Assays of three key mutations reveal that RBP-P is indispensable for the localization of glutelin and prolamine mRNAs in rice endosperm cells.
Abstract
In developing rice (Oryza sativa) endosperm, mRNAs of the major storage proteins, glutelin and prolamine, are transported and anchored to distinct subdomains of the cortical endoplasmic reticulum. RNA binding protein RBP-P binds to both glutelin and prolamine mRNAs, suggesting a role in some aspect of their RNA metabolism. Here, we show that rice lines expressing mutant RBP-P mislocalize both glutelin and prolamine mRNAs. Different mutant RBP-P proteins exhibited varying degrees of reduced RNA binding and/or protein-protein interaction properties, which may account for the mislocalization of storage protein RNAs. In addition, partial loss of RBP-P function conferred a broad phenotypic variation ranging from dwarfism, chlorophyll deficiency, and sterility to late flowering and low spikelet fertility. Transcriptome analysis highlighted the essential role of RBP-P in regulating storage protein genes and several essential biological processes during grain development. Overall, our data demonstrate the significant roles of RBP-P in glutelin and prolamine mRNA localization and in the regulation of genes important for plant growth and development through its RNA binding activity and cooperative regulation with interacting proteins.
INTRODUCTION
mRNA localization, the process by which mRNAs are transported to specific subcellular compartments for localized translation, is an essential and universal mechanism that efficiently drives protein targeting in eukaryotic and prokaryotic organisms (Nevo-Dinur et al., 2011; Medioni et al., 2012; Blower, 2013; Weis et al., 2013; Tian and Okita, 2014). The targeting of mRNA expedites the accumulation of the coded proteins to specific cell compartments and is an underlying mechanism responsible for cell polarity, cell patterning, and cell-fate determination (Medioni et al., 2012; Blower, 2013; Tian and Okita, 2014). As such, mRNA localization is prominently observed in developing oocytes and early embryos, structurally polarized fibroblasts and neuron cells, and budding yeast (Jansen, 2001; Medioni et al., 2012; Weis et al., 2013; Tian and Okita, 2014). In Drosophila melanogaster embryos, 71% of the 3000 transcripts examined were found to be specifically localized in distinct subcellular patterns (Lécuyer et al., 2007). In yeast, more than 20 mRNAs have been reported to specifically localize to daughter cells during vegetative growth (Gonsalvez et al., 2005).
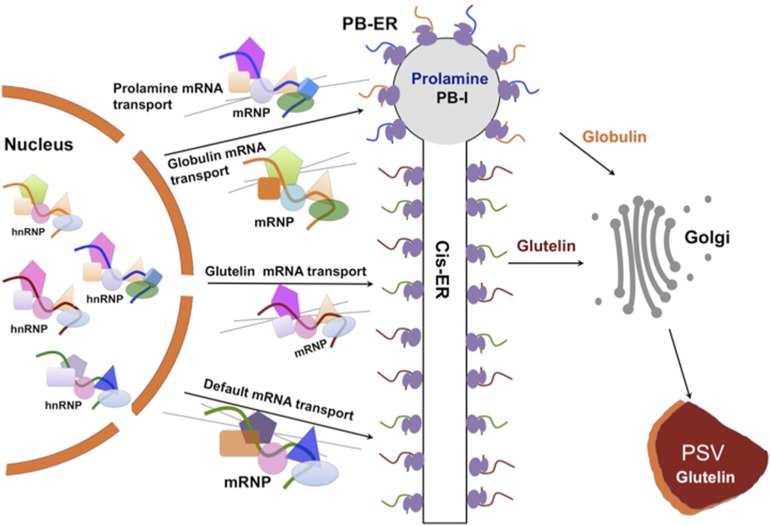
The localization of RNAs to specific intracellular destinations is also evident in higher plants. Developing rice (Oryza sativa) grains synthesize and store two major types of storage proteins, glutelin and prolamine, in endosperm cells. Although both storage protein types are synthesized on the rough endoplasmic reticulum (ER), they are packaged into separate endomembrane compartments. Prolamines are retained as intracisternal inclusions in the ER lumen where they form ER-derived protein body I (PB-I), whereas glutelins are exported to the Golgi and subsequently transported to the protein storage vacuoles (PSVs) to form PB-II (Tian and Okita, 2014). These protein deposition processes are facilitated by the localization of the mRNAs on distinct ER subdomains (Figure 1): Prolamine mRNAs are concentrated on the protein body ER (PB-ER), whereas glutelin mRNAs are localized on adjoining cisternal-ER (Cis-ER) (Choi et al., 2000; Crofts et al., 2005; Doroshenk et al., 2012; Tian and Okita, 2014).
Figure 1.
Model of mRNA Localization to the ER Subdomains in Rice Endosperm Cells.
The localization of mRNAs is initiated in the nucleus. Zip code and other cis-elements of transcribed mRNAs are recognized and bound by relevant RBPs in the nucleus, forming RNP complexes. Upon export to the cytoplasm, RNP complexes are remodeled, i.e., selective RBPs and accessory proteins are removed while others are added, enabling their transport via the cytoskeleton to their destination sites at the cortical ER where RNAs are translated, stored, or processed. Three transport pathways have been identified in developing rice endosperm based on the localization of storage protein mRNAs. Prolamine and globulin mRNAs are targeted to the protein body-ER (PB-ER) that delimits the prolamine intracisternal inclusions, while glutelin mRNAs are transported and localized to adjoining Cis-ER. In addition, there is a default RNA transport pathway to the Cis-ER.
RNA localization is mediated by cis-sequence elements, or zip codes, that are recognized by specific trans-factors. The cis-sequences/zip codes are usually located in the 3′ untranslated region (UTR) of the mRNA but can also be found in the coding region (Jansen, 2001; Andreassi and Riccio, 2009; Tian and Okita, 2014). In the case of rice storage protein mRNA localization, multiple zip codes are found in the coding sequence and 3′UTR (Hamada et al., 2003b; Andreassi and Riccio, 2009; Washida et al., 2009; Tian and Okita, 2014). The zip code sequences for both prolamine and glutelin mRNAs are AU-rich (Hamada et al., 2003a; Washida et al., 2009). Removing these zip code sequences results in the mislocalization of the corresponding mRNAs, while their addition specifically targets reporter RNAs to either the PB-ER (prolamine zip codes) or Cis-ER locations (glutelin zip codes) (Hamada et al., 2003b; Andreassi and Riccio, 2009; Washida et al., 2009; Tian and Okita, 2014).
The zip code sequences are recognized and bound by specific RNA binding proteins (RBPs), which together with other accessory proteins form a large ribonucleoprotein (RNP) complex. The RNP complex is initially formed in the nucleus and is remodeled (i.e., undergoes loss and addition of RBPs and accessory proteins) several times to accommodate the various steps encompassing RNA localization. These steps include nuclear export, translation suppression, transport to and anchoring at the destination site in the cytoplasm, and translation activation (Figure 1). Consistent with this multistep process, 15 prolamine RNA binding proteins were isolated via affinity chromatography using a prolamine zip code sequence as bait (Crofts et al., 2010). Five of those RBPs, RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q, coassemble into three zip code RNA binding complexes to target prolamine mRNAs to their final location (Yang et al., 2014). By contrast, a single glutelin RNA binding protein, RBP-P, has been identified through northwestern blot analysis using glutelin zip code RNA (Doroshenk et al., 2014). As RBP-P was also found in the collection of prolamine zip code-bound RBPs, it is likely involved in both prolamine and glutelin gene expression and RNA localization (Doroshenk et al., 2014).
Here, we provide direct evidence that RBP-P plays an essential role in the localization of glutelin and prolamine mRNAs. Our studies show that mutations in RBP-P result in the mislocalization of both glutelin and prolamine mRNAs on the cortical ER, which is likely due to reduced RNA binding affinity and/or the interruption of protein-protein interactions. Moreover, partial loss of RBP-P function results in a broad deficiency in plant growth and development, indicating that this RNA binding protein plays multiple roles in RNA metabolism and, in turn, gene regulation.
RESULTS
Mutations in RBP-P Result in Mislocalization of Glutelin and Prolamine mRNAs
RBP-P was identified as a putative RNA binding protein that binds to the zip code sequences from both glutelin and prolamine RNAs (Crofts et al., 2010; Doroshenk et al., 2014). It is encoded by the rice gene LOC_Os01g16090, which contains a 1473-bp coding sequence devoid of introns (Figure 2A). The coded RBP-P protein is modular in structure and contains two RNA recognition motifs (RRMs) bound by N-terminal peptides rich in alanine (A) or glutamic acid (E) residues and a C-terminal region rich in glycine (G) residues (Figure 2A; Supplemental Figure 1). Sequence alignment (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of orthologs from other species shows that RBP-P belongs to the UBA2 family of proteins (Supplemental Figure 1). Arabidopsis thaliana AtUBA2a and AtUBA2b, which share 50% identity with RBP-P, have been found to interact with the oligouridylate binding protein 1 (UBP1)-associated protein (Lambermon et al., 2002), and together this multiprotein complex is reported to function in pre-mRNA maturation and RNA stability in plants (Lambermon et al., 2000). Although the two RRM domains are highly conserved among these orthologous proteins, the N and C termini are highly variable (Supplemental Figure 1), suggesting the function of these proteins may have diverged during evolution.
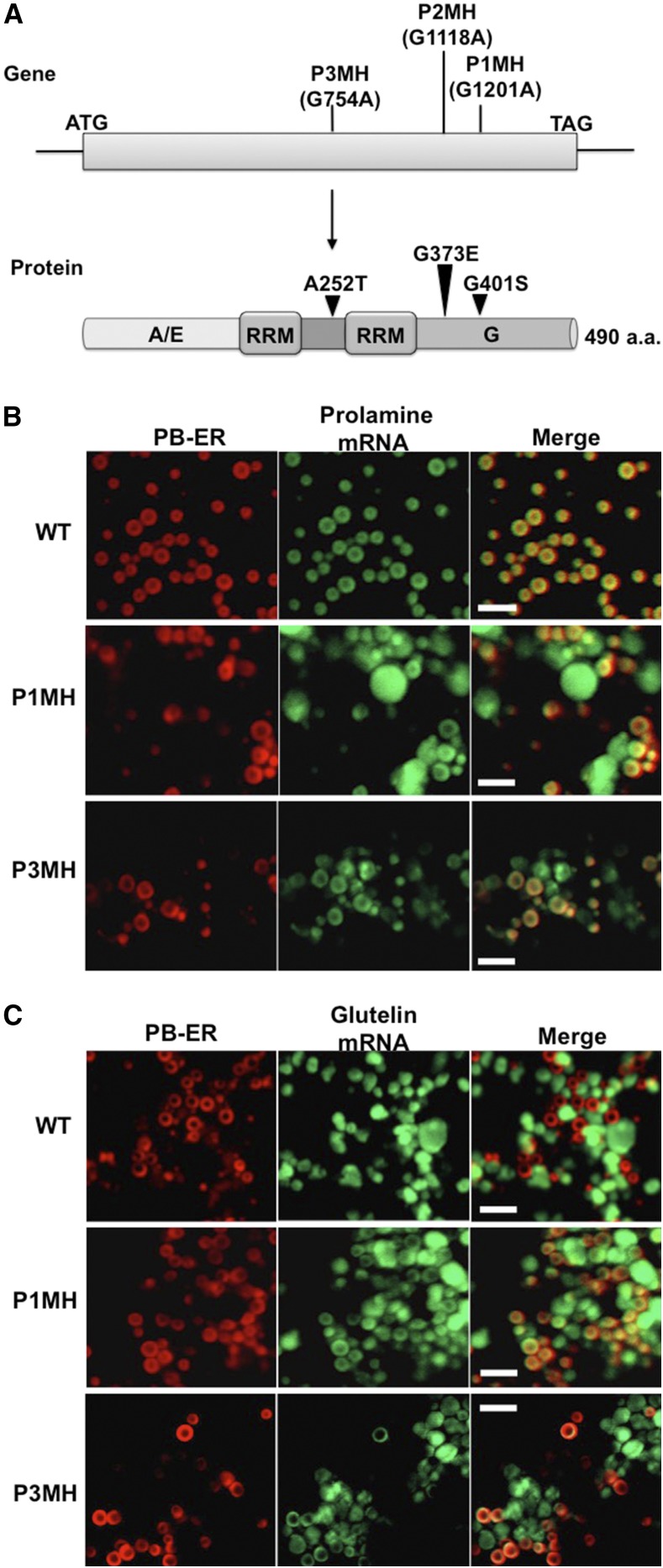
Figure 2.
Mutations of RBP-P Confer Partial Mislocalization of Prolamine and Glutelin mRNAs.
(A) Schematic representation of RBP-P point mutation sites in P1MH, P2MH, and P3MH mutants. A/E, alanine (A) and glutamic acid (E) enrichment in the N-terminal domain; G, glycine (G) enrichment in the C-terminal domain. aa, amino acids.
(B) and (C) Results of in situ RT-PCR to assess the location of prolamine (B) and glutelin (C) mRNAs to subdomains of the cortical-ER in wild-type and RBP-P mutant lines. In situ RT-PCR was performed directly on rice grain sections in the presence of Alexa-488-UTP (green) to label mRNAs. PB-ER was stained using Rhodamine B dye (red). Note that prolamine and glutelin mRNAs are localized to the PB-ER and Cis-ER, respectively, in the wild type. In P1MH and P3MH, the location of both mRNA species is disrupted and instead distributed to both PB-ER and Cis-ER. Bar = 5 µm.
TILLING studies identified three rice lines containing point mutation sites within the RBP-P gene (http://www.shigen.nig.ac.jp/rice/oryzabase). G-to-A replacements at nucleotide positions 754, 1118, and 1202 resulted in nonsynonymous amino acid substitutions A252T, G373E, and G401S in P3MH, P2MH, and P1MH mutant lines, respectively (Figure 2A; Supplemental Figure 2). The A252T mutation site in P3MH lies within the linker sequence between the two RRM domains, while the G373E and G401S substitutions in P2MH and P1MH, respectively, are located in the glycine-rich C-terminal region.
To provide direct evidence that RBP-P is involved in rice glutelin and prolamine mRNA localization, in situ RT-PCR was performed on developing rice grains of the wild type and two of the RBP-P mutants to determine the spatial distribution of glutelin and prolamine mRNAs on the cortical ER (Choi et al., 2000). P2MH was not studied as it exhibited aberrant flower development (Figure 3) leading to infertile panicles. In wild-type tissue, prolamine mRNAs are targeted to the PB-ER membranes that delimit the prolamine intracisternal inclusion granules, while glutelin mRNAs are distributed to the adjoining Cis-ER. As shown in Figure 2B, distribution of prolamine mRNAs are restricted to the PB-ER in wild-type endosperm cells, while prolamine mRNAs are partially mislocalized to the Cis-ER in P1MH and P3MH mutants. Similarly, glutelin mRNAs are partially mislocalized in P1MH and P3MH mutants, as they are observed on the PB-ER as well as their normal location on Cis-ER. These results indicate that two independent mutations in RBP-P mediate the mislocalization of glutelin and prolamine mRNAs in developing rice endosperm cells. Hence, RBP-P is essential for proper localization of prolamine and glutelin mRNAs to the PB-ER and Cis-ER, respectively.
Figure 3.
Phenotype of P2MH Carrying a G373E Mutation in the RBP-P Gene.
(A) Morphology of the wild type, P2MH (homozygous P2 mutant), and P2N (background genotype carrying normal RBP-P isolated from the heterozygous P2 mutant) during vegetative growth. The P2MH mutant is dwarf, has chlorophyll-deficient leaves, and produces fewer tillers than the wild type.
(B) Direct comparison of the leaf blades of P2MH and P2N with the wild type. Box shows an enlarged image of a region of the P2MH leaf.
(C) P2MH mutant showed late development of reproductive organs. When grains produced by wild-type and P2NH plants started to mature (turn brown), P2MH just began to “flower.”
(D) and (E) Comparison of the tassels from the wild type, P2N, and P2MH. (E) is an enlarged view of the area enclosed in a rectangle in (D).
(F) Severe flowering defect in P2MH. Due to the abnormal development of flower structure, P2MH failed to produce grains. Bar = 1 cm.
Proper mRNA localization is required for efficient packaging of storage proteins to their destination site in the endomembrane system. Prolamine polypeptides are locally translated on the PB-ER to form PB-I (Figure 1). Glutelin polypeptides are synthesized on the Cis-ER but exported to the Golgi and then transported to PSV (Figure 1). To investigate whether glutelin and prolamine proteins are mistargeted from their destination due to their mRNA mislocalization, we performed electron microscopy studies to investigate the distribution of the storage proteins in wild-type and RBP-P mutant endosperm cells (Figure 4). While PSVs are irregularly shaped, darkly stained granules due to their high electron density, PB-Is are more spherical in shape with light uniform staining and are surrounded by rough ER containing ribosomes (Figures 4A to 4F). The structures of PB-I and PSV in P1MH and P3MH endosperm cells were not significantly different from those of the wild type (Figures 4A to 4F).
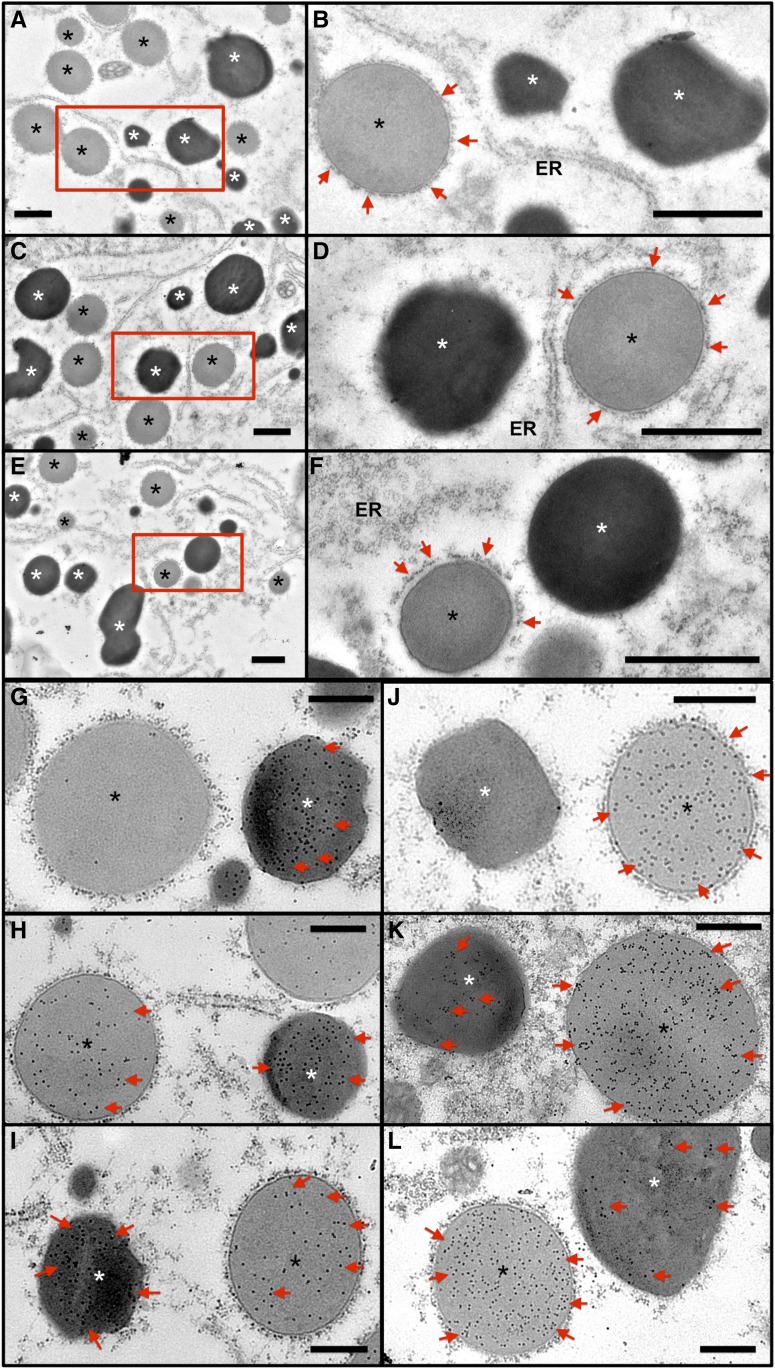
Figure 4.
Distribution of Glutelin and Prolamine Proteins in PB-I and PSV of P1MH and P3MH Endosperm Cells Compared with That of the Wild Type.
(A) to (F) Ultrastructure of PB-I and PSV in wild-type ([A] and [B]), P1MH ([C] and [D]), and P3MH ([E] and [F]) endosperm cells. (B), (D), and (F) are enlargements of the boxed regions in (A), (C), and (E). Bar = 1 μm. Red arrows denote rough ER surrounding PB-I. White asterisks indicate PSV, and black asterisks indicate PB-I.
(G) to (L) Immunolabeling of glutelin ([G] to [I]) and prolamine ([J] to [L]) proteins using monospecific antibodies and 15-nm gold particle-conjugated secondary antibodies. (G) and (J), wild type; (H) and (K), P1MH; (I) and (L), P3MH. Red arrows denote gold particle labeling. Note that in wild-type endosperm cells, glutelin and prolamine are dominantly detected in PSV and PB-I, respectively, although a few gold particles are evident due to slight background nonspecific labeling. Bar = 500 nm.
To obtain more detailed insight into protein body structure, immunocytochemical studies at the electron microscopy level were conducted. In wild-type endosperm, immunogold labeling analysis showed that glutelin was only detected in PSV though a few gold particles were also observed on PB-I due to nonspecific background labeling (Figure 4G). In P1MH and P3MH endosperm cells, however, glutelin proteins were also readily detected in PB-I in addition to their normal accumulation in PSV (Figures 4H and 4I). A similar situation was also obtained for prolamine. While prolamine proteins are exclusively localized in PB-I in the wild type (Figure 4J), prolamine proteins were observed in both PB-I and PSV in P1MH and P3MH endosperm cells (Figures 4K and 4L). Overall, partial mistargeting of glutelin and prolamine mRNAs results in the mislocalization of their coded protein products, i.e., glutelin and prolamine proteins are partially deposited in PB-I and PSV, respectively.
G373E and A252T Mutations Significantly Reduce RNA Binding Activities in Vitro
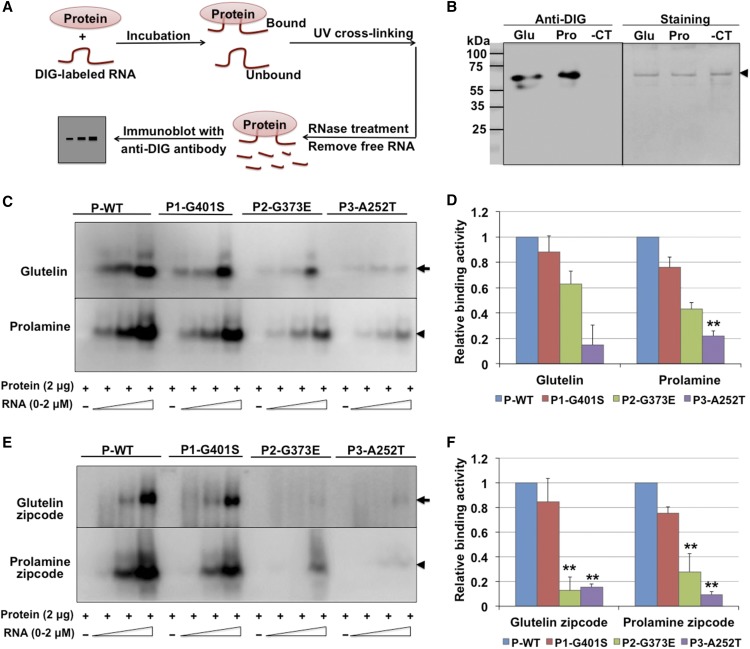
We employed an in vitro RNA-protein UV-cross-linking assay (Doroshenk et al., 2014) to assess the binding activity of RBP-P to glutelin and prolamine RNAs. Recombinant RBP-P and DIG-labeled RNAs were incubated together to form the binding complex and then exposed to UV light to cross-link the bound RNA and protein. The unbound RNAs were removed by RNase treatment and the RNA-protein complexes analyzed by immunoblotting using anti-DIG antibody (Figure 5A). Consistent with our previous findings (Crofts et al., 2010; Doroshenk et al., 2014), RBP-P displays high binding affinity to glutelin and prolamine RNAs compared with the RNA control (Figure 5B).
Figure 5.
In Vitro Binding Activities of Wild-Type and Mutant RBP-P to Glutelin and Prolamine mRNAs.
(A) Simplified representation of the RNA-protein UV-cross-linking assay.
(B) Binding activity of wild-type RBP-P to glutelin (Glu) and prolamine (Pro) mRNAs compared with the negative RNA control (-CT, RNA transcribed from cloning vector). Left panel: Immunoblot with anti-DIG antibody to detect the bound RNAs. Right panel: Coomassie blue-stained gel to show cross-linked RBP-P. Arrowhead indicates the position of RBP-P protein on SDS-PAGE gel.
(C) and (E) Binding activities of wild type (P-WT) and mutant RBP-P lines (P1-G401S, P2-G373E, and P3-A252T) to full-length glutelin and prolamine RNAs (C) and zip code (E) sequences. Arrow and arrowheads in indicate the mobility of glutelin and prolamine RNA bound RBP-P, respectively.
(D) and (F) Relative binding affinities of mutant RBP-Ps to glutelin and prolamine RNAs full-length (D) and zip code (F) sequences compared with wild-type RBP-P as assessed by ImageJ analysis of immunoblot results depicted in (C) and (E). The results represent the means ± se of duplicate determinations. *P value of two-tailed t test < 0.05; **P value of two-tailed t test < 0.01.
To determine whether the nonsynonymous mutations in the three RBP-P variants affected RNA binding, 2 µg of recombinant wild-type and mutant RBP-P proteins was incubated with increasing amounts of glutelin and prolamine RNAs for in vitro RNA-protein binding analyses (Figure 5C). In contrast to wild-type RBP-P, the amount of bound glutelin RNA was significantly less for P2-G373E (63% relative affinity to the wild type) and almost abolished for P3-A252T (15%), while the binding of P1-G401S to glutelin RNA was not significantly affected (Figures 5C and 5D). The same RNA binding properties were also evident when prolamine RNAs were used as the substrate. P1-G401S maintained 76% of the binding affinity to prolamine RNA, but the affinity of P2-G373E and P3-A252T was dramatically reduced to 43% and 22% relative affinity, respectively, of wild-type RBP-P (P-WT) (Figures 5C and 5D). The reduction in RNA binding activity by P2-G373E and P3-A252T indicates that residues G373 and A252 are important for RNA binding by RBP-P.
RBP-P strongly binds to the zip code RNA elements located in the 3′UTR of glutelin RNAs (Doroshenk et al., 2014), and we next assessed whether the mutant RBP-Ps were still capable of binding this RNA sequence. In contrast to the ability of wild-type RBP-P to bind glutelin and prolamine zip code RNAs, P2-G373E and P3-A252T exhibited diminished binding to these zip code RNA sequences (Figures 5E and 5F), while P1-G401S showed only a minor reduction in binding. These results further support the critical role of residues G373 and A252 in RNA binding. By contrast, G401 is not essential for RNA binding, which may be due to its location far downstream from the RNA binding RRM motifs (Figure 2A; Supplemental Figure 1).
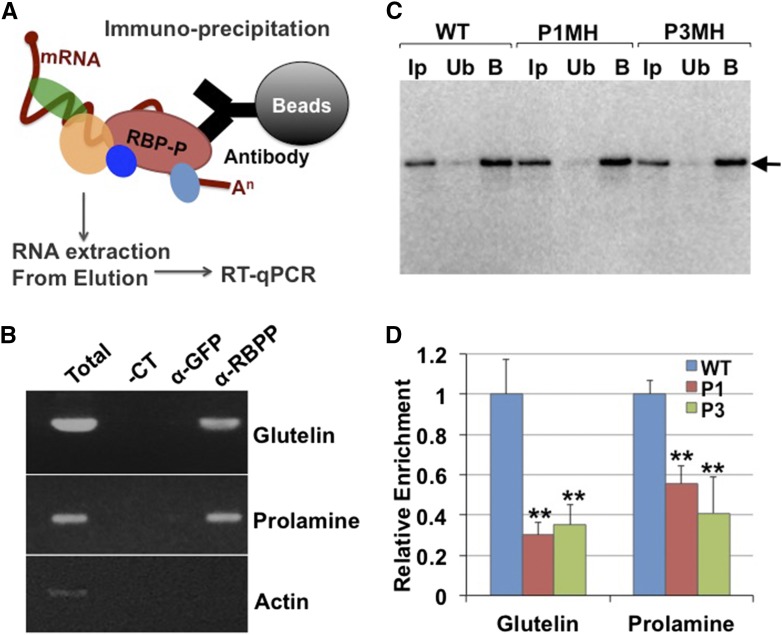
Mutant RBP-P Exhibits Reduced Affinity for Glutelin and Prolamine mRNAs in Vitro
To further characterize the RNA binding properties of the RBP-P mutants, we investigated the in vivo binding activities by RNA-immunoprecipitation (RNA-IP) (Figure 6). Extracts were prepared from developing grain sections from wild-type and mutant plant lines pretreated with formaldehyde to preserve the in vivo RNA-protein complexes. The extracts were then incubated with RBP-P antibody immobilized to protein A/G agarose beads to capture RBP-P complexes. RNA was subsequently isolated from the IP and subjected to RT-PCR (Figures 6A and 6B). Compared with the negative control using anti-GFP antibody, glutelin, and prolamine mRNAs were enriched in the wild-type IPs generated by the RBP-P antibody (Figure 6B). To assess the RNA binding properties of P1MH and P3MH mutants, anti-RBP-P antibody was immobilized onto agarose beads and an equal volume of conjugated beads was used for RNA-IP experiments of wild-type, P1MH, and P3MH extracts to ensure the same amount of RBP-P was captured by the beads in all three rice lines (Figure 6C). As shown in Figure 6D, the relative amount of glutelin mRNAs captured by RBP-P antibody in P1MH and P3MH mutants was significantly reduced to 30% and 35% of wild-type levels, respectively. Similarly, prolamine mRNAs were also less enriched in IPs generated with P1MH and P3MH compared with wild-type grain extracts, exhibiting reduced levels of 55% and 41%, respectively (Figure 6D). These results indicate that the amounts of glutelin and prolamine mRNAs bound to RBP-P are substantially lower in P1MH and P3MH endosperm cells than in the wild type. The reduced association of RBP-P in P3MH further confirms the crucial role of the A252 residue in the binding activities to glutelin and prolamine mRNAs. Replacement of A252 with Thr also partially disrupted their normal localization on the cortical-ER (Figure 2).
Figure 6.
In Vivo Binding Activities of Wild-Type and Mutant RBP-P to Glutelin and Prolamine mRNAs.
(A) Simplified representation of RNA-IP procedure.
(B) Agarose gel-resolved cDNA products synthesized from RNAs associated with IPs generated by anti-RBP-P (α-RBPP) or anti-GFP (α-GFP). Total: PCR using cDNA synthesized from starting material for IP. −CT: Negative control using water as template.
(C) Immunoblot of RNA-IP products from the wild type, P1MH, and P3MH using anti-RBP-P. Note that similar amounts of RBP-P were captured by IP of wild-type, P1MH, and P3MH grain extracts. Ip, input; Ub, unbound fraction; B, elution of bound fraction from IP. Arrowhead indicates the position of RBP-P on SDS-PAGE gel.
(D) The relative enrichment of glutelin and prolamine mRNAs in IPs generated by anti-RBP-P antibodies of wild-type, P1, and P3 developing grain extracts. *P value of two-tailed t test < 0.05; **P value of two-tailed t test < 0.01.
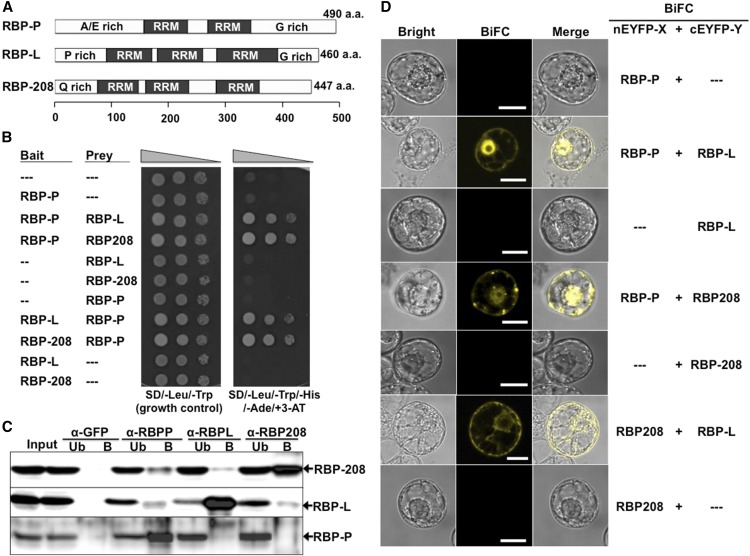
RBP-L and RBP-208 Are Interacting Partners of RBP-P
Although the mutation in P1-G401S had only a minor effect on in vitro RNA binding (Figures 5C to 5F), it did significantly reduce the binding activity in vivo (Figure 6D). A possible reason for this discrepancy is that the G401S replacement interfered with the interactions of RBP-P with other proteins that are involved in RNA binding and/or ribonucleoprotein complex formation. To address this, we first identified the interacting partners of RBP-P through a yeast two-hybrid (Y2H) approach. The RBP-P gene was cloned at the fusion site of the GAL4 DNA binding domain (BD) of the pGBK plasmid to serve as bait for screening rice cDNA sequences cloned at the GAL4 activation domain (AD) of the pGAD plasmid. Yeast cells growing in the absence of histidine (−His/+3-AT) and adenine (−Ade), indicative of the activation of reporter genes, were selected as positive colonies. A total of 14 protein candidates were isolated (Supplemental Table 1) and two, RBP-L and RBP-208, were selected for further study based on their previous characterization as cytoskeleton-associated RNA binding proteins (Doroshenk et al., 2009; Crofts et al., 2010). Both RBP-L and RBP-208 contain three RRMs (Figure 7A). RBP-L also contains a proline-rich (P-rich) N terminus and a glycine-rich (G-rich) C terminus, while the N terminus of RBP-208 is glutamine (Q) rich (Figure 7A). RBP-L exhibits significant homology to the RBP-45 family proteins of Arabidopsis and tobacco (Nicotiana tabacum) (Supplemental Figure 3) (Lorković et al., 2000; Peal et al., 2011), while RBP-208 shows significant identity to the three UBP1 proteins from Arabidopsis, AtUBP1a, AtUBP1b, and AtUBP1c (Lambermon et al., 2000) (Supplemental Figure 4). The Y2H results indicating an interaction between RBP-P and RBP-208 are consistent with other studies in that RBP-P is a homolog of Arabidopsis UBA2 family proteins, which have been found to bind to UBP1 (Lambermon et al., 2000).
Figure 7.
Identification of RBP-L and RBP-208 as Interacting Partners of RBP-P.
(A) Schematic structure of RBP-P, RBP-L, and RBP-208. A/E rich, alanine and glutamic acid-rich domain; G-rich, glycine-rich domain; P-rich, proline-rich domain; Q-rich, glutamine-rich domain. a.a., amino acids.
(B) Interactions of RBP-P with RBP-L and with RBP-208 as revealed by Y2H assay. Yeast cells were cotransfected with pGBK and pGAD constructs carrying the corresponding genes and grown on SD/-Leu/-Trp or SD/-Leu/-Trp/-His/-Ade/+ 3-AT.
(C) Co-IP analyses using affinity-purified antibodies specific against each RBP. Input, starting rice grain lysate; Ub, unbound fraction; B, elution of bound fraction from IP.
(D) In vivo interactions among RBP-P, RBP-L, and RBP-208 as revealed by BiFC analysis in BY-2 protoplasts. —, Original empty vector as negative control. Bars = 20 μm.
The Y2H-based interactions of RBP-P with RBP-L and RBP-208 were further studied to exclude the possibility of false-positive interactions. Both RBP-L and RBP-208 exhibited stringent two-hybrid interactions with RBP-P (Figure 7B). Immunoprecipitation (IP) using affinity-purified antibodies of each RBP was further performed to confirm their interaction in developing rice grains (Figure 7C). Each RBP antibody precipitated its corresponding RBP antigen, while no binding was detected using an anti-GFP antibody as a negative control. RBP-L and RBP-208 were both detected in the bound elution sample using anti-RBP-P antibody, indicating that RBP-P interacts with RBP-L and RBP-208 in rice endosperm cells. IP using either anti-RBP-L or anti-RBP-208 antibody revealed the presence of RBP-208 and RBP-L, respectively, in the bound fraction, suggesting an interaction between these two proteins. Interestingly, no detectable RBP-P was observed in immunoprecipitates generated using anti-RBP-L or anti-RBP-208 antibodies, which may indicate RBP-L and RBP-208 are not accessible to their respective antibodies when they are bound to RBP-P. Overall, these results suggest that RBP-L and RBP-208 interact to form multiprotein complexes in the presence or absence of RBP-P.
To further verify the interactions of RBP-P to RBP-L and RBP-208 in vivo, bimolecular fluorescence complementation (BiFC) assays were performed (Figure 7D). The three proteins were fused to C- or N-terminal-EYFP (cEYFP or nEYFP) for transient coexpression in BY-2 protoplasts. Negative controls were performed using each RBP construct and the complementary empty vector, which did not exhibit any YFP signal (Figure 7D). Protoplasts expressing nEYFP-RBP-P and cEYFP-RBP-L displayed strong fluorescence in the nucleus and a diffuse distribution in the cytoplasm, indicating that RBP-P interacts with RBP-L. RBP-208 was also observed to interact with RBP-P, although the associated fluorescence patterns were distinct from that observed for the RBP-P-RBP-L complex, with bright YFP signals exhibiting speckle-like complexes in the nucleus and foci-like structures in the cytoplasm (Figure 7D). We also observed that RBP-L interacted with RBP-208 in the cytoplasm, which confirmed their interaction detected by co-IP. The distribution of their complex in the cytoplasm differed from the others, indicating the presence of a distinct complex formed by RBP-L and RBP208. The absence of the complex containing RBP-L and RBP-208 in the nucleus suggests that this complex does not participate in the formation of RNA transcripts.
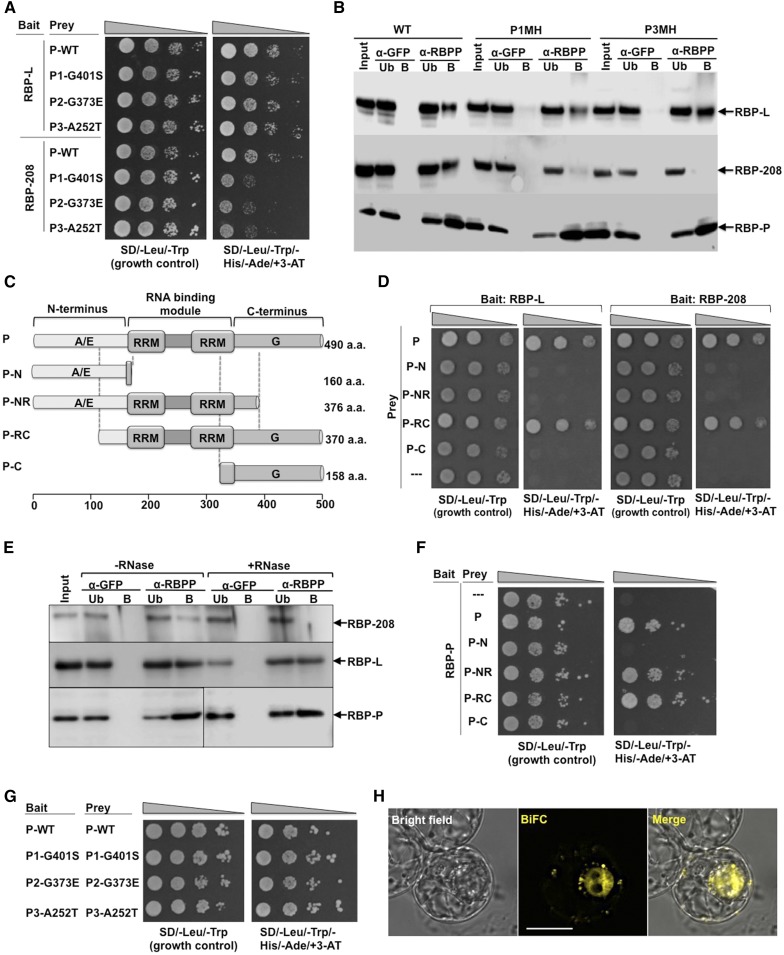
Both A252T and G401S Mutations Affect the Interaction of RBP-P to RBP-208
The capacity of RBP-L and RBP-208 to interact with wild-type and mutant RBP-P was assessed by Y2H analysis. The mutant RBP-P constructs were cloned into the pGAD vector as prey for mating with RBP-L or RBP-208 constructs as bait (Figure 8A). When assessed by growth on control and selection plates, mutations within RBP-P did not affect the interaction with RBP-L, as the colonies grew at similar rates to that of wild-type RBP-P. Colonies coexpressing RBP-208 and mutant RBP-P proteins P1-G401S, P2-G373E, and P3-A252T, however, grew much slower than the wild type, suggesting that the interaction between RBP-P and RBP-208 was weakened by these mutations.
Figure 8.
Protein-Protein Interactions of RBP-P with Itself, RBP-L, and RBP-208.
(A) and (B) The effect of mutations (G401S, G373E, and A252T) on RBP-P interactions with RBP-L and RBP-208 as revealed by Y2H (A) and co-IP analyses (B) compared with the wild type. Note the retardation of growth of yeast clones carrying RBP-208 and mutant RBP-Ps (A) and the absence of RBP-208 in anti-RBP IPs (B). Input, starting rice grain lysate; Ub, unbound fraction; B, elution of the bound fraction from IP; α-GFP and α-RBPP, samples from anti-GFP and anti-RBP-P columns, respectively. Arrows indicate the presence of RBP-L, RBP-208, and RBP-P in various immunoprecipitates formed by anti-RBP-P but not by anti-GFP.
(C) Schematic structure of truncated RBP-P protein. aa, amino acids.
(D) Interactions of RBP-L (left panel) and RBP-208 (right panel) with intact and truncated RBP-P as viewed by Y2H analyses.
(E) Co-IP using anti-RBP-P antibody to detect its interaction with RBP-L and RBP-208 in developing rice grain extracts pretreated with or without RNase. Arrows denote RBP-P, RBP-L, and RBP-208 in immunoblot analyses.
(F) Identification of binding sites responsible for homodimerization of RBP-P. Note that dimer formation requires the two RRM domains, as interaction is seen with P-NR and P-RC but not with P-N or P-C.
(G) Dimerization analysis of wild-type and mutant RBP-P forms viewed by Y2H.
(H) BiFC analysis reveals the in vivo dimerization of RBP-P. nEYFP-RBP-P complemented with cEYFP-RBP-P for coexpression in BY-2 protoplasts. Bar = 20 μm.
To further explore the capacity of RBP-P to interact with RBP-208 and RBP-L, we assessed the association of these RBPs in IPs generated using anti-RBP-P and extracts from wild-type, P1MH, and P3MH rice grains (Figure 8B). While the normal interaction of RBP-P and RBP-L was maintained in mutant grain extracts, RBP-208 was nearly undetectable in anti-RBP-P-generated IPs obtained from both P1MH and P3MH extracts. These results validate those obtained by Y2H where the interaction between RBP-208 and the RBP-P variant forms P1MH, and P3MH is nearly abolished. By contrast, P1MH and P3MH interacted normally with RBP-L, which is in agreement with the Y2H results.
The C-Terminal and RRM Domains of RBP-P Are Required for RBP-L and RBP-208 Association
To obtain additional information on how mutations in RBP-P affect its interactions with RBP-L and RBP-208, we prepared truncated forms of RBP-P for Y2H analysis to identify the binding site of RBP-P to RBP-L and RBP-208 (Figures 8C and 8D). The truncated P-N and P-C contain N-terminal residues 1 to 160 and C-terminal residues 334 to 490, respectively, of RBP-P. P-NR and P-RC are missing the C- and N-terminal regions, respectively (Figure 8C). As shown in Figure 8D, both RBP-L and RBP-208 interacted with full-length RBP-P and P-RC, but not with P-N, P-NR, or P-C. The lack of interaction with P-N, but with P-RC, indicates that the binding of RBP-P to RBP-L or RBP-208 does not require the N-terminal region of RBP-P. The interaction of P-RC, but not of P-NR or P-C, indicates that both the RRM domains and C-terminal end of RBP-P are required to bind to RBP-L and RBP-208.
As RRM motifs are RNA binding modules, we asked whether the interaction was dependent on RNA. To address this, developing grain extracts were treated with RNase and then incubated with anti-RBP-P antibody to determine whether those interactions are RNase-sensitive. Under these conditions, RBP-208 was not associated with RBP-P (Figure 8E), suggesting that the interaction of RBP-P and RBP-208 only occurs when bound to RNA. By contrast, the interaction of RBP-L and RBP-P is independent of RNA as RNase treatment had no effect on their capacity to interact.
RBP-P Can Form a Homodimer
In our Y2H analysis, we found that RBP-P could self-assemble to form a homodimer (Figure 8F). Further studies on the dimer binding sites of RBP-P via Y2H with truncated RBP-P confirmed that RBP-P might form a dimer through its RRM motifs as RBP-P interacted with NR and RC domains but not N and C termini (Figure 8F). As amino acid substitutions may also cause structural changes affecting dimer formation, we assessed whether dimer formation of the RBP-P variant forms P1-G401S, P2-G373E, and P3-A252T was affected. Y2H results showed that growth of yeast cells carrying dimers of P1-G401S, P2-G373E, or P3-A252T was not suppressed compared with wild-type RBP-P (Figure 8G), suggesting that dimer formation was unaffected. We further tested dimer formation by BiFC analysis in vivo and found that RBP-P fused with cEYFP and nEYFP in complementation to form a complex yielding fluorescence signals in both the nucleus and cytoplasm (Figure 8H). The signal of the RBP-P dimer from the nucleus also displayed a speckle-like pattern similar to that seen earlier for the RBP-P/RBP-208 complex (Figure 7D).
RBP-P Mutants Exhibit Growth Defects
The three RBP-P mutant lines exhibit multiple growth defects. This was most conspicuous for P2MH, which exhibited a more severe phenotype than P1MH and P3MH. P2MH plants were dwarfed and their leaves chlorophyll deficient (Figure 3). This mutant line also produced about one-fourth the number of tillers than normal and exhibited delayed panicle emergence and flowering (Table 1). Moreover, P2MH produced structurally abnormal, sterile florets (Figure 3). These growth defects were not evident in a segregating P2N line, which contained a homozygous wild-type RBP-P gene (Table 1).
Table 1. Analysis and Comparison of Plant Growth among Wild-Type and RBP-P Mutant Lines and Their Segregate Wild-Type RBP-P Siblings.
| Genotype | Mature Plant Height (cm)a | Days to Floweringa | No. of Tillers per Planta | Spikelet Fertility (%)a | Grain Weight (mg)b |
|---|---|---|---|---|---|
| Wild type | 68.2 ± 3.5 | 65.2 ± 0.8 | 14.5 ± 2.0 | 99.0 ± 0.5 | 23.3 ± 1.0 |
| P1Nc | 65.6 ± 3.9 | 70.8 ± 1.9** | 12.2 ± 1.7* | 69.1 ± 9.6** | 22.3 ± 1.7** |
| P1MH | 65.4 ± 3.8 | 86.2 ± 3.2** | 12.7 ± 3.2 | 33.6 ± 5.4** | 15.9 ± 1.4** |
| P2Nd | 67.4 ± 3.6 | 65.6 ± 1.5 | 14.7 ± 1.1 | 90.0 ± 1.5** | 22.8 ± 1.5 |
| P2MH | 48.0 ± 1.8** | 73.6 ± 2.1** | 3.6 ± 0.9** | ND | ND |
| P3Ne | 68.8 ± 1.8 | 69.8 ± 1.6** | 17.8 ± 1.4* | 80.3 ± 7.4** | 20.6 ± 1.0** |
| P3MH | 70.8 ± 3.4 | 72.1 ± 2.9** | 22.3 ± 1.4** | 64.4 ± 5.0** | 19.1 ± 1.56** |
ND, not determined due to the unavailability of grains from P2MH line. Each of the traits in mutants was compared to the wild type using the Student’s t test. *P value < 0.05; **P value < 0.01.
Data are shown as the mean ± sd (n = 10).
Data are shown as the mean ± sd (n = 100).
Mutant line carrying the normal RBP-P gene, which segregated from heterozygous P1MH.
Mutant line carrying the normal RBP-P gene, which segregated from heterozygous P2MH.
Mutant line carrying the normal RBP-P gene, which segregated from heterozygous P3MH.
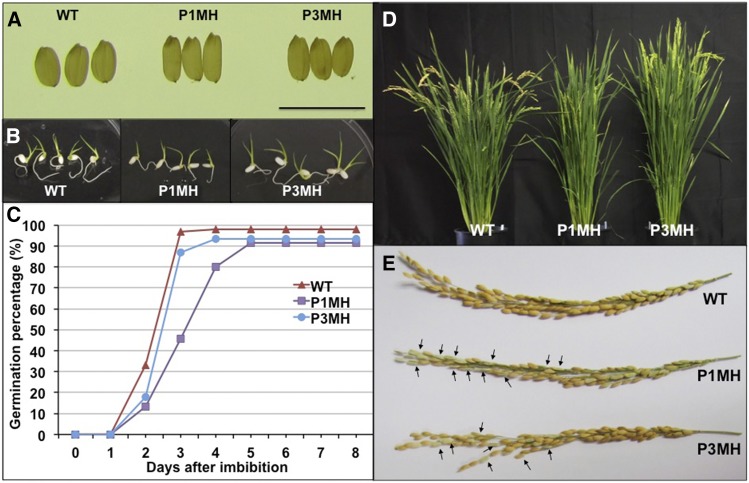
The defects exhibited by P1MH and P3MH lines were less severe than seen for P2MH. Both lines grew slower, flowered later, and produced smaller grains (Table 1). Both lines also exhibited lower spikelet fertility, with P1MH showing much lower fertility than P3MH (Table 1; Figure 9E). While P1MH produced a normal number of tillers, P3MH produced 50% more tillers than the wild type. Differences were also evident at the germination stage where P3MH plants were similar to the wild type, while germination of P1MH mutant grains was delayed by 2 d (Figures 9B and 9C). By contrast, homozygous RBP-P wild-type lines, P1N and P3N, which segregated from P1MH and P3MH in the F2 population, respectively, exhibited growth properties much more similar to normal plants (Table 1).
Figure 9.
RBP-P Mutants Exhibit Several Growth Defects.
(A) Morphology of wild-type, P1MH, and P3MH grains. Bar = 1 cm.
(B) Germinated grains after 5 d of imbibition.
(C) Germination rate of wild-type, P1MH, and P3MH grains. Grain germination was monitored daily for 8 d after initiation and grains were considered germinated when the radicle was extended more than 1 cm.
(D) P1MH and P3MH mutants showed a late flowering phenotype. At around 85 d of growth, mature grains could be observed on wild-type plants, while P3MH and P1MH were in the blooming and heading stages, respectively.
(E) Spikelet fertility was lower in P1MH and P3MH mutants than in the wild type. Arrows indicate some empty glumes in P1MH and P3MH. More detailed information can be found in Table 1.
RBP-P Has a Broad Function in Gene Regulation
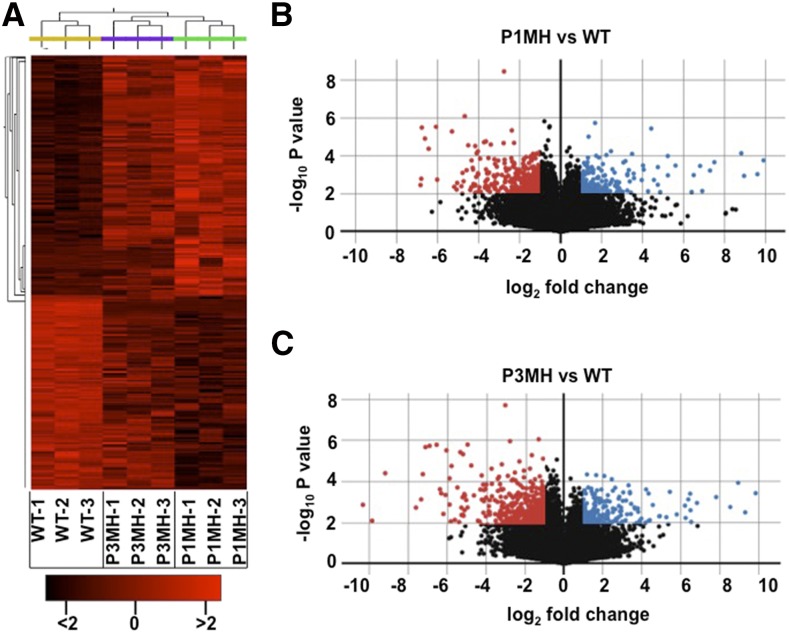
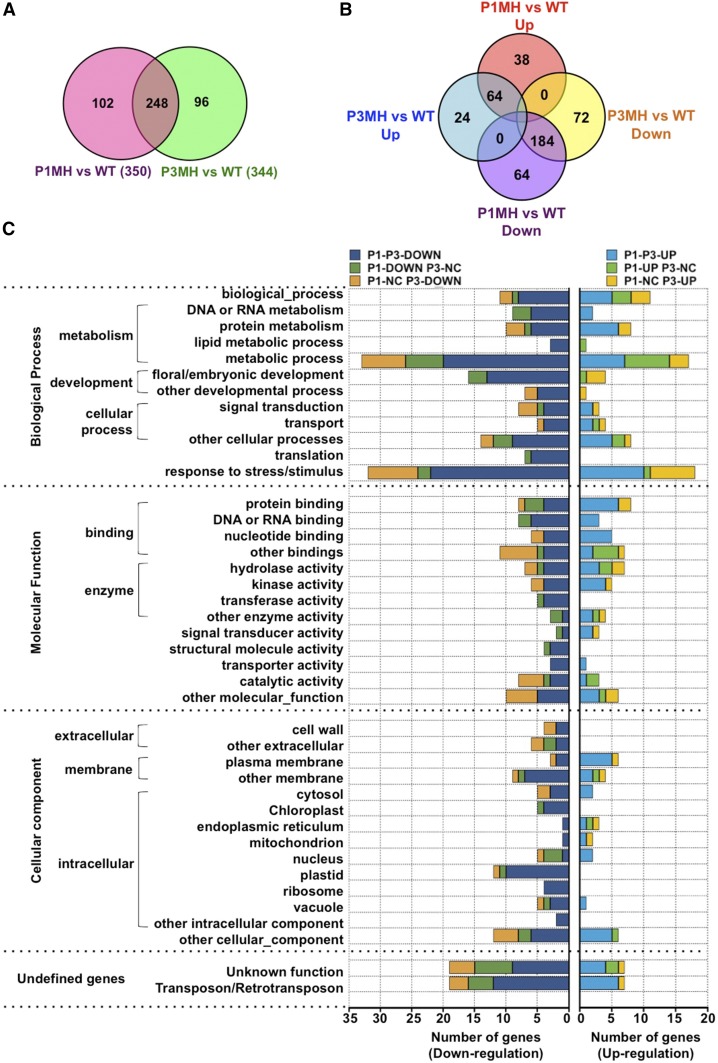
To investigate the effect of RBP-P on gene regulation in detail, we performed high-throughput sequencing and data analysis of triplicate samples of RNA extracted from developing grains of the wild type, P1MH, and P3MH. A correlation-based clustered heat map of the nine samples readily showed that P1MH and P3MH RNAs were clustered into groups distinct from the wild type (Figure 10). When compared with wild-type expression levels, 350 and 344 genes (log2 fold changes greater than 1, P value < 0.01) were differentially expressed in P1MH and P3MH mutant lines, respectively (Figures 10B 10C ,and11A; Supplemental Data Sets1 and 2). Of these differentially expressed RNAs, 248 genes were common in both P1MH and P3MH (Figure 11A; Supplemental Data Sets 2 and 3). A total of 248 and 256 genes were downregulated in P1MH and P3MH, respectively, including a shared set of 184 genes by the two mutants (Figure 11B; Supplemental Data Sets 2 and 3). In the 126 upregulated genes, 64 genes were shared by P1MH and P3MH (Figure 11B). We classified these differentially expressed genes into six groups, down- or upregulated in both P1MH and P3MH and down- or upregulated in only P1MH or P3MH (Supplemental Data Set 3).
Figure 10.
Gene Expression Profile Presented as a Clustered Heat Map and Volcano Plot.
(A) Clustered heat map showing the genotype grouping of wild type and RBP-P mutants, P1MH and P3MH.
(B) and (C) Volcano plots of differentially expressed genes in P1MH (B) and P3MH (C) compared with the wild type. The horizontal lines represent log2 fold changes, and the vertical lines represent P value (−log10). Differentially expressed genes were defined as having a log2 fold change of >1 and a P value of <0.01 (−log10 > 2). Upregulated and downregulated genes are represented by blue and red dots, respectively.
Figure 11.
Differentially Expressed Genes in RBP-P Mutants as Revealed by RNA-Seq Studies.
(A) and (B) Venn diagrams showing the number of differentially expressed genes (A) and down- or upregulated genes (B) in P1MH and P3MH compared with the wild type.
(C) GO classification of differentially expressed genes based on biological process, molecular function, and cellular component. Detailed information is available in Supplemental Data Sets 2 to 9.
Further analysis (Figure 11C; Supplemental Data Sets 4 to 9) was performed on the identified genes to classify the target biological processes, molecular functions, and cellular components regulated by RBP-P. This analysis showed that genes affected by the RBP-P mutations belonged to several essential biological processes. More than 70 genes are involved in the stress response, with 53 downregulated and 23 upregulated. These include OsIAA6 (LOC_Os01g53880) (Jung et al., 2015), NBS-LRR-type disease resistance proteins (LOC_Os06g06400 and LOC_Os02g02670) (van Ooijen et al., 2008), OsRR6 type-A response regulator (LOC_Os04g57720) (Hirose et al., 2007), NHL25 (LOC_Os01g59680) (Varet et al., 2002), CHASE domain-containing protein (LOC_Os12g26940) (von Schwartzenberg et al., 2016), expansin (LOC_Os06g41700) (Marowa et al., 2016), and defensin family proteins (LOC_Os11g45360, LOC_Os12g12230, and LOC_Os07g41290) (Carvalho et al., 2011).
Furthermore, 31 genes are linked to flower and embryo development (Supplemental Data Set 6), including several with well-documented roles in flower development and outgrowth such as the “no apical meristem protein” (Cheng et al., 2012), FRIGIDA (Shindo et al., 2005), CHASE domain containing protein (Lee et al., 2014; von Schwartzenberg et al., 2016), GRF-INTERACTING FACTOR1 (Lee et al., 2014), transcriptional corepressor LEUNIG (Conner and Liu, 2000), and calmodulin binding protein (Golovkin and Reddy, 2003). Genes classified using GOSlim molecular function ontology terms were highly represented by protein binding and DNA/RNA binding proteins. These DNA/RNA binding proteins are coded by 14 genes involved in transcription, including many transcription factors, and 20 genes with known DNA/RNA binding activity such as two pentatricopeptide repeat (PPR) proteins, two zinc finger family proteins, and two RRM domain-containing proteins (Supplemental Data Set 5). In the cellular component classification, genes encoding plastid and membrane associated proteins dominated. Sixteen genes are assigned to the chloroplast component, including 11 ribosomal proteins and 2 translation initiation factors (Supplemental Data Set 7).
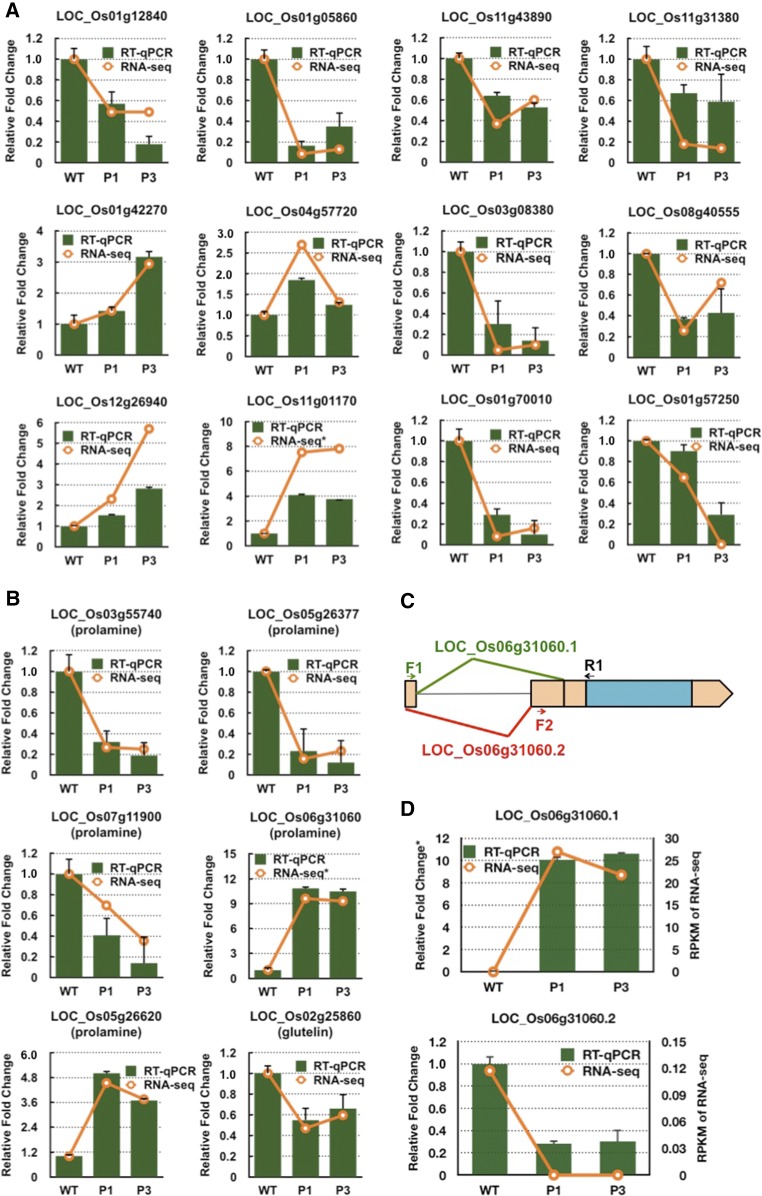
Several differentially expressed genes were selected for RT-qPCR analysis to verify the reliability of the RNA-seq data (Figure 12). LOC_Os11g43890, a WD domain, a G-beta repeat domain-containing protein (designated RBP10), and LOC_Os01g12840 (RBP213) were previously identified as putative RBPs involved in mRNA localization in rice endosperm cells (Crofts et al., 2010; Doroshenk et al., 2012). LOC_Os01g05860, a PPR repeat domain-containing protein, belongs to a large family of RNA binding proteins involved in RNA processing and maturation (Git and Standart, 2002; Lunde et al., 2007; Glisovic et al., 2008; Bailey-Serres et al., 2009; Lorković, 2009; Lee and Kang, 2016). These three putative RBP genes are significantly suppressed in both RBP-P mutants based on RNA-seq and RT-qPCR results. The “no apical meristem protein” (LOC_Os11g31380), which possesses DNA binding activity, is a critical transcriptional regulator in several developmental processes including root, leaf, and floral development (Cheng et al., 2012). Its expression was also reduced by RBP-P mutations and confirmed by RT-qPCR. Four additional genes involved in flower/embryo development, including downregulated LOC_Os03g08380 (ABC transporter) and LOC_Os08g40555 (ATPase) and upregulated LOC_Os01g42270 (transcriptional corepressor LEUNIG) and LOC_Os12g26940 (CHASE domain containing protein), each showed the same expression pattern as assessed by RNA-seq data. Genes falling within the cell component classification that were chosen for RT-qPCR analysis include ribosomal protein LOC_Os01g70010, which was suppressed in both mutants, and glycine-rich cell wall protein LOC_Os01g57250, which was significantly downregulated in the P3MH mutant. LOC_Os04g57720, a OsRR6 type-A response regulator involved in the stress response, tiller outgrowth, and flower development (Hirose et al., 2007) showed elevated expression in P1MH. The expression patterns exhibited by these selected genes were consistent with those of RNA-seq analysis, confirming the reliability of the RNA-seq data.
Figure 12.
RT-qPCR Validation of Differentially Expressed Genes and Expression of Storage Proteins in Rice Grains.
(A) and (B) Expression levels of selected genes differentially expressed in P1MH and P3MH. Green columns indicate the results of RT-qPCR, and orange lines show the data from RNA-seq. y axis, fold change relative to that of the wild type. The relative fold changes in the RNA-seq data for LOC_01g01170 and LOC_Os06g31060 panels are based on log2 fold changes.
(C) Schematic representation of the two splicing forms of the prolamine gene, LOC_Os06g31060. The location of the intron is denoted by a thin black line. The two exons involved in alternative splicing are highlighted in orange blocks. F1, F2, and R1 show the location of the designed primers for detection of the two splicing forms.
(D) RT-qPCR to detect the relative expression of LOC_Os06g31060.1 and LOC_Os06g31060.2 compared with the wild type. Right y axis indicates the RPKM data from RNA-seq. Relative fold change in the LOC_Os06g31060.1 panel is based on a log2 fold change. The error bars in (A), (B), and (D) represent the se of the mean from three technical repeats of RT-qPCR.
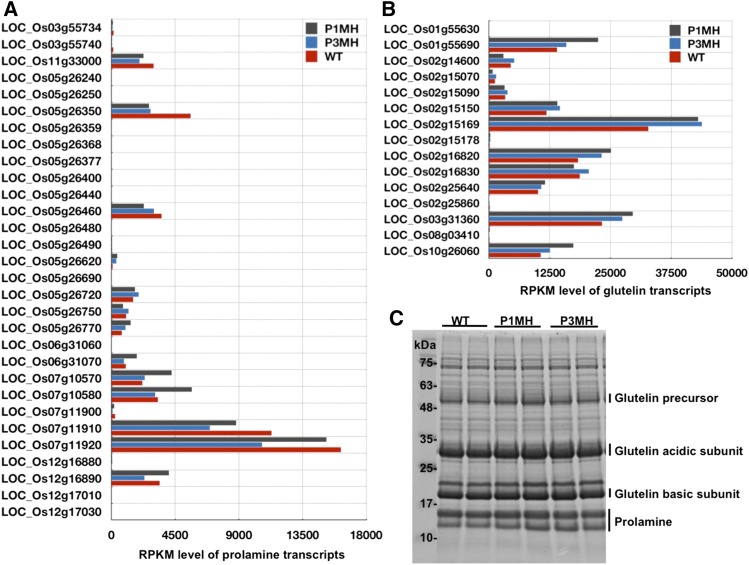
The expression levels of individual members of the glutelin and prolamine families were differentially regulated as well. Three downregulated (LOC_Os03g55740, LOC_Os05g26377, and LOC_Os07g11900) and two upregulated (LOC_Os06g31060 and LOC_Os05g26620) prolamine genes and one slightly downregulated glutelin gene (LOC_Os02g25860) were evident in both mutants based on RNA-seq and RT-qPCR analyses (Figure 12B; Supplemental Data Set 4), while the expression of other prolamine and glutelin genes was not significantly altered in either RBP-P mutant line (Supplemental Data Set 4). The expression of the prolamine gene LOC_Os06g31060 was significantly activated in both P1MH and P3MH (Figure 12B). We found that LOC_Os06g31060 has two alternative splicing forms and the difference in splicing between the two forms resides in the two alternative exons located in the 5′UTR (Figure 12C). Two pairs of primers to amplify the corresponding splicing forms were designed for RT-qPCR to verify the RNA-seq result (Figures 12C and 12D). Compared with the wild type, the expression of splicing variant 1, LOC_Os06g31060.1, was found to be significantly increased and consistent with the RNA-seq data, while variant 2, LOC_Os06g31060.2, was downregulated, suggesting splicing variant 1 is the dominant form. Mutations in RBP-P altered the alternative splicing events of this prolamine gene.
Glutelin and prolamine are encoded by multigene families, and although they account for most of the total protein found in rice grains (up to 80% for glutelin and 5–10% for prolamine), not all gene members within the two families are expressed at high levels (Figures 13A and 13B). Some transcripts, such as LOC_Os12g17030 (prolamine) and LOC_Os01g55630 (glutelin), could only be detected with low RPKM (<10) in the wild type and RBP-P mutants, while others reached extremely high RPKM, such as prolamine LOC_Os07g11920 (>10,000) and glutelin LOC_Os02g15169 (>30,000) (Figures 13A and 13B; Supplemental Data Set 4). The six prolamine and glutelin genes regulated by RBP-P mutations are not highly expressed members in the prolamine and glutelin gene families. Therefore, the expression changes of those genes had no significant effect on the abundance of prolamine and glutelin proteins in the two RBP-P mutants. This is also reflected by the total storage protein profile shown in Figure 13C, which showed that the accumulation level of prolamine and glutelin proteins was not significantly altered in either RBP-P mutant.
Figure 13.
Expression Level of Prolamine and Glutelin in Developing Rice Grains.
(A) and (B) Expression level of prolamine (A) and glutelin (B) transcripts in wild-type, P1MH, and P3MH grains as revealed by RPKM values.
(C) Total protein profile to reveal the expression of prolamine and glutelin proteins in wild-type, P1MH, and P3MH grains. Glutelin precursor, acidic and basic subunits, and prolamine are indicated. Note that the total accumulation levels of glutelin and prolamine are not altered in either RBP-P mutant.
DISCUSSION
RBP-P Is Required for Proper Subcellular Localization of Both Glutelin and Prolamine mRNAs
Developing rice grains are an ideal model for studying RNA localization in plants due to the asymmetric distribution of the storage protein RNAs on the structurally polarized cortical-ER. Sorting of glutelin and prolamine to different endomembrane storage sites depends on the localization of their mRNAs to distinct cortical-ER domains, the Cis-ER and PB-ER, respectively (Doroshenk et al., 2012; Tian and Okita, 2014). In addition to facilitating the subsequent location of its encoded protein product, the targeting of prolamine and glutelin mRNAs to specific ER subdomains also prevents potentially deleterious interactions between proteins (Crofts et al., 2005; Doroshenk et al., 2012; Washida et al., 2012; Tian and Okita, 2014).
Two RNA binding proteins have been found to specifically bind prolamine and glutelin mRNA and function in localization of these mRNAs. One is OsTudor-SN (Sami-Subbu et al., 2001; Okita and Choi, 2002; Wang et al., 2008), previously called Rp120, which consists of four tandem staphylococcus nuclease (SN)-like domains (4SN module) followed by a C-terminal region containing a Tudor domain and truncated SN (TSN module). Mutations of OsTudor-SN resulted in mislocalization of both prolamine and glutelin mRNAs (Chou et al., 2017). While the 4SN module exhibits RNA binding activity, the TSN module binds to multiple RNA binding proteins. The two independent modules likely cooperate in mRNA localization (Chou et al., 2017).
The other RNA binding protein, RBP-P, contains two RRM domains with unique peptide domains bordering the RRM module. RBP-P was initially found in a collection of RNA binding proteins that were specifically captured by the prolamine zip code and later as a potential candidate binding protein to the glutelin zip code RNA (Crofts et al., 2010; Doroshenk et al., 2014). Although RBP-P possesses binding affinity to both prolamine and glutelin RNAs (Crofts et al., 2010; Doroshenk et al., 2014), direct evidence addressing the involvement of RBP-P in the localization of prolamine and glutelin mRNAs was lacking. Here, we show that the localization of glutelin and prolamine mRNAs on the Cis-ER and PB-ER, respectively, is dependent on RBP-P. Rice cell lines P1MH (G401S) and P3MH (A252T) expressing variant RBP-P proteins possessing nonsynonymous mutations show partial mislocalization of glutelin and prolamine mRNAs. In both mutant lines, glutelin mRNAs are also distributed to the PB-ER, while prolamine mRNAs are also found on the Cis-ER. These observations indicate that RBP-P is required for the restricted localization of storage protein mRNAs at their proper intracellular locations on the cortical-ER.
We evaluated the RNA binding activities of wild-type RBP-P and the three mutant proteins, P1(G401S), P2(G373E), and P3(A252T). The in vitro binding activity of the RBP-P variant P3(A252T) exhibited very little in vitro binding activity to glutelin and prolamine RNAs, especially to their zip code sequences (Figure 5). A reduction in RNA binding was also observed for P2(G373E), while P1(G401S) displayed nearly equivalent in vitro binding activity as the wild type. Although the in vitro RNA binding activity of the recombinant P1(G401S) was not significantly impacted by the mutation, the in vivo binding affinity of native P1(G401S) showed more than a twofold reduction in binding of both glutelin and prolamine RNAs (Figure 6D). These differences between the in vitro and in vivo RNA binding properties of recombinant and native RBP forms are likely due to other factors such as the interaction of native RBP-P with other RNA binding proteins, which may include RBP-L and RBP-208.
Structural Features of RBP-P
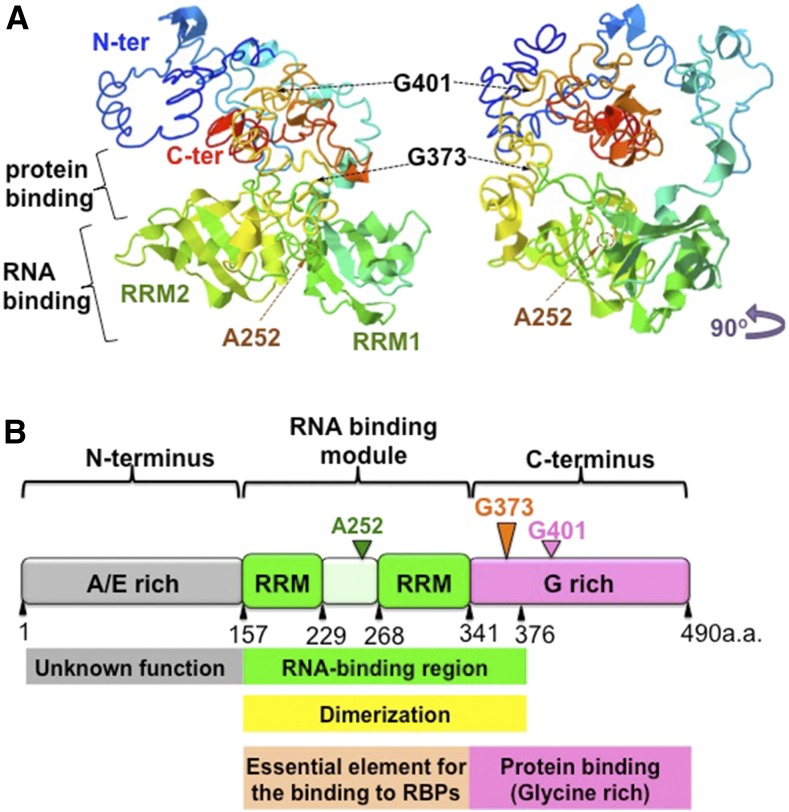
The RRM is the most abundant RNA binding motif in all life kingdoms (Maris et al., 2005). The motif is usually composed of 70 to 90 amino acids exhibiting typical βαββαβ topology with a four-stranded antiparallel β-sheet packed against two α-helices (Figure 14A; Supplemental Figure 1; Maris et al., 2005). The four-stranded β-sheet is the main surface recognizing and interacting with RNA. RRM is a highly conserved domain, and the conservation is highly restricted to the two consensus conserved sequences, RNP1 and RNP2, located in β3 and β1 sheets, which are directly involved in RNA binding (Figure 14A; Supplemental Figure 1; Maris et al., 2005). Any amino acid changes in the highly conserved RRM sequence usually largely alter its binding preference and specificity to various RNAs (Maris et al., 2005). However, one RRM motif can only bind to two or three nucleotides (Maris et al., 2005). The evolution of unique N- and C-terminal regions, as well as the acquisition of multiple RRM motifs, allowed RRM-containing proteins to recognize longer nucleotide sequences and increased their binding affinity (Maris et al., 2005; Lunde et al., 2007). The unique N- and C-terminal regions as well as the RRM domains interact with other RNA binding proteins or accessory proteins forming larger multiprotein complexes, which collectively enhance the sequence specificity and strength of RNA binding.
Figure 14.
Hypothetical Function and Structure of the Multiple Domains of RBP-P.
(A) Predicted structure of RBP-P. Right panel is a 90° left rotation view of the left panel. The two RRM motifs, RRM1 and RRM2, are labeled. The residues Gly-401, Gly-373, and Ala-252 are indicated by arrows.
(B) Schematic representation of the structure and function of the RRM domain and the N and C terminus in RBP-P. The N terminus of RBP-P contains the Ala-rich and Glu-rich motifs of unknown function. The RNA binding module consists of two RRM domains, ∼80 amino acids in length, the interdomain linker, and a short sequence in the C terminus. This protein module is responsible for RNA binding activity as well as dimerization with another RBP-P molecule or other proteins, such as RBP-L. The glycine-rich C terminus is involved in protein-protein interactions.
The nonsynonymous mutation A252T, located between the two RRM domains, disrupted the binding of RBP-P to its target glutelin and prolamine mRNAs in vitro and in vivo (Figures 5 and 6). Sequence alignment of RBP-P with its putative orthologs shows that the two RRMs in those proteins are highly conserved while the sequence that links the two RRMs displays considerable sequence diversity (Supplemental Figure 1). Despite the sequence divergence in the linker region, A252 and P253 are strictly conserved among these proteins (Supplemental Figure 1). The conservation of these residues, especially A252, suggests that these two amino acids likely play a critical role in supporting the RNA binding activity of the two RRMs.
Replacement of Gly-373 by glutamic acid (P2MH, G373E) disrupted both the capacity of in vitro RNA binding activity (Figure 5) and protein interaction with RBP-208 as viewed by Y2H (Figure 8, RBP-208). G373 is part of a conserved motif GYG shared by RBP-P and its orthologs (Supplemental Figure 1). Based on the predicted structure of RBP-P (Figure 14A), the GYG motif forms a loop located close to a coil structure formed by residues 240 to 245 of the RRM linker sequences. This arrangement may stabilize the spatial structure of the conjoining double RRMs for RNA binding and the adjoining C terminus for protein interaction. The mutation of G373E may impede loop formation that, in turn, impairs RNA binding and protein interaction of RBP-P to RBP-208.
The substitution of Gly-401 by Ser (P1MH, G401S) affected the interaction of RBP-P with RBP-208 and in vivo RNA binding but not in vitro RNA binding activity. The reduction in in vivo RNA binding is likely due to the loss of protein interaction with RBP-208 and possibly other RNA binding proteins. Compared with the Gly-373 site, Gly-401 is located away from the RNA binding domain (Figure 14). Conversely, the exposure of Gly-401 on the surface of the RBP-P structure affords it high accessibility to interact with other proteins and is likely the basis for why the G401S mutation affects the interaction with RBP-208. Although Gly-401 is not highly conserved, it is associated with the Gly-rich 401GXXGG405 motif. This motif has been found in several enzymes (Bocharov et al., 2012; Rajagopalan et al., 2013; Verma et al., 2015) and has been suggested to drive dimerization of helices (Bocharov et al., 2012; Remorino and Hochstrasser, 2012; Rajagopalan et al., 2013). Whether the GXXGG motif is a novel peptide facilitating protein-protein interaction deserves further investigation.
Based on these results, we propose a model (Figure 14B) on the structure-function of RBP-P. While the function of the N-terminal region containing peptides rich in Ala or Glu remains to be determined, the region containing the two RRMs is the main RNA binding module and the distal C-terminal segment is prominent in protein-protein interaction. The short glycine-rich sequence immediately following the second RRM supports the function of both RNA binding and protein-protein interaction, while the more distal glycine-rich region near the C terminus is likely the major contributor to protein-protein interaction.
Multiple Complexes Formed by RBP-P Are Likely Involved in mRNA Localization of Storage Proteins
Our results show that RBP-P is capable of forming multiple protein complexes. It can assemble with itself to form a homodimer and can interact with RBP-L and RBP-208. Homodimer formation of RBP-P is dependent only on the two RRM domains. Interactions of RBP-P with RBP-L or RBP-208 require the C-terminal region of RBP-P in addition to the RRM domain (Figure 8D), although the interactive properties are distinct between these two RNA binding proteins. The interaction of RBP-208 with RBP-P is disrupted by the two nonsynonymous mutations, A252T and G401S, as well as by RNase treatment (Figure 8). By contrast, the interaction of RBP-P with RBP-L is not affected by these mutations and is independent of RNA. Hence, the association of RBP-208 is very sensitive to small changes in RBP-P structure and is dependent on the presence of RNA. These results suggest that protein-protein interaction of RBP-P is achieved by three mechanisms: RRM-mediated RBP-P homodimerization; RRM- and C-terminal-mediated interaction; and RNA-dependent, glycine-rich C-terminal-mediated protein-protein interaction (Figure 14B).
RBP-P was found to be located in both the nucleus and cytoplasm (Doroshenk et al., 2014), an observation consistent with its multiple roles during transcription and in RNA localization. In this study, localization of RBP-P dimer and the complexes formed by RBP-P with RBP-L and/or RBP-208 was also observed in both the nucleus and cytoplasm (Figure 7). As shown in other studies (Cheng et al., 2003; Riera et al., 2006; Dias et al., 2010; Meyer et al., 2015; Wu et al., 2016), the speckle-like signal of RBP-P observed in the nucleus is consistent with a possible role in transcription and RNA processing events. Evidence for the latter role is supported by the differential splicing event of the prolamine transcript LOC_Os06g31060 in mutant lines where significant upregulation of one splicing variant and downregulation of the alternative splicing event was observed (Figure 12). These differential splicing events may result from the failure of RBP-P to interact with RBP-208 in the RBP-P mutant lines. In addition, we identified transcription factor TFIIS, a modular factor in RNA polymerase II preinitiation complexes during transcription (Schweikhard et al., 2014), as an interacting partner of RBP-P through Y2H screening (Supplemental Table 1). This interaction with TFIIS infers a potential role of RBP-P in transcription.
The participation of RBP-P with RBP-L and RBP-208 in posttranscriptional RNA processing events is supported by the suggested roles of the Arabidopsis orthologs. Arabidopsis UBP1, the ortholog of RBP-208, plays multiple roles in RNA splicing and mRNA stability (Lambermon et al., 2000). UBA2a, the ortholog of RBP-P, was reported to be involved in RNA stability through interaction with UBP1 (Lambermon et al., 2000), while RBP-45, the ortholog of RBP-L, was reported to be a structural and functional-related protein of UBP1 and involved in some unknown steps of RNA maturation distinct from the role of UBP1 (Lambermon et al., 2000; Lorković et al., 2000; Lewandowska et al., 2004; Wachter et al., 2012). Overall, our results in rice and those reported for the Arabidopsis RNA binding proteins support the view that RBP-P, together with RBP-L and RBP-208, play multiple roles in gene regulation, including RNA splicing, maturation, and mRNA stability. Indeed, mutations of RBP-P altered the expression of several glutelin and prolamine genes (Figure 12; Supplemental Data Set 4) including the previously discussed prolamine splicing variant LOC_Os06g31060.
Based on the available information on the complexes formed by these three RNA binding proteins, we propose a model of multiple complexes involved in RNA-protein assembly during RNA localization. RNA localization is a multistep process that is initiated in the nucleus and terminated at the cortical-ER. RBP-P, a protein bound to both prolamine and glutelin zip code RNAs, acts as a scaffold for the binding of other proteins, as evidenced by its interaction with itself, RBP-L and RBP-208, to form multiple complexes (Figures 7 and 8). These complexes in the nucleus and cytoplasm likely reflect the distinct processes involving transcription, splicing, export from the nucleus, intracellular transport, anchoring at the destination site, translation, storage, and turnover, all of which are required for the precise control of mRNA localization. In the nucleus, RBP-P interacts with RBP-L and RBP-208, although it remains to be determined whether they form two independent complexes or a single complex containing all three RNA binding proteins. Irrespective of whether there is a single or multiple complexes, it is likely that they participate in the different steps of gene expression through interaction with other proteins and/or regulation factors. The RBP-P dimer and RBP-P/RBP-208 complexes may be involved in RNA splicing, which is supported by speckle-like signals from those complexes in the nucleus as well as alternative splicing of a prolamine gene caused by the failure of mutant RBP-P to coassemble with RBP-208. RBP-P may also regulate other steps of RNA maturation or expression of a different set of genes through interaction with RBP-L. After export from the nucleus to the cytoplasm, the complexes may undergo remodeling to form different RNA-protein assemblies for cytoskeleton-mediated RNA transport to their destination. Except for the complexes formed by RBP-P, a separate complex may be formed by RBP-L and RBP208 in the absence of RBP-P, which is supported by the distinct distribution pattern of the RBP-L and RBP208 complex in BiFC analysis (Figure 7D). This complex is not involved in RNA events in the nucleus but may participate in mRNA transport and localization through interaction with other RNA binding proteins. All three RNA binding proteins mentioned in this study were previously identified from a cytoskeleton-enriched fraction in developing rice grains (Doroshenk et al., 2009; Crofts et al., 2010), supporting the involvement of these RBPs in cytosolic mRNA localization events.
It is worth mentioning that the expression of several transcription factors, RNA binding proteins, and PPR domain-containing proteins was also suppressed in the RBP-P mutants (Supplemental Data Set 5), which may further regulate the expression and/or localization of glutelin and prolamine. We found that two of these RNA binding proteins, LOC_Os01g12840 (RBP-213) and LOC_Os11g43890 (RBP-10), which were previously identified as putative proteins involved in RNA localization (Doroshenk et al., 2009), were also downregulated in P1MH and P3MH mutants (Figure 12). Unlike most RNA binding proteins, these two proteins are devoid of any known RNA binding motifs. Hence, they likely function as protein binding factors during mRNP complex assembly. For example, RBP-10 contains a WD domain and G-beta repeats. The WD domain contains 4 to 16 repeating units, which can serve as a rigid scaffold for protein-protein interaction (Stirnimann et al., 2010). Proteins containing WD40 domains are implicated in several processes including signal transduction and transcription regulation (van Nocker and Ludwig, 2003). Whether these RBPs help to regulate the expression and localization of glutelin and prolamine mRNAs deserves further study.
Mislocalization of Glutelin and Prolamine mRNAs Disrupts the Transport of the Corresponding Proteins
The consequence of mislocalization of glutelin and prolamine mRNAs is the mistargeting of the corresponding proteins from their normal intracellular sites of accumulation in the endomembrane system. Mislocalization of glutelin mRNAs on the PB-ER resulted in the retention of the locally synthesized glutelin within PB-I. Similarly, prolamine polypeptides synthesized from its mislocalized mRNAs on the Cis-ER were exported and transported to PSV (Figure 4). This result further substantiates our hypothesis that the final deposition of storage proteins in rice endosperm cells is controlled by mRNA localization (Washida et al., 2009).
The retention of the mistargeted glutelin proteins in PB-I is not due to a lack of membrane trafficking to the Golgi as this ER subdomain rapidly exports newly synthesized globulin polypeptides to the Golgi apparatus for further packaging to the PSV (Washida et al., 2009). The most likely explanation for the retention of glutelin polypeptides within PB-I is that newly synthesized mistargeted glutelin polypeptides do not fold correctly into their normal tertiary conformation as this process requires protein disulfide isomerase, PDIL1-1. This molecular chaperone is absent or at reduced levels in PB-I and is concentrated in the Cis-ER lumen (Satoh-Cruz et al., 2010). The inability to fold properly into its native tertiary structure prevents the newly synthesized glutelin polypeptide from achieving competency for ER export to the Golgi, thereby enabling their retention and accumulation within PB-I.
The transport of prolamine proteins, synthesized by mislocalized RNAs to the Cis-ER, to PSV indicates that these proteins are capable of being exported from the ER. This condition was also seen for the maize (Zea mays) 10-kD δ-zein, which was also transported to the PSV when its RNA was misdirected from the PB-ER to Cis-ER (Washida et al., 2009). The retention of these proteins within PB-I is likely due to their ability to interact with each other to form larger complexes that exclude them from being exported from the ER. This protein-protein interaction is mediated by BiP, which has been demonstrated to bind to nascent, newly synthesizing prolamine chains as well as intact prolamine polypeptides (Li et al., 1993). Consistent with its role in prolamine folding and protein interactions, this molecular chaperone is substantially enriched within PB-I compared with adjoining Cis-ER (Li et al., 1993).
Involvement of RBP-P in Gene Regulation of Several Biological Processes
The main impetus for this study was to elucidate the role of RBP-P in the transport and localization of storage protein RNAs to the PB-ER and Cis-ER. Like other RNA binding proteins, RBP-P likely plays multiple roles in RNA metabolism and, hence, is involved in many essential biological processes during plant development (Doroshenk et al., 2014; Tian and Okita, 2014). These multiple functions are reflected in the temporal and spatial expression patterns of RBP-P, which is constitutively present in multiple organs and tissues throughout the life cycle of the rice plant (Supplemental Figure 5). The phenotypic properties displayed by the RBP-P mutant lines, P1MH and P3MH (Table 1), are consistent with this RNA binding protein having multiple functions. Both mutant lines grew slower, as reflected by the delayed panicle emergence and flowering time, and exhibited lower fertility and smaller grains. Although the RBP-P mutant lines were generated by treatment of fertilized egg cells by N-methyl-N-nitrosourea treatment (Satoh-Cruz et al., 2010) and, hence, contain multiple mutations, genetic segregation analysis indicated that many of the defects observed in these TILLING lines are likely due to the mutant RBP-P gene. This is particularly apparent for P2MH, which is chlorophyll deficient, dwarfed, and has aberrant sterile flowers. These defects are largely reversed in the homozygous RBP-P normal P2N line, which segregated from P2MH in the F2 population (Figure 3). Likewise, near normal growth and developmental traits in two independent segregate lines, P1N and P3N, further supports the role of RBP-P in these different processes.
Many of the changes in gene expression seen in the P1MH and P3MH rice lines are a consequence of mutations in RBP-P. Of the 344 to 350 genes differentially expressed in these two mutant lines, ∼250 genes were common in these two mutant lines. Although the alterations in expression of the 100 or so genes unique to P1MH or P3MH may be products of mutations generated by chemical modification in other genes, these differences could also be due to the specific effects mediated by the RBP-P mutations. Specifically, the A252T replacement in the RRM linker region and G401S substitution in the C-terminal G-rich region may alter different functions of RBP-P. We attempted to directly test this hypothesis by complementing P1MH and P3MH mutant lines with the wild-type RBP-P gene via Agrobacterium tumefaciens-mediated transformation. In both instances, however, P1MH and P3MH produced poorly growing calli, which were not amenable to transformation and plant regeneration.
In addition to their suggested role in RNA processing in the nucleus and later RNA transport and localization in the cytoplasm, RBP-P and its interacting partner RBP-208 are likely involved in other activities as suggested by studies in Arabidopsis. The Arabidopsis RBP-208 putative ortholog, UBP1, is a core component in the stress granule (SG) assembly and stress response machinery via its ability to selectively sequester translationally repressed RNAs into aggregated SGs (Sorenson and Bailey-Serres, 2014). Its interacting partners, the UBA2 family proteins (related to RBP-P), respond to abscisic acid treatment and mechanical wounding, and their constitutive overexpression led to the activation of stress-associated genes (Lambermon et al., 2000; Riera et al., 2006; Bove et al., 2008; Kim et al., 2008). Similar to the properties of UBP1 and UBA2 family proteins in the stress response, RBP-P may also respond to stress and function as the interacting partner of RBP-208 to regulate genes involved in the stress response.
Mutations in RBP-P affect rice reproduction, especially at the flowering and grain development stages. This is readily evident for the P2MH plant line, which produced structurally abnormal, sterile florets (Figure 3). Flowering in the P1MH and P3MH rice lines was also delayed and the grains were smaller than those of the wild type. Consistent with these phenotypic changes, 31 genes involved in these developmental processes were differentially regulated in P1MH and P3MH mutants (Figure 11; Supplemental Data Set 6). Most notably was the GRF-interacting factor 1 (LOC_Os03g52320), which is required for cell specification maintenance during reproductive organ development in Arabidopsis (Lee et al., 2014), and the “no apical meristem protein” (LOC_Os11g31380), which is essential for floral organ identity (Cheng et al., 2012). These genes are suppressed in P1MH and P3MH mutants, which may postpone and disrupt normal development of flowers and grains in rice.
P1MH showed defects in growth and reproductive development, i.e., slower growth and reduced tiller number, distinct from P3MH. One possible explanation for these dissimilarities is the differences in expression of OsRR6 (LOC_Os04g57720) between these two RBP-P mutant lines. OsRR6 is a negative regulator of cytokinin signaling and its overexpression in transgenic lines results in a dwarf phenotype with a reduced branching pattern, abnormal flowers, and a reduction in the number of spikelets (Hirose et al., 2007). The expression of this gene is activated 2.7-fold in P1MH, but not significantly changed in P3MH (Figure 12; Supplemental Data Set 3).
Another interesting gene set that is differentially expressed between the wild-type and RBP-P mutant lines is those involved in chloroplast development (Figure 11; Supplemental Data Set 7). Sixteen genes associated with chloroplast development, including translation initiation factors and ribosomal proteins, were depressed in both P1MH and P3MH mutants. While P1MH and P3MH did not exhibit any abnormalities in chloroplast development, the P2MH line harboring the G373E mutation was chlorophyll deficient (Figure 3). These results suggest that RBP-P plays an essential role in chloroplast development.
Overall, our study showed that RBP-P is an essential factor in the localization of storage protein RNAs to specific subdomains of the cortical-ER. Such a role likely occurs as early as in the nucleus where nuclear-localized RBP-P (Figures 7Dand8H) recognizes the zip code RNA sequences and, together with RBP-L and RBP-208, processes the newly synthesized RNA into a mature transcript competent for export to the cytoplasm. Once in the cytoplasm, the RNP complex containing RBP-P is transported to the cortical-ER where it is localized to the PB-ER or Cis-ER and the mRNA is translated. A partial loss of function of this RNA binding protein disrupts RNA localization. Due to its multiple roles in RNA processing, RBP-P has a broad function in plant growth and is indispensable for normal plant development. In addition, given their close homology to Arabidopsis UBA2a and UBP1, RBP-P and RBP-208 are likely components of stress granules and may be essential for tolerance to biotic and abiotic stress.
METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa japonica variety TC65) RBP-P mutant lines, P1MH, P2MH, and P3MH, originally named RBP-1-MH, RBP-2-MH, and RBP-3-MH, were initially identified and grown at Kyushu University, Japan. Homozygous mutant lines were then grown at Washington State University for three generations to obtain uniformly stable plant lines. Homozygous RBP-P wild-type lines, P1N, P2N, and P3N, which segregated from heterozygous P1MH, P2MH, and P3MH in the F2 population, were also collected as background mutation lines. Wild-type (O. sativa japonica variety TC65) and mutant lines were grown in walk-in growth chambers with a diurnal cycle of 12 h light/12 h dark at 27°C. Lighting was provided by a mixture of high-pressure sodium and metal halide lamps at a lighting intensity (canopy height) of 400 to 700 µmol m−2 s−1 depending on the chamber utilized. For the germination assay, 100 grains were sterilized with 20% (v/v) commercial bleach (ProPower Germicidal Ultra Bleach), rinsed with distilled water five times, and then germinated in a Petri dish with daily changes of water. Grains were considered germinated when the radicle attained a length of at least 1 cm.
IP Assay
IP was performed as previously described (Yang et al., 2014) with a slight modification of the extraction buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, pH 8.0, 0.5% [v/v] Nonidet P-40, 0.1 mM DTT, 100 units/mL SUPERase In [Ambion], 0.0.8 µM aprotinin, 40 µM bestatin, 14 µM E-64, 20 µM leupeptin, and 15 µM pepstatin A in DMSO) and 1 mM PMSF.
Y2H Analysis
Y2H analysis using RBP-P as a bait to identify interacting proteins from a rice cDNA library was performed as described previously (Yang et al., 2014). Yeast mating between RBP-P and candidate transformants was performed to confirm the interactions between the candidates and RBP-P. Double transformants expressing RBP-P and candidate interacting proteins were grown on synthetic dropout (SD) growth media without leucine and tryptophan (SD/-Leu/-Trp) and further screened on SD/-Leu/-Trp/-His/-Adenine medium (SD medium without leucine, tryptophan, histidine, and adenine) supplemented with 3 mM 3-amintriazole (3-AT) (SD/-Leu/-Trp/-His/-Adenine/+ 3-AT) to verify their interaction.
In Situ RT-PCR
In situ RT-PCR was performed as previously described (Chou et al., 2017). The distribution of prolamine and glutelin RNAs coded by LOC_Os07g10570 and LOC_Os01g55690, respectively, was evaluated (Chou et al., 2017).
RNA-Protein Binding Analysis
In vitro RNA-protein UV-cross-linking assay and RNA-IP were performed as previously described (Doroshenk et al., 2014). Following the RNA-IP, RT-qPCR was performed using an iTaq Universal SYBR Green Supermix Kit (Bio-Rad Laboratories) and a Bio-Rad CFX real-time PCR detection system. For qPCR, 1 μg RNA obtained from anti-RBP-P generated IP was subjected to cDNA synthesis using M-MuLV Reverse Transcriptase (NEB). As the anti-GFP antibody control precipitated much less RNA than did the anti-RBP-P antibody, the same ratio of RNA sample to anti-RBP-P antibody precipitation was used for cDNA synthesis. One microliter of 1000-fold diluted cDNA obtained from RNA-IP was used as template for RT-qPCR. PCR conditions were 95°C for 2 min, 40 cycles of 95°C for 10 s, and 60°C for 1 min. Melting curves were performed to analyze primer dimer formation and specificity of primers. Relative fold changes of mRNA transcripts based on triple technical repeats of qPCR were analyzed using Bio-Rad CFX Manager software.
Vector Construction and Production of Antibodies
The full-length cDNAs of RBP-P, RBP-L, and RBP-208 were cloned from wild-type rice cDNA and subcloned into pET30a for recombinant His-tagged protein expression and purification by cobalt (Co2+) immobilized metal chelate affinity beads (Thermo Fisher Scientific). His-tagged GFP protein was also prepared as abovementioned. The purified His-tagged RBP-P, RBP-L, RBP-208, and GFP proteins were used for immunizing New Zealand White rabbits for antibody production. The cDNAs of mutated RBP-P (P1, P2, and P3) were cloned from RBP-P mutants, P1MH, P2MH, and P3MH, and subcloned into pET30a for recombinant protein expression and Y2H vectors for detection of changes in protein-protein interaction.
BiFC Analysis
RBP-P, RBP-L, and RBP-208 cDNA sequences were cloned into pSAT1-nEYFP-C1 or pSAT1-cEYFP-C1-B (Arabidopsis Biological Resource Center), respectively, to generate N- or C-terminal of EYFP fusion protein for BiFC analysis.
BY-2 suspension cells were cultured as described previously (Smertenko et al., 2010). BY-2 cells were protoplasted by digesting the cell walls with 1% (w/v) cellulase (Onozuka RS; PhytoTechnology Laboratories), 0.05% (w/v) pectolyase (Seishin Pharmaceutical), 0.2% (w/v) Driselase (Sigma-Aldrich) in 20 mM KCl, 10 mM CaCl2, 20 mM MES hydrate, and 0.5 M sucrose (pH 5.7) at room temperature for 3 h. The protoplasts were washed in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 5 mM glucose, pH 5.8 to 6.0) and suspended in MMM solution (15 mM MgCl2, 0.1% MES, and 0.5 M mannitol, pH 5.8), and then resuspended at a density of 2 × 106 cells/mL. For transformation, 300 μL of BY-2 protoplasts was incubated with 30 to 40 μg of total plasmid DNA and an equal volume of PEG solution [40% PEG 4000, 0.4 M Mannitol, and 0.1 M Ca(NO3)2, pH 8 to 9]. After incubation at room temperature for 15 to 20 min, 10 mL W5 solution was added, mixed, and centrifuged at 100g for 5 min at room temperature to remove PEG solution. The transformed protoplasts were finally resuspended in 1 mL protoplast culture medium (4.3 g/L MS salts, 0.4 M sucrose, 500 mg/L MES hydrate, 750 mg/L CaCl2, and 250 mg/L NH4NO3, pH 5.7) and cultured at 26°C for 16 h before observation. The fluorescence and light images were observed using a Zeiss LSM 510 Meta confocal microscope.
RNA-Seq and Data Processing
The 10- to 14-d-old developing grains of the wild type andP1MH and P3MH mutants were dehulled to remove the outer fibrous lemma and palea and subjected to total RNA extraction using Plant RNA Reagent (Invitrogen) as previously described (Doroshenk et al., 2014). Purified RNA samples from developing grains from three independent plants for each rice genotype comprise the three biological replicates analyzed by next-generation RNA-seq. RNA-seq libraries were constructed using a ScriptSeq v2 RNA-Seq Library Preparation Kit (Illumina) following the manufacturer’s instructions and sequenced on an Illumina HiSeq instrument. RNA-seq reads were trimmed and mapped to the rice genome database (http://rice.plantbiology.msu.edu/index.shtml) using the CLC genomics workbench (Qiagen). Only unique reads were mapped to the rice genome and normalized as RPKM. Raw data of RPKM of all genes are listed in Supplemental Data Set 1. Genes with a log2 fold change > 1 and a P value lower than 0.01 were defined as differentially expressed genes (Figure10). GOslim annotation was retrieved from http://rice.plantbiology.msu.edu/index.shtml and categorized based on Arabidopsis GO slim terms (https://www.arabidopsis.org) with slight modifications. Some undefined genes were manually categorized based on putative function.
Verification of RNA-Seq by RT-qPCR
Total RNA extraction, cDNA synthesis, and quantitative PCR were performed as mentioned above. The relative expression of target genes was calculated based on the 2−ΔΔCт value with normalization to actin or ubiquitin mRNA levels. All primer sequences are listed in Supplemental Table 2.
Transmission Electron Microscopy
Transmission electron microscopy samples were prepared as previously described (Tian and Sun, 2011). Briefly, the developing grains from the wild type, P1MH, and P3MH were collected at 10 to 14 d after flowering, fixed in 4% (v/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde at 4°C overnight, dehydrated with 30, 50, 70, 80, 90, and 100% ethanol solution, and embedded in LR White resin. Immunogold labeling on ultrathin sections of each samples was performed using antiglutelin and antiprolamine antibodies at 1:200 dilution and gold-coupled secondary antibodies at 1:50. After poststaining with 2% uranyl acetate and Reynold’s lead citrate, the ultrasections were examined on a FEI Tecnai G2 20 Twin transmission electron microscope coupled to a FEI Eagle 4k CCD camera equipped with a 200-kV LaB6 electron source.
Protein Structure Prediction
The structure of RBP-P protein was predicted by online software I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Zhang, 2008; Roy et al., 2010; Yang et al., 2015).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under NCBI accession numbers shown in the legends of Supplemental Figures 1, 3, and 4.
Supplemental Data
Supplemental Figure 1. Protein sequence alignment of RBP-P with its orthologs from other species.
Supplemental Figure 2. Screening of homozygous mutants of P1MH and P3MH.
Supplemental Figure 3. Protein sequence alignments of RBP-L with RBP-45 family proteins from other plant species.
Supplemental Figure 4. Protein sequence alignments of RBP-208 with its UBP1 family proteins from other plant species.
Supplemental Figure 5. Expression profile of RBP-P in various organs and tissues.
Supplemental Table 1. List of interacting partners of RBP-P identified by yeast two hybrid.
Supplemental Table 2. Sequence information of primers used in the study.
Supplemental Data Set 1. Expression of all genes from wild type, P1MH, and P3MH mutant revealed by RNA sequencing analysis.
Supplemental Data Set 2. Summary information of differentially expressed genes in P1MH and P3MH compared with the wild type.
Supplemental Data Set 3. Detailed information of up- and downregulated genes in P1MH and/or P3MH mutants.
Supplemental Data Set 4. Expression of glutelin and prolamine genes.
Supplemental Data Set 5. Differentially expressed transcription/translation factors and putative RNA binding proteins.
Supplemental Data Set 6. Differentially expressed putative genes involved in floral and embryonic development.
Supplemental Data Set 7. Differentially expressed putative genes involved in chloroplast.
Supplemental Data Set 8. Differentially expressed putative genes involved in stress response.
Supplemental Data Set 9. Raw Data of GOslim assignment obtained from MSU Rice genome database.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Oliver Oviedo from Los Alamos National Laboratory, Los Alamos, NM, for his valuable suggestions on RNA-seq data analysis. This work was supported by grants by the National Science Foundation (MCB-1444610 and IOS-1701061), by WSU CAHNRS USDA-NIFA Projects 0590 and 0119, and by the Japan Society for the Promotion of Science. Preparation of the manuscript took place while T.W.O. was serving as a program director at the National Science Foundation.
AUTHOR CONTRIBUTIONS
L.T. designed the research and conducted most of the experiments and data analysis. H.-L.C. performed the in situ RT-PCR experiment. L.Z. contributed to immunoprecipitation studies. S.-K.H constructed the vectors for recombinant protein expression of mutant RBP-P. S.R.S. performed RNA-seq. K.A.D. identified RBP-P in a previous study. T.K. provided rice mutants. T.W.O. supervised the research. L.T. and T.W.O. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Andreassi C., Riccio A. (2009). To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 19: 465–474. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Sorenson R., Juntawong P. (2009). Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 14: 443–453. [DOI] [PubMed] [Google Scholar]
- Blower M.D. (2013). Molecular insights into intracellular RNA localization. Int. Rev. Cell Mol. Biol. 302: 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov E.V., Mineev K.S., Goncharuk M.V., Arseniev A.S. (2012). Structural and thermodynamic insight into the process of “weak” dimerization of the ErbB4 transmembrane domain by solution NMR. Biochim. Biophys. Acta 1818: 2158–2170. [DOI] [PubMed] [Google Scholar]
- Bove J., Kim C.Y., Gibson C.A., Assmann S.M. (2008). Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 67: 71–88. [DOI] [PubMed] [Google Scholar]
- Carvalho Ade O., Gomes V.M. (2011). Plant defensins and defensin-like peptides - biological activities and biotechnological applications. Curr. Pharm. Des. 17: 4270–4293. [DOI] [PubMed] [Google Scholar]
- Cheng X., Peng J., Ma J., Tang Y., Chen R., Mysore K.S., Wen J. (2012). NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol. 195: 71–84. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Kato N., Wang W., Li J., Chen X. (2003). Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev. Cell 4: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.B., Wang C., Muench D.G., Ozawa K., Franceschi V.R., Wu Y., Okita T.W. (2000). Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767. [DOI] [PubMed] [Google Scholar]
- Chou H.L., Tian L., Kumamaru T., Hamada S., Okita T.W. (2017). Multifunctional RNA binding protein OsTudor-SN in storage protein mRNA transport and localization. Plant Physiol. 175: 1608–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J., Liu Z. (2000). LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts A.J., Washida H., Okita T.W., Satoh M., Ogawa M., Kumamaru T., Satoh H. (2005). The role of mRNA and protein sorting in seed storage protein synthesis, transport, and deposition. Biochem. Cell Biol. 83: 728–737. [DOI] [PubMed] [Google Scholar]
- Crofts A.J., Crofts N., Whitelegge J.P., Okita T.W. (2010). Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 231: 1261–1276. [DOI] [PubMed] [Google Scholar]
- Dias A.P., Dufu K., Lei H., Reed R. (2010). A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 1: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshenk K.A., Crofts A.J., Morris R.T., Wyrick J.J., Okita T.W. (2009). Proteomic analysis of cytoskeleton-associated RNA binding proteins in developing rice seed. J. Proteome Res. 8: 4641–4653. [DOI] [PubMed] [Google Scholar]
- Doroshenk K.A., Crofts A.J., Morris R.T., Wyrick J.J., Okita T.W. (2012). RiceRBP: A Resource for Experimentally Identified RNA Binding Proteins in Oryza sativa. Front. Plant Sci. 3: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshenk K.A., Tian L., Crofts A.J., Kumamaru T., Okita T.W. (2014). Characterization of RNA binding protein RBP-P reveals a possible role in rice glutelin gene expression and RNA localization. Plant Mol. Biol. 85: 381–394. [DOI] [PubMed] [Google Scholar]
- Git A., Standart N. (2002). The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA 8: 1319–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (2003). A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc. Natl. Acad. Sci. USA 100: 10558–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez G.B., Urbinati C.R., Long R.M. (2005). RNA localization in yeast: moving towards a mechanism. Biol. Cell 97: 75–86. [DOI] [PubMed] [Google Scholar]
- Hamada S., Ishiyama K., Choi S.B., Wang C., Singh S., Kawai N., Franceschi V.R., Okita T.W. (2003a). The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15: 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Ishiyama K., Sakulsingharoj C., Choi S.B., Wu Y., Wang C., Singh S., Kawai N., Messing J., Okita T.W. (2003b). Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Makita N., Kojima M., Kamada-Nobusada T., Sakakibara H. (2007). Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48: 523–539. [DOI] [PubMed] [Google Scholar]
- Jansen R.P. (2001). mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2: 247–256. [DOI] [PubMed] [Google Scholar]
- Jung H., Lee D.K., Choi Y.D., Kim J.K. (2015). OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 236: 304–312. [DOI] [PubMed] [Google Scholar]
- Kim C.Y., Bove J., Assmann S.M. (2008). Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytol. 180: 57–70. [DOI] [PubMed] [Google Scholar]
- Lambermon M.H., Simpson G.G., Wieczorek Kirk D.A., Hemmings-Mieszczak M., Klahre U., Filipowicz W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19: 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambermon M.H., Fu Y., Wieczorek Kirk D.A., Dupasquier M., Filipowicz W., Lorković Z.J. (2002). UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Mol. Cell. Biol. 22: 4346–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187. [DOI] [PubMed] [Google Scholar]
- Lee K., Kang H. (2016). Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 39: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Wynn A.N., Franks R.G., Hwang Y.S., Lim J., Kim J.H. (2014). The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386: 12–24. [DOI] [PubMed] [Google Scholar]
- Lewandowska D., Simpson C.G., Clark G.P., Jennings N.S., Barciszewska-Pacak M., Lin C.F., Makalowski W., Brown J.W., Jarmolowski A. (2004). Determinants of plant U12-dependent intron splicing efficiency. Plant Cell 16: 1340–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu Y., Zhang D.Z., Gillikin J.W., Boston R.S., Franceschi V.R., Okita T.W. (1993). Rice prolamine protein body biogenesis: a BiP-mediated process. Science 262: 1054–1056. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14: 229–236. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Wieczorek Kirk D.A., Klahre U., Hemmings-Mieszczak M., Filipowicz W. (2000). RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6: 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde B.M., Moore C., Varani G. (2007). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F.H. (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272: 2118–2131. [DOI] [PubMed] [Google Scholar]
- Marowa P., Ding A., Kong Y. (2016). Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 35: 949–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C., Mowry K., Besse F. (2012). Principles and roles of mRNA localization in animal development. Development 139: 3263–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Koester T., Staiger D. (2015). Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins implicated in the splicing process. Biomolecules 5: 1717–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K., Nussbaum-Shochat A., Ben-Yehuda S., Amster-Choder O. (2011). Translation-independent localization of mRNA in E. coli. Science 331: 1081–1084. [DOI] [PubMed] [Google Scholar]
- Okita T.W., Choi S.B. (2002). mRNA localization in plants: targeting to the cell’s cortical region and beyond. Curr. Opin. Plant Biol. 5: 553–559. [DOI] [PubMed] [Google Scholar]
- Peal L., Jambunathan N., Mahalingam R. (2011). Phylogenetic and expression analysis of RNA-binding proteins with triple RNA recognition motifs in plants. Mol. Cells 31: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S., Teter S.J., Zwart P.H., Brennan R.G., Phillips K.J., Kiley P.J. (2013). Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat. Struct. Mol. Biol. 20: 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remorino A., Hochstrasser R.M. (2012). Three-dimensional structures by two-dimensional vibrational spectroscopy. Acc. Chem. Res. 45: 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M., Redko Y., Leung J. (2006). Arabidopsis RNA-binding protein UBA2a relocalizes into nuclear speckles in response to abscisic acid. FEBS Lett. 580: 4160–4165. [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sami-Subbu R., Choi S.B., Wu Y., Wang C., Okita T.W. (2001). Identification of a cytoskeleton-associated 120 kDa RNA-binding protein in developing rice seeds. Plant Mol. Biol. 46: 79–88. [DOI] [PubMed] [Google Scholar]
- Satoh-Cruz M., Crofts A.J., Takemoto-Kuno Y., Sugino A., Washida H., Crofts N., Okita T.W., Ogawa M., Satoh H., Kumamaru T. (2010). Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol. 51: 1581–1593. [DOI] [PubMed] [Google Scholar]
- Schweikhard V., Meng C., Murakami K., Kaplan C.D., Kornberg R.D., Block S.M. (2014). Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc. Natl. Acad. Sci. USA 111: 6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M.J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A.P., Deeks M.J., Hussey P.J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123: 3019–3028. [DOI] [PubMed] [Google Scholar]
- Sorenson R., Bailey-Serres J. (2014). Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann C.U., Petsalaki E., Russell R.B., Müller C.W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35: 565–574. [DOI] [PubMed] [Google Scholar]
- Tian L., Okita T.W. (2014). mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr. Opin. Plant Biol. 22: 77–85. [DOI] [PubMed] [Google Scholar]
- Tian L., Sun S.S.M. (2011). A cost-effective ELP-intein coupling system for recombinant protein purification from plant production platform. PLoS One 6: e24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Ludwig P. (2003). The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G., Mayr G., Kasiem M.M., Albrecht M., Cornelissen B.J., Takken F.L. (2008). Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 59: 1383–1397. [DOI] [PubMed] [Google Scholar]
- Varet A., Parker J., Tornero P., Nass N., Nürnberger T., Dangl J.L., Scheel D., Lee J. (2002). NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol. Plant Microbe Interact. 15: 608–616. [DOI] [PubMed] [Google Scholar]
- Verma V.V., Gupta R., Goel M. (2015). “Phylogenetic and evolutionary analysis of functional divergence among Gamma glutamyl transpeptidase (GGT) subfamilies”. Biol. Direct 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwartzenberg K., Lindner A.C., Gruhn N., Šimura J., Novák O., Strnad M., Gonneau M., Nogué F., Heyl A. (2016). CHASE domain-containing receptors play an essential role in the cytokinin response of the moss Physcomitrella patens. J. Exp. Bot. 67: 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A., Rühl C., Stauffer E. (2012). The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 3: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Washida H., Crofts A.J., Hamada S., Katsube-Tanaka T., Kim D., Choi S.B., Modi M., Singh S., Okita T.W. (2008). The cytoplasmic-localized, cytoskeletal-associated RNA binding protein OsTudor-SN: evidence for an essential role in storage protein RNA transport and localization. Plant J. 55: 443–454. [DOI] [PubMed] [Google Scholar]
- Washida H., Kaneko S., Crofts N., Sugino A., Wang C., Okita T.W. (2009). Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol. 50: 1710–1714. [DOI] [PubMed] [Google Scholar]
- Washida H., Sugino A., Doroshenk K.A., Satoh-Cruz M., Nagamine A., Katsube-Tanaka T., Ogawa M., Kumamaru T., Satoh H., Okita T.W. (2012). RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J. 70: 471–479. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Schleiff E., Zerges W. (2013). Protein targeting to subcellular organelles via MRNA localization. Biochim. Biophys. Acta 1833: 260–273. [DOI] [PubMed] [Google Scholar]
- Wu Z., et al. (2016). RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. Plant Cell 28: 55–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Crofts A.J., Crofts N., Okita T.W. (2014). Multiple RNA binding protein complexes interact with the rice prolamine RNA cis-localization zipcode sequences. Plant Physiol. 164: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]