β-Cyclocitral, generated in the chloroplast under high-light stress, mitigates photooxidative stress by recruiting the xenobiotic response involving SCL14, TGAII, and ANAC102 transcription factors.
Abstract
When exposed to unfavorable environmental conditions, plants can absorb light energy in excess of their photosynthetic capacities, with the surplus energy leading to the production of reactive oxygen species and photooxidative stress. Subsequent lipid peroxidation generates toxic reactive carbonyl species whose accumulation culminates in cell death. β-Cyclocitral, an oxidized by-product of β-carotene generated in the chloroplasts, mediates a protective retrograde response that lowers the levels of toxic peroxides and carbonyls, limiting damage to intracellular components. In this study, we elucidate the molecular mechanism induced by β-cyclocitral in Arabidopsis thaliana and show that the xenobiotic detoxification response is involved in the tolerance to excess light energy. The involvement of the xenobiotic response suggests a possible origin for this pathway. Furthermore, we establish the hierarchical structure of this pathway that is mediated by the β-cyclocitral-inducible GRAS protein SCARECROW LIKE14 (SCL14) and involves ANAC102 as a pivotal component upstream of other ANAC transcription factors and of many enzymes of the xenobiotic detoxification response. Finally, the SCL14-dependent protective mechanism is also involved in the low sensitivity of young leaf tissues to high-light stress.
INTRODUCTION
Plants often encounter light intensities that exceed their photosynthetic capacities, due to unfavorable environmental conditions that prevent a good match between absorbed light energy and carbon metabolism (Ort, 2001). The photosynthetic electron transport chain uses molecular oxygen as an electron carrier, generating biologically damaging molecules such as reactive oxygen species (ROS), peroxides, and radicals (Apel and Hirt, 2004; Asada, 2006; Li et al., 2009). In particular, triplet excited chlorophylls, whose lifetime increases under excess light conditions, can transfer excitation energy directly to oxygen resulting in the formation of singlet oxygen (1O2) (Krieger-Liszkay et al., 2008; Triantaphylidès and Havaux, 2009). Besides its toxic effects, 1O2 can trigger a specific signaling cascade, leading to programmed cell death or to acclimation (Wagner et al., 2004; Gadjev et al., 2006; Ramel et al., 2013a; Chan et al., 2016). Nevertheless, due to a high reactivity and short lifetime (∼100 ns in biological tissues), direct involvement of 1O2 in retrograde signaling is unlikely; rather, signaling may originate in the oxidation of preferential targets, which then act as mediators.
Carotenoids are efficient 1O2 physical quenchers (Frank and Cogdell, 1996) which are sometimes oxidized by 1O2 at the level of photosystem II, generating, among other products, the retrograde signaling mediators β-cyclocitral (β-cc) and dihydroactinidiolide (Ramel et al., 2012b; Havaux, 2014; Shumbe et al., 2014, 2017). β-cc is generated in the chloroplast and its basal level (∼50 ng g−1 fresh leaf weight) triples during high-light stress, while dihydroactinidiolide concentration (∼5 ng g−1) increases almost 10-fold, suggesting chronic production of 1O2 during photosynthesis and a dramatic increase under stress conditions (Ramel et al., 2012b). Interestingly, treatment of plants with exogenous β-cc or dihydroactinidiolide increases their internal leaf concentrations to levels comparable with the ones measured under stress conditions (∼180 and 45 ng g−1, respectively) and elicit a genetic response leading to acclimation to high-light stress (Ramel et al., 2012b; Shumbe et al., 2014).
A striking feature of the gene regulation induced by β-cc in Arabidopsis thaliana is the induction of various detoxification mechanisms, among which the induction of several GSTs and UDP-glycosyltransferases (Ramel et al., 2012b) that also participate in phase II of the xenobiotic detoxifying process (Sandermann, 1992). In fact, in plants, ectopic reactive chemicals are inactivated by a set of detoxifying enzymes that modify and eliminate these compounds in three phases: modification, conjugation, and compartmentalization (Sandermann, 1992; Riechers et al., 2010). While the conjugation phase has been reported to be induced under many different stresses, the modification phase has only been characterized more recently and its involvement in physiological responses is still unclear (Mueller et al., 2008; Riechers et al., 2010; Ramel et al., 2012c). In particular, in the modification phase, the GRAS protein SCARECROW LIKE14 (SCL14) and the glutaredoxin GRX480/ROXY19 compete for binding with the TGAII transcription factors and mediate the activation or the inhibition, respectively, of the detoxification response (Ndamukong et al., 2007; Fode et al., 2008; Köster et al., 2012; Huang et al., 2016).
In this work, we identify a SCL14-dependent xenobiotic detoxification response to a physiological condition, rather than an artificial stimulus. Furthermore, we decode its role in the β-cc-induced retrograde signaling that occurs under high light stress, and we show that SCL14-dependent detoxification is necessary for the resilience of Arabidopsis plants to photooxidative stress. In this work, we enrich our understanding of the SCL14-dependent pathway by showing the hierarchical relationship between several players and make the link between the xenobiotic detoxification pathway and its physiological role in detoxifying toxic reactive carbonyl species (RCS). Finally, we show that SCL14 mediates part of the intrinsic resistance of young leaves to excessive light.
RESULTS
β-cc Elicits an Excessive Light-Like Response Involving the TGA and MYC Transcription Factors
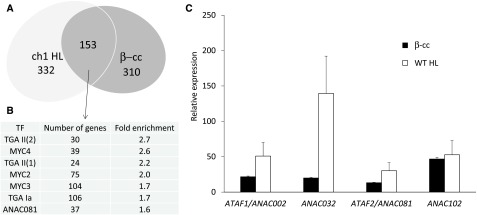
The comparison of the genetic response of 5-week-old Arabidopsis plants treated with β-cc (100 μL β-cc for 4 h; http://urgv.evry.inra.fr/CATdb, Project CEA10-03_Cyclocitral) with the response of 1O2-overproducing ch1 mutant plants exposed to excessive light (1200 μmol photons m−2 s−1 at 10°C for 24 h, Project CEA10-02_Light) (Ramel et al., 2013b) revealed a 30% overlap of the genes modified more than 1.5 fold in log2 values (Figure 1A). The overlap suggests the existence of common mechanisms to cope with photooxidative damage, elicited by β-cc and during enhanced production of 1O2.
Figure 1.
β-cc Elicits a SCL14-Like Response Involving the TGA and MYC Transcription Factors.
(A) Number of genes whose expression increases or decreases more than 1.5-fold in log2 values compared with the respective controls in the transcriptome of β-cc-treated wild-type plants (4 h treatment, β-cc) and of the ch1 mutant line under high-light stress (ch1 HL, 1200 μmol photons m−2 s−1 at 10°C for 24 h).
(B) Transcription factors cis element enrichment in the regulatory regions (−1000 + 50 bp from the starting codon) of the common genes.
(C) ANAC002, ANAC032, ANAC081, and ANAC102 expression levels (fold on control levels) in wild-type plants exposed to β-cc or under excessive light stress (HL), measured by RT-qPCR. Every value showed a significant difference when tested against CTRL conditions (P < 0.01). Error bars = +sd between the four technical replicates from pools of three plants per treatment. Two full experimental replicates.
The regulatory regions of the 153 genes in the common cluster (−1000 to +50 bp from the starting codon) show an enrichment of the MYC (MYC2, 3, and 4) and of the TGA (class I and II) transcription factor binding sites (AthaMap gene analysis; Steffens et al., 2004) (Figure 1B). MYC transcription factors are activators of the jasmonic acid response, while TGA transcription factors are well known players in the salicylic acid response and in detoxifying mechanisms (Chini et al., 2007; Kesarwani et al., 2007; Mueller et al., 2008). Moreover, the interaction of SCL14 with the TGA transcription factors induces the expression of a subset of the detoxification genes, mainly belonging to the modification phase of the xenobiotic detoxification pathway (Fode et al., 2008; Köster et al., 2012). Altogether, the MYC and TGA transcription factors mediate the response to xenobiotics and their concerted action is required for the complete activation of the response. In fact, as highlighted in the analysis of the expression of the phase I enzyme cytochrome P450 CYP81D11 (Köster et al., 2012), only the activation of both the MYC and TGA pathways leads to the maximum induction of the latter gene.
β-cc and Excessive Light Induce SCL14-Regulated ANAC Transcription Factors
Among the modified genes identified in the aforementioned transcriptomes, the expression levels of the SCL14-regulated ANAC002 and ANAC032, and of the related ANAC081 and ANAC102 genes (Fode et al., 2008; Ratnakaran, 2014) have been further analyzed by RT-qPCR in control and β-cc-treated wild-type plants and in plants after excessive light stress (1500 μmol photons m−2 s−1 at 7°C for 24 h). This analysis confirmed the induction of the four ANAC genes by β-cc (20, 20, 13, and 50 times the control levels, respectively), further pointing to ANAC102 as the most induced ANAC transcription factor by the β-cc treatment (Figure 1C).
Furthermore, the stronger induction of ANAC002, ANAC032, and ANAC081 under excessive light (50, 30, and 130 times the control values, respectively) suggests the coexistence of several molecular signals able to induce this pathway generated by high light stress. The origin of the pathway induced by xenobiotics is still unclear, and several mechanisms elicit the conjugation phase of the xenobiotic response (Riechers et al., 2010). Among these, the response to oxylipins, such as jasmonic acid, and lipid-derived reactive electrophilic compounds like 12-oxo-phytodienoic acid and phytoprostanes (Mueller and Berger, 2009; Riechers et al., 2010), with all these species being present in excessive light stress (Mano et al., 2005; Mano, 2012; Farmer and Mueller, 2013). Conversely, here, we highlight the activation of the modification phase of the xenobiotic response under physiological conditions.
β-cc Induces a SCL14-Regulated Response
β-cc enhances the expression of the four ANAC transcription factors involved in the SCL14-dependent response to xenobiotics; therefore, we wanted to test the involvement of the TGA-SCL14 regulation in the β-cc response. The TGA family of transcription factors and the corresponding cis-element as-1 are well-described transcriptional control mechanisms in plants (Katagiri et al., 1989; Redman et al., 2002). The as-1 element, from the CAMV 35S viral promoter, and the ocs element, from the bacterial octopine synthase promoter, in the form of TGACG sequences in the promoter, are mainly activated by the TGA II transcription factors, under auxin and salicylic acid mediated stimuli (Lam and Lam, 1995). as-1-bound TGA transcription factors recruit the GRAS protein SCL14 to the transcription site, activating several genes inducible by xenobiotics, which contribute to the protection of plants (Mano et al., 2005; Kotchoni et al., 2006; Fode et al., 2008; Turóczy et al., 2011; Mano, 2012).
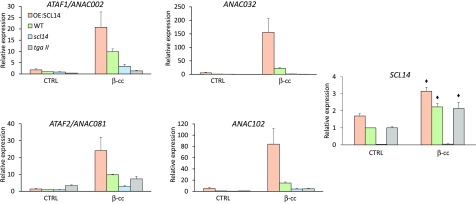
To determine whether the SCL14-dependent xenobiotic detoxification pathway is implicated in the genetic response induced by β-cc, we analyzed ANAC002, 032, 081, and 102 expression levels in the tgaII and scl14 mutant lines and in the SCL14-overexpressing line (OE:SCL14) by RT-qPCR after the β-cc treatment. These analyses showed that β-cc induction of the ANAC transcription factors is weakened or impeded in the tgaII and scl14 mutant lines (70 to 90% lower), while it is strengthened in the SCL14 overexpressing lines (200 to 800% higher) (Figure 2).
Figure 2.
β-cc Induces SCL14 and the Downstream Response.
ANAC002, ANAC032, ANAC081, ANAC102, and SCL14 expression levels (relative to wild-type control levels [WT CTRL], which are set to 1) in wild-type, scl14, tgaII, and OE:SCL14 plants under control conditions or exposed to β-cc, measured by RT-qPCR. ANAC genes induced by β-cc in the OE:SCL14, scl14, and tgaII plants showed a significant difference when tested against β-cc induction in the wild type (P < 0.01). The black diamonds indicate expression levels significantly different from SCL14 expression levels in the corresponding control condition (P < 0.01). Error bars = +sd between the four technical replicates from pools of three plants per genotype per treatment.
In the response to salicylic acid, TGAII transcription factors can interact with NPR1 (Després et al., 2000; Zhou et al., 2000; Fan and Dong, 2002). We tested an eventual competition between NPR1 and SCL14 on the induction of the ANAC genes regulated by TGA 2, 5, and 6 by analyzing ANAC002, 032, 081, and 102 expression levels in the npr1 mutant lines by RT-qPCR after the β-cc treatment. The genetic response of the four SCL14-dependent ANAC transcription factors, ANAC002, ANAC032, ANAC081, and ANAC102, to β-cc was similar in the npr1-1 mutant and in the wild type, suggesting no competition between the two TGA-regulating proteins under these conditions (Supplemental Figure 1).
The participation of SCL14 in the response to exogenous artificial molecules is well known, and many xenobiotics are able to induce the SCL14-dependent detoxification pathway (De Veylder et al., 1997; DeRidder et al., 2002; Fode et al., 2008; Riechers et al., 2010; Skipsey et al., 2011; Taylor et al., 2013). On the contrary, β-cc is an endogenous molecule showing a SCL14-dependent response. This suggests that the SCL14-regulated response is more general than the detoxification of xenobiotics and could play a role in the response to changes in natural environments.
β-cc Induces SCL14
As the SCL14-dependent response was enhanced by the β-cc treatment, we analyzed the expression level of SCL14 itself. We confirm that SCL14 is not expressed in the scl14 mutant, neither in control conditions nor in plants treated with β-cc (Figure 2). Then, we show that SCL14 is expressed 1.5 times more in the OE:SCL14 than in the wild type in control conditions, in line with the protein levels previously described (Fode et al., 2008). Although being under the control of a 35S constitutive promoter, SCL14 transcript levels exhibited a relatively small increase in the OE:SCL14 line, suggesting the intervention of a posttranscriptional regulation. Finally, β-cc treatment upregulated SCL14 expression both in wild-type and in the OE:SCL14 lines (Figure 2). Remarkably, the faint “overexpression” present in the OE:SCL14 was able to mediate a marked increase in the response of ANAC genes to β-cc (200 to 800% higher in the OE:SCL14 than in the wild type). Therefore, the increase in SCL14 gene expression observed in the wild type in response to β-cc (in the same range as the SCL14 overexpressor in the absence of β-cc treatment), together with the concomitant activation of the MYC pathway (Figure 1B), could explain the substantial effects of the apocarotenoid on ANAC transcription factors (Figure 2) and on genes encoding detoxifying enzymes (below; Figure 5) (Köster et al., 2012).
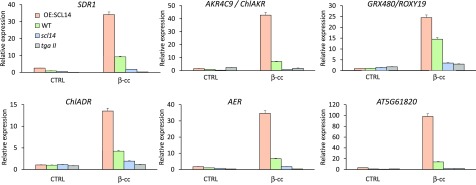
Figure 5.
β-cc Induces a SCL14-Dependent Xenobiotic-Detoxification Response.
SDR1, ChlADR, AKR4C9, AER, GRX480/ROXY19, and AT5G61820 expression levels (relative to wild-type control levels [WT CTRL], which are set to 1) in wild-type, scl14,tgaII, and OE:SCL14 plants under control conditions or exposed to β-cc, measured by RT-qPCR. Every gene induced by β-cc in the OE:SCL14, scl14, and tgaII plants showed a significant difference when tested against β-cc induction in the wild type (P < 0.01). Error bars = +sd between the four technical replicates from pools of three plants per genotype per treatment.
SCL14 Mediates Plant Resilience to Excessive Light
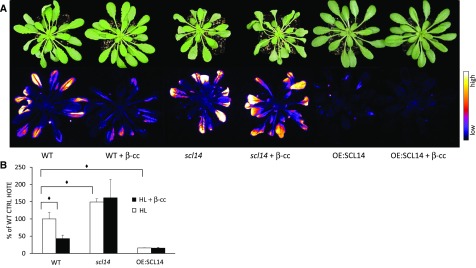
SCL14 regulates the genetic response of plants treated with β-cc; consequently, we wondered about the role of SCL14 under excessive light stress, which causes endogenous increases of β-cc in a much more complex metabolic response. Therefore, we analyzed the behavior of the scl14 knockout mutant and the OE:SCL14 overexpressor under excessive light stress (1500 μmol photons m−2 s−1 at 7°C for 24 h) following pretreatment with β-cc or with water (100 μL for 4 h). Lipid peroxidation was used as a marker of photooxidative damage and analyzed by image quantification of plant autoluminescence, derived from the spontaneous decomposition of lipid peroxides (Birtic et al., 2011), and by HPLC-UV quantification of HOTEs (hydroxy octadecatrienoic acid isomers) derived from the oxidation of linolenic acid (Montillet et al., 2004; Birtic et al., 2011). scl14 plants showed stronger leaf bleaching after 24-h exposure to stress conditions compared with wild-type plants, as well as more intense lipid-peroxidation-related luminescence (Figure 3A). By contrast, OE:SCL14 plants showed almost no detrimental effects due to the stress conditions. The sensitive and resistant behaviors of scl14 and OE:SCL14 lines were confirmed by the quantification of HOTE in leaf samples of the stressed plants, showing 1.5-time higher HOTE levels in the mutant line and five times lower HOTE levels in the overexpressing line compared with the wild type (Figure 3B).
Figure 3.
scl14 Mutant Lines Are Not Able to Acquire β-cc-Induced Resistance to Excessive Light.
(A) Leaf bleaching (top panel) and lipid peroxidation, monitored by autoluminescence imaging (bottom of panel), of wild-type, scl14, and OE:SCL14 plants pretreated with β-cc or with water. The color palette shows signal intensity from low (dark blue) to high (white) values.
(B) HPLC-UV quantification of the HOTE normalized to the wild-type control (CTRL). The black diamonds indicate significant differences with P < 0.05. Error bars = +sd between the four biological replicates from leaves of three plants per genotype per treatment. Two full experimental replicates.
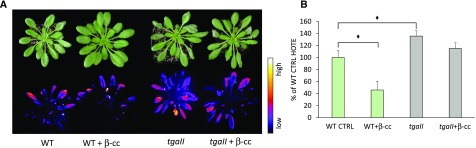
SCL14 has been shown to interact with TGA 2, 5, and 6 transcription factors and to coregulate at least a subset of the genes presenting the TGA binding as-1 motif in their promoter (Fode et al., 2008). We consequently evaluated the role of the TGA 2, 5, and 6 transcription factors in the response to excessive light by exposing the tgaII triple mutant line to stress conditions following pretreatment with β-cc or with water.tgaII plants showed stronger leaf bleaching after 24-h exposure to stress conditions compared with wild-type plants, as well as more intense lipid peroxidation-related luminescence (Figure 4A). The sensitive behaviors of the tgaII line was confirmed by the quantification of HOTE in leaf samples of the stressed plants, showing 1.5 times higher HOTE levels in the mutant line compared with the wild type (Figure 4B).
Figure 4.
tgaII Mutant Lines Are Sensitive to Excessive Light and Unresponsive to β-cc.
(A) Leaf bleaching (top panel) and lipid peroxidation, monitored by autoluminescence imaging (bottom panel), of wild-type and oftgaII mutant plants pretreated with β-cc or with water. The color palette shows signal intensity from low (dark blue) to high (white) values.
(B) HPLC-UV quantification of the HOTE normalized on wild-type control (CTRL). The black diamonds indicate significant differences with P < 0.05. Error bars = +sd between the four biological replicates from leaves of three plants per genotype per treatment. Two full experimental replicates.
scl14 Mutant Lines Are Not Able to Acquire β-cc-Induced Resistance to Excessive Light
A 4-h β-cc pretreatment of wild-type plants is sufficient to induce a genetic response leading to acclimation to excessive light stress (Ramel et al., 2012b; Shumbe et al., 2017). The acclimated status is indicated by a lower accumulation of lipid peroxides under excessive light stress (<50% compared with untreated plants), revealed by plant autoluminescence and HPLC-UV HOTE quantification (Ramel et al., 2012b). The treatment of plants with β-cc, sufficient to protect wild-type plants from photooxidative stress (Figures 3A and 3B), was ineffective on scl14 mutant lines. β-cc-treated scl14 plants showed comparable leaf bleaching and autoluminescence to untreated samples (Figure 3A). In addition, the quantification of HOTE confirmed the inability of scl14 mutant plants to acquire the β-cc-induced acclimation to excessive light (Figure 3B). The lack of β-cc-induced protection in the scl14 mutant line highlights SCL14 as central actor in the β-cc signaling network leading to acclimation. These results demonstrate that SCL14 is not only necessary for the proper genetic response to β-cc and excessive light but that xenobiotic detoxification is necessary both for light-induced acclimation to photooxidative stress and for β-cc-induced acclimation.
Similarly as in scl14, the treatment of tgaII plants with β-cc was ineffective. In fact, β-cc-treated tgaII plants showed comparable leaf bleaching and autoluminescence to untreated samples (Figure 4A). In addition, the quantification of HOTE confirmed the inability of tgaII mutant plants to acquire the β-cc-induced acclimation to excessive light (Figure 4B). Results of Figure 4 show that the tgaII mutant phenocopies the scl14 mutant at the whole plant level (Figure 3).
β-cc Induces a SCL14-Dependent Xenobiotic Detoxification Response
The response to β-cc is, at least in part, dependent on SCL14, but the SCL14-dependent xenobiotic response is an integrated network involving many molecular targets. Among these, four ANAC transcription factors (ANAC 2, 32, 81, and 102) and families of modifying enzymes, like cytochrome P450s, short-chain dehydrogenases/reductases (SDRs), monooxygenases, 2-alkenal reductases (AERs), aldo-keto reductases (AKRs), and aldehyde dehydrogenases (ALDH), take part in the modification phase of the xenobiotic response (Fode et al., 2008). The analysis of the transcriptome of β-cc-treated plants suggests that many of these genes are also induced by β-cc, and we further verified by RT-qPCR the expression of a selection of genes: ChlADR (CHLOROPLASTIC ALDEHYDE REDUCTASE; AT3G04000), SDR1 (AT4G13180), AER (AT5G16970), AKR4C9 (CHLOROPLASTIC ALDO-KETO REDUCTASE [ChlAKR]; AT2G37770), GRX480/ROXY19 (AT1G28480), and a gene of unknown function but strongly induced by β-cc (AT5G61820). These analyses showed that induction of the selected genes by β-cc is weakened or impeded in the tgaII and scl14 mutant lines (70 to 99% lower), while it is strengthened in the SCL14 overexpressing lines (150 to 1000% higher) (Figure 5). Therefore, we can confidently conclude that β-cc is able to induce a xenobiotic-like response (Sandermann, 1992; Kreuz et al., 1996; Riechers et al., 2010).
Furthermore, these families of enzymes are of particular interest when we consider lipid peroxidation occurring under excessive light stress, especially in the chloroplast. In this stress, lipid peroxides mainly derive from the linoleic (18∶2), linolenic (18∶3), and roughanic (16∶3) polyunsaturated fatty acids (Montillet et al., 2013). These primary hydroperoxy or hydroxy fatty acids can generate aldehydes, oxo-acids, epoxydes, and cyclized compounds such as jasmonates or phytoprostanes (Mano et al., 2005; Montillet et al., 2013). Several of these compounds are reactive carbonyls (Mano, 2012) that can specifically be anabolized by the SDR, AER, AKR, and ALDH enzyme families, exactly the same as those induced by the xenobiotic response (Mano et al., 2005; Yamauchi et al., 2011; Mano, 2012). Furthermore, chloroplasts are the main sites of lipid peroxidation under excessive light stress, and at least two of the enzymes induced by β-cc, ChlADR and ChlAKR, can detoxify RCS directly at the primary location of production, as they are expressed in the stroma (Yamauchi et al., 2011).
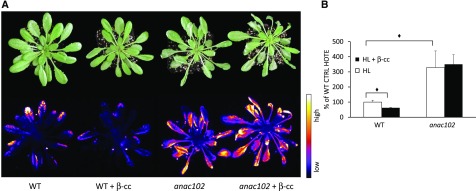
anac102 Mutant Lines Are Sensitive to Excessive Light and Unresponsive to β-cc
ANAC102 was the most induced SCL14-dependent ANAC transcription factor by the treatment with β-cc. Therefore, we analyzed anac102 knockout mutant plants, pretreated with β-cc or with water, under excessive light stress (1500 μmol photons m−2 s−1 at 7°C for 24 h). Five-week-old anac102 plants showed stronger leaf bleaching after 24-h exposure to stress conditions compared with wild-type plants, as well as enhanced lipid-peroxide-dependent autoluminescence (Figure 6A). The photosensitive behavior of anac102 was confirmed by the quantification of HOTEs in leaf samples of the stressed plants, which showed 3 times higher HOTE levels in the mutant line compared with the wild type (Figure 6B). Furthermore, β-cc-treated anac102 plants showed comparable leaf bleaching and autoluminescence as untreated samples (Figure 6A), indicating that β-cc signaling was incomplete in this mutant. In addition, HOTE quantification confirmed the inability of anac102 mutant plants to acquire the β-cc-induced acclimation to excessive light (Figure 2B). The lack of β-cc-induced protection in the anac102 mutant line implies that ANAC102 participates in the β-cc retrograde signaling downstream of SCL14.
Figure 6.
anac102 Mutant Lines Are Sensitive to Excessive Light and Unresponsive to β-cc.
(A) Leaf bleaching (top panel) and lipid peroxidation, monitored by autoluminescence imaging (bottom panel), of wild-type and of anac102 mutant plants pretreated with β-cc or with water. The color palette shows signal intensity from low (dark blue) to high (white) values.
(B) HPLC-UV quantification of the HOTE normalized to wild-type control (CTRL). The black diamonds indicate significant differences with P < 0.05. Error bars = +sd between the three biological replicates from leaves of three plants per genotype per treatment. Two full experimental replicates.
β-cc-Induced SCL14-Dependent Detoxification Response Is Blocked in anac102
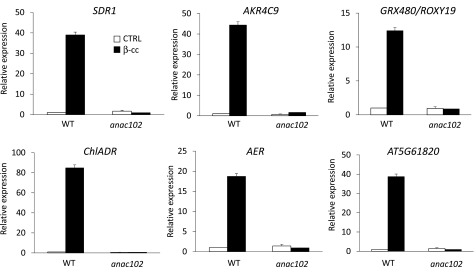
We verified by RT-qPCR the expression of SCL14-dependent genes, ChlADR, SDR1, AER, AKR4C9 (ChlAKR), GRX480/ROXY19, and AT5G61820, in wild-type and anac102 mutant lines. These analyses showed that β-cc induction of the selected genes is weakened or impeded in the anac102 mutant (90 to 99% lower) (Figure 7). These results place ANAC102 downstream of SCL14, as previously shown (Fode et al., 2008), but upstream of the analyzed detoxifying enzymes.
Figure 7.
β-cc-Induced SCL14-Dependent Detoxification Response Is Blocked in anac102.
SDR1, ChlADR, AKR4C9, AER, GRX480/ROXY19, and AT5G61820 expression levels (relative to wild-type control levels [WT CTRL], which were set as 1) in wild-type and anac102 plants under control conditions or exposed to β-cc, measured by RT-qPCR. Every gene induced by β-cc in the anac102 plants showed a significant difference when tested against β-cc induction in the wild type (P < 0.01). Error bars = +sd between the four technical replicates from pools of three plants per genotype per treatment.
ANAC102 Is Upstream of ANAC002, ANAC032, and ANAC081
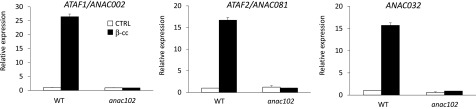
To better elucidate the molecular pathway downstream of SCL14 in the response to β-cc, we analyzed the expression levels of ANAC002, ANAC032, and ANAC081 in the wild type and in the anac102 mutant line. The analyses showed that β-cc induction of the three SCL14-regulated ANAC transcription factors was completely blocked in the anac102 mutant line (Figure 8). This finding reveals transcriptional control of ANAC002, ANAC032, and ANAC081 transcription factors by ANAC102 in the SCL14-dependent pathway. By these analyses, we have shown not only that ANAC102 controls the induction of the detoxification enzymes, previously reported at the same level of ANAC102 (Fode et al., 2008), but also that ANAC transcription factors follow a hierarchical order in β-cc retrograde signaling.
Figure 8.
ANAC102 Is Upstream of ANAC002, ANAC032, and ANAC081.
ANAC002, ANAC032, and ANAC081 expression levels (relative to wild-type control levels [WT CTRL], which were set as 1) in wild-type and anac102 plants under control conditions or exposed to β-cc, measured by RT-qPCR. Every gene induced by β-cc in anac102 plants showed a significant difference when tested against β-cc induction in the wild type (P < 0.01). Error bars = +sd between the four technical replicates from pools of three plants per genotype per treatment.
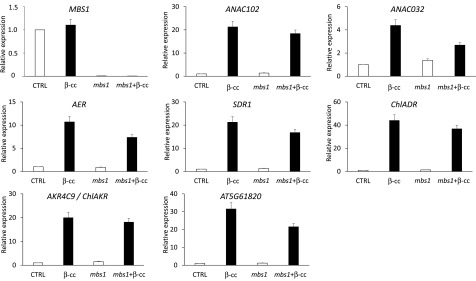
SCL14-Dependent Detoxification Pathway Is Independent of MBS1
β-cc elicits the SCL14-dependent xenobiotic response pathway to mediate the phototolerance to excessive light. However, it was recently demonstrated that β-cc retrograde signaling depends on the MBS1 protein (Shumbe et al., 2017). We then wondered whether the two pathways are interdependent or operate in parallel. By analyzing the genetic response of ANAC032, ANAC102, ChlADR, SDR1, AER, AKR4C9 (ChlAKR), and AT5G61820 to β-cc in the mbs1 mutant line (Figure 9), we found that all those marker genes of detoxification were equally induced or had a limited induction, in the mutant line compared with the wild type. Based on the normal induction of the detoxification response in the mbs1 mutant, we must hypothesize the coexistence of at least two mechanisms downstream of β-cc: the MBS1-independent detoxification response controlled by SCL14 and a pathway controlled by MBS1. Nevertheless, under the rather severe light stress conditions used here (sudden transfer of plants from low light to high light), both pathways appear to be required for β-cc-induced phototolerance.
Figure 9.
SCL14-Dependent Detoxification Pathway Is Independent of MBS1.
MBS1, ANAC102, ANAC032, AER, SDR1, ChlADR, AKR4C9, and AT5G61820 expression levels (relative to wild-type control levels [CTRL], which were set as 1) in wild-type and mbs1 plants under control conditions or exposed to β-cc, measured by RT-qPCR. MBS1 expression in the mutant is significantly lower than in the wild type (P < 0.01). ANAC102, ANAC032, AER, SDR1, ChlADR, AKR4C9, and AT5G61820 expression levels after β-cc treatment are significantly different from the relative control level in wild-type or mbs1 mutant plants (P > 0,01). Error bars = +sd between the four technical replicates from pools of three plants per genotype per treatment. Two full experimental replicates.
ANAC102 Is Particularly Induced in Young Leaves
We generated stable Arabidopsis transgenic lines carrying the full ANAC102 (AT5G63790) gene (−2041 +1124), including the putative 5′ regulatory region and both introns and exons, coding for the ANAC102 protein fused to the β-glucuronidase reporter. By histochemical assay, we show that the ANAC102 reporter is present at low levels under physiological conditions (Figure 10; Supplemental Figure 2), being detectable only in the conductive tissues of young leaves. On the contrary, after the treatment with β-cc, the reporter strongly accumulated in the full limb of young leaves as well as in the conductive tissues of mature leaves (Figure 10). We further verified that the strong accumulation of ANAC102 in the β-cc-treated samples is due to an upregulation of gene expression rather than a stabilization of the ANAC102-β-glucuronidase fusion protein. Samples treated with the protein synthesis inhibitor cycloheximide did not show the accumulation of the reporter (Supplemental Figure 2). In the light of the positive effect of ANAC102 and SCL14 on the resilience to excessive light, the higher accumulation of ANAC102 in young leaves suggests that the younger leaf tissues could be more resistant to this stress than the old, mature tissues.
Figure 10.
ANAC102 Is Particularly Induced in Young Leaves.
(A) Schematization of the ANAC102 translational reporter.
(B) Histochemical analyses of the translational reporter after 4 h treatment with water (CTRL) or β-cc and overnight development of the staining. Two full experimental replicates.
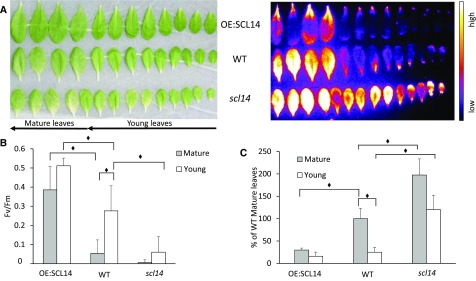
Young Leaves Are More Resistant to Excessive Light Than Mature Leaves, and This Difference Is Dependent on SCL14
As shown in Figures 3A and 6A, young leaves in the center of the rosette do not show lipid-peroxide-derived luminescence after high light stress, suggesting a high tolerance to photooxidation. Using whole plants, resistance of younger tissues to excessive light could depend both on biological factors like systemic acquired acclimation (Rossel et al., 2007; Carmody et al., 2016) and on technical factors like the heterogeneity of irradiance and leaf temperature. To determine whether young leaves are intrinsically resistant to excessive light, we cut young and mature leaves from several plants and placed them on a flat surface covered with water, where irradiance and temperature during the stress are more homogeneous (1100 μmol photons m−2 s−1 at 4°C for 16 h). Furthermore, the cut-leaf system excludes the eventual participation of the systemic and long-distance signaling to the high-light tolerance. By this analysis, under more controlled conditions, we feel confident to confirm the higher resilience of young leaves to excessive light compared with mature leaves by an organ autonomous mechanism. In fact, not only autoluminescence after the stress was much lower in young leaves than in mature leaves (Figure 11A), but also the quantification of HOTE confirmed this difference (75% less HOTE in young leaves) (Figure 11C), and Fv/Fm was higher in young leaves, indicating lower photoinhibition of the photosynthetic apparatus (Figure 11B).
Figure 11.
The Resilience of Young Leaves to Excessive Light Depends on SCL14.
(A) Leaf bleaching (on the left) and lipid peroxidation monitored by autoluminescence imaging (on the right) of wild-type, scl14, and OE:SCL14 detached leaves after high-light stress.
(B) Maximum quantum yield of PSII photochemistry determined by the Fv/Fm chlorophyll fluorescence ratio of wild-type, scl14, and OE:SCL14 mature and young detached leaves after high-light stress.
(C) HPLC-UV quantification of the HOTE normalized to wild-type mature leaves. Black diamonds indicate significant differences (P < 0.05). Error bars = +sd between the three biological replicates of pools of four mature leaves or 10 young leaves. Two full experimental replicates.
Leaves can acclimate to excessive light through molecular, anatomical, and physiological changes (Oguchi et al., 2003; Kouřil et al., 2013). In particular, younger leaves can better resist to photoinhibition than older leaves due to a higher plasticity allowing a faster redesign of their anatomy and photosynthetic apparatus, optimizing them to the new light conditions (Sims and Pearcy, 1992; Bielczynski et al., 2017). Furthermore, many pathways can increase the photoprotective capacity of younger leaves such as a higher capacity to accumulate ascorbate peroxidase and superoxidase dismutase (Moustaka et al., 2015). Both enzymes are part of the ROS scavenging system, allowing lower oxidative damage upstream of lipid peroxidation.
To discern the involvement of SCL14 in the response of young leaves to high light, we tested leaves cut from 5-week-old scl14 mutant plants or SCL14-overexpressing lines. The excessive light treatment induced lower photooxidative stress in OE:SCL14 leaves compared with the wild type, as shown by a lower autoluminescence intensity. On the contrary, scl14 leaves showed much stronger luminescence both in mature and young leaves (Figure 11A). These results were also confirmed by the HPLC-UV quantification of HOTE, showing higher HOTE levels in scl14 mature and young leaves compared with wild-type mature and young leaves (Figure 11C). In addition, young leaves of the scl14 mutant line showed a weaker intrinsic resistance to high light compared with young tissues of the wild type. While in the wild type we found a strong decrease in lipid peroxidation (75% less HOTE) in young leaves relative to mature leaves, we found only a partial protection (40% less HOTE) of young tissues compared with mature ones in the scl14 mutant (Figure 11C). Furthermore, HOTE levels found in mature leaves of OE:SCL14 lines were much lower than in wild-type leaves (25% of HOTE levels present in wild-type mature leaves) and comparable to the ones found in young wild-type leaves. In addition, reduced photodamage of the photosynthetic apparatus were observed in OE:SCL14 lines as shown by the high Fv/Fm chlorophyll fluorescence ratio values (Figure 11B), while greater photodamage was observed in scl14 mutant leaves. Considering these results, we can conclude that the SCL14-dependent response participates in the resilience of young leaves to excessive light, in parallel to their higher photoprotective capacities.
SCL14-Dependent Detoxification Pathway Acts on Toxic RCS Rather Than on ROS Accumulation
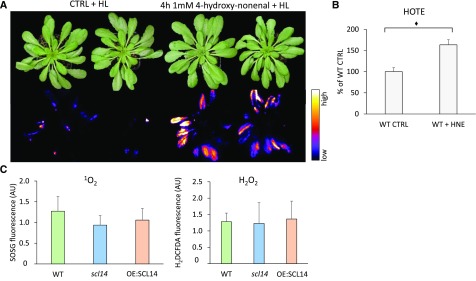
In Figures 3 and 11, we show higher lipid peroxidation in the scl14 mutant line compared with the wild type, and a marked resistance of the OE:SCL14 line. The enzymes, whose transcriptional levels are controlled by SCL14, take part in the modification phase of the xenobiotic detoxification response (Sandermann, 1992; Fode et al., 2008). These enzymes are able to detoxify toxic RCS, mainly aldehydes, to less reactive carboxylates or alcohols (Mano et al., 2005; Yamauchi et al., 2011; Mano, 2012). RCSs are not merely markers of oxidative stress, but rather active factors that can deplete glutathione pools and then exacerbate stress effects (Mano, 2012). We tested this idea in our stress conditions by pretreating wild-type plants with 1 mM 4-hydroxy-nonenal (HNE), a well-known RCS detoxified by AER (Mano et al., 2005; Mano, 2012). As shown in Figure 12A, a 4-h treatment with HNE worsened plant fitness against excessive light stress and amplified oxidative damage. In fact, treated plants showed higher autoluminescence and increased HOTE level after high light stress compared with control plants (Figure 12A). We can therefore confirm that the higher peroxidation levels found in the scl14 mutant line can be due to inefficient RCS scavenging that leads to amplified oxidative damage. On the contrary, the lower peroxidation found in the OE:SCL14 lines is compatible with an enhanced detoxification response and, thus, with reduced RCS levels and consequent oxidative damage. For the HNE treatment, for example, the AER gene is induced 5 times more in the OE:SCL14 line than in the wild type by the β-cc treatment, while it is induced 4 times less in the scl14 mutant line than in the wild type (Figure 5). In addition, we also analyzed ROS accumulation in those mutant or transgenic lines by means of fluorescent dies. OE:SCL14, wild-type, and scl14 plants were put in stress conditions for 4 h and then infiltrated with the 1O2 specific probe SOSG (Singlet Oxygen Sensor Green) or with the general ROS probe H2DCFDA, mainly responsive to H2O2. The quantification of the fluorescence emitted by the ROS-activated probes, showed no differences in ROS accumulation between the three genotypes (Figure 12C). Altogether, these results suggest that the sensitivity of scl14 line and the tolerance of the OE:SCL14 lines to excessive light are likely due to a different accumulation of RCS rather than differential ROS accumulations.
Figure 12.
SCL14-Dependent Detoxification Pathway Acts on Toxic RCS Rather Than on ROS Accumulation.
(A) Leaf bleaching (top panel) and lipid peroxidation, monitored by autoluminescence imaging (bottom panel), of wild-type plants pretreated with 1 mM HNE or with water. The color palette shows signal intensity from low (dark blue) to high (white) values.
(B) HPLC-UV quantification of the HOTE normalized to the wild-type control (CTRL). The black diamonds indicate significant differences with P < 0.05. Error bar = +sd between the four biological replicates from leaves of three plants per treatment.
(C) SOSG or H2DCFDA fluorescence in wild-type, scl14, or OE:SCL14 plants tested measured after 4 h of high-light stress. Error bars = +sd between fluorescence deriving from five leaves per plant. Two plants per genotype per treatment.
DISCUSSION
For more than 25 years, it was known that plants are able to detoxify xenobiotic compounds explaining, for example, different sensitivities to herbicides (Sandermann, 1992; Kreuz et al., 1996; Riechers et al., 2010). Artificial molecules that enter the cells can elicit a detoxification process that inactivates them by a three-phase mechanism. First, hydroxylases, reductases (i.e., AER, AKRs, and SDRs), cytochrome P450 monooxygenases, or peroxidases introduce or modify reactive side groups. In the second phase, these modified exogenous compounds are conjugated to sugar moieties or glutathione by either glycosyl transferases or glutathione S-transferases. Finally, the conjugates are transported to the vacuole or to the apoplast (Sandermann, 1992). The exact origin of the pathway elicited by xenobiotics is still unclear, but several mechanisms can participate in the response (Riechers et al., 2010). Among these, xenobiotics elicit a genetic modification partially overlapping the response to oxylipins, such as jasmonic acid, or reactive electrophilic lipid-derived compounds, like 12-oxo-phytodienoic acid and phytoprostanes (Mueller and Berger, 2009; Riechers et al., 2010).
The concentration of oxylipins, carbonyls, and RCS, generated by enzymatic activities and by ROS-dependent oxidation, increases under many stresses (Mano, 2012; Farmer and Mueller, 2013; Roach et al., 2017). In fact, ROS, lipid peroxides, and RCS, corresponding to the α,β-unsaturated aldehydes and ketones derived from lipid hydroperoxides, are critical cell-damaging agents in plants under environmental stresses, which can lead to cell death (Mano, 2012). These compounds characterize the oxidative response and they can be specifically inactivated by families of enzymes of the detoxification pathway, such as AER, AKRs, SDRs, and ALDHs (Mano, 2012). The beneficial role of the latter enzymes for plant tolerance to environmental constraints has been reported in several plant species (Mano et al., 2005; Kotchoni et al., 2006; Stiti et al., 2011; Turóczy et al., 2011; Mano, 2012)
The β-carotene oxidation by-product β-cc, generated under photooxidative stress, is the first identified nonartificial compound that can induce the SCL14-dependent xenobiotic response, exploiting this pathway to confer resistance to excessive light. Furthermore, we show that this response is independent of MBS1 signaling but equally necessary for coping with high-light stress and for the β-cc-induced resistance to high light. We identified the SCL14-dependent xenobiotics detoxification in response to a physiological condition, rather than an artificial stimulus. In fact, plants lacking the TGAII-regulative factor SCL14, or the TGA2, 5, and 6 transcription factors themselves (Figure 4), became sensitive to photooxidative stress. We propose that the enhanced cell death and lipid peroxide accumulation found in these lines is due to a stunted induction of the detoxification response downstream of SCL14 and ANAC102. In fact, high RCS levels, obtained by a pretreatment with 4-hydroxy-nonenal, made the plant more sensitive to high-light conditions. On the contrary, the overexpression of SCL14 that permits a stronger induction of the detoxification response was sufficient to confer increased resistance to photooxidative stress. As under excessive light and considering that plants suffer excessive light on a daily basis (Ort, 2001), it would be worth testing the overexpression of this regulative factor under other stresses and in other plant species to verify the possible impact of enhancing the detoxification response on field yields, like previously described for single enzymes downstream of SCL14 (Mano et al., 2005; Kotchoni et al., 2006; Stiti et al., 2011; Turóczy et al., 2011; Mano, 2012).
Finally, we can state that the SCL14-dependent-xenobiotic response occurs during photooxidative stress and that it participates in the β-cc-induced resistance to excessive light and to the intrinsic resistance of young tissues to this stress. Therefore, we propose a mechanism in which photooxidation under excessive light generates toxic RCS metabolites and increases β-cc concentration. β-cc enhances SCL14 expression, likely increasing the number of interactions between SCL14 and the as1-bound TGA transcription factors that, together with the activation of the MYC-dependent signaling pathway (Figure 1), induces strong activation of the xenobiotic detoxification response (Figure 13) (Köster et al., 2012). In fact, the concerted action of the TGA and the MYC transcription factors is required for the complete activation of the detoxification response (Köster et al., 2012). Furthermore, the limited induction of SCL14 by β-cc, similar to the overexpression levels found in the OE:SCL14 line, could explain why we detected no competition between SCL14 and NPR1, one of the many interacting protein partners of the TGAII transcription factors (Després et al., 2000; Zhou et al., 2000; Fan and Dong, 2002). In fact, the genetic response of the four SCL14-dependent ANAC transcription factors, ANAC002, ANAC032, ANAC081, and ANAC102, to β-cc was similar in the npr1-1 mutant and in the wild type (Supplemental Figure 2)
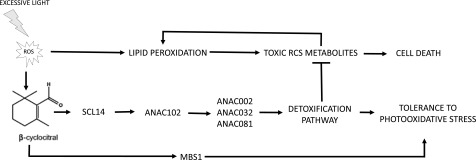
Figure 13.
β-cc Mediates Resilience to Photooxidative Stress via the SCL14-Dependent Xenobiotic Response.
Photooxidation under excessive light stress generates toxic RCS metabolites and increases β-cc concentration. β-cc induces the expression of SCL14 leading to enhanced expression of ANAC102 and finally a strong activation of the xenobiotic detoxification response. In this response, ANAC102 is upstream of ANAC002, ANAC032, and ANAC081 expression and consequently of the enzymes controlled by these transcription factors. Furthermore, the strong induction of the AER, AKR, ALDH, and SDR enzymes and of the glucosyl and glutathione transferases in the xenobiotic detoxification pathway assures the elimination of the RCS produced under stress conditions. Reducing RCS accumulation limits the positive feedback on lipid peroxidation and leads to tolerance rather than cell death.
In this work, we enrich the SCL14-dependent pathway by showing the hierarchical relationship between the several players and make the link between the response to xenobiotics and its physiological role in detoxifying toxic RCS, which have a negative impact under stress conditions. More specifically, we show that among all the elements regulated by SCL14, ANAC102 is a master regulator of the downstream response. Accordingly, ANAC002, ANAC032, and ANAC081 were unresponsive to β-cc in the anac102 mutant line (Figure 8).
Under excessive light, SCL14, TGAII, and likely MYC transcription factors mediate the induction of ANAC102. This transcription factor was shown to be indispensable for coping with high light and to mediate the induction of ANAC002, ANAC032, and ANAC081 and, consequently, of the downstream enzymes (Ratnakaran, 2014). Finally, the strong induction of the AER, AKR, ALDH, and SDR enzymes and of the glucosyl and glutathione transferases (Ramel et al., 2012b) assures the elimination of RCS produced under stress conditions. RCSs are toxic for the cell and their accumulation leads to a reinforced accumulation of lipid peroxides under excessive light (Figure 12) (Mano, 2012). The activation of this detoxification mechanism precludes cell death owing to lower accumulation of toxic compounds and depicts a novel process by which β-cc enhances plant performance under photooxidative stress (Figure 13). It is likely that the SCL14-dependent pathway of Figure 13 is part of a more complex mechanism of phototolerance, which could involve regulation of phytohormones such as jasmonic acid. Indeed, acclimation of the 1O2-overproducing ch1 Arabidopsis mutant is associated with a block of the biosynthesis of this phytohormone in high light (Ramel et al., 2013b). Further studies will have to clarify the links between the β-cc-induced detoxification pathway shown here and hormonal regulations.
Chloroplasts are the main sites of lipid peroxidation under excessive light stress, and at least two of the enzymes induced by β-cc, the stroma-localized ChlADR and ChlAKR, can detoxify RCS directly at the primary location of production. The perfect correspondence of the toxic metabolites produced under oxidative stress with the target of the enzymatic families that compose the xenobiotic response let us hypothesize a possible explanation for the origin of this pathway. In other words, the exceptional diversity of reactive damaging molecules generated under photooxidative stress could have led to the evolution of a general mechanism of detoxification that mediates the inactivation of several reactive and potentially toxic compounds (Huang et al., 2016).
In this work, we further deciphered the intrinsic resistance of young tissues to excessive light. Younger leaves can better resist photoinhibition than older leaves due to a higher plasticity (Sims and Pearcy, 1992; Bielczynski et al., 2017) and to higher capacity to accumulate ascorbate peroxidase and superoxidase dismutase (Moustaka et al., 2015). In addition, high light triggers both autonomous signals required for direct HL and ROS perception and nonautonomous distal systemic acquired acclimation (Rossel et al., 2007; Carmody et al., 2016). Altogether, these mechanisms may participate in the tolerance to photooxidative stress in young leaves under our stress conditions. In fact, we were unable to observe photooxidation in the young leaves of whole plants (Figures 3, 6, and 12), and we had to increase stress conditions, by prolonging the stress to 48 h, to observe a limited peroxidation and photoinhibition in wild-type young leaves (Supplemental Figure 3). Wild-type young leaves are resistant to high light both when whole plants or cut leaves were put under stress conditions, suggesting autonomous mechanisms are prevalent in the tolerance. On the other hand, young scl14 leaves were drastically more sensitive to high light than the wild type in the detached leaves experiments, while only limitedly more sensitive when we analyzed whole plants (Supplemental Figure 3). This suggests an additional role for the systemic acquired acclimation upstream of SCL14. In fact, when systemic signaling is excluded, SCL14 is pivotal for reducing peroxidation, while other mechanisms are induced in young leaves in a systemic way that could limit RCS formation by reducing ROS concentrations (Matsuo et al., 2015; Carmody et al., 2016).
To sum up, we propose a mechanism elicited by β-cc to achieve plant tolerance to high light, in parallel with the previously described MBS1-mediated protection against singlet oxygen. The SCL14-dependent detoxification is linked to a physiological process rather than to the resistance to artificial or synthetic xenobiotics. The induction of the many enzymes belonging to the first phase of the detoxification can now be correlated with the increase in the concentration of their substrates, namely oxidized lipids (e.g., HOTEs) and derived metabolites. Finally, we highlight a more complete and integrated view on the processes happening downstream of photooxidation, which are schematized in Figure 13.
METHODS
Plant Growth and Stress Treatment
The wild type (ecotype Col-0) and anac102 (SALK_030702C) were obtained from the Nottingham Arabidopsis Stock Centre (Arabidopsis.info) (Christianson et al., 2009), scl14, the triple mutant tgaII (tga 2x5x6), and the SCL14-overexpressing Arabidopsis thaliana lines were kindly provided by the Christiane Gatz laboratory (Gottingen, Germany) (Fode et al., 2008). The npr1 mutant line was kindly provided by Gerit Bethke of the Glazebrook lab (St. Paul, MN). All the lines were grown for 5 weeks in short-day conditions (8 h/16 h, day/night) under a moderate PFD of ∼150 µmol photons m−2 s−1 provided by HQI metal halide bulbs (Osram), controlled temperature (22°C/18°C, day/night), and a relative air humidity of 65%. Photooxidative stress was applied by subjecting at least three plants of wild-type, anac102, scl14, tgaII, npr1, and OE:SCL14 lines per experiment, to 1500 µmol m−2 s−1 PFD, 7°C/18°C temperature day/night, respectively, and 380 ppm CO2 in a growth chamber (Ramel et al., 2012b; Havaux, 2014). Alternatively, for the photooxidative stress on detached leaves, mature and young leaves were cut from 5-week old plants of the different lines and placed on a flat and wet surface. Leaf age was defined by leaf position (mature, leaves 5–11; young, leaves >14) (Mousavi et al., 2013). Stress conditions were imposed by placing the leaves in a cold chamber (6°C) under a PFD of 1100 µmol photons m−2 s−1 for 16 h. β-cc treatment was performed by placing plants in a transparent airtight Plexiglas box and by applying defined volumes (50 or 100 µL) of pure β-cc on cotton balls in the Plexiglas boxes (Ramel et al., 2012b; Shumbe et al., 2014, 2017). As a control, β-cc was replaced with water. The Plexiglas boxes were thoroughly sealed and placed in a growth chamber under controlled conditions of light and temperature (50 µmol photons m−2 s−1 and 22°C) for 4 h. β-cc was obtained from Sigma-Aldrich.
The treatment with 4-HNE was performed by spraying plants with 1 mM 4-HNE in water and putting the plant in sealed Plexiglas boxes for 4 h. As a control, 4-HNE was replaced with water. Then high-light stress conditions were imposed, as described above.
The translational reporter line ANAC102:ANAC102-GUS was obtained by cloning the full AT5G63790 gene (−2041 +1124), including the putative 5′ regulative region and both introns and exons, in frame to the uidA gene in the pBGWFS7 Gateway entry vector (Karimi et al., 2002) (Supplemental Table 1). The vector was transformed in Agrobacterium tumefaciens C58C1 strain and wild-type plants were transformed by floral dip.
Quantification of Lipid Peroxidation and Imaging
Lipids were extracted from 0.3 to 0.5 g of leaves, deriving from a pool of three plants for each condition, then frozen in liquid nitrogen. The leaves were ground in an equivolume methanol/chloroform solution containing 5 mM triphenylphosphine and 1 mM 2,6-tert-butyl-p-cresol (5 mL g−1 fresh weight) and 1 M citric acid (2.5 mL g−1 fresh weight), using an Ultra-Turrax blender. 15-HEDE was added as an internal standard to a final concentration 100 nmol g−1 fresh weight and mixed properly. After centrifugation at 700 rpm and 4°C for 5 min, the lower organic phase was carefully taken out with the help of a glass syringe and transferred into a 15-mL glass tube. The syringe was rinsed with ∼2.5 mL chloroform and emptied in the tube containing the upper organic phase. The process was repeated, and the lower layer was again collected and pooled with the first fraction. The solvent was evaporated under N2 gas, at 40°C. The residues were recovered by 1.25 mL absolute ethanol and 1.25 mL of 3.5 n NaOH and hydrolyzed at 80°C for 30 min. The ethanol was evaporated under N2 gas at 40°C for ∼10 min. After cooling to room temperature, pH was adjusted to 4 to 5 by adding 2.1 mL 1 M citric acid. Hydroxy fatty acids were extracted with hexane/ether 50/50 (v/v). The organic phase of three samples for each condition was analyzed by straight phase HPLC-UV, as previously described (Montillet et al., 2004). ROS-induced and LOX-mediated HOTE isomers (9-, 12-, 13-, and 16-HOTE derived from the oxidation of the main fatty acid, linolenic acid) were quantified based on the 15-HEDE internal standard (Montillet et al., 2004). Lipid peroxidation was also visualized in whole plants by autoluminescence imaging. Stressed plants were dark adapted for 2 h, and the luminescence emitted from the spontaneous decomposition of lipid peroxides was captured by a highly sensitive liquid N2-cooled CCD camera, as previously described (Birtic et al., 2011). One exemplificative plant was analyzed for each condition, and experiments were repeated at least twice. The images were treated using Image J software (NIH)
PSII Photochemical Activity
Chlorophyll fluorescence from intact leaves was measured with a PAM-2000 fluorometer (Walz), as described previously (Ramel et al., 2012b). The maximum quantum yield of PSII was determined by the Fv/Fm ratio, measured in dark-adapted intact leaves. Chlorophyll fluorescence imaging was done with a laboratory-built instrument described by Johnson et al. (2009).
RNA Isolation and RT-qPCR
Total RNA was isolated from 150 mg leaves, deriving from a pool of the shoots of three plants for each condition, using the Nucleospin RNA plant kit (Macherey-Nagel). The concentration was measured on a NanoDrop2000 (Thermo Fisher Scientific). First-strand cDNA was synthesized from 3 µg total RNA using the PrimeScript Reverse Transcriptase kit (Takara). RT-qPCR was performed on a Lightcycler 480 real-time PCR system (Roche). Three microliters of a reaction mixture comprising SYBR Green I Master (Roche), 10 µM each of forward and reverse primers, and water, was added to 2 μL of a 50-fold diluted cDNA sample in a 384-well plate. Each condition was represented by four technical replicates. The PCR program used was: 95°C for 10 min, then 45 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 15 s. Primers for all genes examined (Supplemental Table 1) were designed using the Primer-BLAST software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). PROFILIN-1 and CYCLOPHYLIN-5 were used as reference genes for the normalization of gene expression levels.
β-Glucuronidase Histochemical Assay
Histochemical staining was performed on 5-week-old plants of the ANAC102-GUS translational reporter. Samples, belonging to two transgenic lines, were analyzed for β-glucuronidase activity by observing the specific blue staining. Samples were incubated overnight at 37°C in the reaction medium [1 mM X-Gluc, 0.05% Triton X-100, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6 3× H2O, 10 mM EDTA, and 50 mM sodium phosphate buffer, pH 7.0].
Detection of ROS Production
1O2 or H2O2 production was measured in attached leaves using the SOSG or the H2DCFDA fluorescent probe (Invitrogen) (Flors et al., 2006), as previously described (Ramel et al., 2012a). Leaves were infiltrated with 100 mM probe by a 1-mL syringe, without needle. Plants were exposed for 4 h to a PFD of 1500 μmol m-2 s−1 at 7°C then infiltrated and put further 30 min under stress conditions. SOSG fluorescence was then measured using a Perkin-Elmer spectrofluorometer (LS 50B) at 515 nm with a 475-nm excitation. H2DCFDA fluorescence was measured at 525 nm with a 490-nm exciting light beam.
Statistics
All experiments were performed at least on three biological replicates, and the images represent typical examples. Each experiment included the corresponding independent control. The values are represented as the means + sd. The statistical significance was tested using Student’s t test (two-tailed, unequal variances).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MBS1 (AT3G02790), SCL14 (AT1G07530), ANAC102 (AT5G63790), ANAC002 (AT1G01720), ANAC032 (AT1G77450), and ANAC081 (AT5G08790).
Supplemental Data
Supplemental Figure 1. β-cc induction of ANAC genes is independent of NPR1.
Supplemental Figure 2. Effect of cycloheximide on ANAC102 induction by β-cc.
Supplemental Figure 3. The resilience of young leaves to excessive light depends on SCL14.
Supplemental Table 1. Primers used in the work.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the Phytotec platform (CEA Cadarache) for growing plants under control and stress conditions. We also thank C. Gatz (Gottingen, Germany) for the kind gift of seeds of the tgaII and of the scl14 knockout mutant and the SCL14 overexpressor. We thank G. Bethke (St.Paul, Minnesota) for the kind gift of the npr1 mutant seeds. Finally, we thank X. Johnson (CEA Cadarache) for help with chlorophyll fluorescence imaging and for careful reading of the manuscript. This work was supported by a grant from the French National Research Agency (ANR project SLOSAM, 14-CE02-0010-02).
AUTHOR CONTRIBUTIONS
S.D. and M.H. conceived the study. S.D. performed most experiments. B. K. performed HOTE quantifications. S.D. and M.H. analyzed the data and wrote the article.
References
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielczynski L.W., Łącki M.K., Hoefnagels I., Gambin A., Croce R. (2017). Leaf and plant age affects photosynthetic performance and photoprotective capacity. Plant Physiol. 175: 1634–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtic S., Ksas B., Genty B., Mueller M.J., Triantaphylidès C., Havaux M. (2011). Using spontaneous photon emission to image lipid oxidation patterns in plant tissues. Plant J. 67: 1103–1115. [DOI] [PubMed] [Google Scholar]
- Carmody M., Crisp P.A., d’Alessandro S., Ganguly D., Gordon M., Havaux M., Albrecht-Borth V., Pogson B.J. (2016). Uncoupling high light responses from singlet oxygen retrograde signaling and spatial-temporal systemic acquired acclimation. Plant Physiol. 171: 1734–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.X., Phua S.Y., Crisp P., McQuinn R., Pogson B.J. (2016). Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 67: 25–53. [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Christianson J.A., Wilson I.W., Llewellyn D.J., Dennis E.S. (2009). The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149: 1724–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRidder B.P., Dixon D.P., Beussman D.J., Edwards R., Goldsbrough P.B. (2002). Induction of glutathione S-transferases in Arabidopsis by herbicide safeners. Plant Physiol. 130: 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290. [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Van Montagu M., Inzé D. (1997). Herbicide safener-inducible gene expression in Arabidopsis thaliana. Plant Cell Physiol. 38: 568–577. [DOI] [PubMed] [Google Scholar]
- Fan W., Dong X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E., Mueller M.J. (2013). ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 64: 429–450. [DOI] [PubMed] [Google Scholar]
- Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U., Mullineaux P.M., Nonell S., Wilson M.T., Baker N.R. (2006). Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57: 1725–1734. [DOI] [PubMed] [Google Scholar]
- Fode B., Siemsen T., Thurow C., Weigel R., Gatz C. (2008). The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20: 3122–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank H.A., Cogdell R.J. (1996). Carotenoids in photosynthesis. Photochem. Photobiol. 63: 257–264. [DOI] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M. (2014). Carotenoid oxidation products as stress signals in plants. Plant J. 79: 597–606. [DOI] [PubMed] [Google Scholar]
- Huang L.-J., Li N., Thurow C., Wirtz M., Hell R., Gatz C. (2016). Ectopically expressed glutaredoxin ROXY19 negatively regulates the detoxification pathway in Arabidopsis thaliana. BMC Plant Biol. 16: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Vandystadt G., Bujaldon S., Wollman F.A., Dubois R., Roussel P., Alric J., Béal D. (2009). A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth. Res. 102: 85–93. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Katagiri F., Lam E., Chua N.-H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340: 727–730. [DOI] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Thurow C., Kruse K., Meier A., Iven T., Feussner I., Gatz C. (2012). Xenobiotic- and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol. 159: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni S.O., Kuhns C., Ditzer A., Kirch H.H., Bartels D. (2006). Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 29: 1033–1048. [DOI] [PubMed] [Google Scholar]
- Kouřil R., Wientjes E., Bultema J.B., Croce R., Boekema E.J. (2013). High-light vs. low-light: Effect of light acclimation on photosystem II composition and organization in Arabidopsis thaliana. Biochim. Biophys. Acta 1827: 411–419. [DOI] [PubMed] [Google Scholar]
- Kreuz K., Tommasini R., Martinoia E. (1996). Old enzymes for a new job. Herbicide detoxification in plants. Plant Physiol. 111: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Fufezan C., Trebst A. (2008). Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 98: 551–564. [DOI] [PubMed] [Google Scholar]
- Lam E., Lam Y.K. (1995). Binding site requirements and differential representation of TGF factors in nuclear ASF-1 activity. Nucleic Acids Res. 23: 3778–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260. [DOI] [PubMed] [Google Scholar]
- Mano J. (2012). Reactive carbonyl species: their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 59: 90–97. [DOI] [PubMed] [Google Scholar]
- Mano J., Belles-Boix E., Babiychuk E., Inzé D., Torii Y., Hiraoka E., Takimoto K., Slooten L., Asada K., Kushnir S. (2005). Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase. Detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol. 139: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Johnson J.M., Hieno A., Tokizawa M., Nomoto M., Tada Y., Godfrey R., Obokata J., Sherameti I., Yamamoto Y.Y., Böhmer F.D., Oelmüller R. (2015). High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol. Plant 8: 1253–1273. [DOI] [PubMed] [Google Scholar]
- Montillet J.L., et al. (2013). An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet J.L., Cacas J.L., Garnier L., Montané M.H., Douki T., Bessoule J.J., Polkowska-Kowalczyk L., Maciejewska U., Agnel J.P., Vial A., Triantaphylidès C. (2004). The upstream oxylipin profile of Arabidopsis thaliana: a tool to scan for oxidative stresses. Plant J. 40: 439–451. [DOI] [PubMed] [Google Scholar]
- Mousavi S.A.R., Chauvin A., Pascaud F., Kellenberger S., Farmer E.E. (2013). GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426. [DOI] [PubMed] [Google Scholar]
- Moustaka J., Tanou G., Adamakis I.D., Eleftheriou E.P., Moustakas M. (2015). Leaf age-dependent photoprotective and antioxidative response mechanisms to paraquat-induced oxidative stress in arabidopsis thaliana. Int. J. Mol. Sci. 16: 13989–14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M.J., Berger S. (2009). Reactive electrophilic oxylipins: pattern recognition and signalling. Phytochemistry 70: 1511–1521. [DOI] [PubMed] [Google Scholar]
- Mueller S., Hilbert B., Dueckershoff K., Roitsch T., Krischke M., Mueller M.J., Berger S. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A.A., Thurow C., Fode B., Zander M., Weigel R., Gatz C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50: 128–139. [DOI] [PubMed] [Google Scholar]
- Oguchi R., Hikosaka K., Hirose T. (2003). Does the photosynthetic light-acclimation need change in leaf anatomy? Plant Cell Environ. 26: 505–512. [Google Scholar]
- Ort D.R. (2001). When there is too much light. Plant Physiol. 125: 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Cuiné S., Triantaphylidès C., Ravanat J.-L., Havaux M. (2012a). Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158: 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012b). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 109: 5535–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Sulmon C., Serra A.A., Gouesbet G., Couée I. (2012c). Xenobiotic sensing and signalling in higher plants. J. Exp. Bot. 63: 3999–4014. [DOI] [PubMed] [Google Scholar]
- Ramel F., Ksas B., Akkari E., Mialoundama A.S., Monnet F., Krieger-Liszkay A., Ravanat J.-L., Mueller M.J., Bouvier F., Havaux M. (2013a). Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25: 1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Ksas B., Akkari E., Mialoundama A.S., Monnet F., Krieger-Liszkay A., Ravanat J.-L., Mueller M.J., Bouvier F., Havaux M. (2013b). Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25: 1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakaran N. (2014). Identification of the Role of Arabidopsis ATAF-Type NAC Transcription Factors in Plant Stress and Development. PhD dissertation (Georg-August University School of Science; ). [Google Scholar]
- Redman J., Whitcraft J., Johnslon C., Arias J. (2002). Abiotic and biotic stress differentially stimulate as-1 element activity in Arabidopsis. Plant Cell Rep. 21: 180–185. [Google Scholar]
- Riechers D.E., Kreuz K., Zhang Q. (2010). Detoxification without intoxication: herbicide safeners activate plant defense gene expression. Plant Physiol. 153: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach T., Baur T., Stöggl W., Krieger-Liszkay A. (2017). Chlamydomonas reinhardtii responding to high light: a role for 2-propenal (acrolein). Physiol. Plant. 161: 75–87. [DOI] [PubMed] [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandermann H., Jr (1992). Plant metabolism of xenobiotics. Trends Biochem. Sci. 17: 82–84. [DOI] [PubMed] [Google Scholar]
- Shumbe L., Bott R., Havaux M. (2014). Dihydroactinidiolide, a high light-induced β-carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis. Mol. Plant 7: 1248–1251. [DOI] [PubMed] [Google Scholar]
- Shumbe L., D’Alessandro S., Shao N., Chevalier A., Ksas B., Bock R., Havaux M. (2017). METHYLENE BLUE SENSITIVITY 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of β-cyclocitral. Plant Cell Environ. 40: 216–226. [DOI] [PubMed] [Google Scholar]
- Sims D.A., Pearcy R.W. (1992). Response of leaf anatomy and photosynthetic capacity in Alocasia macrorrhiza (Araceae) to a transfer from low to high light. Am. J. Bot. 79: 449. [Google Scholar]
- Skipsey M., Knight K.M., Brazier-Hicks M., Dixon D.P., Steel P.G., Edwards R. (2011). Xenobiotic responsiveness of Arabidopsis thaliana to a chemical series derived from a herbicide safener. J. Biol. Chem. 286: 32268–32276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens N.O., Galuschka C., Schindler M., Bülow L., Hehl R. (2004). AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 32: D368–D372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiti N., Missihoun T.D., Kotchoni S.O., Kirch H.-H., Bartels D. (2011). Aldehyde dehydrogenases in Arabidopsis thaliana: Biochemical requirements, metabolic pathways, and functional analysis. Front. Plant Sci. 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor V.L., Cummins I., Brazier-Hicks M., Edwards R. (2013). Protective responses induced by herbicide safeners in wheat. Environ. Exp. Bot. 88: 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C., Havaux M. (2009). Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 14: 219–228. [DOI] [PubMed] [Google Scholar]
- Turóczy Z., Kis P., Török K., Cserháti M., Lendvai A., Dudits D., Horváth G.V. (2011). Overproduction of a rice aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol. Biol. 75: 399–412. [DOI] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., op den Camp R., Kim C., Landgraf F., Lee K.P., Wursch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Hasegawa A., Taninaka A., Mizutani M., Sugimoto Y. (2011). NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 286: 6999–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-M., Trifa Y., Silva H., Pontier D., Lam E., Shah J., Klessig D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13: 191–202. [DOI] [PubMed] [Google Scholar]