Analysis of a spo11-1 hypomorphic allele in Arabidopsis showed that reducing the number of meiotic DSBs resulted in a profound redistribution of meiotic crossovers on a genome-wide scale.
Abstract
Meiotic recombination generates genetic diversity and ensures proper chromosome segregation. Recombination is initiated by the programmed formation of double-strand breaks (DSBs) in chromosomal DNA by DNA Topoisomerase VI-A Subunit (SPO11), a topoisomerase-like enzyme. Repair of some DSBs leads to the formation of crossovers (COs). In most organisms, including plants, the number of DSBs greatly exceeds the number of COs and which DSBs become CO sites is tightly controlled. The CO landscape is affected by DNA sequence and epigenome features of chromosomes as well as by global mechanisms controlling recombination dynamics. The latter are poorly understood and their effects on CO distribution are not well elucidated. To study how recombination dynamics affects CO distribution, we engineered Arabidopsis thaliana plants to carry hypomorphic alleles of SPO11-1. Two independent transgenic lines showed ∼30% and 40% reductions in DSB numbers, which were commensurate with the dosage of the SPO11-1 transcript. The reduction in DSB number resulted in proportional, although smaller, reductions of the number of COs. Most interestingly, CO distribution along the chromosomes was dramatically altered, with substantially fewer COs forming in pericentromeric chromosome regions. These results indicate that SPO11 activity, and the resulting DSB numbers are major factors shaping the CO landscape.
INTRODUCTION
Meiotic recombination is the primary mechanism generating new genetic variation in plants. Recombination is initiated by formation of programmed double-strand breaks (DSBs) in chromosomal DNA by a complex of proteins that in plants includes two topoisomerase-like proteins, SPO11-1 and SPO11-2, as well as several accessory proteins (Edlinger and Schlögelhofer, 2011; Vrielynck et al., 2016). The DSBs are then processed by another protein complex named MRE11-RAD50-NBS1, which removes SPO11 from DNA and resects the DSBs, creating single-stranded DNA overhangs (Mimitou and Symington, 2009). The overhangs are coated by two recombination proteins RAD51 and DMC1 (Kurzbauer et al., 2012; Brown et al., 2015), which promotes their invasion into double-stranded DNA of the corresponding regions on the homologous chromosome (Hunter and Kleckner, 2001). Eventually, meiotic recombination results in the formation of two recombination products, crossovers (COs) and non-crossovers (Hollingsworth and Brill, 2004; Rockmill et al., 2013).
In most species, COs represent only a small fraction of recombination products. In Arabidopsis thaliana, there are around 150 to 250 DSBs, based on various estimates, formed per meiosis (Vignard et al., 2007; Serrentino and Borde, 2012). However, DSB repair results in the formation of only ∼10 COs (Higgins et al., 2004; Chelysheva et al., 2007; Osman et al., 2011). There are two distinct CO types (Zalevsky et al., 1999; de los Santos et al., 2003; Wang et al., 2016), class I and class II, which are formed by different protein complexes (Mercier et al., 2005; Holloway et al., 2008). MLH1 and MLH3 are the key proteins required for the formation of class I COs, whereas class II CO formation involves MUS81 and MMS4.
A number of factors affect CO distribution along chromosomes. Most of them are related to DNA sequence and epigenome features of the chromosome regions preferred as sites of recombination events (Giraut et al., 2011; Choi et al., 2013; Wijnker et al., 2013; Shilo et al., 2015; Kianian et al., 2018). CO distribution is also shaped by globally operating mechanisms of recombination dynamics affecting how many and which DSBs are repaired as COs. How these mechanisms operate and how they impact CO landscape is not clear. At least three such mechanisms exist: CO assurance, CO interference, and CO homeostasis. CO assurance ensures formation of at least one obligatory CO per homologous chromosome pair, which is required for proper chromosome orientation at metaphase I and chromosome segregation at anaphase I (Jones and Franklin, 2006). Interference affects CO distribution by preventing CO formation in close proximity to each other (Zhang et al., 2014). However, only some COs are subject to interference. Class I COs, which constitute ∼85% of all Arabidopsis COs, are sensitive to interference (Drouaud et al., 2013). In contrast, class II COs, which are the remaining 15%, are interference-insensitive (Henderson, 2012). Homeostasis affects CO distribution by maintaining a stable CO number even if the DSB number changes widely (Martini et al., 2006). However, although CO homeostasis is robust in yeast and mouse (Martini et al., 2006; Cole et al., 2012), its effects in plants appear to be limited (Sidhu et al., 2015).
To study how recombination dynamics affects CO distribution, we characterized Arabidopsis lines carrying hypomorphic alleles of SPO11-1 and showing decreased numbers of meiotic DSBs. While these changes did not result in major defects in DSB repair, chromosome synapsis, or segregation, we discovered that the SPO11-1 hypomorphs exhibited dramatic alterations of CO distribution patterns, primarily due to significant decreases in CO formation in pericentromeric regions of chromosomes. These data indicate that SPO11 activity is a major determinant of CO landscape.
RESULTS
Hypomorphic Alleles of SPO11-1 Exhibit Reduced DSB Numbers
To study how DSB dynamics affects CO landscape, we expressed a wild-type copy of the SPO11-1 gene in the spo11-1 mutant background and selected hypomorphic lines exhibiting incomplete complementation. To do this, plants of the Col-0 ecotype heterozygous for the spo11-1-3 allele (Sanchez-Moran et al., 2007) were transformed with a construct containing the full-length SPO11-1 gene together with its native promoter. Then, 20 randomly selected T1 plants homozygous for the spo11-1-3 mutation were selected and propagated to the T2 generation.
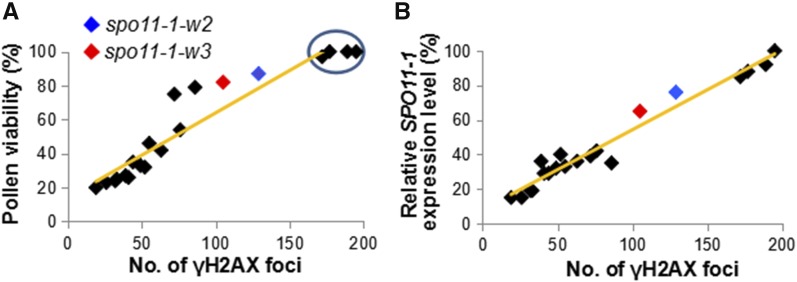
The T2 lines displayed a broad range of defects in pollen viability, ranging from severe sterility with 20% of pollen viability to a complete fertility (i.e., 100% of pollen viability) (Figure 1A; Supplemental Table 1). The decreased pollen viability phenotypes were stable, remaining constant for at least four generations. To quantify the number of meiotic DSBs produced in the transgenic lines, we used an anti-γH2AX antibody, which detects phosphorylation of the H2AX histone variant near DSB sites (Kurzbauer et al., 2012). Lines without obvious pollen viability defects (i.e., exhibiting pollen viability higher than 97%) showed ∼172 to 195 γH2AX foci per meiocyte (highlighted by a circle in Figure 1A). These numbers were similar to the ones reported in literature for wild-type Arabidopsis plants (Vignard et al., 2007; Serrentino and Borde, 2012). In contrast, DSB numbers in lines with reduced pollen viability were significantly lower (Figure 1A; Supplemental Table 1) and proportional to the decrease in pollen viability (r = 0.94) and the SPO11-1 transcript levels examined using real-time RT-PCR (r = 0.93) (Figure 1B; Supplemental Table 1).
Figure 1.
Most Arabidopsis Lines Carrying the SPO11-1 Transgene Exhibit Incomplete Complementation of the spo11-1-3 Mutation.
(A) The relationship between the number meiotic DSBs, measured by the number of γH2AX foci, and pollen viability. Pollen viability was quantified using at least 10 anthers for each line. γH2AX foci were quantified using immunolocalization experiments on ten leptotene cells for each line. Lines without obvious pollen viability defects are indicated by a circle.
(B) The relationship between the number of γH2AX foci and the level of SPO11-1 transcript. The transcript levels of SPO11-1 in were examined using reverse transcription quantitative PCR with the ACTIN2 gene utilized as an internal standard.
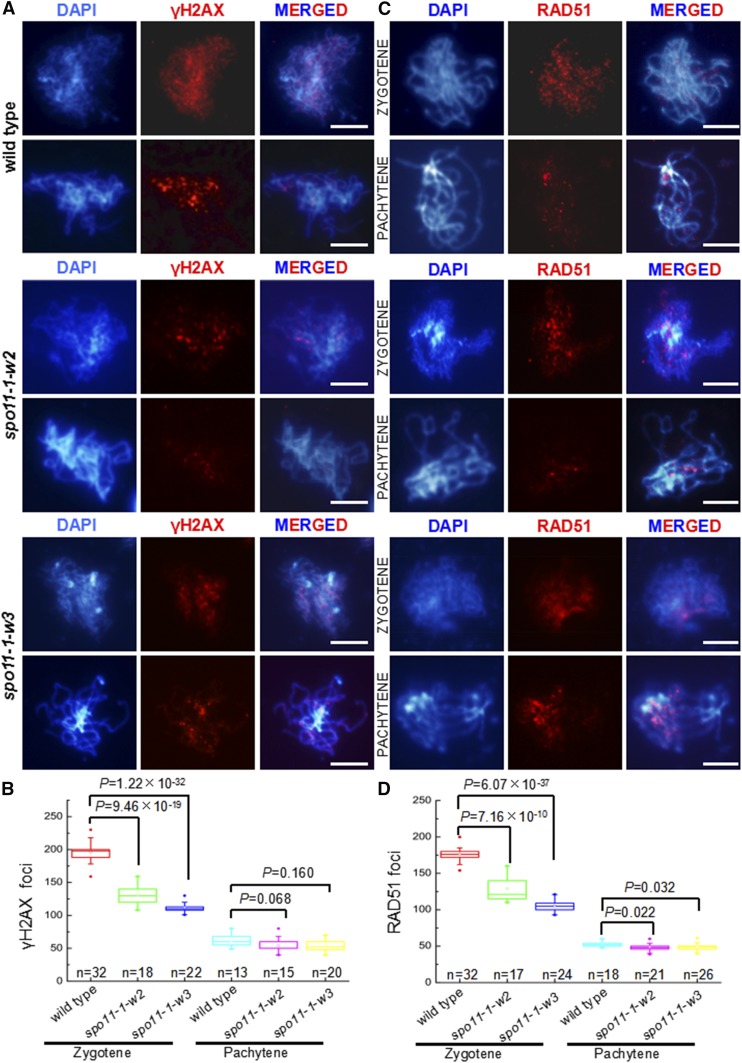
For further studies, we selected two of the transgenic lines with mild to moderate decreases in pollen viability (Supplemental Figure 1), spo11-1-w2, which exhibited an average of 129 γH2AX foci per meiocyte in zygotene, and spo11-1-w3, which had an average of 105 γH2AX foci per meiocyte (Figures 2A and 2B). The γH2AX focus numbers in both lines were significantly lower (Figure 2B) than the average of 196 γH2AX foci in wild-type Col-0 plants (Figures 2A and 2B). The DSB number reductions were confirmed by immunolocalizing the RAD51 protein, which marks DSB sites and facilitates the first step of DSB repair (Pradillo et al., 2014). At zygotene, there was an average of 129 RAD51 foci in spo11-1-w2 and 105 foci in spo11-1-w3 (Figures 2C and 2D), which represented statistically significant decreases (Figure 2D) compared with the average of 176 foci in wild-type Col-0 (Figures 2C and 2D). Overall, the γH2AX and RAD51 immunolocalization assays were in agreement, showing an ∼30% DSB number decrease in spo11-1-w2 and an ∼40% decrease in spo11-1-w3.
Figure 2.
Meiocytes in the spo11-1-w2 and spo11-1-w3 Lines Show Reduced Numbers of Meiotic DSBs.
(A) Analysis of meiotic DSBs formation using the anti-γH2AX antibody (red) in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants. Bars = 5 μm.
(B) Numbers of γH2AX loci in zygotene and pachytene in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants. Data are presented as box plots. Results of the two-tailed Student’s t test are shown.
(C) Analysis of meiotic DSBs formation using the anti-RAD51 antibody (red) in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants. Bars = 5 μm.
(D) Numbers of RAD51 loci in zygotene and pachytene in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants. Data are presented as box plots. Results of the two-tailed Student’s t test are shown.
In summary, these data showed that by transforming spo11-1 mutants with wild-type SPO11-1, we were able to generate a series of transgenic lines exhibiting varying levels of SPO11-1 expression, which resulted in reduced numbers of DSBs relative to wild type.
SPO11-1 Hypomorphs Do Not Show Abnormalities in Synapsis
Previous studies have shown that near-complete elimination of DSBs in Arabidopsis using null spo11 mutations resulted in severe defects in chromosome synapsis, including delayed and incomplete installation of ZIPPER1 (ZYP1), the central element protein of the synaptonemal complex (Grelon et al., 2001; Stacey et al., 2006; De Muyt et al., 2007).
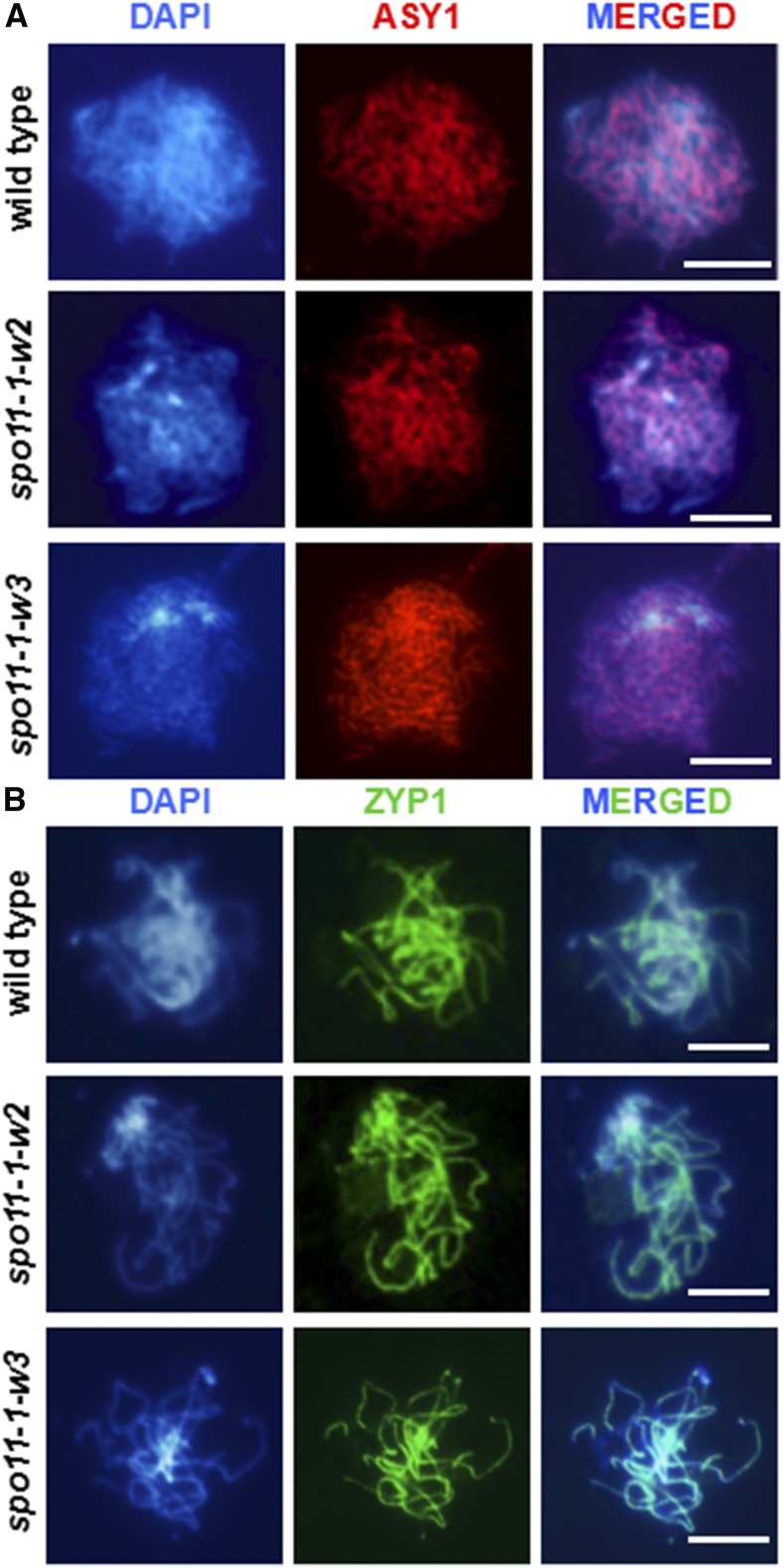
Consequently, we conducted immunolocalization analyses of ZYP1 (Higgins et al., 2005) along with ASY1, a protein associated with the lateral element of the synaptonemal complex (Armstrong et al., 2002). Cytological observations with both antibodies did not reveal detectable differences in spo11-1-w2 or spo11-1-w3 compared with wild-type Col-0 plants (Figure 3). Thus, it appears that the mild reductions in DSB number in the hypomorphic lines did not result in serious synapsis defects.
Figure 3.
Synaptonemal Complex Formation Is Not Affected in spo11-1-w2 and spo11-1-w3 Meiocytes.
(A) Immunolocalization of the ASY1 protein (red) in wild-type, spo11-1-w2, and spo11-1-w3 male meiocytes in zygotene. Bars = 10 μm. In each line, at least 10 meiocytes were examined at each stage.
(B) Immunolocalization of the ZYP1 protein (green) in wild-type, spo11-1-w2, and spo11-1-w3 male meiocytes in pachytene. Bars = 10 μm. In each line, at least 10 meioctytes were examined at each stage.
Chromosome Appearance, Bivalent Formation, and Chromosome Segregation Appear Normal in spo11-1-w2 and spo11-1-w3
To examine whether decreasing DSB numbers in the SPO11-1 hypomorphic lines resulted in changes in chromosome dynamics, we examined chromosomes from leptotene to telophase II in spo11-1-w2 and spo11-1-w3 using 4′,6-diamidino-2-phenylindole (DAPI) staining. We found that chromosome appearance from leptotene to telophase II in spo11-1-w2 and spo11-1-w3 was indistinguishable from the wild-type Col-0 (Supplemental Figure 2). Chromosome segregation in anaphase I and II were also similar to the wild type. Altogether, these data indicated that decreasing DSB numbers in the SPO11-1 hypomorphic lines did not results in easily detectable defects in chromosome appearance or behavior.
Centromere Interactions Do Not Appear Affected in SPO11-1 Hypomorphic Lines
To examine whether decreasing DSB numbers disrupted the SPO11-dependent interactions of centromeres during early meiotic prophase I (Ronceret et al., 2009; Da Ines et al., 2012), we examined centromere behavior using fluorescence in situ hybridization (FISH) with a centromere-specific probe. These analyses did not uncover obvious differences between spo11-1-w2 and spo11-1-w3 and wild-type Col-0 plants (Supplemental Figure 3), indicating that interactions of centromeric regions are not altered in the hypomorphic lines in a major way.
There Are No Severe Defects in DSB Repair in SPO11-1 Hypomorphic Lines
It has been reported that mutants with dramatically reduced numbers of RAD51 foci exhibit defective DSB repair, manifested by high numbers of RAD51 foci persisting well into pachytene (Su et al., 2017). To investigate if this was the case in the Arabidopsis SPO11-1 hypomorphs, we examined γH2AX and RAD51 foci numbers in pachytene. As criteria to identify pachytene meiocytes, we used chromosome appearance, and their presence as bivalents, as well as the pairing status of centromere regions detected using FISH. We found that the γH2AX and RAD51 foci numbers in pachytene in both spo11-1-w2 and spo11-1-w3 were similar to those in wild-type Col-0 plants (Figures 2C and 2D). These data suggested that the more modest DSB reductions exhibited by the two hypomorphic lines have not resulted in severe defects in DSB repair.
CO Numbers Are Reduced in spo11-1-w2 and spo11-1-w3
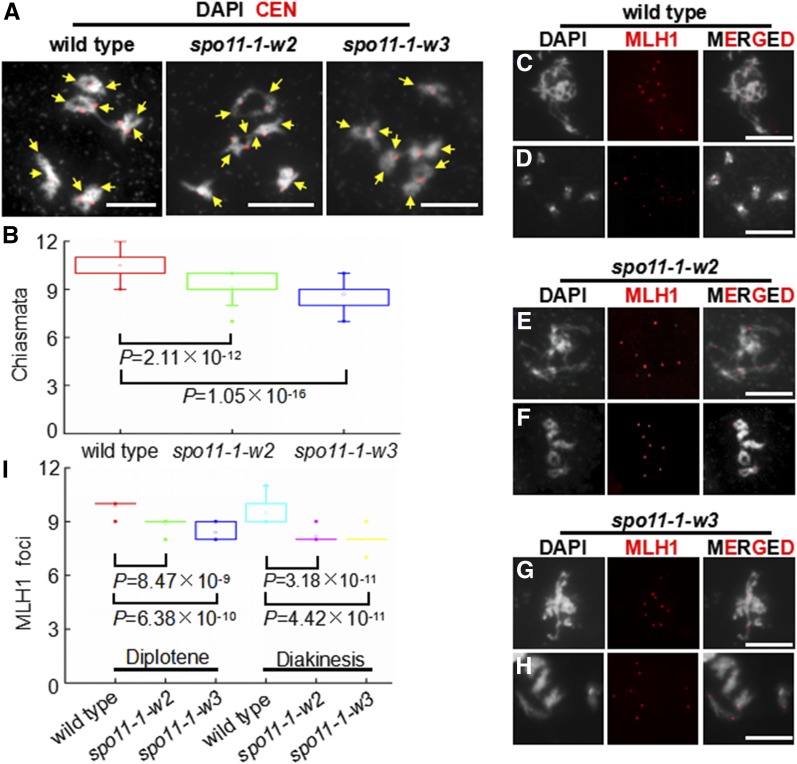
To study how reducing DSB numbers affected recombination dynamics, we examined CO numbers. To quantify all COs, we counted chiasmata, cytological structures that are sites of COs and are visible under the microscope in late meiotic prophase I (Sanchez-Moran et al., 2002; Sidhu et al., 2015). In diakinesis, we found, on average, 9.1 chiasmata in spo11-1-w2 (n = 45) and 8.7 chiasmata in spo11-1-w3 (n = 45) (Figures 4A and 4B). This number indicated statistically significant CO number reductions in both lines (Figure 4B) compared with wild-type Col-0, which showed, on average, 10.5 chiasmata per meiocyte (n = 45) (Figures 4A and 4B). The wild-type numbers were consistent with the previously reported numbers of 9 to 11 COs in Arabidopsis meiosis (Higgins et al., 2004; Chelysheva et al., 2007; Giraut et al., 2011; Osman et al., 2011).
Figure 4.
Cytological Analysis of Crossovers in Wild-Type, spo11-1-w2, and spo11-1-w3 Meiocytes.
(A) Chiasma analysis in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants in diakinesis. White = chromosomes stained with DAPI. Red = centromeres visualized using a centromere-specific FISH probe. Yellow arrows point to chiasmata. Bars = 10 μm.
(B) Chiasma numbers in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants. Results of the two-tailed Student’s t test are shown.
(C) to (H) Localization of the MLH1 protein foci (red) in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants at diplotene ([C], [E], and [G]) and diakinesis ([D], [F], and [H]). Bars = 10 μm.
(I) Numbers of MLH1 foci in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants. Results of the two-tailed Student’s t test are shown.
To specifically examine class I COs, we scored chromosomal foci of MLH1. In wild-type Col-0 meiocytes, there was an average of 9.9 MLH1 foci in diplotene (n = 19) and 9.5 foci in diakinesis (n = 22) (Figures 4C, 4D, and 4I). In contrast, in spo11-1-w2, we found on average of 8.9 MLH1 foci in diplotene (n = 15) and 8.2 foci in diakinesis (n = 17) (Figures 4E, 4F, and 4I), whereas in spo11-1-w3, there were on average 8.4 MLH1 foci in diplotene (n = 18) and 8.0 foci in diakinesis (n = 19) (Figures 4G to 4I). At both stages, the differences between the transgenic lines and wild type were statistically significant (Figure 4I).
Altogether, the CO analyses indicated that decreasing the DSB numbers in the SPO11-1 hypomorphic lines resulted in reductions in the numbers of COs. The CO number decreases were proportional, albeit smaller, to the DSB number decreases and were primarily due to reductions in class I CO formation.
CO Distributions Are Altered in spo11-1-w2 and spo11-1-w3
In addition to examining the CO number, we also investigated CO distribution by assessing recombination rates in several intervals genome-wide. To do this, spo11-1-3 mutant plants (in the Col-0 ecotype background) were backcrossed nine times to the Landsberg erecta (Ler) ecotype. The resulting spo11-1-3 (Ler) plants were crossed to the spo11-1-w2 and spo11-1-w3 hypomorphic lines, which also carried homozygous spo11-1-3 mutations and were in the original Col-0 background. CO rates were assessed in male meiosis in the hybrid plants by using them to pollinate wild-type Col-0 plants and genotyping the resulting BC1 progeny (see Methods for more information). For genotyping, we used 30 pairs of insertion/deletion (InDel) markers, which were distributed fairly uniformly across four of the five Arabidopsis chromosomes (Supplemental Figure 4). Chromosome 3 was excluded from the analyses as it harbors the SPO11-1 locus (Grelon et al., 2001) and the hybrid plants needed to be homozygous for the spo11-1-3 mutation.
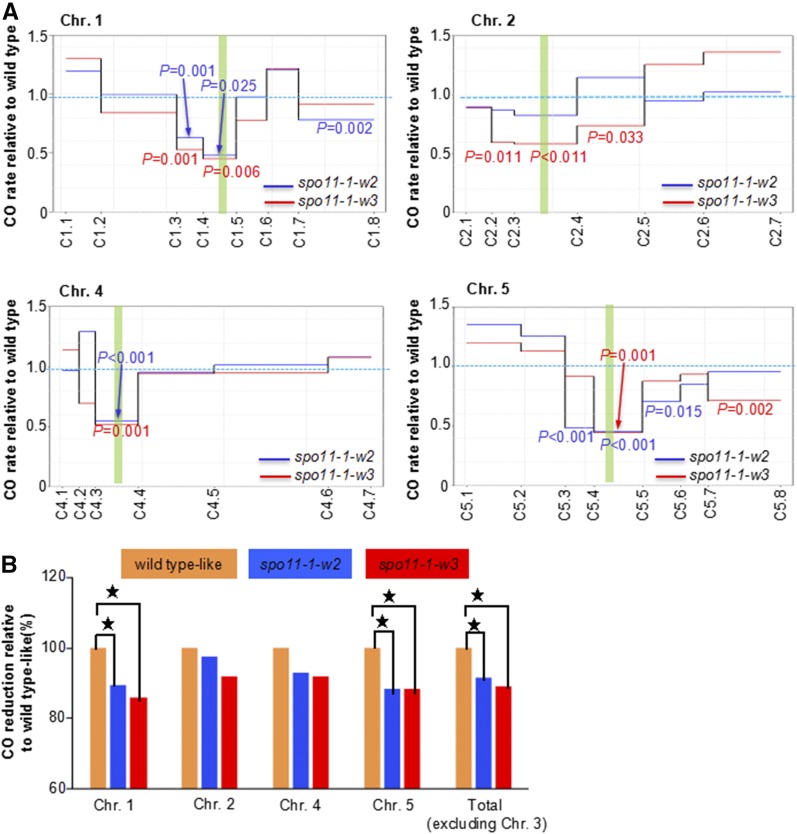
In the spo11-1-w2 population, we genotyped 346 plants homozygous for spo11-1-3, along with 357 plants with heterozygous SPO11-1/spo11-1-3 genotype from a corresponding sib population control (see Methods). For spo11-1-w3, 327 mutant plants and 316 control-sib-population plants were genotyped. We found striking differences in chromosome-wide recombination patterns between the two SPO11-1 hypomorphic populations and their control sibs. Both hypomorphic populations exhibited significantly fewer COs in pericentromeric regions on all chromosomes (Figure 5A). On chromosomes 1, 4, and 5, the decreases in centromeric bins were as much as 50%. On the other hand, CO rates on chromosome arms in spo11-1-w2 and spo11-1-w3 plants were mostly similar or slightly higher than in the control populations, exceeding 25% in only a few cases (Figure 5A). They were also generally not significantly different between the mutant and control populations.
Figure 5.
CO landscapes in male meiosis in spo11-1-w2 and spo11-1-w3.
(A) Patterns of CO distribution on individual chromosomes. Green vertical bars represent centromeres. Solid lines represent CO frequency of each interval in spo11-1-w2 (blue) and spo11-1-w3 (red) relative to sib population control. Probability values according to the Z-test are shown for intervals in which CO rates in spo11-1-w2 (blue) and spo11-1-w3 (red) differ significantly from those in sib population control.
(B) CO reductions on each chromosome in spo11-1-w2 and spo11-1-w3 populations relative to the sib population control. Asterisks indicate statistically significant differences according to the Student’s t test.
In addition to the changes in individual intervals, we also detected overall CO rate decreases on entire chromosomes (Figure 5B). The overall decreases were substantially larger on chromosomes 1 and 5, and the smallest on chromosome 2. These differences could be related to the genetic length of chromosomes; chromosomes 1 and 5 are the longest and chromosome 2 is the shortest of Arabidopsis chromosomes.
Altogether, genome-wide analyses of CO rates in populations carrying hypomorphic alleles of SPO11-1 reveled significant changes in CO distribution patterns, primarily caused by substantial decreases in CO numbers in pericentromeric chromosome regions.
DISCUSSION
Examination of recombination patterns in Arabidopsis plants carrying hypomorphic alleles of SPO11-1 indicated that reduced SPO11 activity, and the resulting change in meiotic DSB numbers, affected CO landscape. Other than CO distribution, reducing DSB numbers by as much as 40% did not majorly disturb chromosome behavior.
T-DNA integration events can be associated with translocations, deletions, or other types of chromosomal aberrations (Clark and Krysan, 2010; Crismani and Mercier, 2013), which may be capable of affecting the CO landscape. However, several lines of evidence imply that the phenotypes we observed were not caused by chromosomal abnormalities. (1) Very similar CO distribution patterns were exhibited by two independently derived transgenic lines. (2) Changes in CO patterns were observed on all four chromosomes, whereas chromosomal aberrations would be expected to only affect a single (deletions, duplications, and inversions) or two chromosomes (translocations). (3) Chromosomal aberrations would result in the appearance of linkage between markers that are unlinked in wild-type plants (Clark and Krysan, 2010), which we have not found. (4) Aberrations such as inversions and translocations would result in the formation of anaphase bridges, which we have not detected (Supplemental Figures 2 and 3). (5) Heterozygous translocations and inversions would also result in a close to 50% pollen viability decrease (Clark and Krysan, 2010; Crismani and Mercier, 2013), which would be much more severe than the one we observed (Supplemental Figure 1). (6) Finally, the mutant and control populations used for CO distribution analyses only differed by the presence of homozygous spo11-1-3 alleles (see Methods). The spo11-1-3 mutation has been used in numerous previous studies (Panoli et al., 2006; Stacey et al., 2006; Hartung et al., 2007; Sanchez-Moran et al., 2007; De Storme et al., 2013) but was not reported to result in CO pattern alterations, which indicates that presence of the mutation alone would not be sufficient to cause the CO distribution changes we observed. Based on all the evidence, we conclude that it is highly unlikely that the changes in CO patterns resulted from chromosomal abnormalities rather than altered SPO11-1 expression.
DSB Number, Chromosome Interactions, and CO Homeostasis
We found that reducing DSB numbers by as much as 40% did not result in detectable defects in chromosome behavior or cause DSB repair defects. Several studies have reported chromosome interaction defects in mutant plants exhibiting very severe DSB number reductions (Grelon et al., 2001; Pawlowski et al., 2003; Stacey et al., 2006; De Muyt et al., 2007; Hartung et al., 2007; Miao et al., 2013; Ji et al., 2016; Vrielynck et al., 2016). Null mutants in Arabidopsis SPO11-1 and SPO11-2 genes exhibited absence of chromosome pairing and synapsis (Grelon et al., 2001; Stacey et al., 2006; Hartung et al., 2007). In contrast, the DSB number reductions in spo11-1-w2 and the spo11-1-w3 did not result in detectable defects in chromosome behavior. These observations suggest that there might be a numerical threshold of DSBs that are required to ensure normal chromosome interactions.
The DSB number reductions in the two hypomorphic lines were accompanied by significant, albeit smaller, decreases in CO numbers. These results shed new light on the functioning of CO homeostasis in Arabidopsis. The existence of CO homeostasis has been proposed in budding yeast and mouse (Martini et al., 2006; Cole et al., 2012). However, in maize (Zea mays), CO control is robust only at the low bound of the CO number to provide CO assurance (Sidhu et al., 2015). After the requirement of one CO per chromosome pair is met, the CO number becomes linearly related to the DSB number, indicating absence of CO homeostasis. In Arabidopsis, mutants in FASCIATA 1 (FAS1), a gene encoding one of the three subunits of Chromatin Assembly Factor 1 (CAF-1) exhibit increased DSB formation, whereas the CO number remained the same as in the wild type (Varas et al., 2015). However, as the most likely cause of the DSB number increase in the fas1 mutants was a chromatin structure alteration, it cannot be excluded that the chromatin defect affected DSB formation and CO formation independently, leading to parallel increases of both DSBs and COs. In contrast to Varas et al. (2015), we detected a change in CO number accompanying a proportional change in the DSB number, which implies that the extent of homeostasis in Arabidopsis is limited. If one assumes that five obligate COs need to be formed in each Arabidopsis meiocyte to ensure correct chromosome segregation and that the remaining COs are directly proportional to the DSB number, the expected CO numbers would be 8.8 per meiocyte in spo11-1-w2 and 8.3 in spo11-1-w3. These numbers are not significantly different from the observed numbers. It should be noted that our conclusions are based on a reduction of DSB number below the wild-type level, whereas Varas et al. (2015) examined a DSB number increase over the wild-type level. These two directions of DSB number change could trigger different responses. For example, a threshold mechanism may exist to prevent CO number increase to avoid genome instability.
Interestingly, the changes in the CO number in spo11-1-w2 and spo11-1-w3 were mostly due to changes in the number of class I COs, as measured by the eventual number of MLH1 foci present at diakinesis. Although we cannot exclude that class II COs were not affected at all, because their numbers are relatively small compared with class I COs, we conclude that the overall CO number change was not predominantly due to class II COs.
DSB Number and Recombination Dynamics
Our most unexpected finding was that reductions of DSB numbers resulted in dramatic alterations of CO distribution patterns, with substantially fewer COs formed in pericentromeric chromosome regions. Studies of recombination dynamics have suggested that early forming DSBs are more likely to be repaired as COs (Kauppi et al., 2013). However, lowering SPO11-1 expression would be unlikely to alter where on chromosomes the first DSBs were formed. Instead, we hypothesize that decreasing DSB numbers changed their densities on chromosomes, which affected recombination dynamics. Alternatively, a possible mechanism for such interaction may be provided by studies of COs on sex chromosomes in mouse. Guaranteeing the formation of a CO in the relatively short pseudoautosomal region of sex chromosomes requires higher DSB densities than the ones present on autosomes (Kauppi et al., 2011). Hence, DSB density may be a major factor determining the ability of specific chromosome regions to become CO sites. Such mechanism could be responsible for the CO number decrease in pericentromeric regions in the SPO11-1 hypomorphs. Meiotic chromosome interactions in Arabidopsis initiate with pairing of centromeres (Ronceret et al., 2009; Da Ines et al., 2012). CO formation in pericentromeric regions may take place during these centromere-initiated early pairing interactions. Lower DSB densities could result in fewer COs being formed near centromeres, which allows chromosome arms to become CO sites. The latter further decreases CO formation in pericentromeric regions due to CO interference (Jones and Franklin, 2006)
It is very notable that the patterns of CO redistribution in spo11-1-w2 and spo11-1-w3 were similar to the CO patterns reported in the Arabidopsis ddm1 and met1 mutants exhibiting reduced DNA methylation (Colomé-Tatché et al., 2012; Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012; Yelina et al., 2012) (Supplemental Figure 5). CO rate decreases in pericentromeric regions in these mutants were unexpected as DNA methylation loss in these normally highly methylated regions should have resulted in increased recombination. Consequently, it has been proposed that decreasing DNA methylation levels must have altered chromatin structure of whole chromosomes, making the already open chromatin in distal regions even more accessible to recombination than in wild type (Colomé-Tatché et al., 2012; Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012; Yelina et al., 2012).
Altogether, our data indicate that SPO11 activity is a major determinant of CO distribution. We propose that its interplay with chromatin modification patterns shapes CO landscape. Similarities between the CO distribution patterns in the SPO11-1 hypomorphic lines and in the met1 as well as ddm1 mutants suggest that the magnitude of the effect of recombination dynamics on CO distribution is similar to the impact of chromatin structure.
In addition to furthering the understanding of factors that shape CO landscape, our data provide a unique example of how genetic diversity in species and populations can be modulated by expression levels of a single gene. These findings may also be useful for the efforts to modify CO patterns in crops for more efficient breeding.
METHODS
Growing and Genotyping spo11-1-3 Mutant Plants
Seeds of Arabidopsis thaliana Columbia ecotype (Col-0, CS3879) and the SALK spo11-1-3 mutant line (SALK_146172) were obtained from the ABRC. The seeds were surface-sterilized using 70% ethanol for 1 min, followed by 50% bleach for 10 min, and washed four times with sterilized water for 1 min each time. They were then germinated on half-strength Murashige and Skoog (MS) agar medium containing 1% sucrose and grown in a growth chamber with a photoperiod of 16 h light (300 micromoles of quanta per meter squared per second) at 22°C and 8 h dark at 20°C for 7 d. Seedlings were transferred to soil and grown under the same conditions. The methods of DNA extraction and genotyping were described previously (He et al., 2009). PCR primers used for genotyping are listed in Supplemental Table 2.
Complementation of the spo11-1-3 Mutant Using SPO11-1 Native Promoter Driving a Full-Length SPO11-1 Coding Sequence Fragment
The Arabidopsis SPO11-1 promoter (from −1067 to −1 bp) was amplified from genomic DNA using primers SPO11-1L and SPO11-1R that included SacI and NcoI restriction sites, respectively. The amplified fragment was cloned into the pGEM-T vector (Promega). Once the cloning accuracy was verified by sequencing, the promoter sequence was subcloned into pCAMBIA1305 to generate the transformation construct. The full-length coding sequence of SPO11-1 was cloned into the pGEM-T vector using primers SPO11-2L and SPO11-2R with NcoI and PmlI restriction sites and then subcloned into ProSPO11-1:GUS to generate the ProSPO11-1:SPO11-1 construct by replacing the GUS fragment. Sequences of all primers used in construct generation are listed in Supplemental Table 2. The ProSPO11-1:SPO11-1 construct was transferred into Agrobacterium tumefaciens strain GV3101 and subsequently transformed into spo11-1-3 heterozygous plants through the floral dip method (Clough and Bent, 1998). Transgenic plants were selected using 50 ng/μL hygromycin. Twenty spo11-1-3 homozygous plants with a range of pollen viability were selected for subsequent analysis.
RNA Extraction and RT-qPCR
RNA was extracted from young buds using an RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized from 5 μg of total RNA using the ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs) following the manufacturer’s instructions. Quantitative real-time PCR was conducted using the ABI 7500 real-time PCR system (Applied Biosystems) according to the manufacturer’s instructions. PCR amplifications were performed in a total volume of 20 μL. Reaction mixtures contained 10 μL Taq Universal SYBR Green Supermix (Bio-Rad), 0.5 μL 10 ng/μL of forward and reverse primers (SPO11-1-3L and SPO11-1-3R), and 2 μL of diluted cDNA. The PCR program was as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. The relative expression of SPO11-1 in different transgenic plants was first normalized using the expression level of ACTIN2 as an internal control and further calculated using the ratio of the expression in transgenic plants relative to wild-type plants. Data shown were the averages of three independent experiments. Primer sequences are listed in Supplemental Table 2.
Pollen Viability Analysis
Pollen viability was examined using Alexander staining (Alexander, 1969).
Chromosome Spreading
Meiotic chromosome spreads were performed as described by Ross et al. (1996). Chromosomes were stained by DAPI (Vector Lab) and examined under a fluorescence microscope.
FISH
FISH was performed as described by Furner et al. (1998) with some modifications. After the fixation step and spreading, the slides were treated with 70% deionized formamide in 2× SSC for 5 min at 95°C, and dehydrated using a 70 to 100% ethanol series. Approximately 10 μL hybridization buffer containing 1 μL probe was applied to the slides, which were hybridized for 3 h at 37°C in a humid chamber. Then, slides were washed in 2× SSC four times and counterstained using DAPI. An oligonucleotide 5′-GGTTGCGGTTTAAGTTCTTATACTCAATCATACACATGAC-3′ labeled with FITC at the 5′end was used for centromere detection (Ronceret et al., 2009). Images were obtained using a Zeiss Axio Imager A2 microscope and examined in Photoshop CS3 (Adobe Systems). FISH analysis was conducted following a previously published procedure (Han et al., 2009).
Immunostaining
Immunostaining was conducted as described previously (Wang et al., 2012). Primary antibodies used for immunofluorescence experiment were diluted as follows: 1:100 for ZYP1 (Wang et al., 2012) and MLH1 (Chelysheva et al., 2010), 1:300 for ASY1 (Wang et al., 2012), and 1:200 for γH2AX (Miao et al., 2013) and RAD51 (Wang et al., 2012). Images were collected using a Zeiss Axio Imager A2 microscope (Zeiss) and processed in Photoshop 5.0 (Adobe Systems). The Rad51 and γH2AX foci in all lines were counted and statistically analyzed using Image Tool version 3.0 (Collins, 2007). The Image tool 3.0 developed by UTHSCSA (University of Texas Health Science Center, San Antonio, TX) was used to quantify the number of foci. Images sharing the same size and resolution were expanded into full view so that foci could be easily observed. Foci were identified manually and counted automatically using the “Analysis-Count and Tag” feature.
CO Pattern Analysis
To examine CO distribution in the spo11-1-w2 population, Ler/Col spo11-1-3/spo11-1-3 spo11-1-w2/+ hybrid plants and their Ler/Col SPO11-1/spo11-1-3 spo11-1-w2/+ sibs were identified and crossed as males to wild-type Col plants to create BC1 progeny. An identical crossing scheme was used to create BC1 progeny using the spo11-1-w3 transgenic line. To maintain the transgenic construct, the seeds of transgenic plants were germinated on the MS medium containing hygromycin. To examine CO patterns, we used 30 InDel PCR markers distributed roughly uniformly across four Arabidopsis chromosomes (Supplemental Table 2). Genotyping data were uploaded into MapDisto (http://mapdisto.free.fr/) (Lorieux, 2012) for the recombination ratio calculation as previously described (Mirouze et al., 2012).
Accession Numbers
Sequence data from this article can be found at https://www.arabidopsis.org under accession number AT3G13170 (SPO11-1).
Supplemental Data
Supplemental Figure 1. Pollen viability in wild-type, spo11-1-w2, and spo11-1-w3 plants.
Supplemental Figure 2. Meiosis progression in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 plants (n = 10 for each line at each stage).
Supplemental Figure 3. Analysis of chromosome dynamics in male meiocytes in wild-type, spo11-1-w2, and spo11-1-w3 using a centromere FISH probe (red) (n = 10 for each line at each stage).
Supplemental Figure 4. Locations of makers used for CO rate analysis.
Supplemental Figure 5. Comparison of the effects of met1, spo11-1-w2, and spo11-1-w3 on CO distribution.
Supplemental Table 1. Pollen viability, number of γH2AX foci, and relative expression of SPO11-1 in each transgenic line.
Supplemental Table 2. Primers used in the study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank members of our laboratories for helpful assistance and discussion during research and manuscript preparation. This work was supported by grants from the National Natural Science Foundation of China to Y.H. (Grants 31471172 and 31671277) and Y.W. (Grants 31570314 and 31870293), by the State Key Laboratory of Genetic Engineering, Fudan University, and Rijk Zwaan to Y.W., and by the U.S. National Science Foundation to W.P.P. (IOS-1025881 and IOS-1546792).
AUTHOR CONTRIBUTIONS
Y.H., Y.W., and W.P.P. designed the research. M.X. and L.J. performed plant transformation, transgene expression analyses, and pollen viability analyses. J.W. performed cytological experiments. S.W. constructed plasmids for plant transformation. M.X., J.W., L.J., M.W., Y.W., and Y.H. analyzed the data. M.W. conducted statistical analyses of CO patterns. M.X., Y.H., Y.W., and W.P.P. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Grubb J., Zhang A., Rust M.J., Bishop D.K. (2015). Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet. 11: e1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Gendrot G., Vezon D., Doutriaux M.P., Mercier R., Grelon M. (2007). Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Grandont L., Vrielynck N., le Guin S., Mercier R., Grelon M. (2010). An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129: 143–153. [DOI] [PubMed] [Google Scholar]
- Choi K., Zhao X., Kelly K.A., Venn O., Higgins J.D., Yelina N.E., Hardcastle T.J., Ziolkowski P.A., Copenhaver G.P., Franklin F.C., McVean G., Henderson I.R. (2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.A., Krysan P.J. (2010). Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 64: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cole F., Kauppi L., Lange J., Roig I., Wang R., Keeney S., Jasin M. (2012). Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14: 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T.J. (2007). ImageJ for microscopy. Biotechniques 43: 25–30. [DOI] [PubMed] [Google Scholar]
- Colomé-Tatché M., et al. (2012). Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. USA 109: 16240–16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W., Mercier R. (2013). Identifying meiotic mutants in Arabidopsis thaliana. Methods Mol. Biol. 990: 227–234. [DOI] [PubMed] [Google Scholar]
- Da Ines O., Abe K., Goubely C., Gallego M.E., White C.I. (2012). Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet. 8: e1002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M. (2003). The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Vezon D., Gendrot G., Gallois J.L., Stevens R., Grelon M. (2007). AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 26: 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Zamariola L., Mau M., Sharbel T.F., Geelen D. (2013). Volume-based pollen size analysis: an advanced method to assess somatic and gametophytic ploidy in flowering plants. Plant Reprod. 26: 65–81. [DOI] [PubMed] [Google Scholar]
- Drouaud J., Khademian H., Giraut L., Zanni V., Bellalou S., Henderson I.R., Falque M., Mézard C. (2013). Contrasted patterns of crossover and non-crossover at Arabidopsis thaliana meiotic recombination hotspots. PLoS Genet. 9: e1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlinger B., Schlögelhofer P. (2011). Have a break: determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 62: 1545–1563. [DOI] [PubMed] [Google Scholar]
- Furner I.J., Sheikh M.A., Collett C.E. (1998). Gene silencing and homology-dependent gene silencing in Arabidopsis: genetic modifiers and DNA methylation. Genetics 149: 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L., Falque M., Drouaud J., Pereira L., Martin O.C., Mézard C. (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7: e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon D., Gendrot G., Pelletier G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Gao Z., Birchler J.A. (2009). Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Wurz-Wildersinn R., Fuchs J., Schubert I., Suer S., Puchta H. (2007). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Mawhinney T.P., Preuss M.L., Schroeder A.C., Chen B., Abraham L., Jez J.M., Chen S. (2009). A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 60: 679–690. [DOI] [PubMed] [Google Scholar]
- Henderson I.R. (2012). Control of meiotic recombination frequency in plant genomes. Curr. Opin. Plant Biol. 15: 556–561. [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Armstrong S.J., Franklin F.C.H., Jones G.H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Brill S.J. (2004). The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J.K., Booth J., Edelmann W., McGowan C.H., Cohen P.E. (2008). MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4: e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Ji J., Tang D., Shen Y., Xue Z., Wang H., Shi W., Zhang C., Du G., Li Y., Cheng Z. (2016). P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice. Proc. Natl. Acad. Sci. USA 113: 10577–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.H., Franklin F.C.H. (2006). Meiotic crossing-over: obligation and interference. Cell 126: 246–248. [DOI] [PubMed] [Google Scholar]
- Kauppi L., Barchi M., Baudat F., Romanienko P.J., Keeney S., Jasin M. (2011). Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., Keeney S. (2013). Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 27: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian P.M.A., et al. (2018). High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9: 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbauer M.-T., Uanschou C., Chen D., Schlögelhofer P. (2012). The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24: 2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M. (2012). MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30: 1231–1235. [Google Scholar]
- Martini E., Diaz R.L., Hunter N., Keeney S. (2006). Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Levy A.A. (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: E981–E988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Jolivet S., Vezon D., Huppe E., Chelysheva L., Giovanni M., Nogué F., Doutriaux M.P., Horlow C., Grelon M., Mézard C. (2005). Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3,whereas the other one is not. Curr. Biol. 15: 692–701. [DOI] [PubMed] [Google Scholar]
- Miao C., Tang D., Zhang H., Wang M., Li Y., Tang S., Yu H., Gu M., Cheng Z. (2013). Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. (2009). DNA end resection: many nucleases make light work. DNA Repair (Amst.) 8: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M., Lieberman-Lazarovich M., Aversano R., Bucher E., Nicolet J., Reinders J., Paszkowski J. (2012). Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K., Higgins J.D., Sanchez-Moran E., Armstrong S.J., Franklin F.C. (2011). Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 190: 523–544. [DOI] [PubMed] [Google Scholar]
- Panoli A.P., Ravi M., Sebastian J., Nishal B., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I. (2006). AtMND1 is required for homologous pairing during meiosis in Arabidopsis. BMC Mol. Biol. 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski W.P., Golubovskaya I.N., Cande W.Z. (2003). Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradillo M., Varas J., Oliver C., Santos J.L. (2014). On the role of AtDMC1, AtRAD51 and its paralogs during Arabidopsis meiosis. Front. Plant Sci. 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Lefrançois P., Voelkel-Meiman K., Oke A., Roeder G.S., Fung J.C. (2013). High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet. 9: e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A., Doutriaux M.P., Golubovskaya I.N., Pawlowski W.P. (2009). PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc. Natl. Acad. Sci. USA 106: 20121–20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K.J., Fransz P., Jones G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4: 507–516. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Armstrong S.J., Santos J.L., Franklin F.C., Jones G.H. (2002). Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J.L., Jones G.H., Franklin F.C. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino M.E., Borde V. (2012). The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp. Cell Res. 318: 1347–1352. [DOI] [PubMed] [Google Scholar]
- Shilo S., Melamed-Bessudo C., Dorone Y., Barkai N., Levy A.A. (2015). DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell 27: 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu G.K., Fang C., Olson M.A., Falque M., Martin O.C., Pawlowski W.P. (2015). Recombination patterns in maize reveal limits to crossover homeostasis. Proc. Natl. Acad. Sci. USA 112: 15982–15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey N.J., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48: 206–216. [DOI] [PubMed] [Google Scholar]
- Su H., Cheng Z., Huang J., Lin J., Copenhaver G.P., Ma H., Wang Y. (2017). Arabidopsis RAD51, RAD51C and XRCC3 proteins form a complex and facilitate RAD51 localization on chromosomes for meiotic recombination. PLoS Genet. 13: e1006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas J., Sánchez-Morán E., Copenhaver G.P., Santos J.L., Pradillo M. (2015). Analysis of the relationships between DNA double-strand breaks, synaptonemal complex and crossovers using the Atfas1-4 mutant. PLoS Genet. 11: e1005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignard J., Siwiec T., Chelysheva L., Vrielynck N., Gonord F., Armstrong S.J., Schlögelhofer P., Mercier R. (2007). The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 3: 1894–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrielynck N., Chambon A., Vezon D., Pereira L., Chelysheva L., De Muyt A., Mézard C., Mayer C., Grelon M. (2016). A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351: 939–943. [DOI] [PubMed] [Google Scholar]
- Wang C., et al. (2016). The role of OsMSH4 in male and female gamete development in rice meiosis. J. Exp. Bot. 67: 1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng Z., Huang J., Shi Q., Hong Y., Copenhaver G.P., Gong Z., Ma H. (2012). The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 8: e1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., et al. (2013). The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina N.E., et al. (2012). Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 8: e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen A.J., Duffy J.B., Kemphues K.J., Villeneuve A.M. (1999). Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liang Z., Hutchinson J., Kleckner N. (2014). Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 10: e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]