtained from the Maize Genetics Cooperation Stock Center, and they were originally employed for a different purpose. All seeds appeared normal and were propagated by self-pollination. One self-pollinated ear segregated normal and opaque seeds at a 3:1 ratio, whereas the other fourteen did not display any kernel phenotype (Figure 1A). We did not expect that UFMu06493 seeds would produce an opaque phenotype, and we eventually verified that the opacity was not genetically linked to the gene that we meant to study, indicating that this phenotype is caused by an independent mutation. Because we are interested in maize endosperm development, we continued to follow up our studies on this mutant. This opaque mutant was designated ocd1-1 (see below).
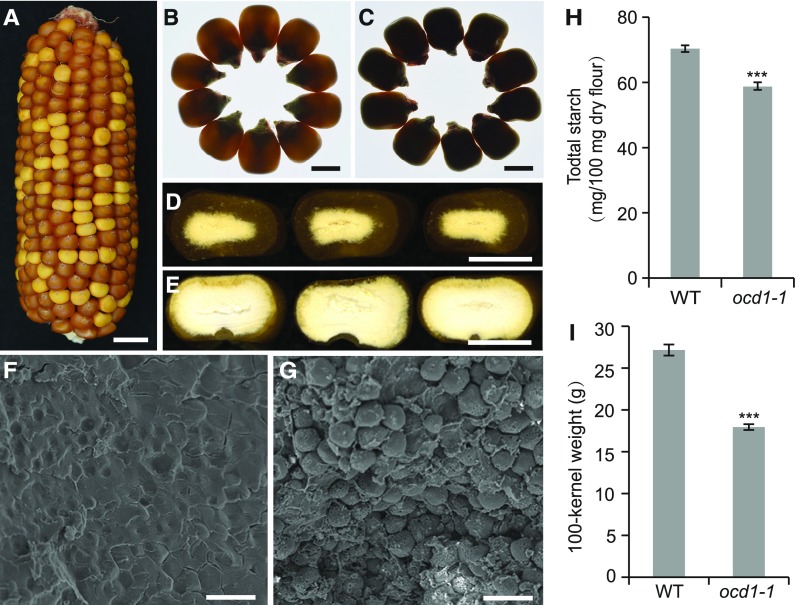
Figure 1.
Kernel Phenotypes of the Wild Type and ocd1-1 Mutant.
(A) A self-pollinated ear segregating opaque mutants.
(B) and (C) Mature kernels of the wild type (B) and ocd1-1 (C) observed in a light box.
(D) and (E) Cross sections of mature wild-type (D) and ocd1-1 (E) kernels.
(F) and (G) Scanning electron micrographs of the peripheral area of the mature wild-type (F) and ocd1-1 (G) endosperm.
(H) Quantification of the starch content in the wild-type and ocd1-1 mature seeds.
(I) 100-kernel weight of wild type and ocd1-1.
Bars = 1 cm in (A), 0.5 cm in (B) to (E), and 20 µm in (F) and (G). Error bars show sd from three biological replicates. ***P < 0.001 as determined by Student’s t tests.
We propagated the normal seeds from the segregating ear for several generations (F2, F3, and F4). In each generation, the normal and opaque seeds from the segregating ears segregated at a 3:1 ratio, indicating that this phenotype resulted from a single gene mutation. Normal seeds lacking spotting (spots are indicative of active transposition) from a nonsegregating ear were used as the wild type in the following experiments. When observed on the light box, the mutant seeds (F3) appeared completely nonvitreous (Figures 1B and 1C). The outer region of the wild-type endosperm cross section is vitreous (Figure 1D), whereas the ocd1-1 endosperm is entirely starchy (Figure 1E). Scanning electron microscopy observation showed that the starch granules in the outer region of the endosperm were likely embedded in a proteinaceous matrix in the wild-type seeds (Wu and Messing, 2010a) but were naked and loosely packed in the ocd1-1 seeds (Figures 1F and 1G). We then determined the starch content in dry seeds and kernel weight. The wild type contained 70.3 mg of starch per 100 mg of dry seed flour, and the starch content was comparatively reduced by 16.5% in ocd1-1 (58.7 mg per 100 mg) (Figure 1H). The 100-kernel weight of ocd1-1 was only 17.9 g, which is 33.9% lower than that of the wild type (27.1 g) (Figure 1I). These results suggest that the reduced starch content and insufficient endosperm filling are responsible for the decreased kernel weight in ocd1-1.
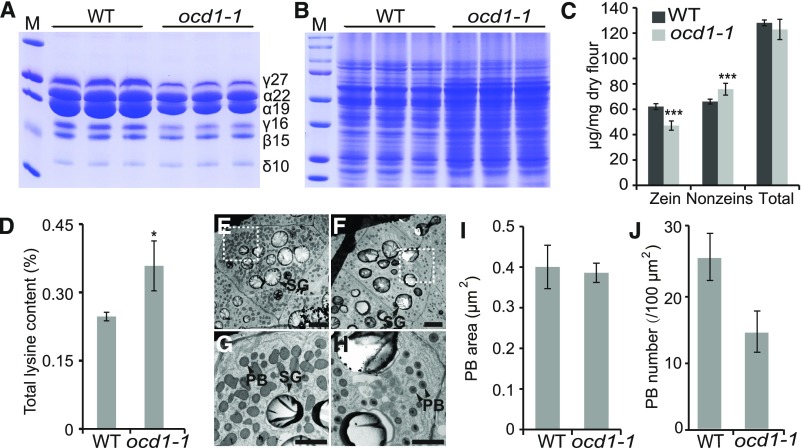
Zein proteins account for more than 60% of the total endosperm proteins and previous studies showed that endosperm opacity is often related to zein synthesis or organization in protein bodies (PBs; Larkins and Hurkman, 1978; Holding et al., 2007; Wu and Messing, 2010b; Zhang et al., 2015). To test this distribution, zein proteins extracted from wild-type and ocd1-1 seeds were analyzed by SDS-PAGE. As shown in Figure 2A, the accumulation of all zein proteins was decreased, and the 19-kD α-zein and 27-kD γ-zein were most affected in ocd1-1. Because of proteome rebalancing, the synthesis of other proteins (nonzeins) increased in ocd1-1 to compensate for the decreased levels of zeins (Figure 2B). Quantitative measurements revealed that the zein levels were significantly decreased and nonzein levels were increased in ocd1-1, which resulted in the total protein content not being apparently altered compared with that of the wild type (Figure 2C). Remarkably, the total lysine content, which is deficient in zein proteins, was significantly enhanced in ocd1-1 seeds (Figure 2D).
Figure 2.
Protein Contents and PBs in the Wild-Type and ocd1-1 Endosperm.
(A) and (B) SDS-PAGE analysis of zein (A) and nonzein (B) proteins in the wild type and ocd1-1. Total protein loaded in each lane was equal to 200 μg of maize flour. The size of each zein protein band is indicated beside it. M, protein markers in (A) from top to bottom correspond to 37, 25, 20, 15, and 10 kD and in (B) correspond to 250, 150, 100, 75, 50, 37, 25, and 20 kD.
(C) and (D) Quantification of zein, nonzein, total protein (C), and total lysine (D) contents in wild-type and ocd1-1 mature seeds.
(E) and (F) Ultrastructure of the 20-DAP endosperms of the wild type (E) and ocd1-1 (F). Bars = 10 μm.
(G) and (H) Enlarged images of boxed areas in (E) and (F). Bars = 1 μm.
(I) and (J) PB size (I) and number (J) in the 20-DAP endosperms of the wild type and ocd1-1.
Error bars in (C) and (D) show the sd calculated from at least three biological replicates. For the PB size and number analysis, more than 20 endosperm cells in the wild type and ocd1-1 were calculated. *P < 0.05 and ***P < 0.001. SG, starch granular.
Consistent with the protein accumulation patterns, an RT-qPCR assay (Supplemental Figure 1) revealed that the transcript levels of most zein genes (especially those encoding the z1A, z1B, and z1D 19-kD α-zeins) were markedly decreased in the ocd1-1 endosperm at 20 days after pollination (DAP), when the storage products were actively synthesized for endosperm filling. However, transcripts of the gene encoding the 27-kD γ-zein were not downregulated, indicating that its protein reduction in ocd1-1 is not regulated at the transcriptional level (Supplemental Figure 1). PBs derived from the rough endoplasmic reticulum included the organelle for the synthesis and storage of zein proteins. A transmission electron microscopy analysis of the developing endosperm at 20 DAP determined that the size of the PBs was not apparently altered in ocd1-1, although the number of PBs was significantly fewer compared with that in the wild type (Figures 2E to 2J).
In addition to defects in the endosperm, the ocd1-1 mutant displayed abnormalities in development, with ocd1-1 seeds exhibiting a dramatic reduction in germination rate (only 30%) in the greenhouse while the wild-type seeds germinated normally (90%) (Supplemental Figure 2A). The ocd1-1 seedlings were obviously smaller and presented a shorter root length than the wild type (Supplemental Figures 2B to 2D). The ocd1-1 plants were difficult to reproduce. These observations suggest that the ocd1-1 mutation has pleiotropic effects on plant development.
Cloning of the Mutant Gene
We created an F2 population with B73 and ocd1-1 for map-based cloning. Taking advantage of single-nucleotide polymorphism markers and 21 individuals, the ocd1-1 locus was located on chromosome 8L (Figure 3A). We then used SSR (simple sequence repeat) and Indel (insertion and deletion) markers for a fine mapping. Using 597 individuals, the Ocd1 gene was narrowed down to a 210-kb region between markers SSR17.80 and SSR18.21 (Figure 3A). Based on the B73 genome in the MaizeGDB database, four protein-coding genes (GRMZM2G080722, GRMZM2G080521, GRMZM2G175253, and GRMZM2G175171) were annotated in this interval (Figure 3A). PCR amplification and sequencing determined that GRMZM2G175171 from the mutant bears a 1.4-kb insertion in the first exon (+297 bp relative to the start codon) and the other three genes were identical to their wild-type counterparts. A sequence BLAST revealed that this 1.4-kb DNA fragment was a Mutator element (Mu8) (Figure 3B). To determine the relationship between the phenotype and genotype, four primers were designed. The P1 and P2 primers were specific to GRMZM2G175171, whereas 810F and 810R were specific to Mu8. Because of the Mu8 insertion in GRMZM2G175171, the combinations of P1+810R and P2+801F were expected to specifically amplify the sequences flanking the left and right junction sites, respectively. Based on this designation, if a homozygous seed for this insertion was examined via PCR, then the primer pairs P1+810R and P2+801F but not P1+P2 would generate the correct band. Our results revealed that all examined opaque seeds (n = 48) were homozygotes for this insertion, whereas wild-type seeds (n = 48) from a nonsegregating ear had no insertion in this gene. A representative PCR amplification is shown in Figure 3C. These results confirmed that the disrupted GRMZM2G175171 is associated with the mutant phenotype.
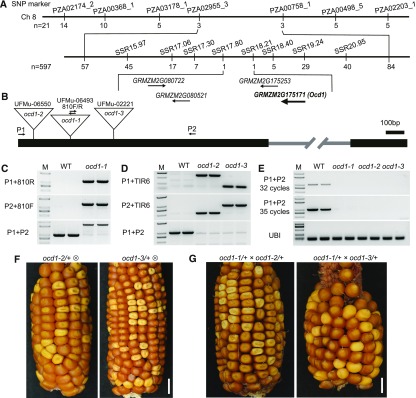
Figure 3.
Map-Based Cloning and Genetic Confirmation of ocd1.
(A) Fine mapping of ocd1 using the F2 population of ocd1 and B73. The numbers under each bar indicate the number of recombinants identified by the corresponding molecular marker.
(B) Schematic representation of the Ocd1 gene structure with mutant alleles indicated. The black bold line and gray line indicate exons and introns, respectively. The triangles indicate Mu insertions in the three alleles. The indicated primers were used to characterize the mutant as shown in (C) to (E).
(C) Identification of the Mu insertion in ocd1-1. The primers were used as indicated in (B).
(D) Identification of the Mu insertion in ocd1-2 and ocd1-3. The suggested TIR6 was used as the insertion-specific primer.
(E) RT-PCR analysis of the Ocd1 expression. The Ubi gene was used as an internal control.
(F) Self-pollinated ears of ocd1-2/+ (left panel) and ocd1-3/+ (right panel). Both ears segregate opaque seeds. Bars = 1 cm.
(G) Ears of a heterozygous ocd1-1/+ plant pollinated by ocd1-2/+ (left panel) and ocd1-3/+ (right panel) pollen. Bars = 1 cm.
To further confirm that this mutation is responsible for the opaque phenotype, we performed a reverse genetic screen for additional Mu-inserted alleles. Two stocks, UFMu-06550 and UFMu-02221, were identified to bear a Mu insertion in the first exon of GRMZM2G175171, creating two additional mutant alleles (thus designated ocd1-2 and ocd1-3). PCR amplification and sequencing positioned the Mu in ocd1-2 and ocd1-3 at +94 and +540 bp, respectively (Figure 3B). Ears from self-pollinated heterozygous plants of the two alleles segregated normal and opaque seeds at a 3:1 ratio (Figures 3F). We determined the genotypes of 48 opaque seeds from each of the self-pollinated ocd1-2/+ and ocd1-3/+ and found that they were all homozygous for the Mu insertion (the primer TIR6 recommended by MaizeGDB was based on the terminal inverted repeats in the Mu element) (Figure 3D). The RNA transcripts of Ocd1 were detected in the wild type but not in the ocd1-1, ocd1-2, and ocd1-3 plants with the P1 and P2 primers (Figure 3E), indicating that these three knock-out alleles are not functional.
We then analyzed the zeins and nonzeins in ocd1-2 and ocd1-3 endosperms and revealed that they displayed similar protein accumulation patterns to that in ocd1-1 (Supplemental Figure 3). To perform the allelic test, the pollen of heterozygous ocd1-2/+ and ocd1-3/+ plants was pollinated to the ear of ocd1-1/+. The resulting ears segregated normal and opaque seeds at a 3:1 ratio (Figure 3G). These genetic data showed that GRMZM2G175171 is the gene responsible for the ocd1 phenotype.
Ocd1 Is Generally Expressed
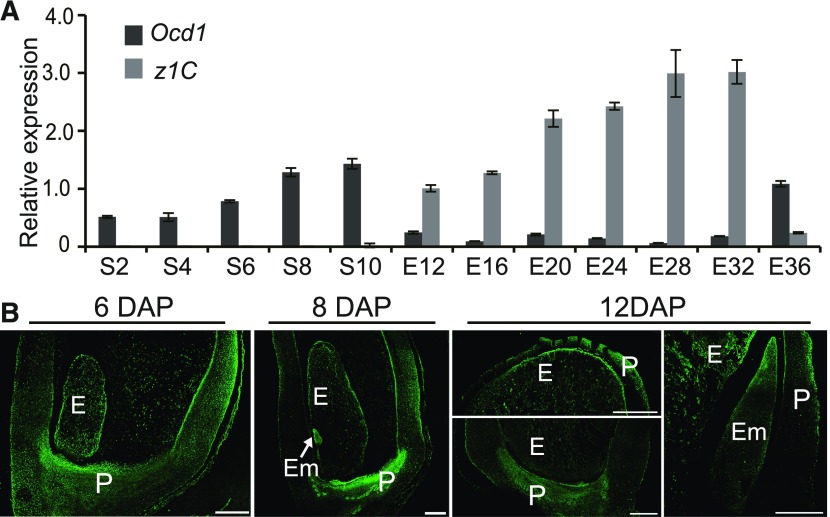
According to the maize eFP browser (http://www.maizegdb.org/gene_center/gene/GRMZM2G175171) and published RNA-seq data (Chen et al., 2014), Ocd1 is widely expressed in maize tissues (Supplemental Figure 4A). Indeed, real-time PCR detected transcripts of Ocd1 in different tissues at varying levels (Supplemental Figure 4B). We further investigated the expression profile of Ocd1 during seed development. Its expression was gradually increased and reached a peak at 10 DAP. Thereafter, the endosperm was easily separated from the embryo and pericarp. Basically, the levels of the Ocd1 transcripts decreased throughout endosperm development beginning from 12 to 32 DAP. At 36 DAP, Ocd1 expression increased again to a level comparable to that before 12 DAP (Figure 4A) (Chen et al., 2014). As a comparison, the endosperm-specific z1C genes encoding the 22-kD α-zein proteins were investigated. They began to express from 10 DAP and reached a maximum level at 32 DAP. At 36 DAP, their transcripts decreased to a basal level.
Figure 4.
The Transcript and Protein Expression Pattern of Ocd1 during Seed Development.
(A) RT-qPCR analysis of Ocd1 expression during seed development. z1C was used as an endosperm-specific marker. All expression levels are normalized to that of Ubi. Three replicates for each RNA sample were made and error bars represent ±sd.
(B) Immunofluorescent assay of the OCD1 distribution during seed development.
E, endosperm; Em, embryo; P, pericarp, S, seed. Bars = 500 μm.
To understand the OCD1 distribution pattern during seed development, the polyclonal antiserum was raised against the OCD1 protein. The specificity of the antibodies was tested by immunoblot. The single primary band representing the OCD1 protein was present in the wild type but missing in ocd1-1 (Supplemental Figure 5). Immunofluorescence experiments showed that the OCD1 protein was detected in the endosperm, embryo, and pericarp from 6 to 12 DAP seeds (Figure 4B). It appears that the OCD1 protein accumulated at a higher level in the peripheral region of the endosperm cells.
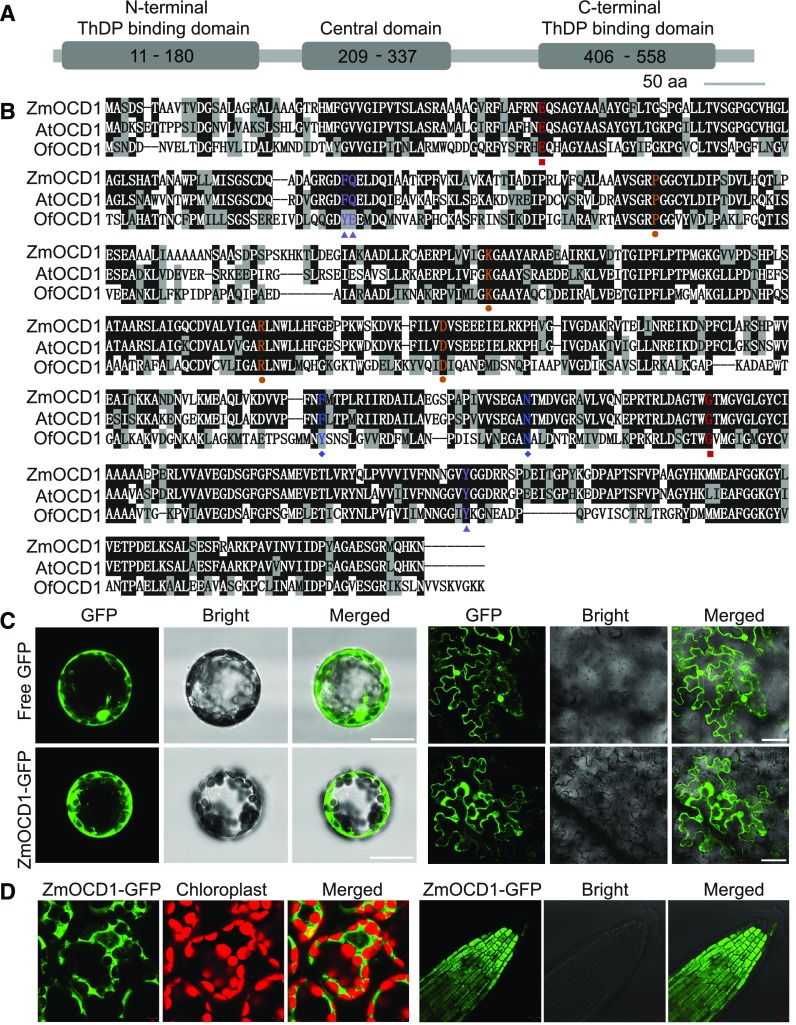
Domain and Sequence Analysis of the ZmOCD1 Protein
Ocd1 is composed of two exons and one intron, and it is predicted to encode a 575-amino acid protein. Pfam searches (http://pfam.xfam.org/search) using the OCD1 amino acid sequence indicated that this protein contains three conserved domains, an N-terminal thiamine pyrophosphate (ThDP) binding domain, a central ThDP and magnesium (Mg) ion binding domain, and a C-terminal ThDP binding and catalytic activity domain (Figure 5A). To infer the function of the OCD1 protein, public databases (NCBI) were searched using BLASTP with its protein sequence as a query. This search identified OCD1 homologs in Arabidopsis (AtOCD1) and O. formigenes (OfOCD1). ZmOCD1 shares 83% and 62% similarity with AtOCD1 and OfOCD1, respectively.
Figure 5.
Domain, Sequence, and Subcellular Localization of ZmOCD1.
(A) Schematic of the ZmOCD1 protein with conserved domains indicated. aa, amino acids.
(B) Alignment of the ZmOCD1 protein and its homologs from Arabidopsis and O. formigenes. The key amino acids for enzyme functions are indicated by different colored shapes. Red squares, ThDP binding sites; orange circles, ADP binding sites; blue diamonds, Mg binding sites; and purple triangles, active-site residues.
(C) Transient expression of the free 35S:GFP and 35S:ZmOCD1-GFP fusion (left panel) in Arabidopsis mesophyll protoplast cells and in tobacco leaf epidermal cells (right panel). Bars = 25 µm.
(D) Subcellular localization of ZmOCD1 in 35S:ZmOCD1-GFP transgenic plants. The GFP fluorescence in cotyledon (left panel) and root tip (right panel) cells from the T2 plants was observed.
A structural analysis of OfOCD1 has characterized the sites required for ThDP, ADP and Mg binding, and other active site residues (Berthold et al., 2005). We compared the OCD1 amino acids and showed that the ThDP binding sites and ADP binding sites are identical in OfOCD1, AtOCD1, and ZmOCD1 (Figure 5B). The two Mg binding sites and the three active site residues of OfOCD1 were identical or replaced with the similar amino acids in Arabidopsis and maize (Figure 5B). The high similarity and conservation of key amino acid sites in prokaryotic and eukaryotic OCD proteins suggests that ZmOCD1 may function as an oxalyl-CoA decarboxylase in oxalate degradation.
ZmOCD1 Is Located in the Cytoplasm
To determine its subcellular localization, the ZmOCD1 protein was fused to GFP. Free GFP was used as a control. Both gene cassettes were driven by the constitutive 35S promoter. The resulting constructs were first transiently expressed in Arabidopsis leaf mesophyll protoplasts. In contrast to the signal from 35S:GFP, which was detected both in nuclei and the cytoplasm, the signal from 35S:ZmOCD1-GFP appeared to be restricted to the cytoplasm (Figure 5C). Similar cytosol localization of 35S:ZmOCD1-GFP was observed in tobacco (Nicotiana benthamiana) leaves, which were injected with Agrobacterium tumefaciens harboring this construct (Figure 5C). To further determine ZmOCD1 localization, stable transgenic Arabidopsis plants were generated with this construct. Confocal observation of cotyledon and root tip cells from the T2 plants showed that ZmOCD1 was most likely a cytoplasmic protein as well (Figure 5D). It is still intriguing to investigate whether a small portion of ZmOCD1 is able to locate to mitochondria or peroxisomes.
ZmOCD1 Catalyzes the Breakdown of Oxalyl-CoA into Formyl-CoA and CO2
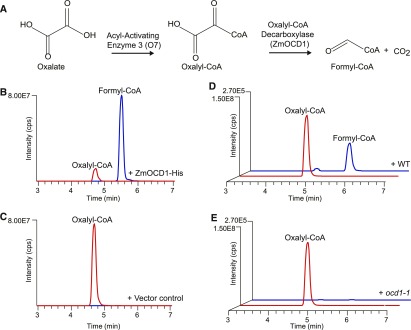
AAE3 catalyzes the reaction that ligates oxalate and CoA to form oxalyl-CoA, which becomes the substrate for oxalyl-CoA decarboxylase (Figure 6A). To investigate whether ZmOCD1 indeed functions as an oxalyl-CoA decarboxylase in the oxalate degradation pathway, a 6×histidine (His)-tagged ZmOCD1 (ZmOCD1-His) was constructed and purified in Escherichia coli. The ZmOCD1-His and the purified vector control proteins (Supplemental Figure 6A) were examined for the ability to degrade oxalyl-CoA. Due to the lack of commercially available oxalyl-CoA, we chemically synthesized this compound from acyl chloride through several steps of reactions as previously described (Quayle, 1962). The resulting oxalyl-CoA was determined to have a precise molecular weight (m/z+ 840.1, positive mode) and corresponding fragmentation patterns (Supplemental Figures 6B and 6C) via liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) analysis. The purified ZmOCD1-His and vector control proteins were incubated with the synthesized oxalyl-CoA and cofactors. After incubation for 15 min, the products were subjected to LC-MS/MS analysis. In addition to the peak corresponding to the substrate oxalyl-CoA (m/z+ 840.1), ZmOCD1-His produced a new peak (m/z+ 796.1, positive mode) representing a lower molecular mass of formyl-CoA (Figure 6B). LC-MS/MS analysis of the new peak product revealed a fragmentation pattern consistent with formyl-CoA (Supplemental Figure 6D). The weight difference between the two peaks is equal to the loss of a CO2 (m/z 44.0), suggesting degradation of oxalyl-CoA into formyl-CoA and CO2 by ZmOCD1-His. However, the incubation with the vector control only showed a peak that corresponded to the molecular mass (m/z+ 840.1) of the substrate oxalyl-CoA rather than the product formyl-CoA (m/z+ 796.1), indicating that catalysis did not occur (Figure 6C).
Figure 6.
Biochemical Function of Recombinant ZmOCD1.
(A) Partial pathway for oxalate degradation. The first two steps of oxalate degradation are proposed to be catalyzed by O7 and ZmOCD1.
(B) and (C) In vitro enzyme activity analysis. Purified recombinant ZmOCD1 (B) or the empty vector control (C) was incubated with oxalyl-CoA and the cofactors.
(D) and (E) In vivo enzyme activity analysis. 20-DAP endosperm extracts of the wild type (D) or ocd1-1 (E) were incubated with oxalyl-CoA and the cofactors.
Product formation was analyzed by LC-MS/MS. The relative intensities of the LC-MS/MS transitions of oxalyl-CoA (m/z+ = 840.1, red line) and formyl-CoA (m/z+ = 796.1, blue line) are shown. The LC-MS/MS analyses were performed under the positive mode (M/H)+.
We next assayed the oxalyl-CoA decarboxylase activity in vivo by using homogenates from wild-type and ocd1-1 endosperms at 20 DAP (Supplemental Figure 6E). The reactions were performed as described above. When the wild-type endosperm extract was added, the peaks corresponding to the substrate oxalyl-CoA and product formyl-CoA were detected, although the amount of formyl-CoA was much lower than that produced by ZmOCD1-His (Figure 6D). In contrast, no formyl-CoA was detected by incubation with the ocd1-1 endosperm extract, indicating that the extract from the ocd1-1 endosperms is deficient in the oxalyl-CoA decarboxylase activity (Figure 6E). These biochemical data collectively demonstrated that ZmOCD1 is the oxalyl-CoA decarboxylase that catalyzes the breakdown of oxalyl-CoA into formyl-CoA and CO2.
Maize O7 is a homolog of AtAAE3, which functions as an oxalyl-CoA synthetase. o7 showed an opaque endosperm as observed in ocd. Maize o7 was cloned earlier than aae3 in Arabidopsis (Miclaus et al., 2011; Wang et al., 2011), although the biochemical function of O7 had not been experimentally confirmed. We also expressed and purified the O7 protein (Supplemental Figure 6A) and performed the enzymatic analysis using oxalate as the substrate. The LC-MS/MS analysis showed that the O7 protein produced a new peak corresponding to the molecular mass of oxalyl-CoA (m/z− 838.1, negative mode). The production was proportional to the supplied oxalate concentration (Supplemental Figure 7A). LC-MS/MS also revealed a fragmentation pattern consistent with oxalyl-CoA (Supplemental Figure 7B). Based on these analyses, we concluded that ZmOCD1 functions downstream of O7 for oxalate degradation in maize.
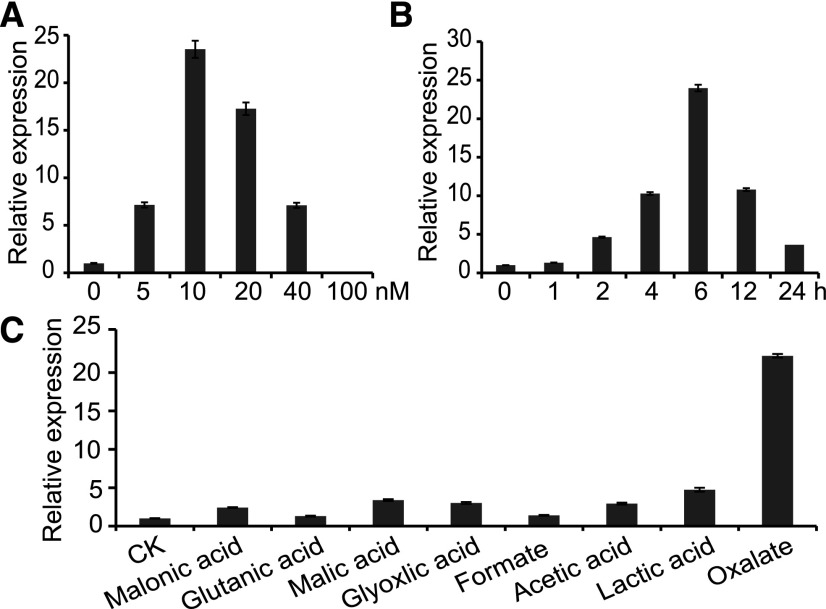
Ocd1 Is Specifically Induced by Oxalate
To investigate whether Ocd1 responds to exogenous oxalate treatment, wild-type seeds were germinated and grown vertically on a moist paper bag. Seven-day-old seedlings were water-cultured in the following treatments. First, we tested the induction of Ocd1 with different oxalate concentrations for 6 h. As shown in Figure 7A, the expression of root Ocd1 was greatly induced by external oxalate treatments, although 10 mM oxalate led to maximum induction. Higher oxalate concentrations may result in more severe stress or toxicity to plant cells and as a consequence, they may suppress gene expression. Second, we tested induction with 10 mM oxalate in a time-course study. The RNA transcripts of Ocd1 gradually increased over time and reached a maximum at 6 h. The expression decreased thereafter (Figure 7B). Third, we tested the specificity of induction with different organic acids (10 mM), including formate, acetic acid, lactate, and oxalate. Water was used as a negative control. Our results showed that the oxalate-induced expression reached a level more than 20-fold higher than that in the control, whereas other inductions were not comparable to that of oxalate (Figure 7C). These analyses demonstrated that Ocd1 specifically responds to oxalate treatment.
Figure 7.
Oxalate-Induced Expression of Ocd1 in Roots.
(A) Ocd1 expression is induced by different oxalate concentrations.
(B) Ocd1 expression is induced by different times.
(C) Ocd1 expression is induced by different organic acids.
The wild-type roots were treated with different conditions as indicated. All data were normalized relative to the expression of Ubi. The values are indicated as the means ± sd of three independent treatments.
Oxalate Is Overaccumulated in ocd1 and o7 Mutants
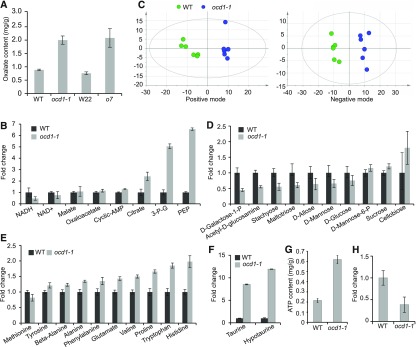
Due to lack of oxalyl-CoA decarboxylase in ocd1 mutants (Figure 6E), we inferred that they had a defect in oxalate metabolism. To test this inference, oxalate contents were measured in the mature wild-type and ocd1-1 dry seeds by a coupled enzyme reaction. The seeds o7 and its wild-type counterpart (W22) were also analyzed. The oxalate content in o7 (4.6 mg of oxalate/g dry flour) lacking ZmAAE3 was ∼2-fold higher than that in the wild type (2.2 mg of oxalate/g dry flour). We also observed that ocd1-1 accumulated 2-fold higher oxalate levels than the wild type (Figure 8A). These results suggest that oxalate degradation was disrupted in the o7 and ocd1-1 mutants.
Figure 8.
Oxalate Contents and Metabolome Analysis in the Wild Type and ocd1.
(A) Measurement of oxalate contents in mature wild-type, W22, o7, and ocd1-1 seeds. Error bars represent ±sd from three biological replicates.
(B) Significant fold changes of metabolites in wild-type and ocd1-1 20-DAP endosperms revealed by targeted metabolomics analyses.
(C) Principal component analysis of nontargeted metabolomes. Metabolites were extracted from six individual endosperms at 20 DAP and examined under positive and negative mode.
(D) Relative contents of variant saccharides with significant fold changes in ocd1-1 compared to the wild type.
(E) Relative contents of variant amino acids with significant fold changes in ocd1-1 compared to the wild type.
(F) Relative contents of taurine and hypotaurine in the wild type and ocd1-1.
(G) Concentrations of ATP in the wild type and ocd1-1. Levels of ATP were quantified enzymatically.
(H) Relative content of indole acetic acid in the wild type and ocd1-1.
Levels of metabolites (except for ATP) in wild type were designated 1. The error bars indicate ±sd from six biological replicates.
Metabolome Alterations in the ocd1 Endosperm
To test the metabolic alterations caused by the Ocd1 mutation, we first performed a targeted metabolomics analysis with 20-DAP endosperms by LC-MS/MS. In this assay, a total of 25 metabolites involved in the energy metabolism process were identified, among which eight displayed a significant change in their contents (Supplemental Data Set 1). The levels of NAD+ and NADH, the oxidized and reduced forms of nicotinamide adenine dinucleotide, were reduced by 24.1% and 53.4%, respectively, compared with those in the wild type (Figure 8B). Citrate, the first intermediate product produced in the tricarboxylic acid (TCA) cycle, was increased by 1.4-fold compared with that in the wild type. Two other TCA cycle intermediates (malate and oxaloacetate) were also increased. Phosphoenolpyruvate (PEP) and 3-phospho-glycerate (3-P-G) from the glycolysis pathway showed the highest elevation among the six increased metabolites. Cyclic adenosine monophosphate (cyclic AMP) was elevated by 28% in ocd1-1 compared with that in the wild type (Figure 8B). However, some other TCA cycle and glycolic metabolites such as fumarate, succinate, and pyruvate, were not observed to have an apparent alteration in ocd1-1 (Supplemental Data Set 1).
Due to the limited number of standard chemicals for the targeted metabolomics analysis, we further performed a nontargeted analysis using the LC-MS/MS (Supplemental Data Set 2). Endosperms of the wild type and ocd1-1 at 20 DAP were harvested for the extraction of metabolites. Principal component analysis showed that the six biological replicates of each sample were clustered into one group under the positive and negative model (Figure 8C). The statistical analysis identified multiple significant changed metabolites in the ocd1-1 endosperm. The contents of PEP and 3-P-G in ocd1-1 were remarkably enhanced compared with those in the wild type. The fold changes were similar with those in the targeted metabolomics analysis. Levels of multiple saccharides and their derivatives such as d-glucose, d-galactose 1-phosphate, stachyose, maltotriose, d-allose, d-mannose, and N-acetyl-d-glucosamine showed a significantly reduced level, while sucrose, d-mannose-6-phosphate, and cellobiose in ocd1 were elevated compared with the levels in the wild type (Figure 8D). Eight free amino acids (aspartate, tyrosine, alanine, phenylalanine, glutamate, valine, tryptophan, and histidine) were also elevated in the mutant endosperm whereas methionine was reduced by 18% (Figure 8E). Several other small molecular metabolites also showed altered contents. Taurine, a supplement for human health, and its biosynthetic intermediate hypotaurine, were ranked as the top two increased metabolites in ocd1-1 (Figure 8F). The energy charge (ATP) was also enhanced in ocd1-1, which displayed a more than 2-fold increase compared with that in the wild type (Figure 8G). By contrast, the plant hormone auxin (indole-3-acetic acid) showed more than 60% decrease in ocd1-1 (Figure 8H). These results collectively demonstrated that a large perturbation in the metabolome is involved in multiple pathways in the ocd1-1 endosperm.
DISCUSSION
Discovery of Genes for Oxalate Catabolism in Plants
Oxalate is a widespread organic acid that is synthesized by organisms ranging from prokaryotic to eukaryotic kingdoms. Due to its relatively strong acidity and chelating ability, excessive accumulation of this acid is toxic in vivo and negatively impacts cell growth and organ development. Plants have evolved several pathways to metabolize oxalate. The best characterized pathway is via oxalate oxidase, which directly converts oxalate into CO2 and H2O2 (Lane et al., 1993), although certain species, such as Arabidopsis, do not present oxalate oxidase activity. An alternative pathway for the degradation of oxalate has been proposed. This pathway involves four enzymesoxalyl-CoA synthetase, oxalyl-CoA decarboxylase, formyl-CoA hydrolase, and formate dehydrogenase. Maize o7 is a classic mutant that exhibits an opaque phenotype in the endosperm. The o7 mutant was first described nearly half a century ago and characterized as having significantly more lysine due to a general reduction in zein levels and a compensatory increase in nonzeins. Two other high-lysine mutants were characterized at that time, o2 and fl2. o2 was determined to encode a bZIP transcription factor regulating most zein gene expression (Schmidt et al., 1992), and fl2 was found to be caused by a defective signal peptide on a 22-kD α-zein protein (Coleman et al., 1997). By a simple implication, the involvement of O7 in the synthesis of zein proteins was inferred. The o7 gene was eventually cloned by two independent labs (Miclaus et al., 2011; Wang et al., 2011). BLAST searches using the O7 protein sequence indicated that O7 belonged to the acyl-CoA synthetase superfamily and presented the greatest similarity to AAE3 based on the phylogenetic tree (Wang et al., 2011). An analysis of amino acids and metabolites suggested that O7 might affect amino acid biosynthesis, although the exact function had not been elaborated. Later, the Arabidopsis homolog AtAAE3 was cloned and shown to encode an oxalyl-CoA synthetase, which catalyzes the ligation of oxalate and CoA into oxalyl-CoA (Foster et al., 2012). The aae3 mutants lacked oxalyl-CoA synthetase activity and accumulated a 3-fold higher level of oxalate than the wild type. This finding represented a breakthrough insight into oxalate metabolism in plants. The final step involved in formate catabolism catalyzed by formate dehydrogenase has also been reported (Suzuki et al., 1998; Lou et al., 2016b).
In this study, we isolated an opaque mutant, ocd1. Based on map-based cloning and allelic complementation, the causal gene was determined to be GRMZM2G175171, which was predicted to encode an oxalyl-CoA decarboxylase and therefore designated Ocd1. It was proposed that oxalyl-CoA decarboxylase catalyzed the degradation of oxalyl-CoA, the product of AAE3, into formyl-CoA and CO2 (Foster et al., 2012, 2016; Foster and Nakata, 2014). A series of evidence supported the function of ZmOCD1 as an oxalyl-CoA synthetase. First, although ZmOCD1 shared only 42% identity with the characterized OfOCD1, the functional key sites required for ThDP, ATP, and Mg binding, and other active site residues were highly conserved (Figure 5A). Second, the expression of Ocd1 was specially induced by oxalate (Figure 7). Third, we experimentally demonstrated that the recombinant OCD1 protein catalyzed the breakdown of oxalyl-CoA into formyl-CoA and CO2 (Figure 6B). Finally, no oxalyl-CoA decarboxylase activity was detected and oxalyl-CoA failed to be degraded in the ocd1-1 endosperms (Figure 6E).
In human and rat, the OCD1 homolog (42% and 41% identity, respectively) was named 2-hydroxyacyl-CoA lyase (2-HPCL). OCD1 and 2-HPCL display several similarities including the ability to bind Mg and ThDP as cofactors and produce the same product, formyl-CoA. However, OCD1and 2-HPCL used different substrates for catalysis. 2-HPCL catalyzes the carbon-carbon bond cleavage of 3-methyl-branched fatty acids and 2-hydroxy straight-chain fatty acids, such as phytanic acid, during α-oxidation (Foulon et al., 1999; Casteels et al., 2007). A possible explanation for the diverged function between ZmOCD1 and 2-HPCL is their different subcellular localization. 2-HPCL is localized in the peroxisome (Foulon et al., 1999), whereas ZmOCD1 is mainly, if not all, located to the cytoplasm (Figures 5C and 5D). Moreover, humans have evolved a special strategy to dispose of oxalate by exploiting the gut bacteria O. formigenes (Stewart et al., 2004; Siener et al., 2013), which degrades oxalate via the same AAE3- and OCD1-mediated pathway.
Developmental Defects and Reduced Endosperm Storage Reserves in ocd1 Mutants
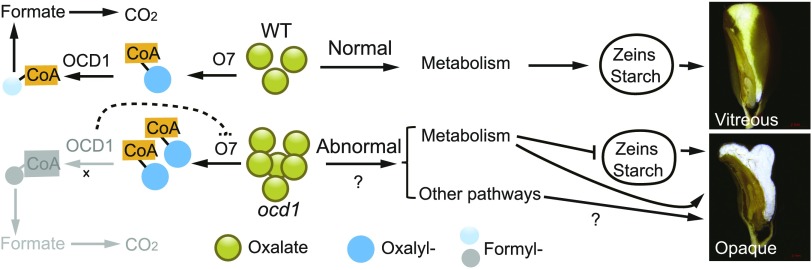
The ocd1 seeds were first characterized based on their opaque phenotype (Figures 1A to 1C). In the wild-type kernel, the outer region of the endosperm is vitreous and the inner area is starchy (Figure 1D). In the vitreous endosperm, spaces between starch granules are tightly filled with a proteinaceous matrix primarily composed of PBs (Wu and Messing, 2010a). PBs represent the organelle for the synthesis and storage of zein proteins. Each subfamily of zeins is proposed to play a different role in PB formation. The γ- and β-zeins are expressed slightly earlier than the α- and δ-zeins and likely initiate PB formation, whereas the α- and δ-zeins are subsequently deposited into the inner region of the PBs to enlarge their size. Reduced zein synthesis and opaque (floury) phenotype have been observed in several maize mutants. The classic opaque mutant o2 affects the transcription of most zein genes (Schmidt et al., 1992; Zhang et al., 2015; Yang et al., 2016). In fl3, the contents of the storage protein zeins were decreased and the endosperm was floury (Li et al., 2017). In addition, the direct suppression of γ-, β-, or α-zein expression resulted in a reduced PB number or size, and opaque phenotype was subsequently produced in the endosperm (Wu and Messing, 2010b; Guo et al., 2013). Similar to o7, ocd1 accumulated overtly decreased amounts of zein proteins (Figure 2A). ocd1 appears to have a differential effect on zein gene expression. As displayed via SDS-PAGE, different zein proteins were not evenly decreased, with 19-kD α-zeins and 27-kD γ-zein the most affected (Figure 2A). However, in contrast to other zein genes, RNA transcripts encoding 27-kD γ-zein were not correspondingly reduced (Supplemental Figure 1), suggesting that ocd1 affects its expression only at the translational level. There were fewer PBs in o7 and ocd1 than in the wild type (Figure 2J; Wang et al., 2011). Therefore, the opaque phenotype in ocd1 is closely related to the reduction of zein proteins and PB number, although the mechanism by which ocd1 affects the synthesis of zein proteins remains unclear. It is unlikely that deficiency in oxalyl-CoA decarboxylase or consequent overaccumulation of oxalate has a specific or direct effect on zein protein synthesis, since the starch content was also reduced in ocd1-1 seeds (Figure 1H). As a strong metal chelator, the overaccumulation of oxalate may cause disrupted metal homeostasis and in turn affect functions of different organelles and zein protein and starch synthesis in the ocd1 endosperm.
Although O7 functions in the same pathway and acts upstream of ZmOCD1, o7 appeared to have a different phenotype compared with ocd1. The penetrance and expressivity of the opaque phenotype in o7 is genetic-background dependent. When o7 was outcrossed to other lines, the ratio of opaque kernels on ears generated by self-pollination was often much lower than the expected 25% (Miclaus et al., 2011). However, the deviation from Mendelian segregation was not observed in ocd1. Although o7 presents poor seed germination, like ocd1, the mature plants flower and have a seed setting rate comparable to that of the wild type. In contrast to the three null ocd1 alleles (Figure 3E), which all resulted from a Mu insertion in the first exon near the N terminus, the o7-ref allele bore a 12-bp in-frame deletion in the coding region, which significantly reduced rather than abolished the accumulation of the o7 protein. A second, Ds-tagged o7 allele had the insertion near the C-terminal end. However, whether the two alleles were weak alleles that had a partial function was not clarified.
Mutation in ocd1 Affects the Endosperm Metabolome
The endosperm is a tissue with highly active metabolic pathways that are mainly dedicated to synthesis of storage proteins and starch. We showed that the ocd1 endosperm was impaired in several pivotal metabolic pathways, such as glycolysis and the TCA cycle. Glycolysis is a series of reactions catalyzed by several enzymes, which convert glucose into pyruvate and produce ATP for living cells. Our targeted and nontargeted metabolomics analysis revealed that PEP and 3-P-G in the ocd1-1 endosperm, two critical intermediates for glycolysis, accumulated to levels that were 5- and 6-fold higher than that of the wild type (Figure 8B), suggesting that the glycolysis in ocd1 is subject to a large perturbation. The TCA cycle catalyzes the oxidation of acetyl-CoA into CO2 and generates ATP and the reducing agent NADH, which can be used in numerous other biochemical reactions. Oxaloacetate, a precursor for oxalate, is also a metabolic intermediate in the TCA cycle. We observed an increased level of oxaloacetate and citrate in ocd1 (Figure 8B), suggesting the TCA cycle is substantially altered. It is likely that overaccumulation of oxalate in ocd1 causes a feedback response in the TCA cycle. Moreover, transferring the increased ATP in ocd1 for the synthesis of storage products does not appear to be possible because its kernel weight is much less than that of the wild type. The altered levels of NADH and NAD+ suggest that the redox state of the ocd1-1 endosperm was imbalanced, which may in turn affect other metabolic reactions that use NADH as cofactors.
Pyruvate phosphate dikinase (PPDK) reversibly converts AMP, pyrophosphate, and phosphoenolpyruvate to orthophosphate, pyruvate, and ATP. Mutations in pdk1 and pdk2 (encoding PPDK) resulted in essentially doubling of the steady state concentration of multiple glycolytic intermediates including PEP and hexose phosphates (Lappe et al., 2018). The complete PPDK knockout seeds exhibited an opaque phenotype. However, both starch and zein contents in PPDK deficiency seeds were not apparently affected. In this scenario, it is possible that the opaque phenotype is independent of the reduction in zein (Lappe et al., 2018). The OCD1 deficiency results in large metabolic perturbations in the endosperm (Figure 8) and consequently may change the quality of the endosperm cells as they mature. Although the zein protein reduction is a direct causal factor for the opaque phenotype (Wu and Messing, 2010a, 2011, 2017; Guo et al., 2013; Zhang et al., 2015), it is likely that a combination of altered metabolite fluxes, at least indirectly, contribute to the opacity in the ocd1 endosperm.
In summary, our study cloned a gene that encodes the oxalyl-CoA decarboxylase in maize. We demonstrated that this enzyme acts downstream of O7 and catalyzes the breakdown of oxalyl-CoA into formyl-CoA and CO2, which filled the gap in the second step of oxalate catabolism in plants. Our results showed that lack of ZmOCD1 leads to failure to break down oxalyl-CoA, which may impose negative feedback on the function of O7. This negative feedback results in excessive accumulation of oxalate in the ocd1 endosperm. We also showed that buildup of oxalate causes dramatic perturbations in a wide variety of metabolites. Since the changes are so extreme, identifying direct connections between the oxalate accumulation, altered metabolites, storage synthesis, and the final seed phenotype is challenging. We propose that these changes may somehow directly lead to the opaque phenotype or indirectly through affecting the synthesis of storage zein proteins and starch in the ocd1 endosperm (Figure 9). It will be important to investigate the mechanisms by which the oxalate levels affect metabolism in these complex ways. Oxalate has also been reported to be implicated in the infection of fungal pathogens and in the tolerance to lead and Al toxicity in some plant species (Yang et al., 2000, 2008; Guimarães and Stotz, 2004; Dong et al., 2008). It will be also interesting to investigate the functions of O7 and ZmOCD1 in the metal stress and pathogen resistance in the future.
Figure 9.
Proposed Model for ZmOCD1 Function and the Opaque Phenotype.
Due to the loss of function of oxalyl-CoA decarboxylase in ocd1, the reactions downstream of oxalyl-CoA are blocked (in gray). The dashed line indicates putative a feedback inhibition, which results in an excessive accumulation of oxalate in the ocd1 endosperm. The altered metabolites in the ocd1 endosperm may cause reduced synthesis of zeins and starch. The former directly affects the vitreous endosperm formation and the latter leads to the kernel weight loss. Dramatic metabolome perturbations, as well as other unidentified pathways, may contribute to the opaque phenotype in ocd1 kernels in a way that is independent of the zein protein decrease.
METHODS
Genetic Materials
The maize (Zea mays) mutants were obtained from the Maize Genetics Cooperation stock center under the accession numbers UFMu06493 or mu1051305 (ocd1-1), UFMu-06550 or mu1054504 (ocd1-2), and UFMu-02221 or mu1016386 (ocd1-3). The o7 seeds were obtained from Joachim Messing’s lab at Rutgers University as previously described (Miclaus et al., 2011). These genetic materials were planted in Shanghai, China in summer and Sanya, China in winter. For the germination assay, the seeds were grown in the phytotron at the Institute of Plant Physiology and Ecology, Shanghai, China. Tobacco (Nicotiana benthamiana) and Arabidopsis thaliana plants were grown in a growth chamber under a day/night regime of 16/8 h at a temperature of 20 to 25°C.
Phenotype Observations
For the light transmission assay, the wild-type and ocd1 seeds were placed on a medical light box and images were directly taken by a camera. For the cross-section observations, the seeds were latitudinally cut and placed on the platform of the stereoscope and observed via light microscopy. The scanning and transmission electron microscopy methods were previously described (Zhang et al., 2015). For the germination assay, 100 wild-type and cod1-1 seeds were planted and the germination ratio was calculated at 10 d after sowing. Twenty wild-type and cod1-1 plants were measured for root length and plant height. The 100-kernel weight of wild type and ocd1-1 (isolated from segregating ears) was quantified by a balance. The above experiments were repeated three times and similar results were obtained.
Measurement of Proteins and Starch
Five mature wild-type and ocd1 seeds from three individual ears were ground into fine flour using steel beads. Three biological replicates for each were performed. One hundred milligrams of flour for each sample was used for extraction of zein and nonzein proteins. Briefly, 100 mg of flour was incubated with 1 mL of zein extraction buffer (70% ethanol, 2% 2-mercaptoethanol [v/v], 3.75 mM sodium borate, pH 10, and 0.3% SDS) in a 2-mL tube at room temperature. The mixture was centrifuged at 13,000 rpm for 15 min, and then 100 μL of the supernatant was transferred to a new tube containing 10 μL of 10% SDS. The mixture was dried using a Concentrator plus (Eppendorf) and then dissolved in 100 μL of distilled water. For the nonzein extraction, the above precipitation was extracted by zein buffer three additional times and vortexed with 1 mL of double-distilled water. After centrifugation at 13,000 rpm for 15 min, the supernatant containing the nonzein proteins was transferred to a new tube. SDS-PAGE was performed to analyze the accumulation patterns of zein and nonzein proteins in the wild type and ocd1 mutants. Protein quantification was performed as described previously (Zhang et al., 2015) using the BCA protein assay kit (Pierce).
The same flours used in the storage protein measurements were used for the total starch content analysis. After filtering with an 80-mesh nylon screen and drying at 37°C for 3 d, the starch content of each sample was measured with the Total Starch Assay Kit (K-TSTA; Megazyme) according to the manufacturer’s procedure.
Map-Based Cloning of ocd1
Normal seeds from segregating ears (homozygotes or heterozygotes for the Ocd1 allele) were planted. All plants were numbered. Each plant was self-pollinated and crossed to B73 as male at the same time. If a self-pollinated ear segregated mutant seeds, its corresponding cross with B73 was saved and planted in the next season to generate the F2 population. The endosperms of mutant kernels from segregating ears at 20 DAP were harvested for genomic DNA extraction and then genotyping. Chromosome localization of ocd1 was performed using KASP markers (LGC), and the fine mapping was performed using SSR or Indel markers. The primers for the molecular markers are listed in Supplemental Table 1.
RNA Extraction and RT-qPCR Analysis
Tissues were collected at the time indicated and kept at −80°C until use. Prior to a TRIzol extraction, the fine ground seed or endosperm flour was treated with extraction buffer (50 mM Tris, pH 8.0, 150 mM LiCl, 5 mM EDTA, pH 8.0, and 1% SDS) and 1:1 phenol-chloroform three times. Other tissues were directly extracted using TRIzol reagent (Invitrogen). The RNA samples were purified with the RNeasy Mini Kit (Qiagen). Total RNA (5 μg) was used for reverse transcription with a Superscript III first-strand kit (Invitrogen). The resulting cDNA was diluted to 20 ng/μL, and real-time PCR was performed with SYBR Green (TAKARA) on a Bio-Rad CFX-96 thermocycler. The fold changes in gene expression were calculated using the ΔΔCt (cycle threshold) values. The maize Ubiquitin gene was used as an internal control. All primers used in the study are listed in Supplemental Table 1.
Subcellular Localization Analysis
For the GFP or RFP fusion, ZmOCD1 or AtAAE3 cDNA was cloned into the modified binary vector pCAMBIA1300-35S-GFP (driven by the 35S promoter) using a recombination method. The vector of pCAMBIA1300-35S-GFP was used as a free GFP control. The resulting 35S:GFP and 35S:ZmOCD1-GFP constructs were transiently expressed in Arabidopsis protoplasts via the PEG-CaCl2 method directly and tobacco leaf via the Agrobacterium tumefaciens strain GV3101 as described (Yang et al., 2016). The 35S: ZmOCD1-GFP plasmid was also transformed into Arabidopsis (Col-0) plants by Agrobacterium-mediated transformation. The GFP fluorescence was observed using an Olympus confocal microscope system.
Immunoblot and Immunofluorescence Assays
The fragment (111–311 amino acids) of ZmOCD1 protein was used as antigen and the antibodies were prepared in rabbits by Abclonal of China. Total protein from wild type and mutant endosperms was extracted with the lysis buffer (90 mM HEPES, pH 8.0, 1 mM EDTA, pH 8.0, 10% [v/v] glycerol, and 5mM DTT). Proteins were separated by a 4 to 20% SDS-PAGE gel and transferred electrophoretically to PVDF membrane (Bio-Rad). The membrane was then incubated with antibodies and visualized using Tanon-5200 system. The dilutions of antibodies against OCD1 and ACTIN (Abclonal) were 1:2000 and 1:5000, respectively.
Wild-type seeds at 6, 8, and 12 DAP were fixed overnight in 4% paraformaldehyde in 0.1 M PBS (pH 7.5). Sections (10 µm) were cut out and blocked with 5% BSA (BSA) for 1 h at room temperature. Then the sections were incubated with OCD1 antibodies (diluted at 1:100 with 0.1 M PBS containing 0.1% BSA) overnight at 4°C. After rinsing three times in PBS (5 min each), the samples were immersed in the goat anti-rabbit IgG (H+L) Alexa Fluor 488 (Yeasen; 1:200 dilution). Fluorescence was observed via a confocal laser scanning microscope (LSM880; Zeiss).
Determination of the ZmOCD1 Enzyme Activity
The coding sequences of O7 and ZmOCD1 were amplified and inserted into the expression vector pTOLO-EX2 vector. The constructs were transformed into Escherichia coli DH5α cells for DNA sequencing. After verification by sequencing, these constructs were transformed into E. coli BL21 (Rosetta strain). Target proteins were induced by 1 mM IPTG overnight at 18°C. After the SDS-PAGE analysis, the resulting tagged fusion proteins were purified using Ni-NTA affinity chromatography. The relative purity of the proteins was evaluated and the purified proteins were quantified via SDS-PAGE and A280 absorbance using BSA as a standard, respectively.
To determine the ZmOCD1 enzyme activity, oxalyl-CoA was chemically synthesized as previously described (Quayle, 1962). The reaction system included 50 mM Tris buffer (pH 7.5), 6.6 mM BSA, 0.8 mM MgCl2, 20 mM thiamine pyrophosphate, oxalate-CoA, and 3 μg of ZmOCD1 protein in a total volume of 100 μL. For in vivo enzyme activity assay, 20-DAP wild-type and ocd1-1 endosperms were extracted with the lysis buffer (90 mM HEPES, pH 8.0, 1 mM EDTA, pH 8.0, 10% [v/v] glycerol, and 5 mM DTT). Twenty microliters of endosperm extracts from the wild type and ocd1-1 was added to prepare the reaction system as listed above. After incubation at 37°C for 15 min, the reactions were terminated with an equal volume of methanol and were subjected to LC-MS/MS analysis.
To determine the O7 function, a 2× reaction buffer was first prepared that contained 0.2 M Tris-HCl (pH 8.0), 4 mM DTT, 10 mM ATP, 20 mM MgCl2, 0.8 mM NADH, and 2 mM phosphoenolpyruvate. The reaction system included 300 μL of 2× reaction buffer, 10 units each of myokinase, pyruvate kinase, and lactate dehydrogenase, 6 μg of O7 protein, and oxalate (Sigma-Aldrich) at different concentrations (200, 400, and 600 µM) and CoA (1:1) in a final volume of 600 μL. The reaction was incubated at 37°C for 30 min and analyzed via LC-MS/MS analysis.
LC-MS/MS was performed via a Q-Exactive Quadrupole Orbitrap mass spectrometer (Thermo) coupled to an Acquity Ultra Performance LC system (Waters). An Acquity UPLC HSS T3 column (2.1 × 100 mm, 1.8 um) was used at 30°C. Samples (5 μL) were injected at 10°C. Positive and negative modes were used for the analysis of OCD1 and O7 activity. Ammonium formate (positive mode) or water (negative mode) and acetonitrile were used as mobile phases A and B, respectively. The flow rate was 0.3 mL/min. The gradient of mobile phase B was as follows: 0 min, 0%; 10 min, 20%; 12 min, 90%; and 13.1 min, 0%. The MS system was operated with a HESI ion source in positive and negative mode under the following conditions: capillary voltage, +4.0/−3.0 kV; capillary temperature, 320°C; sheath gas, 35 units; auxiliary gas, 8 units; and auxiliary gas heater temperature, 350°C. The mass spectrometer was run in full MS-ddMS2 mode. The full MS scan used the following settings: resolution, 70,000; AGC target, 3e6; and scan range, 80–1000 m/z. The MS2 scan parameters were as follows: resolution, 17,500; AGC target, 1e5; max IT, 50 ms; and normalized HCD collision energy, 15 and 30 eV.
Measurement of Oxalate and ATP Contents
The oxalate measurement was performed with the oxalate colorimetric assay kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, 50 mg of dry seed flour from wild type and ocd1-1 was homogenized in oxalate assay buffer and incubated for 1 h at room temperature. After centrifugation at 10,000g for 5 min, 15 μL of supernatant was added to a tube with 35 μL of oxalate assay buffer. Then, 2 μL of oxalate converter was added to each sample and incubated at 37°C for 1 h. A master reaction mix (46 μL of oxalate development buffer, 2 μL of enzyme mix, and 2 μL of probe) was added to each of the wells, the wells were mixed using a horizontal shaker, and the reaction was incubated in the dark for 1 h at 37°C. The absorbance of the reaction mixes was measured at 450 nm (A450), and the concentrations were calculated according to a standard curve.
ATP content was measured with an ATP assay kit (Beyotime). For the standard curve, ATP solutions (0.001, 0.003, 0.01, 0.03, 0.1, 0.3, and 1 μM) were added to 100 μL of the substrate solution. The luminescence was recorded in an Illuminometer (Promega) with an integration time of 2 s per well. The 20-DAP endosperm of the wild type and ocd1-1 was mixed well with lysis solution. After centrifugation to remove the endosperm cell debris, 20 μL of supernatant was added to 100 μL of substrate solution. Luminescence was detected as described above, and the concentration was calculated according to the standard curve.
Metabolite Measurements
Metabolites were extracted from six individual endosperms. Endosperm tissues retrieved from −80°C storage were ground into fine powder in liquid nitrogen. Powder (60 mg) was vortexed in a 1 mL solution of methanol/ acetonitrile/water. After sonication for 30 min on ice, the mix was stored at −20°C for 1 h to precipitate proteins. The mix was centrifuged for 15 min (13,000 rpm, 4°C) and dried by a vacuum drying system. A targeted metabolic analysis was performed using an LC-MS/MS system. The dried metabolites were dissolved in 100 μL of acetonitrile/water (1:1, v/v) and centrifuged (13,000 rpm) for 15 min. Electrospray ionization was conducted with an Agilent 1290 Infinity chromatography system and AB Sciex QTRAP 5500 mass spectrometer. NH4COOH4 (15 mM) and acetonitrile were used as mobile phases A and B, respectively. A binary solvent gradient was used as follows: A, NH4COOH; B, 0 to 18 min at 90% to 40% acetonitrile; 18 to 18.1 min at 40% to 90% acetonitrile; and 18.1 to 23 min at 90% acetonitrile. The LC-MS/MS was operated in the negative mode under the following conditions: source temperature, 450°C; ion source gas 1, 45; ion source gas 2, 45; curtain gas, 30; and ion spray voltage floating, −4500 V.
For the nontargeted metabolic analyses, the Agilent 1290 Infinity LC (UHPLC0) and Triple TOF 5600 (AB Sciex) were used. A solvent gradient was used as follows: A solution of 25 mM NH4COOH and 25 mM NH3H20 was used as A and acetonitrile was used as B; the gradient for A was 100% and for B was 0 to 1 min at 95%; 1 to 14 min at 95% to 65%; 14 to 16 min at 65% to 40%; 16 to 18 min at 40%; 18 to 18.1 min at 40% to 95%; and 18.1 to 23 min at 95%. After the UHPLC analysis, the samples were analyzed under positive and negative ion mode MS. The conditions were as follows: ion source gas 1, 60; ion source gas 2, 60; curtain gas, 30; source temperature, 600°C; ion spray voltage floating, ±5500 V; TOF MS scan m/z range, 60 to 1000 D; product ion scan m/z range, 25 to 1000 D; and collision energy, 35 ± 15 eV.
Accession Numbers
Sequence data from this study can be found in GenBank/EMBL data libraries under accession numbers NP_001130784 (ZmOCD1), NP_197240 (AtOCD1), and WP_005881708.1 (OfOCD1).
Supplemental Data
Supplemental Figure 1. Quantitative RT-PCR Measurement of the Expression Levels of α- (22-kD z1C, 19-kD z1A, 19-kD z1B, and 19-kD z1D), γ- (16-kD and 27-kD), δ- (10-kD), and β- (15-kD) Zein Genes in the Wild Type and ocd1.
Supplemental Figure 2. Germination Ratio and Seedling Phenotypes of the Wild Type and ocd1-1.
Supplemental Figure 3. SDS-PAGE Analysis of Zein and Nonzein Proteins in the Wild Type and ocd1 Mutants.
Supplemental Figure 4. Expression Analysis of Ocd1.
Supplemental Figure 5. Specificity of OCD1 Antibodies.
Supplemental Figure 6. SDS-PAGE Analysis of Purified Recombinant and Endosperm Extract Proteins and LC-MS/MS Analysis of the Substrate and Product during Enzymatic Experiments.
Supplemental Figure 7. Biochemical Function of Recombinant O7.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Targeted Metabolic Analysis.
Supplemental Data Set 2. Nontargeted Metabolic Analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This research was supported by the Ministry of Science and Technology of China (2016YFD0100500 to Y.W.), the National Natural Science Foundation of China (31601310 to J.Y. and 91635303 to Y.W.), the Chinese Academy of Sciences (XDPB0401 and XDA08020107 to Y.W.), and a Chinese Thousand Talents Program Grant (to Y.W.). We thank Xiaoyan Gao, Zhiping Zhang, Yuanhong Shan, Wenli Hu, and Yining Liu (Institute of Plant Physiology and Ecology, SIBS, CAS) for technical support.
AUTHOR CONTRIBUTIONS
J.Y. and Y.W. designed the research. J.Y., M.F., C.J., Y.H., and Y.W. performed the experiments. J.Y. and Y.W. analyzed the data and wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Bateman D.F., Beer S.V. (1965). Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology 55: 204–211. [PubMed] [Google Scholar]
- Berthold C.L., Moussatche P., Richards N.G., Lindqvist Y. (2005). Structural basis for activation of the thiamin diphosphate-dependent enzyme oxalyl-CoA decarboxylase by adenosine diphosphate. J. Biol. Chem. 280: 41645–41654. [DOI] [PubMed] [Google Scholar]
- Brzezicha-Cirocka J., Grembecka M., Szefer P. (2016). Oxalate, magnesium and calcium content in selected kinds of tea: impact on human health. Eur. Food Res. Technol. 242: 383–389. [Google Scholar]
- Casteels M., Sniekers M., Fraccascia P., Mannaerts G.P., Van Veldhoven P.P. (2007). The role of 2-hydroxyacyl-CoA lyase, a thiamin pyrophosphate-dependent enzyme, in the peroxisomal metabolism of 3-methyl-branched fatty acids and 2-hydroxy straight-chain fatty acids. Biochem. Soc. Trans. 35: 876–880. [DOI] [PubMed] [Google Scholar]
- Castellaro A.M., Tonda A., Cejas H.H., Ferreyra H., Caputto B.L., Pucci O.A., Gil G.A. (2015). Oxalate induces breast cancer. BMC Cancer 15: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zeng B., Zhang M., Xie S., Wang G., Hauck A., Lai J. (2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.E., Harada E., Wada M., Tsuboi H., Morita Y., Kusano T., Sano H. (2001). Detoxification of cadmium in tobacco plants: formation and active excretion of crystals containing cadmium and calcium through trichomes. Planta 213: 45–50. [DOI] [PubMed] [Google Scholar]
- Coleman C.E., Clore A.M., Ranch J.P., Higgins R., Lopes M.A., Larkins B.A. (1997). Expression of a mutant alpha-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94: 7094–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer J.M., Perakis S.S. (2014). Calcium oxalate contribution to calcium cycling in forests of contrasting nutrient status. For. Ecol. Manage. 334: 64–73. [Google Scholar]
- Davies D.D., Asker H. (1983). Synthesis of oxalic acid by enzymes from lettuce leaves. Plant Physiol. 72: 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Ji R., Guo X., Foster S.J., Chen H., Dong C., Liu Y., Hu Q., Liu S. (2008). Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228: 331–340. [DOI] [PubMed] [Google Scholar]
- Foster J., Nakata P.A. (2014). An oxalyl-CoA synthetase is important for oxalate metabolism in Saccharomyces cerevisiae. FEBS Lett. 588: 160–166. [DOI] [PubMed] [Google Scholar]
- Foster J., Kim H.U., Nakata P.A., Browse J. (2012). A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J., Luo B., Nakata P.A. (2016). An oxalyl-CoA dependent pathway of oxalate catabolism plays a role in regulating calcium oxalate crystal accumulation and defending against oxalate-secreting phytopathogens in Medicago truncatula. PLoS One 11: e0149850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulon V., Antonenkov V.D., Croes K., Waelkens E., Mannaerts G.P., Van Veldhoven P.P., Casteels M. (1999). Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids. Proc. Natl. Acad. Sci. USA 96: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi V.R. (1989). Calcium-oxalate formation is a rapid and reversible process in Lemna minor L. Protoplasma 148: 130–137. [Google Scholar]
- Franceschi V.R., Nakata P.A. (2005). Calcium oxalate in plants: formation and function. Annu. Rev. Plant Biol. 56: 41–71. [DOI] [PubMed] [Google Scholar]
- Franceschi V.R., Schueren A.M. (1986). Incorporation of strontium into plant calcium-oxalate crystals. Protoplasma 130: 199–205. [Google Scholar]
- Gil G.A., Castellaro A.M., Pucci O., Tonda A., Cejas H., Caputto B.L. (2013). Oxalate microcalcifications induce breast cancer tumors. Cancer Res. 73: 1–2. [Google Scholar]
- Guimarães R.L., Stotz H.U. (2004). Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136: 3703–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Yuan L., Chen H., Sato S.J., Clemente T.E., Holding D.R. (2013). Nonredundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm. Plant Physiol. 162: 1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Joosten H.J., Niu W., Zhao Z., Mariano P.S., McCalman M., van Kan J., Schaap P.J., Dunaway-Mariano D. (2007). Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi. J. Biol. Chem. 282: 9581–9590. [DOI] [PubMed] [Google Scholar]
- Heaney R.P., Weaver C.M. (1990). Calcium absorption from kale. Am. J. Clin. Nutr. 51: 656–657. [DOI] [PubMed] [Google Scholar]
- Holding D.R., Otegui M.S., Li B., Meeley R.B., Dam T., Hunter B.G., Jung R., Larkins B.A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner H.T., Kausch A.P., Wagner B.L. (2000). Ascorbic acid: A precursor of oxalate in crystal idioblasts of Yucca torreyi in liquid root culture. Int. J. Plant Sci. 161: 861–868. [Google Scholar]
- Islam M.N., Maeda H., Kawasaki M. (2015). Effect of calcium concentration in growth medium on oxalate content and evaluation of the role of guttation in the regulation of oxalate in eddo. Plant Prod. Sci. 18: 464–470. [Google Scholar]
- Kim K.S., Min J.Y., Dickman M.B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21: 605–612. [DOI] [PubMed] [Google Scholar]
- Kirejczyk J.K., Porowski T., Filonowicz R., Kazberuk A., Stefanowicz M., Wasilewska A., Debek W. (2014). An association between kidney stone composition and urinary metabolic disturbances in children. J. Pediatr. Urol. 10: 130–135. [DOI] [PubMed] [Google Scholar]
- Korth K.L., Doege S.J., Park S.H., Goggin F.L., Wang Q., Gomez S.K., Liu G., Jia L., Nakata P.A. (2006). Medicago truncatula mutants demonstrate the role of plant calcium oxalate crystals as an effective defense against chewing insects. Plant Physiol. 141: 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane B.G., Dunwell J.M., Ray J.A., Schmitt M.R., Cuming A.C. (1993). Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268: 12239–12242. [PubMed] [Google Scholar]
- Lappe R.R., et al. (2018). Functions of maize genes encoding pyruvate phosphate dikinase in developing endosperm. Proc. Natl. Acad. Sci. USA 115: E24–E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins B.A., Hurkman W.J. (1978). Synthesis and deposition of zein in protein bodies of maize endosperm. Plant Physiol. 62: 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuwenhoek A. (1975). Microscopical observations. Philos. Trans. R. Soc. Lond. 10: 380–385. [Google Scholar]
- Li Q., Wang J., Ye J., Zheng X., Xiang X., Li C., Fu M., Wang Q., Zhang Z., Wu Y. (2017). The Maize Imprinted Gene Floury3 Encodes a PLATZ Protein Required for tRNA and 5S rRNA Transcription through Interaction with RNA Polymerase III. Plant Cell 29: 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz E.C., Michet C.J., Milliner D.S., Lieske J.C. (2013). Update on oxalate crystal disease. Curr. Rheumatol. Rep. 15: 340–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H.Q., Fan W., Xu J.M., Gong Y.L., Jin J.F., Chen W.W., Liu L.Y., Hai M.R., Yang J.L., Zheng S.J. (2016a). An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol. 172: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H.Q., Gong Y.L., Fan W., Xu J.M., Liu Y., Cao M.J., Wang M.H., Yang J.L., Zheng S.J. (2016b). A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol. 171: 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Hiradate S., Nomoto K., Iwashita T., Matsumoto H. (1997). Internal detoxification mechanism of Al in hydrangea (identification of Al form in the leaves). Plant Physiol. 113: 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Hiradate S., Matsumoto H. (1998). High aluminum resistance in buckwheat. Ii. Oxalic acid detoxifies aluminum internally. Plant Physiol. 117: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazen A.M.A., El Maghraby O.M.O. (1998). Accumulation of cadmium, lead and strontium, and a role of calcium oxalate in water hyacinth tolerance. Biol. Plant. 40: 411–417. [Google Scholar]
- Miclaus M., Wu Y., Xu J.H., Dooner H.K., Messing J. (2011). The maize high-lysine mutant opaque7 is defective in an acyl-CoA synthetase-like protein. Genetics 189: 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata P.A. (2012). Influence of calcium oxalate crystal accumulation on the calcium content of seeds from Medicago truncatula. Plant Sci. 185-186: 246–249. [DOI] [PubMed] [Google Scholar]
- Nakata P.A. (2015). An assessment of engineered calcium oxalate crystal formation on plant growth and development as a step toward evaluating its use to enhance plant defense. PLoS One 10: e0141982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond B. (1963). Oxalates and ionic equilibria in Australian saltbushes (Atriplex). Nature 198: 503–504. [Google Scholar]
- Park S.H., Doege S.J., Nakata P.A., Korth K.L. (2009). Medicago truncatula-derived calcium oxalate crystals have a negative impact on chewing insect performance via their physical properties. Entomol. Exp. Appl. 131: 208–215. [Google Scholar]
- Prychid C.J., Jabaily R.S., Rudall P.J. (2008). Cellular ultrastructure and crystal development in Amorphophallus (Araceae). Ann. Bot. 101: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle J.R. (1962). Chemical synthesis of oxalyl-coenzyme A and its enzymic reduction to glyxylate. Biochim. Biophys. Acta 57: 398–400. [DOI] [PubMed] [Google Scholar]
- Richardson K.E., Tolbert N.E. (1961). Oxidation of glyoxylic acid to oxalic acid by glycolic acid oxidase. J. Biol. Chem. 236: 1280–1284. [PubMed] [Google Scholar]
- Schmidt R.J., Ketudat M., Aukerman M.J., Hoschek G. (1992). Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4: 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Ma J.F., Kyo M., Iwashita T. (2002). Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 215: 394–398. [DOI] [PubMed] [Google Scholar]
- Siener R., Bangen U., Sidhu H., Hönow R., von Unruh G., Hesse A. (2013). The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int. 83: 1144–1149. [DOI] [PubMed] [Google Scholar]
- Stewart C.S., Duncan S.H., Cave D.R. (2004). Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol. Lett. 230: 1–7. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Itai R., Suzuki K., Nakanishi H., Nishizawa N.K., Yoshimura E., Mori S. (1998). Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol. 116: 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.N., Curhan G.C. (2007). Oxalate intake and the risk for nephrolithiasis. J. Am. Soc. Nephrol. 18: 2198–2204. [DOI] [PubMed] [Google Scholar]
- Volk G.M., Lynch-Holm V.J., Kostman T.A., Goss L.J., Franceschi V.R. (2002). The role of druse and raphide calcium oxalate crystals in tissue calcium regulation in Pistia stratiotes leaves. Plant Biol. 4: 34–45. [Google Scholar]
- Wang G., Sun X., Wang G., Wang F., Gao Q., Sun X., Tang Y., Chang C., Lai J., Zhu L., Xu Z., Song R. (2011). Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Weaver C.M., Martin B.R., Ebner J.S., Krueger C.A. (1987). Oxalic acid decreases calcium absorption in rats. J. Nutr. 117: 1903–1906. [DOI] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2010a). RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol. 153: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2010b). RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol. 153: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2011). Novel genetic selection system for quantitative trait loci of quality protein maize. Genetics 188: 1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2017). Understanding and improving protein traits in maize. In Achieving Sustainable Cultivation of Maize. Vol. 1: From Improved Varieties to Local Applications, Watson D., ed (Cambridge, UK: Burleigh Dodds Science Publishing; ), pp. 115–128. [Google Scholar]
- Yang J.L., Zhang L., Zheng S.J. (2008). Aluminum-activated oxalate secretion does not associate with internal content among some oxalate accumulators. J. Integr. Plant Biol. 50: 1103–1107. [DOI] [PubMed] [Google Scholar]
- Yang J., Ji C., Wu Y. (2016). Divergent transactivation of maize storage protein zein genes by the transcription factors Opaque2 and OHPs. Genetics 204: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.Y., Jung J.Y., Song W.Y., Suh H.S., Lee Y. (2000). Identification of rice varieties with high tolerance or sensitivity to lead and characterization of the mechanism of tolerance. Plant Physiol. 124: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Jiang J., Zhang C., Jiang L., Ye N., Lu Y., Yang G., Liu E., Peng C., He Z., Peng X. (2010). Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 61: 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yang J., Wu Y. (2015). Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindler-Frank E., Honow R., Hesse A. (2001). Calcium and oxalate content of the leaves of Phaseolus vulgaris at different calcium supply in relation to calcium oxalate crystal formation. J. Plant Physiol. 158: 139–144. [Google Scholar]