An amino acid variation within the interaction domain of EARLY FLOWERING3 with the E3 ubiquitin ligase HAF1 strongly affects the variation in heading date among japonica rice accessions.
Abstract
The ubiquitin 26S proteasome system (UPS) is critical for enabling plants to alter their proteomes to integrate internal and external signals for the photoperiodic induction of flowering. We previously demonstrated that HAF1, a C3HC4 RING domain-containing E3 ubiquitin ligase, is essential to precisely modulate the timing of Heading Date1 accumulation and to ensure appropriate photoperiodic responses under short-day conditions in rice (Oryza sativa). However, how HAF1 mediates flowering under long-day conditions remains unknown. In this study, we show that OsELF3 (EARLY FLOWERING3) is the direct substrate of HAF1 for ubiquitination in vitro and in vivo. HAF1 is required for maintaining the circadian rhythm of OsELF3 accumulation during photoperiodic responses in rice. In addition, the haf1 oself3 double mutant headed as late as oself3 plants under long-day conditions. An amino acid variation (L558S) within the interaction domain of OsELF3 with HAF1 greatly contributes to the variation in heading date among japonica rice accessions. The japonica accessions carrying the OsELF3(L)-type allele are found at higher latitudes, while varieties carrying the OsELF3(S)-type allele are found at lower latitudes. Taken together, our findings suggest that HAF1 precisely modulates the diurnal rhythm of OsELF3 accumulation to ensure the appropriate heading date in rice.
INTRODUCTION
Flowering time (heading date) is an important agronomic trait in rice (Oryza sativa) that directly influences successful grain production and variety distribution in various planting regions (Izawa, 2007). Although the molecular basis of flowering time control is well established in Arabidopsis thaliana, a long-day (LD) plant, it remains to be dissected for rice, a facultative short-day (SD) plant. Recent studies using molecular genomic approaches have characterized several regulators involved in photoperiodic flowering in rice. Two distinct pathways control photoperiodic heading date in rice (Turck et al., 2008). The OsGI-Hd1-Hd3a (GIGANTEA-Heading Date1-Hd3a) pathway is evolutionarily conserved between rice and Arabidopsis, which used the corresponding GI-CO-FT (GI-CONSTANS-FLOWERING LOCUS T) pathway (Turck et al., 2008). Unlike CO, which promotes flowering in Arabidopsis, Hd1 promotes flowering under SD conditions but represses flowering under LD conditions in rice (Yano et al., 2000; Ishikawa et al., 2011). Hd1 functions as a flowering repressor through interaction with the transcription factor Ghd7 (Nemoto et al., 2016). Hd5/DTH8 (DAYS TO HEADING8)/Ghd8 encodes a putative HAP3/NF-YB subunit of a CCAAT-box binding transcription factor (Wei et al., 2010; Yan et al., 2011;Fujino et al., 2013). The DTH8/Ghd8-Hd1 complex is required for the transcriptional repression of Hd3a by Hd1 under LDs (Du et al., 2017). Polymorphisms in Hd1 affect the formation of the Hd1-NF-YCs-Hd5/Ghd8 heterotrimer to limit Hd1 function to help rice varieties adapt to high latitudes (Goretti et al., 2017). Another photoperiodic heading pathway is Ghd7-Ehd1-Hd3a/RFT1 (Ghd7-Early heading date1-Hd3a/RICE FLOWERING LOCUS T1), which is unique to rice (Xue et al., 2008; Tsuji et al., 2011; Song et al., 2015). Ghd7 encodes a CCT domain protein that represses photoperiodic expression of Ehd1 under LDs (Xue et al., 2008). Ehd1, encoding a rice-specific B-type response regulator, is a heading promoter that functions independently of Hd1 in SDs (Doi et al., 2004). Although the Ehd1-dependent flowering pathway was regard as being unique to rice, recent investigations have shown that it is strongly linked to the Hd1-dependent pathway. Under LDs, Hd1 is a strong repressor of Ehd1 (Gómez-Ariza et al., 2015; Nemoto et al., 2016; Du et al., 2017; Goretti et al., 2017). Multiple heading regulators fine-tune the expression of Ehd1, which is crucial for rice heading at a suitable time. RID1 (Rice Indeterminate 1)/Ehd2 is fundamental for rice heading by activating Ehd1 expression (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008). OsMADS51, a type I MADS box transcription factor, promotes flowering upstream of Ehd1 under SDs (Kim et al., 2007). Another activator of Ehd1 is Ehd4, which encodes a nucleus-localized CCCH-type zinc finger protein unique to rice (Gao et al., 2013). Several repressors of Ehd1 expression have been identified, including Hd5/DTH8/Ghd8 (Wei et al., 2010; Yan et al., 2011;Fujino et al., 2013), the transcription factor OsLFL1 (Peng et al., 2007), OsCOL4 (CONSTANS-LIKE4) (Lee et al., 2010), and Ghd7 (Xue et al., 2008). The altered expression of the floral repressor gene Ghd7 and the floral promoter gene Ehd1 in response to light establishes critical daylength responses to control florigen gene expression under specific photoperiods (Itoh et al., 2010). Hd16/EL1, encoding a casein kinase I protein, acts as a flowering repressor by directly interacting with and phosphorylating Ghd7 (Hori et al., 2013; Kwon et al., 2014). Ehd3 encodes a nuclear protein with two PHD-finger motifs and represses the expression of Ghd7 particularly under LDs (Matsubara et al., 2011). Circadian clock proteins also regulate the expression of Ghd7. OsELF3 (EARLY FLOWERING3) is required to sustain the robust oscillation through maintaining normal expression of clock-related genes and promotes flowering mainly by repressing Ghd7 expression under LDs (Zhao et al., 2012; Yang et al., 2013).
ELF3 was first identified as a floral suppressor in Arabidopsis (Zagotta et al., 1996). ELF3, a component of the ELF3-ELF4-LUX ARRHYTHMO (LUX) evening complex, plays an important role in modulating light input to the circadian clock (Covington et al., 2001; Thines and Harmon, 2010; Kolmos et al., 2011). Early in the night, the evening complex represses the expression of transcription factor genes PIF4 and PIF5 to inhibit hypocotyl growth (Leivar et al., 2009; Leivar and Quail, 2011; Nusinow et al., 2011). The transcriptional regulator BBX19 acts as an adaptor to recruit ELF3 for degradation by the E3 ligase COP1, dynamically gating the formation of the evening complex and resulting in the repression of PIF4/5 PHYTOCHROME-INTERACTING FACTOR 4/5) expression to promote hypocotyl growth (Wang et al., 2015). ELF3 and PIF4 function as key hubs for integrating hypocotyl elongation in response to temperature (Box et al., 2015; Raschke et al., 2015). ELF3 also functions as a substrate adaptor for COP1-GI interaction, which controls circadian function and photoperiodic flowering by enabling COP1 to degrade GI (Yu et al., 2008). Compared with Arabidopsis, rice contains two orthologs of ELF3 (Fu et al., 2009). OsELF3 (LOC_Os06g05060) plays a dominant role in controlling flowering time in rice (Zhao et al., 2012; Yang et al., 2013). Mutation in OsELF3 causes a delayed heading date only under LDs (Yang et al., 2013). However, the elf3 mutant has an early and photoperiod-insensitive flowering time and defective inhibition of hypocotyl elongation in Arabidopsis (Zagotta et al., 1996), suggesting that OsELF3 might be involved in different flowering pathways in rice. Under LDs, OsELF3 promotes flowering mainly by repressing Ghd7 and OsGI expression. Furthermore, the rhythmic expression patterns of some circadian clock-associated genes, such as OsLHY (LATE ELONGATED HYPOTOTYL), OsPRR1 (PSEUDO-RESPONSE REGULATOR 1), OsPRR37, and OsPRR73, are perturbed in oself3 (Zhao et al., 2012; Yang et al., 2013).
In Arabidopsis, ubiquitin-mediated protein degradation is important for regulating the expression and stability of flowering regulators (Song et al., 2015). The temporal transcriptional and posttranslational regulation of CO coinciding with day-length information is essential for proper photoperiodic flowering (Goretti et al., 2017). Under LDs, blue light activates the complex containing E3 ubiquitin ligase FKF1 and GI, which results in the degradation of the CO transcriptional repressors, CDFs (Sawa et al., 2007; Fornara et al., 2009; Song et al., 2012). FKF1-GI also stabilizes CO protein in the afternoon (Sawa et al., 2007; Song et al., 2012). At least two RING-finger E3 ubiquitin ligases, HOS1 and COP1, are involved in CO protein degradation (Jang et al., 2008; Liu et al., 2008; Lazaro et al., 2012). In the morning, HOS1 restricts the accumulation of CO protein by degrading it (Lazaro et al., 2012). In the dark, the COP1-SPA complex degrades CO protein to prevent flowering in SDs (Jang et al., 2008; Liu et al., 2008; Sarid-Krebs et al., 2015). Although multiple E3 ubiquitin ligases involved in flowering have been characterized in Arabidopsis, only a few have been studied in rice. We previously revealed that HAF1, a C3HC4 RING domain-containing E3 ubiquitin ligase, is essential to precisely modulate the timing of Hd1 accumulation and to ensure an appropriate photoperiodic response under SDs (Yang et al., 2015). Under LDs, the haf1 hd1 double mutant flowers as late as haf1, suggesting that HAF1 may interact with additional components to control heading date under LDs (Yang et al., 2015).
Here, to elucidate how HAF1 participates in heading under LDs in rice, we examined its possible interactions with multiple flowering regulators under LDs, such as Ghd7, Ghd8, Ghd7.1, Ehd4 and OsELF3, in yeast two-hybrid assays. We found that OsELF3, a flowering promoter under LDs, physically interacts with HAF1. Moreover, we show that HAF1 mediates the ubiquitination of OsELF3 protein via the 26S proteasome pathway. Natural variation in the interaction domain of OsELF3 with HAF1 contributes to the geographic distribution of various japonica rice accessions. The haf1 oself3 double mutant has the same heading date as oself3, suggesting that HAF1 influences heading date mainly via OsELF3 under LDs. Thus, the ubiquitin-dependent degradation of OsELF3 by HAF1 contributes to the circadian accumulation of OsELF3 and ensures appropriate heading date under LDs.
RESULTS
OsELF3 Interacts with HAF1 in Yeast Two-Hybrid Assays
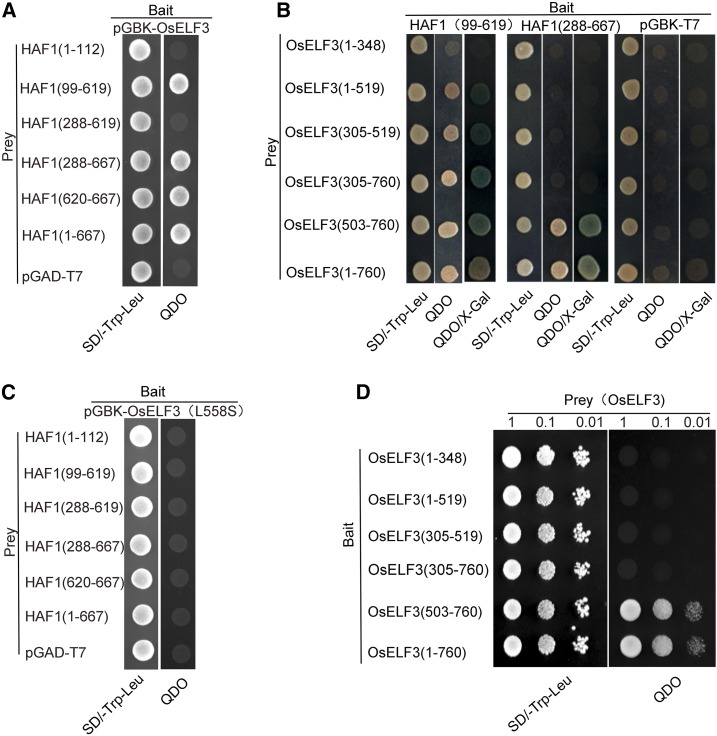
To explore the molecular mechanism underlying how HAF1 regulates heading under LDs, we performed yeast two-hybrid assays to examine the possible physical interactions between HAF1 and OsELF3. Due to the autotranscriptional activation activity of HAF1, truncated fragments of HAF1 (amino acids 1–112, 99–619, 288–619, 288–667, and 620–667) with no transcriptional activation activity (Yang et al., 2015) and full-length HAF1 protein were used to transform yeast cells individually as prey and full-length OsELF3 protein as bait to determine whether HAF1 interacts with OsELF3. The yeast two-hybrid assays showed that constructs containing fragments amino acids 99 to 619 or 288 to 667 of HAF1 and full-length HAF1 protein successfully interacted with OsELF3 (Figure 1A). Considering that the C3HC4 domain is present in the C-terminal region of HAF1 (Yang et al., 2015), its interaction with OsELF3 might occur through the C3HC4 domain of HAF1. We therefore truncated OsELF3 into five fragments (amino acids 1–348, 1–519, 305–519, 305–760, and 503–760) for the interaction assay. The C terminus of OsELF3 containing fragment amino acids 503 to 760 was required for the interaction with HAF1 amino acids 99 to 619 or 288 to 667 (Figure 1B). The substitution of leucine to serine at residue 558 (L558S) in the C terminus of OsELF3 abolished the interaction with HAF1 (Figure 1C). Arabidopsis ELF3 forms a homodimer to regulate its own function (Liu et al., 2001). Our results suggest that OsELF3 also interacts with itself via its C-terminal region (amino acids 503–760) (Figure 1D). Thus, OsELF3 might form a homodimer and interact with HAF1 to perform its function.
Figure 1.
Yeast Two-Hybrid Assay of Interactions between OsELF3 and HAF1.
(A) Interaction analysis of OsELF3 (as bait) with prey constructs containing HAF1 truncated fragments.
(B) Interaction analysis between a series of truncated OsELF3 and HAF1 fragments. HAF1 fragments (amino acids 99–619 or 288–667) interact most strongly with the C-terminal domain of OsELF3 (amino acids 503–760).
(C) The substitution of leucine to serine (L558S) in OsELF3 abolishes its interaction with HAF1.
(D) The C-terminal domain of OsELF3 (amino acids 503–760) is essential for its homodimer formation. Empty pGBK (bait) and pGAD (prey) were used as negative controls. Blue clones in the X-Gal assay and clones that grow on QDO medium indicate protein interactions in the yeast cells. QDO indicates SD-Trp-Leu-His-Ade medium.
OsELF3 Interacts with HAF1 in Vivo and in Vitro
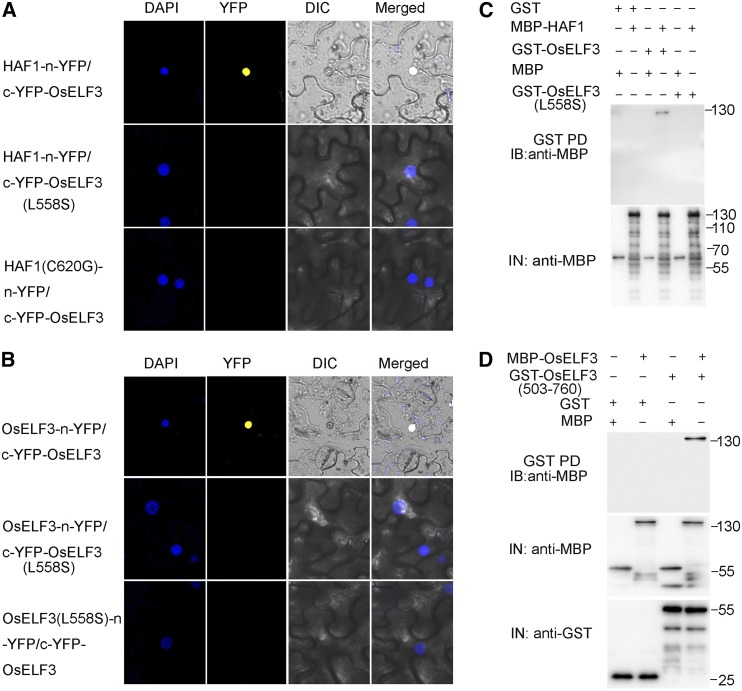
We used bimolecular fluorescence complementation (BiFC) assays to confirm the interaction between OsELF3 and HAF1 in plant cells. Using a transient expression system in wild tobacco (Nicotiana benthamiana) cells, constructs containing HAF1-n-YFP and c-YFP-OsELF3 were cotransformed into the abaxial epidermal cells of wild tobacco leaves. As anticipated, YFP fluorescence was detected in the nuclei of leaf cells, while no fluorescence was observed using mutant fusion proteins c-YFP-OsELF3 (L558S) or HAF1 (C620G)-n-YFP (Figure 2A). These results indicate that OsELF3 interacts with HAF1 in the nucleus in planta. A similar experiment was conducted to verify the formation of OsELF3 homodimers. Strong YFP fluorescence was observed in the nuclei of wild tobacco leaf epidermal cells when OsELF3-n-YFP and c-YFP-OsELF3 were coexpressed (Figure 2B). Although the biological role of the OsELF3 homodimer is unclear, our results confirm that the formation of OsELF3 homodimers occurs in nuclei in planta.
Figure 2.
HAF1 Interacts with OsELF3 in Vivo and in Vitro.
(A) BiFC visualization of the interaction between HAF1 and OsELF3 in the nuclei of N. benthamiana epidermal cells. As negative controls, the substitution of leucine to serine (L558S) of OsELF3 or the substitution of glycine to cysteine (C620G) of HAF1 (HAF1) abolishes the interaction between these proteins.
(B) BiFC showing the homodimer formation of OsELF3. Numbers on the right indicate the molecular masses of marker proteins (the unit is kilodaltons). OsELF3 (L558S) protein was used as a negative control. DAPI, fluorescence of 4′,6-diamino-2-phenylindole; DIC, differential interference contrast.
(C) In vitro pull-down assay demonstrating the direct interaction between OsELF3 and HAF1. MBP-HAF1 was pulled down (PD) by GST-OsELF3 immobilized on glutathione-conjugated agarose beads and analyzed by immunoblotting (IB) using anti-MBP antibody.
(D) In vitro pull-down assay showing that the C-terminal domain (amino acids 503–760) of OsELF3 is required for its homodimer formation.
We further verified the protein interaction of HAF1 with OsELF3 and OsELF3 homodimer formation via a pull-down assay. Maltose binding protein-tagged HAF1 (MBP-HAF1) and GST-tagged OsELF3 (GST-OsELF3) fusion proteins were prepared separately. As shown in Figure 2C, MBP-HAF1 and GST-OsELF3 binding proteins in the pull-down samples with GST beads were detected using anti-MBP antibody. Also, MBP-OsELF3 fusion protein bound to GST-OsELF3 (503–760), while MBP-OsELF3 and GST-OsELF3 (503–760) did not bind to the negative controls GST and MBP, respectively (Figure 2D). Taken together, these results indicate that the C-terminal region of OsELF3 (containing amino acids 503–760) is indispensable for its interaction with HAF1 and OsELF3 homodimer formation.
HAF1 Mediates the Ubiquitination and Proteasomal Degradation of OsELF3
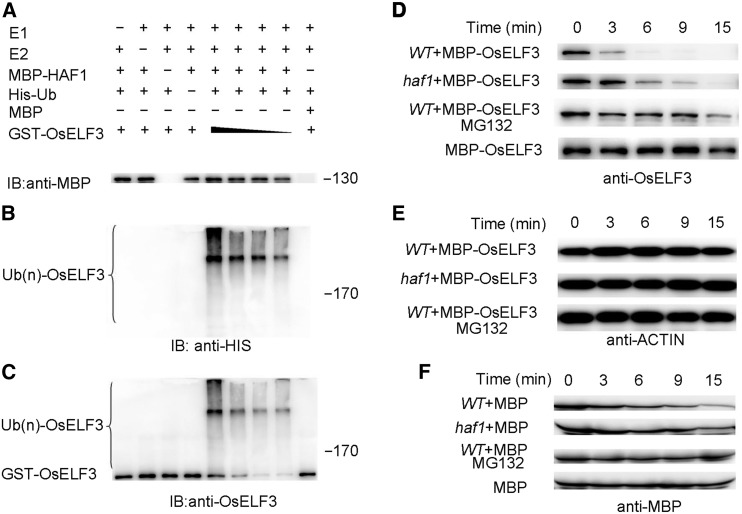
Since HAF1 is a C3HC4 RING domain-containing E3 ubiquitin ligase (Yang et al., 2015), we performed ubiquitination assays to examine the possibility that HAF1 mediates the ubiquitination of OsELF3. We purified the tagged proteins MBP-OsELF3 and GST-OsELF3 in Escherichia coli to test the specificity of the OsELF3 antibody by immunoblot analysis (Supplemental Figures 1A and 1B). With increasing amounts of MBP-OsELF3 and GST-OsELF3 protein, more intense MBP-OsELF3 and GST-OsELF3 bands were detected using the OsELF3 antibody (Supplemental Figures 1C and 1D). Using total protein extracts from wild-type rice variety ‘Zhonghua 11’ (ZH11) or ‘Zhenshan 97’ (ZS97) leaves as samples, a specific band was also detected (Supplemental Figures 1E and 1F). In a sample from a knockdown mutant of OsELF3 (Yang et al., 2013), the intensity of the band was weak (Supplemental Figure 1F). These results indicate that the OsELF3 antibody specifically detects OsELF3. We purified the tagged proteins MBP-HAF1 and GST-OsELF3 for ubiquitination assays in vitro (Figure 3A). In the presence of ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and His-Ub, GST-OsELF3 was found to be ubiquitinated by HAF1. With decreasing amounts of GST-OsELF3, the ubiquitinated GST-OsELF3 bands were less intense, as revealed by immunoblotting using anti-His (Figure 3B) or anti-OsELF3 antibodies (Figure 3C). When MBP-HAF1 was substituted with MBP (negative control), no ubiquitinated GST-OsELF3 band was detected. These results indicate that HAF1 mediates the ubiquitination of OsELF3 in vitro.
Figure 3.
In Vitro Ubiquitination Assays Showing the Degradation of OsELF3 by HAF1.
(A) to (C) HAF1 acts as an E3 ligase to mediate the ubiquitination of OsELF3. GST-OsELF3 ubiquitination assays were performed using MBP-HAF1 (or MBP) as a negative control (A). Ubiquitinated GST-OsELF3 was detected by His antibody (B) and OsELF3 antibody (C). Molecular masses of ubiquitinated GST-OsELF3 in kilodaltons are indicated on the right.
(D) to (F) HAF1 mediates the degradation of OsELF3 in vitro. Ten micrograms of total protein extracts was loaded into each lane, and ACTIN was used as a loading control. MBP was used as a negative control. MG132 is a proteasome inhibitor.
Next, we prepared total protein extracts from wild-type (ZH11) leaves for incubation with purified MBP-OsELF3 in cell-free degradation assays. Total protein was extracted from wild-type and haf1 plants individually. We found that MBP-OsELF3 degraded more slowly during incubation with protein extracts from haf1 plants compared with those from the wild type (Figure 3D). In the presence of the proteasome inhibitor MG132, the rate of MBP-OsELF3 degradation was lower during incubation with the same protein extracts from the wild type (Figure 3D). By contrast, no obvious degradation of rice ACTIN or control MBP protein was observed using the same plant extracts (Figures 3E and 3F). Together, these results further demonstrate that HAF1 is specifically responsible for OsELF3 protein degradation.
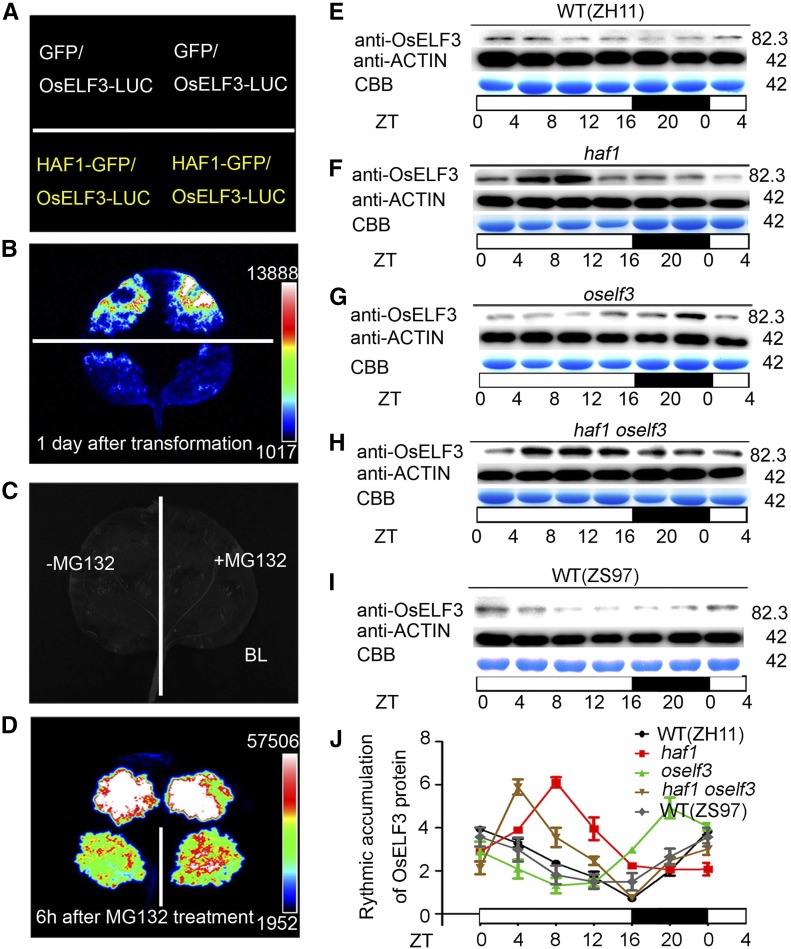
To confirm the ubiquitin-mediated degradation of OsELF3 by HAF1, we performed in vivo protein degradation assay via agroinfiltration in N. benthamiana. As shown in Figure 4A, using GFP protein as a control, GFP-tagged HAF1 (GFP-HAF1) was mixed with OsELF3-LUC (luciferase) and transiently infiltrated into separate areas of a single N. benthamiana leaf. After 1 d of infiltration, we quantified the average LUC activity. Compared with the strong LUC activity in the coexpression control, the coexpression of GFP-HAF1 with OsELF3-LUC led to a sharp decrease in LUC activity (Figure 4B), indicating that HAF1 mediates the degradation of OsELF3 in vivo. When MG132 was added to GFP-HAF1 plus OsELF3-LUC, the fluorescence intensity increased compared with that without MG132 treatment (Figures 4C and 4D), suggesting that HAF1 mediates ubiquitination and targets OsELF3 for degradation via the 26S proteasome pathway.
Figure 4.
In Vivo Ubiquitination Assays Showing the Degradation of OsELF3 by HAF1.
(A) to (D) Relative luciferase activity in wild tobacco leaves coinfiltrated by OsELF3-LUC and HAF1-GFP. GFP was used as a negative control. (A) and (C) show the setup used in (B) and (D), respectively. MG132 inhibits the degradation of OsELF3-LUC by HAF1-GFP. Black represents the weakest luciferase activity, and white represents the strongest luciferase activity.
(E) to (I) Time-course analysis showing rhythmic accumulation of OsELF3 in ZH11 (E), haf1 (F), oself3 (G), haf1 oself3 (H), and ZS97 (I) plants under LD conditions. OsELF3 was detected with anti-OsELF3 antibody. CBB, Coomassie Brilliant Blue staining. Molecular masses of OsELF3 and ACTIN proteins in kilodaltons are indicated on the right. White and black bars represent subjective days and nights, respectively. ZT, Zeitgeber time. ZT = 0 is defined as the time of lights on; numbers represent hours after ZT0 that the sample was harvested.
(J) The relative abundance (rhythmic accumulation) of OsELF3 protein in wild-type (ZH11), haf1, oself3, haf1 oself3, and wild-type (ZS97) plants. Mean and sd (error bars) values were obtained from three biological repeats (each protein sample was obtained from three separate plants).
HAF1 Maintains the Circadian Rhythm of OsELF3 Accumulation
We previously showed that OsELF3 promotes heading date under LDs mainly through maintaining its normal rhythmic expression pattern (Yang et al., 2013). To further investigate the role of HAF1 in OsELF3 accumulation in rice, we compared the diurnal accumulation of OsELF3 in the leaves of haf1, oself3, haf1 oself3, and wild-type plants under LDs. Immunoblot analysis using anti-OsELF3 antibody showed that OsELF3 accumulated to higher levels in the daytime than at night (Figure 4E). OsELF3 accumulated to higher levels in haf1 than in the wild type at most time points examined, with peak expression detected during the daytime (Figure 4F). In oself3, the phase of rhythmic protein expression was shifted backward compared with the wild type (Figure 4G). This pattern of protein oscillation is delayed compared with that observed for OsELF3 transcript (Yang et al., 2013), suggesting that the expression pattern of OsELF3 might be partially controlled by posttranscriptional regulation. Compared with oself3 plants, OsELF3 protein accumulation was higher at most time points in the oself3 haf1 double mutant (Figure 4H). We also examined OsELF3 accumulation in ZS97 leaves; OsELF3 expression showed a similar diurnal rhythm to that in ZH11, but at relatively lower levels (Figure 4I). Combining the data from two additional biological replicates (each protein sample was from three separate plants) (Supplemental Figure 2), we analyzed the rhythmic expression pattern of OsELF3 protein in the wild type (Figure 4E), haf1 (Figure 4F), oself3 (Figure 4G), and haf1 oself3 (Figure 4H) plants. OsELF3 showed a circadian rhythmic expression pattern, with a peak at dawn and a trough at dusk. The OsELF3 accumulation patterns were disturbed in oself3 and haf1. Higher levels of OsELF3 protein were detected in haf1 and oself3 haf1 compared with the wild type and oself3, respectively (Figure 4J). These results indicate that HAF1 is indispensible for maintaining the circadian rhythm of OsELF3 accumulation in rice.
HAF1 Does Not Affect the Transcription of OsELF3
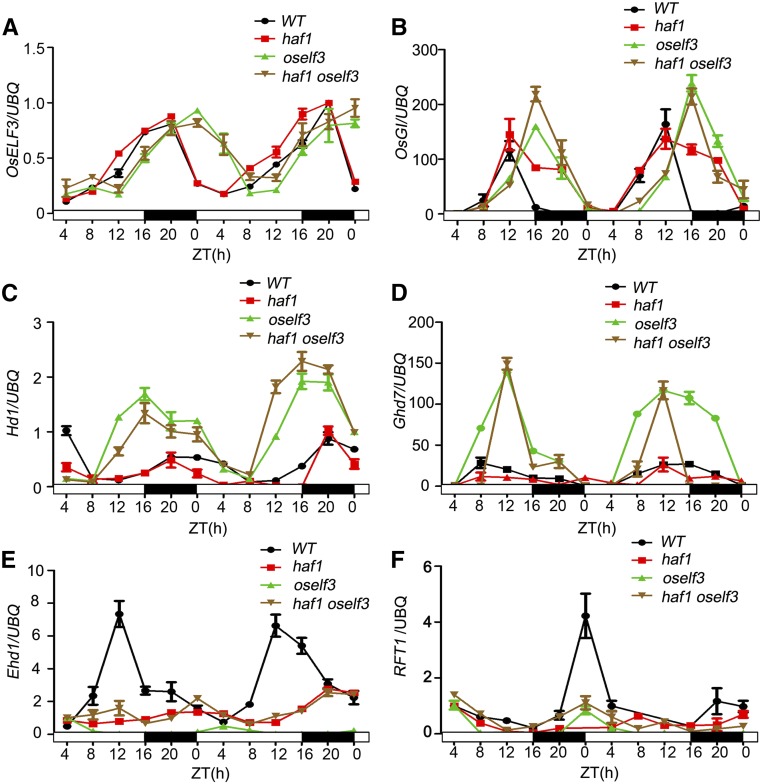
Because OsELF3 promotes flowering only under LDs (Yang et al., 2013), we examined whether HAF1 affects the transcription of OsELF3 and the other flowering regulators under artificial LDs (16 h light/8 h dark). The penultimate leaf blades were harvested every 4 h during a 48-h period from wild-type, haf1, oself3, and haf1 oself3 plants. We monitored the diurnal expression patterns of OsELF3, OsGI, Hd1, Ghd7, Ehd1, and RFT1 by qRT-PCR under LDs. Consistent with our previous investigation (Yang et al., 2013), OsELF3 showed a diurnal expression pattern, but the phase of this diurnal expression pattern was shifted in oself3 (Figure 5A). OsELF3 transcript levels and expression patterns were similar in haf1 plants compared with the wild type. Similar OsELF3 expression patterns were also observed in oself3 and haf1 oself3 plants (Figure 5A). The diurnal expression patterns of the other flowering regulators, including OsGI, Hd1, Ghd7, Ehd1, and RFT1, were consistent with our previous results (Yang et al., 2013). Compared with the wild type, the phase of diurnal expression patterns of OsGI and Hd1 were altered in oself3 (Figures 5B and 5C); the transcript levels of two flowering repressors, Hd1 and Ghd7, were dramatically increased in oself3 (Figures 5C and 5D). However, in the haf1 background, the transcript levels and diurnal expression phases of OsGI, Hd1, and Ghd7 did not obviously differ compared with the wild type or between oself3 and haf1 oself3 plants (Figures 5B to 5D). These results suggest that HAF1 has little effect on the transcription of OsELF3, OsGI, Hd1, and Ghd7. The expression of the flowering activators under LDs, Ehd1 and RFT1, was dramatically suppressed in oself3, haf1, and haf1 oself3 (Figures 5E and 5F), suggesting that HAF1 may participate in an intricate flowering pathway independent of OsELF3 under LDs. Thus, haf1 plants exhibit a delayed heading date (Yang et al., 2015).
Figure 5.
Rhythmic Expression Patterns of Heading Date-Related Genes in Wild-Type, haf1, oself3, and haf1 oself3 Lines under LD Conditions.
Diurnal expression patterns of OsELF3 (A), OsGI (B), Hd1 (C), Ghd7 (D), Ehd1 (E), and RFT1 (F) in different genetic backgrounds. The expression levels are relative to UBQ mRNA. In all panels, the mean of each point is based on the average of three biological repeats (RNA samples were from three separate plants) calculated using the relative quantification method. Error bars indicate standard deviations. White and black bars represent subjective days and nights, respectively. ZT, Zeitgeber time. ZT = 0 is defined as the time of lights on; numbers represent hours after ZT0 that the sample was harvested.
We also compared the expression patterns of circadian clock genes in oself3, haf1, haf1 oself3, and the wild type. OsELF3 had a dramatic effect on the expression of the clock-associated genes OsLHY, OsPRR1, OsPRR37, and OsPRR59 under LDs (Yang et al., 2013). In oself3 plants, the expression level of OsLHY was clearly reduced (Supplemental Figure 3A), whereas the transcript levels of OsPRR1 (Supplemental Figure 3B), OsPRR37 (Supplemental Figure 3C), and OsPRR59 (Supplemental Figure 3D) increased. In the haf1 mutant, HAF1 had no effect on the expression of OsLHY, OsPRR1, OsPRR37, or OsPRR59, which showed identical expression patterns in haf1 and the wild type under LDs (Supplemental Figure 3). Compared with the haf1 oself3 double mutant, the diurnal expression patterns of these genes did not obviously differ in haf1 plants (Supplemental Figure 3). We examined the free-running rhythms of clock-associated genes OsLHY, OsPRR1, OsPRR37, and OsPRR59 under continuous light conditions. These genes maintained the same robust rhythmic expression patterns in haf1 and the wild type and in oself3 and haf1 oself3 plants (Supplemental Figure 4). OsLHY transcript levels were lower in oself3 and haf1 oself3 compared with wild-type and haf1 plants, respectively (Supplemental Figure 4A). However, OsPRR1, OsPRR37, and OsPRR59 transcript levels increased in oself3 and haf1 oself3 plants (Supplemental Figures 4B to 4D). These results confirm the notion that OsELF3 is required for the rhythmic expression of circadian clock genes (Yang et al., 2013). Thus, we propose that HAF1 affects the function of OsELF3 mainly on the protein level but does not affect its diurnal transcription pattern.
OsELF3 Is Genetically Epistatic to HAF1
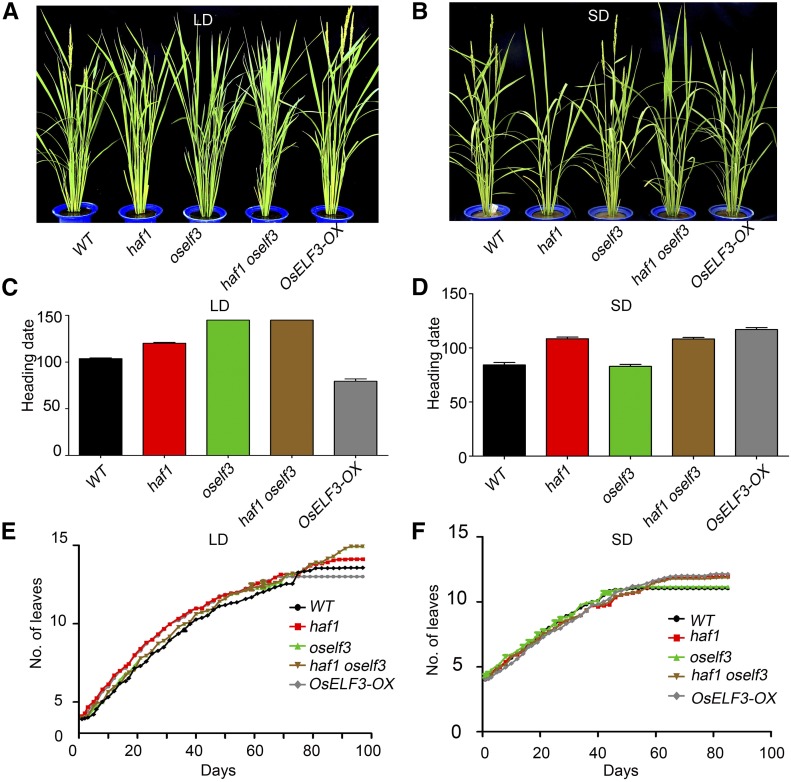
We previously showed that OsELF3 acts as a flowering promoter in LDs. Mutation in OsELF3 resulted in a delayed heading date only under LDs, but not SDs (Yang et al., 2013). However, haf1 showed later heading under both LDs and SDs (Yang et al., 2015). To investigate the genetic relationship between OsELF3 and HAF1, we examined the heading date of the haf1 oself3 double mutant. Under natural LD in summer at Wuhan, haf1 oself3 (116.40 ± 1.86 d) flowered as late as oself3 (116.17 ± 2.02 d), i.e., ∼15 d later than haf1 (101.37 ± 2.96 d) (Supplemental Figure 5). We then surveyed the heading dates of haf1, oself3, haf1 oself3, and OsELF3 overexpression plants (OsELF3-OX) under artificial LDs (16 h light/8 h dark) and SDs (8 h light/16 h dark) (Figure 6). Consistent with previous reports (Yang et al., 2013, 2015), both oself3 (139.73 ± 15.60 d) and haf1 (120.27 ± 5.81 d) plants exhibited delayed heading dates compared with wild-type plants (103.76 ± 5.53 d) under LDs (Figures 6A and 6C). OsELF3-OX plants exhibited a delayed heading date (117 ± 3.65) under SDs and an earlier heading date (79.5 ± 5.00 d) under LDs (Figures 6A to 6D). The leaf emergence rates did not differ among mutant lines and the wild type (Figures 6E and 6F), indicating that the differences in heading date are due to differences in the time of floral induction. The haf1 oself3 double mutant (138.24 ± 15.69 d) headed as late as oself3 plants (139.73 ± 15.60) under LDs (Figures 6A and 6C), indicating that OsELF3 is genetically epistatic to HAF1. We also generated F1 plants by crossing OsELF3-OX with haf1 plants. Under natural LDs in Wuhan, the F1 plants exhibited an early heading date similar to that of OsELF3-OX plants (Supplemental Figure 6). These results suggest that the delayed heading date of haf1 oself3 under LDs is mainly due to the disruption of the diurnal expression pattern of OsELF3 in oself3, not merely to OsELF3 abundance. In addition, the possibility that some flowering repressors degraded by HAF1 under LDs contribute to the delayed heading date in haf1 oself3 plants cannot be excluded.
Figure 6.
Heading Date of the haf1 oself3 Double Mutant under LD and SD Conditions.
(A) and (B) Phenotypes of the wild type, haf1, oself3, and haf1 oself3 at the heading stage under LD (A) and SD (B) conditions.
(C) and (D) Statistics of heading date in wild-type, haf1, oself3, and haf1 oself3 plants under LD (C) and SD (D) conditions. Mean and sd (error bars) values were obtained from at least 20 plants.
(E) and (F) Comparison of leaf emergence rates between wild-type plants and mutant lines under LD (E) and SD (F) conditions.
Interaction between HAF1 and OsELF3 Might Contribute to the Geographic Distribution of Different japonica Varieties
To examine the sequence variation of OsELF3, we analyzed whole genomic data from 979 rice accessions, including 446 wild rice species (Huang et al., 2012) and 533 cultivated rice varieties (Xie et al., 2015), which are distributed worldwide. The coding region of OsELF3 contains 21 single nucleotide polymorphisms (SNPs) that can be classified into 10 haplotypes (Supplemental Figure 7A). The 21 SNPs are present in seven nonsynonymous codons, four synonymous codons, one stop codon, and nine introns. The 10 haplotypes were classified into two major clades: one clade includes Hap1, Hap9, and Hap10, which are mainly represented by Aus accessions; the seven other haplotypes belong to another clade, which was traced to indica and japonica rice (Supplemental Figure 7B). The second SNP, G/A at position 5191, causes an amino acid change from leucine to serine in residue 558 (L558S) at the C terminus of OsELF3 (Supplemental Figure 7C). The OsELF3(L)-type allele is fixed in the cultivated accessions, but all wild and Aus rice varieties carry the OsELF3(S)-type allele (Supplemental Figure 7D). Most indica rice varieties carry the OsELF3(S) type allele, and the frequency of the OsELF3(L)-type allele is only 12.7% in the indica subpopulation. Genome-wide association study (GWAS) of heading date in the japonica population using a linear regression model provided by the FaST-LMM program (Lippert et al., 2011) showed that the second SNP, G/A at position 5191, is significantly associated with heading date (P = 4.95 × 10−8; Supplemental Figure 8). Interestingly, the amino acid change from leucine to serine (L558S) occurs in the interaction region of OsELF3 with HAF1. Yeast two-hybrid and pull-down assays demonstrated that HAF1 interacts with OsELF3(L) protein, but not with OsELF3(S) protein (Figures 1C and 2C). The japonica variety ‘ZH11’ carries the OsELF3(L)-type allele, and indica variety ‘ZS97’ carries the OsELF3(S) type allele. We demonstrated that MBP-OsELF3(L) could be degraded by HAF1 from total protein extracts from ZH11 leaves (Figure 3D). We also prepared total protein extracts from ZS97 leaves for incubation with purified MBP-OsELF3(S) in cell-free degradation assays (Supplemental Figure 9). The degradation of MBP-OsELF(S) showed a similar trend when it was incubated with total protein extracts from ZS97 or haf1 leaves (Supplemental Figure 9A). When MG132 was added to the assay, the protein degradation rate of OsELF3(S) was slower (Supplemental Figure 9B). These results suggest that HAF1 is only responsible for the ubiquitination and degradation of OsELF3(L), but not for the ubiquitination of OsELF3(S).
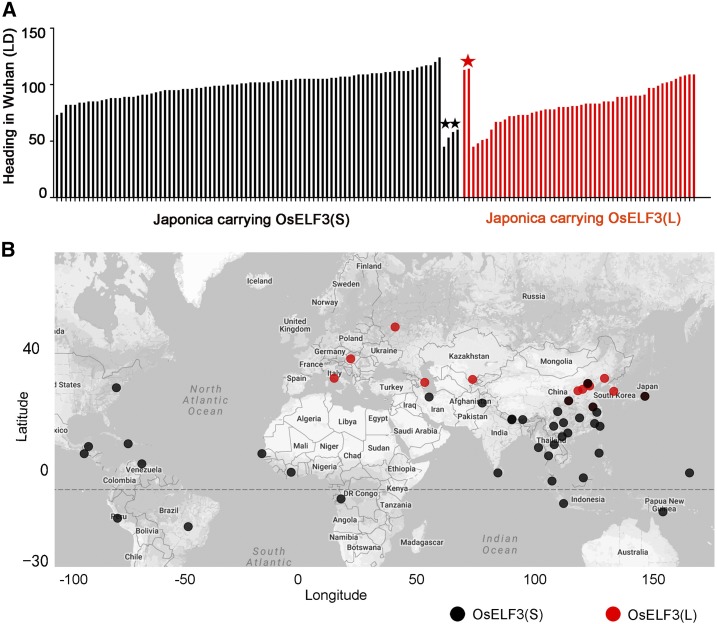
Next, we surveyed the heading dates of 142 japonica accessions (Supplemental Data Set 1) carrying two types of alleles, OsELF3(L) and OsELF(S), in Wuhan, finding that 52 accessions carry the OsELF3(L)-type allele and 90 harbor the OsELF(S)-type allele. With a few exceptions, accessions containing the OsELF3(L) allele flowered earlier than those harboring the OsELF(S) allele, further indicating that the OsELF3(L) protein is functional and may contribute to early flowering in the japonica accessions (Figure 7A). Only four accessions carrying the OsELF(S) allele flowered early, and two accessions carrying the OsELF3(L) allele flowered late. Moreover, japonica accessions carrying the OsELF3(L)/OsELF(S) alleles showed a clear geographic distribution pattern (Figure 7B). The japonica accessions carrying the OsELF3(L) allele are distributed at higher latitudes (between 35.7°N and 55°N), but accessions carrying the OsELF(S) allele are distributed at latitudes below 35.7°N. Thus, the SNP G/A at position 5191 in OsELF3 may be a footprint of natural and artificial selection for geographical distribution during rice breeding.
Figure 7.
Heading Date and Geographical Distribution of japonica Accessions Carrying the OsELF3(S) and OsELF3(L)-Type Alleles.
(A) Heading dates of japonica accessions separately carrying OsELF3(S)- and OsELF3(L)-type alleles in Wuhan in summer 2012. The few outliers are marked with asterisks. Black asterisk indicates early flowering accessions carrying OsELF3(S), and red asterisk indicates late flowering accessions carrying OsELF3(L).
(B) Geographic distribution of accessions carrying the OsELF3(S)-type (black dots) and OsELF3(L)-type (red dots) alleles. Longitude and latitude details for 142 accessions are shown in Supplemental Data Set 1. An Excel file (Supplemental Data Set 1) was converted to text format and imported into R software. A data packet in R software containing longitude and latitude information for each site all over the world enabled every accession to be placed in its proper coordinate on the map.
DISCUSSION
Although we previously showed that HAF1 and OsELF3 play important roles in promoting the heading of rice under LDs (Yang et al., 2013), the molecular mechanism underlying their roles in regulating flowering time was poorly understood. In this study, we showed that OsELF3 is the direct substrate of HAF1 for ubiquitination in vitro and in vivo. The natural variation in OsELF3 affects its interaction with HAF1, indicating that HAF1 mediates the circadian degradation of OsELF3, which may contribute to the variation in geographic distribution of different rice varieties. Thus, OsELF3 accumulation regulated by HAF1 plays a pivotal role in controlling flowering progress under LDs in rice.
Molecular Mechanism of Flowering between OsELF3 and ELF3
Although OsELF3 is regarded as the ortholog of ELF3 in rice, these genes share only 35% sequence identity (Yang et al., 2013). Moreover, ELF3 functions as a flowering repressor in Arabidopsis, as the Arabidopsis elf3 mutant exhibits early and photoperiod-insensitive flowering (Zagotta et al., 1996). Mutation in OsELF3, on the other hand, results in later flowering, and plants overexpressing OsELF3 show early flowering under LDs, indicating that OsELF3 is a flowering activator in rice (Yang et al., 2013). Compared with ELF3 in Arabidopsis, OsELF3 may exert diverse functions by recruiting different components.
ELF3 protein can be functionally characterized into three domains: the N-terminal, middle, and C-terminal domains (Liu et al., 2001; Yu et al., 2008; Herrero et al., 2012). The N-terminal domain is required for the physical interaction of ELF3 with the E3-ligase COP1 (Yu et al., 2008) and the photoreceptor PhyB (Liu et al., 2001). The N-terminal and middle domains of ELF3 participate in the interaction with COP1 and GI separately, which regulate the accumulation pattern of GI to shape its proper functioning in the control of circadian oscillation and photoperiodism (Yu et al., 2008). Additionally, the middle domain is involved in the formation of the ELF4-ELF3-LUX complex, which is required for circadian gating of hypocotyl growth in the evening and regulates leaf senescence by gating jasmonate signaling (Nusinow et al., 2011; Ezer et al., 2017; Zhang et al., 2018). The C-terminal domain of ELF3 does not appear to interact with any protein; it is essential for homodimer formation and for the localization of this protein in nuclear bodies (Liu et al., 2001; Herrero et al., 2012). In rice, the C-terminal domain of OsELF3 is also required for homodimer formation (Figures 1D, 2B, and 2D). Notably, this C-terminal domain mediates the physical interaction of OsELF3 with the E3-ligase HAF1 (Figure 1B). Although both COP1 and HAF1 are RING-finger E3 ligases, COP1 is well characterized and comprises a RING-finger domain, a coiled-coil domain, and seven WD40 repeats (Deng et al., 1992; McNellis et al., 1994); HAF1 contains another domain in addition to the conserved C3HC4-type RING-finger domain (Yang et al., 2015). Furthermore, COP1 and HAF1 interact with the N-terminal domain of ELF3 and the C-terminal domain of OsELF3, respectively. COP1 is involved in light-controlled development and flowering in Arabidopsis (Deng et al., 1991; Xu et al., 2016), but HAF1 has only been characterized as a heading date regulator that functions via the ubiquitination of Hd1 (Yang et al., 2015) and OsELF3 (this study). In Arabidopsis, ELF3 delays flowering by indirectly suppressing the expression of the flowering activator gene CO (Yu et al., 2008). By contrast, OsELF3 promotes heading by suppressing the expression of the key repressor gene, Ghd7, in rice (Matsubara et al., 2012; Yang et al., 2013). These findings suggest that the distinct flowering mechanism of OsELF3 might be more highly evolved in rice compared with that of ELF3 in Arabidopsis.
Natural Variation in OsELF3 Contributes to the Distinct Distribution Patterns of Different Rice Cultivars
Previous studies have suggested that ELF3 performs a highly conserved role in controlling flowering time between Arabidopsis and various crop species (Tajima et al., 2007; Faure et al., 2012; Matsubara et al., 2012; Undurraga et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012; Lu et al., 2017). The polyglutamine repeats in the C terminus of ELF3 affect the function of this protein in the control of circadian rhythms (Tajima et al., 2007), hypocotyl length, and flowering time in Arabidopsis (Undurraga et al., 2012). Analysis of natural variations revealed that the intracellular distribution of ELF3 protein is associated with its role in the circadian clock (Anwer et al., 2014). EARLY MATURITY8 (EAM8) is a barley (Hordeum vulgare) ortholog of Arabidopsis ELF3; various independent eam8 mutations have been selected that facilitated the movement of barley cultivation to high-latitude, short-season environments in Europe (Faure et al., 2012). Two temperate legumes, pea (Pisum sativum) and lentil (Lens culinaris), contain HIGH RESPONSE TO PHOTOPERIOD (HR) orthologs of ELF3; variation in the HR gene is also associated with photoperiod-insensitive early flowering in these two crop species (Weller et al., 2012). Natural variation in the soybean (Glycine max) J (Juvenile) locus, an ortholog of Arabidopsis ELF3, contributes to the long-juvenile trait (associated with high yields) and enables soybean to adapt to tropical regions (Lu et al., 2017). Natural variation in OsELF3 is involved in photoperiodic flowering and the regional distribution of rice varieties (Matsubara et al., 2012). Thus, natural variations in OsELF3 and its orthologs in different species may contribute to regional adaptation and have been selected during domestication.
In this study, we found that a SNP at position 5191 of OsELF3 resulting in an amino acid change from leucine to serine (L558S) is significantly associated with heading date in rice (Supplemental Figures 7C and 8). Further investigation of heading dates in various accessions confirmed that OsELF3(L)-type protein is functional and may contribute to early flowering in japonica accessions (Figure 7A). Our results are consistent with the finding that the amino acid variation at position 558 (S558L) of OsELF3 protein is associated with earlier heading dates (Matsubara et al., 2012). This amino acid variation is located in the interaction region of OsELF3 with HAF1 (Supplemental Figure 7C). Because HAF1 only interacts with the fragment carrying the OsELF3(L)-type protein (Figures 1C and 2C), we speculate that HAF1 may contribute to the natural variation in OsELF3 affecting the geographic distribution of rice accessions. Considering that HAF1 is required for maintaining the circadian rhythm of OsELF3 accumulation in japonica rice varieties (such as ZH11) (Figure 4E), perhaps another E3 ligase is responsible for the rhythmic oscillation of OsELF3 in the varieties carrying the OsELF3(L)-type allele.
HAF1 May Be Involved in Mediating the Degradation of Multiple Flowering Regulators
Protein degradation by the ubiquitin 26S proteasome system plays a critical role in the photoperiodic induction of flowering (Piñeiro and Jarillo, 2013). In Arabidopsis, COP1 is a versatile E3 ligase that mediates the protein degradation of multiple flowering regulators, such as GI, CO, and ELF3 (Liu et al., 2008; Yu et al., 2008; Sarid-Krebs et al., 2015). HAF1, as an essential C3HC4 RING domain-containing E3 ubiquitin ligase, plays a pivotal role in controlling heading date in rice (Yang et al., 2015). We previously demonstrated that HAF1 mediates Hd1 degradation and determines heading date under SDs (Yang et al., 2015). In this study, we demonstrated that HAF1 is essential for modulating the circadian rhythmic accumulation of OsELF3 and controls heading date under LDs. Although the haf1 oself3 double mutant headed as late as oself3 plants, OsELF3-OX plants showed an early heading date under LDs (Figures 6A and 6C), suggesting that OsELF3 may promote flowering, at least in part, independently of HAF1 mediating its degradation. We propose that HAF1 may be involved in mediating the degradation of multiple flowering regulators. OsELF3 promotes flowering mainly through suppressing the expression of flowering repressors Ghd7 and OsGI under LDs (Matsubara et al., 2012; Yang et al., 2013). Further analysis of the flowering repressors downstream of the OsELF3 pathways will be needed to determine whether HAF1 mediates their ubiquitination for degradation via the 26S proteasome system.
METHODS
Plant Materials and Growth Conditions
The Tos17-tagged rice (Oryza sativa) mutant oself3, OsELF3-overexpressing plants (Yang et al., 2013), and T-DNA-tagged mutant haf1 (Yang et al., 2015) were used for genetic analysis. The oself3 and haf1 mutants were generated using O. sativa subsp japonica ‘Zhonghua 11’ (ZH11), which contains the OsELF3(558L) type allele. O. sativa subsp indica ‘Zhenshan 97’ (ZS97) carries the OsELF3(558S)-type allele. To create the haf1 oself3 double mutant, F1 heterozygotes were obtained by crossing the haf1 mutant as the female plant with the oself3 mutant as the pollen donor. The plants were grown in the experimental field at Huazhong Agriculture University in Wuhan, China (30.4°N, 114.2°E) under natural LDs. The plants were grown in controlled growth chambers (B8; Eshengtaihe Ctrl Tech Company) with constant 28°C, 70% humidity under sodium lamp conditions (40,000 lumen) under artificial SDs (8 h light/16 h dark, 28°C) or LDs (16 h light/8 h dark, 28°C). Heading date was recorded as the number of days from germination to the day of panicle emergence from the leaf sheath.
Genotyping of Mutant Plants
Genotyping of wild-type, haf1, oself3, and haf1 oself3 plants was performed by PCR using the following primers: P1, P2, and P3, OsELF3-AS, OsELF3-S, and TRB2 (primers are listed in Supplemental Table 1). PCR reaction system was as follows: 100 ng DNA template, 2.0 μL 10× PCR buffer, 10 mM dNTP, 10 mM primers, 1 unit rTaq polymerase, and double-distilled water to a total volume of 20 μL. PCR was conducted with an initial step of 94°C incubation for 5 min, followed by of 30 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min. Leaf emergence rate was calculated as described by Itoh et al. (1998).
Yeast Two-Hybrid Assay
OsELF3 cDNA used as bait and prey were obtained by RT-PCR from wild-type plants. All truncated fragments of HAF1 (amino acids 1–112, 99–619, 288–619, 288–667, and 620–667) were isolated as previously described (Yang et al., 2015). OsELF3 was divided into five truncated fragments: aa(1–348), aa(1–519), aa(305–519), aa(305–760), and aa(503–760). PCR-amplified cDNAs with restriction sites EcoRI and BamHI were cloned into the pGBK and pGAD vectors (Clontech) (primers are listed in Supplemental Table 2). Mutated OsELF3(L558S) was used as a negative control. Yeast cotransformation and lacZ activity assays were performed following the manual of the Matchmaker Gold Yeast Two-Hybrid System (Clontech). Yeast two-hybrid assays were performed in the absence of adenine, leucine, histidine, and tryptophan in X-α-galactose-containing solid medium.
BiFC Assays
HAF1 and OsELF3 cDNAs were individually cloned into 35S-1301s-nYFP and 35S-1301s-cYFP vectors containing either N- or C-terminal YFP fragments, designated HAF1-n-YFP and c-YFP-OsELF3, respectively. Mutated HAF1(C620G) and OsELF3(L558S) fused separately with N- and C-terminal YFP were used as negative controls, which were designated HAF1(C620G)-n-YFP and c-YFP-OsELF3(L558S), respectively. The method for BiFC assays using Nicotiana benthamiana (wild tobacco) was performed as described by Kudla and Bock (2016). Specific primers used to produce the BiFC constructs are listed in Supplemental Table 2.
Expression of Recombinant Proteins in Bacteria and Purification
Full-length OsELF3 cDNA was inserted into GST fusion vector pGEX-4T-1 (Pharmacia) and a MBP fusion vector pMAL-C2X (primers are listed in Supplemental Table 2). The resulting GST-OsELF3 and MBP-OsELF3 fusion proteins were expressed in BL21-CodonPlus (Stratagene) Escherichia coli cells and purified using glutathione Sepharose 4B (Pharmacia) and amylose resin beads (New England Biolabs). A truncated fragment of the OsELF3 C-terminal domain was inserted into GST fusion vector pGEX-4T-1. MBP/GST-tagged HAF1 protein and MBP/GST alone were described previously (Yang et al., 2015).
Pull-Down Assays
The coding sequences of HAF1 and OsELF3 (amino acids 503–760) were cloned into the pGEX-4T-1 vector to generate the construct GST-HAF1 and GST-OsELF3 (503–760), respectively (primers are listed in Supplemental Table 2). GST-OsELF3(L558S) was used as a negative control. The method use for the in vitro pull-down assays was modified from Yang et al. (2015). Bacterial lysates containing ∼15 mg of GST-OsELF3 or GST-OsELF3(L558S) fusion proteins were mixed with lysates containing ∼30 mg of MBP-HAF1 fusion proteins. Glutathione Sepharose (30 μL; GE Life Sciences) was added to each combined solution, followed by incubation at 4°C for 60 min with rocking. The beads were washed four times with TGH buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, pH 8.0, 1% Triton X-100, 10% glycerol, 1 mM PMSF, and 1× Complete protease inhibitor cocktail [Roche]), and the isolated proteins were further separated on two 10% SDS-PAGE gels and detected by immunoblot analysis with anti-GST antibody (Abmart; lot number M20007L, 1:1000 dilution) and anti-MBP antibody (NEB; lot number E8032L, 1:10,000 dilution), respectively.
In Vitro Ubiquitination Assays
The reaction mixtures (30 μL) for the ubiquitination assays contained 110 ng of E1 (Boston Biochem), 170 ng of E2 (UBCh5a; Boston Biochem), 1 mg of His-ubiquitin (Boston Biochem), and 2 mg of MBP-HAF1 fusion protein with gradually increasing concentrations of GST-OsELF3 fusion protein in reaction buffer containing 50 mM Tris, pH 7.4, 2 mM DTT, 5 mM MgCl2, and 2 mM ATP. After 3 h incubation at 30°C, the reactions were stopped by adding sample buffer, and the mixtures were divided in half and separated in two 10% SDS-PAGE gels. Ubiquitinated GST-OsELF3 was detected using anti-OsELF3 antibody (Abclonal; lot number WG00551D, 1:100 dilution) and anti-HIS antibody (Abmart; lot number M30111L, 1:1000 dilution). Images were visualized using a Tanon-5200 chemiluminescent imaging system (Tanon Science and Technology).
In Vivo Ubiquitination Assay
The coding sequence of OsELF3 was cloned into the PFA2300-LUC vector to generate the construct OsELF3-LUC, and HAF1 cDNA was cloned into the 35s-1300-GFP vector (primers are listed in Supplemental Table 2). Agroinfiltration for the in vivo ubiquitination assay was performed as described by Liu et al. (2010). The above constructs were transformed into Agrobacterium tumefaciens GV3101. Agrobacterium cells carrying the HAF1-GFP and OsELF3-LUC plasmids were infiltrated into N. benthamiana leaves. The corresponding empty GFP vectors were used as the negative controls. The plants were grown in a growth chamber under LD conditions (16 h light and 8 h dark) for 24 h. The infiltrated N. benthamiana leaves were sprayed with luciferin (100 mM) and incubated in the dark for 10 min. The leaves were observed under a low-light cooled CCD imaging apparatus (Tanon Science and Technology). For proteasome inhibition, leaves were infiltrated with 10 mM MgCl2 and 50 μM MG132 (Sigma-Aldrich) solution for 6 h before sample collection. Quantification of LUC activity was performed as described by Zhu et al. (2010) at 24 h after infiltration.
Preparation of the OsELF3 Polyclonal Antibody
To prepare the OsELF3 polyclonal antibody, an 840-bp DNA fragment encoding a 280-amino acid fragment of OsELF3 (residues 170–450) was cloned into the pGEX-4T-1 vector (Novagen). The recombinant protein was expressed in E. coli DE3 cells (Transgen) and purified using glutathione Sepharose 4B (Pharmacia) to produce rabbit polyclonal antibodies (prepared by Abclonal). The antibody (1:100 dilution) was tested by immunoblot analysis using total proteins extracted from ZH11 or ZS97 leaves, and MBP-OsELF3 and GST-OsELF3 were purified from the E. coli cells.
Measuring OsELF3 Levels in Rice
Leaves from 15-d-old seedlings were harvested and ground into a fine powder in liquid nitrogen. A 200-μL aliquot of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, and 1× Complete protease inhibitor cocktail [Roche]) was added to each 100-mg powder sample. The mixture was vortexed and chilled on ice for 5 min. The samples were centrifuged at 13,000 rpm for 20 min at 4°C, and the supernatant was collected and stored at −70°C. An equal amount of rice protein was loaded and separated by SDS-PAGE and stained by Coomassie Brilliant Blue. To detect OsELF3 levels, an equal amount of rice protein was separated on a 10% SDS-PAGE gel and detected by immunoblot analysis with anti-OsELF3 antibody (1:100 dilution). To measure OsELF3 protein levels, 10 μg of total protein extract was loaded into each lane. ACTIN was used as an internal control, with 1 μg total protein extract used to measure ACTIN protein levels and for Coomassie blue staining. For each time point, leaves from three separate plants were sampled. Quantitative analysis for each time point was calculated using Image J software. The parameters for measurement were area, mean gray value, and integrated density. The value for each time point was normalized to ACTIN protein levels. Statistical analysis was performed to determine the mean and sd values from three biological replicates (separate experiments) and three technical replicates (within the same experiment).
Cell-Free Protein Degradation Assay
For the cell-free protein degradation assay, seedlings were grown in a greenhouse for 10 to 14 d under natural daylength conditions. Total proteins were extracted in degradation buffer (25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, and 10 mM ATP). To monitor the degradation of the expressed recombinant MBP-OsELF3, MBP-OsELF3(L558S), and MBP proteins, 200 ng of purified MBP-OsELF3, MBP-OsELF3(L558S), and MBP protein was added to 75 μL of seedlings extracts for the individual assays. The reaction mixtures were incubated at 30°C for the indicated time points. Reactions were stopped with adding 4× SDS sample buffer and boiling at 100°C for 10 min. An equal amount of each reaction was loaded onto a 10% (w/v) SDS-PAGE gel and detected by immunoblot analysis with anti-OsELF3 antibody (1:1000 dilution).
Diurnal Gene Expression Analysis
Rice seedlings were grown for 30 d in the greenhouse under natural daylength conditions. For the LD samples, the plants were transferred to a growth chamber set for LDs (16 h light/8 h dark, 28°C and 50% humidity) and entrained for 5 d. The, penultimate leaves were harvested every 4 h during a 48-h period from wild-type, haf1, oself3, and haf1 oself3 plants. For the free-running samples, the plants were transported to constant light conditions after being entrained for 5 d under LDs. Total RNA was extracted from various plant tissues using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and first-strand cDNA was synthesized from 2.5 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen). qRT-PCR was performed (primers are listed in Supplemental Table 3) with the Applied Biosystems 7500 real-time PCR detection system using SYBR Green Master Mix (Applied Biosystems). The measurements were obtained using the relative quantification method.
GWAS for Heading Date
An association panel consisting of a diverse collection of 156 O. sativa japonica accessions was planted and phenotyped in 2012 in Wuhan under LDs (Xie et al., 2015). Heading date was recorded as the duration of the period from sprouting to heading (panicle emergence from the leaf sheath). The genotype data were obtained from the RiceVarMap database (Zhao et al., 2015). The P values of each SNP within the coding region of OsELF3 protein were calculated, and the most significant SNPs that associated with heading date were identified.
Statistical Analysis
RNA or protein samples used for time-course analysis at each time point were obtained from three separate plants per genotype. For diurnal gene expression analysis, 2−△△CT values were calculated for each ZT, mean values from three biological repeats were used to exhibit differences among genotypes, and error bars indicate sd. For time-course analysis of OsELF3 protein, Image J software was used to calculate the gray value of each band obtained by immunoblotting. Mean values and standard deviations were obtained from three biological and three technical replicates. For statistical analysis of heading date, mean and sd (error bars) from at least 20 plants are indicated on the y axes.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: HAF1, LOC_Os04g55510; OsELF3, LOC_Os06g05060; OsGI, AJ133787; Hd1, LOC_Os06g16370; Ghd7, Os07g15770; Ehd1, AB092506; Hd3a, AB052944; and RFT1, AB062676.
Supplemental Data
Supplemental Figure 1. Detection specificity of the OsELF3 antibody.
Supplemental Figure 2. Biological duplications for time-course detection of OsELF3 protein accumulation.
Supplemental Figure 3. Rhythmic expression patterns of circadian clock genes in the wild type, haf1, oself3, and haf1 oself3 under LDs, as revealed by qRT-PCR analysis.
Supplemental Figure 4. Rhythmic expression patterns of circadian clock genes in the wild type, haf1, oself3, and haf1 oself3 under constant light conditions.
Supplemental Figure 5. Phenotypes of haf1, oself3, and haf1 oself3 under natural LDs in Wuhan.
Supplemental Figure 6. Phenotype of OsELF3-OX/haf1 lines under natural LDs in Wuhan.
Supplemental Figure 7. Haplotype analysis of OsELF3, evolutionary relationship analysis among OsELF3 haplotypes, and frequency analysis of OsELF3(L) and OsELF3(S) alleles.
Supplemental Figure 8. GWAS for heading date in various OsELF3 gene regions.
Supplemental Figure 9. Cell-free degradation assay of MBP-OsELF3(558S) incubated with total protein extracts from ZS97 or haf1 leaves.
Supplemental Table 1. Primers for genotyping analysis.
Supplemental Table 2. Primers for plasmid construction.
Supplemental Table 3. Primers for qRT-PCR.
Supplemental Data Set 1. Regional distribution and heading date information for 142 japonica accessions.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by The National Key Research and Development Program of China (2016YFD0100903), the National Natural Science Foundation of China (31425018, 31370222, and 31630054), the Program for Chinese Outstanding Talents in Agricultural Scientific Research, and the Wuhan Yellow Crane Talent Program. We thank Ying Yang for developing haf1 oself3 mutants.
AUTHOR CONTRIBUTIONS
C.W. and C.Z. conceived this project and designed the research. C.W. supervised the study. C.W., C.Z., Q. P., D.Z., and Y.C. performed the genetic analysis. C.Z., D.F., and M.D. performed the ubiquitin activity analyses. C.Z., Y.Y., W.X., and Y.O. performed the haplotype analysis of OsELF3. C.Z. and X.L. surveyed the heading dates of the japonica accessions. C.W. and C.Z. analyzed the data and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Anwer M.U., Boikoglou E., Herrero E., Hallstein M., Davis A.M., Velikkakam James G., Nagy F., Davis S.J. (2014). Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 3: 02206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box M.S., et al. (2015). ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 25: 194–199. [DOI] [PubMed] [Google Scholar]
- Covington M.F., Panda S., Liu X.L., Strayer C.A., Wagner D.R., Kay S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A., Tian W., Wei M., Yan W., He H., Zhou D., Huang X., Li S., Ouyang X. (2017). The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant 10: 948–961. [DOI] [PubMed] [Google Scholar]
- Ezer D., et al. (2017). The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Turner A.S., Gruszka D., Christodoulou V., Davis S.J., von Korff M., Laurie D.A. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA 109: 8328–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86. [DOI] [PubMed] [Google Scholar]
- Fu C., Yang X.O., Chen X., Chen W., Ma Y., Hu J., Li S. (2009). OsEF3, a homologous gene of Arabidopsis ELF3, has pleiotropic effects in rice. Plant Biol (Stuttg) 11: 751–757. [DOI] [PubMed] [Google Scholar]
- Fujino K., Yamanouchi U., Yano M. (2013). Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor. Appl. Genet. 126: 611–618. [DOI] [PubMed] [Google Scholar]
- Gao H., et al. (2013). Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 9: e1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Ariza J., Galbiati F., Goretti D., Brambilla V., Shrestha R., Pappolla A., Courtois B., Fornara F. (2015). Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 66: 2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretti D., Martignago D., Landini M., Brambilla V., Gómez-Ariza J., Gnesutta N., Galbiati F., Collani S., Takagi H., Terauchi R., Mantovani R., Fornara F. (2017). Transcriptional and post-transcriptional mechanisms limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13: e1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Ogiso-Tanaka E., Matsubara K., Yamanouchi U., Ebana K., Yano M. (2013). Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Aoki M., Kurotani K., Yokoi S., Shinomura T., Takano M., Shimamoto K. (2011). Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics 285: 461–470. [DOI] [PubMed] [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42: 635–638. [DOI] [PubMed] [Google Scholar]
- Itoh J.I., Hasegawa A., Kitano H., Nagato Y. (1998). A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell 10: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58: 3091–3097. [DOI] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.L., Lee S., Kim H.J., Nam H.G., An G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145: 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Herrero E., Bujdoso N., Millar A.J., Tóth R., Gyula P., Nagy F., Davis S.J. (2011). A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell 23: 3230–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Bock R. (2016). Lighting the way to protein-protein interactions: recommendations on best practices for bimolecular fluorescence complementation analyses. Plant Cell 28: 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.T., Yoo S.C., Koo B.H., Cho S.H., Park J.W., Zhang Z., Li J., Li Z., Paek N.C. (2014). Natural variation in Early flowering1 contributes to early flowering in japonica rice under long days. Plant Cell Environ. 37: 101–112. [DOI] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., Jarillo J.A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., et al. (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63: 18–30. [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C.M., Davidson R.I., Heckerman D. (2011). FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835. [DOI] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wagner D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49: 773–779. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Yamanouchi U., Wang Z.X., Minobe Y., Izawa T., Yano M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 148: 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Yamanouchi U., Nonoue Y., Sugimoto K., Wang Z.X., Minobe Y., Yano M. (2011). Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 66: 603–612. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Ogiso-Tanaka E., Hori K., Ebana K., Ando T., Yano M. (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53: 709–716. [DOI] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., Nonoue Y., Yano M., Izawa T. (2016). Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86: 221–233. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., et al. (2008). Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 56: 1018–1029. [DOI] [PubMed] [Google Scholar]
- Peng L.T., Shi Z.Y., Li L., Shen G.Z., Zhang J.L. (2007). Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem. Biophys. Res. Commun. 360: 251–256. [DOI] [PubMed] [Google Scholar]
- Piñeiro M., Jarillo J.A. (2013). Ubiquitination in the control of photoperiodic flowering. Plant Sci. 198: 98–109. [DOI] [PubMed] [Google Scholar]
- Raschke A., et al. (2015). Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 15: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid-Krebs L., Panigrahi K.C., Fornara F., Takahashi Y., Hayama R., Jang S., Tilmes V., Valverde F., Coupland G. (2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 84: 451–463. [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima T., Oda A., Nakagawa M., Kamada H., Mizoguchi T. (2007). Natural variation of polyglutamine repeats of a circadian clock gene ELF3 in Arabidopsis. Plant Biotechnol. 24: 237–240. [Google Scholar]
- Thines B., Harmon F.G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 107: 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Taoka K., Shimamoto K. (2011). Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14: 45–52. [DOI] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594. [DOI] [PubMed] [Google Scholar]
- Undurraga S.F., Press M.O., Legendre M., Bujdoso N., Bale J., Wang H., Davis S.J., Verstrepen K.J., Queitsch C. (2012). Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc. Natl. Acad. Sci. USA 109: 19363–19367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Q., Sarmast M.K., Jiang J., Dehesh K. (2015). The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27: 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J.L., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 109: 21158–21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., You C., Li C., Long T., Chen G., Byrne M.E., Zhang Q. (2008). RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. USA 105: 12915–12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., et al. (2015). Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. USA 112: E5411–E5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhu D., Deng X. (2016). The role of COP1 in repression of photoperiodic flowering. F1000 Fac. Rev. 5: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yan W.H., Wang P., Chen H.X., Zhou H.J., Li Q.P., Wang C.R., Ding Z.H., Zhang Y.S., Yu S.B., Xing Y.Z., Zhang Q.F. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4: 319–330. [DOI] [PubMed] [Google Scholar]
- Yang Y., Peng Q., Chen G.X., Li X.H., Wu C.Y. (2013). OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol. Plant 6: 202–215. [DOI] [PubMed] [Google Scholar]
- Yang Y., Fu D., Zhu C., He Y., Zhang H., Liu T., Li X., Wu C. (2015). The RING-finger ubiquitin ligase HAF1 mediates Heading date 1 degradation during photoperiodic flowering in rice. Plant Cell 27: 2455–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., Sasaki T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta M.T., Hicks K.A., Jacobs C.I., Young J.C., Hangarter R.P., Meeks-Wagner D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10: 691–702. [DOI] [PubMed] [Google Scholar]
- Zakhrabekova S., et al. (2012). Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. USA 109: 4326–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Wei H., Li N., Tian W., Chong K., Wang L. (2018). Circadian evening complex represses Jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11: 326–337. [DOI] [PubMed] [Google Scholar]
- Zhao H., Yao W., Ouyang Y., Yang W., Wang G., Lian X., Xing Y., Chen L., Xie W. (2015). RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res. 43: D1018–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Huang X., Ouyang X., Chen W., Du A., Zhu L., Wang S., Deng X.W., Li S. (2012). OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One 7: e43705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang Y., Li R., Song X., Wang Q., Huang S., Jin J.B., Liu C.M., Lin J. (2010). Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 61: 223–233. [DOI] [PubMed] [Google Scholar]