CLAVATA3 INSENSITIVE RECEPTOR KINASEs function with BARELY ANY MERISTEM 1/2 and RECEPTOR-LIKE PROTEIN KINASE2 to regulate early anther development in Arabidopsis.
Abstract
Appropriate cell division and differentiation ensure normal anther development in angiosperms. BARELY ANY MERISTEM 1/2 (BAM1/2) and RECEPTOR-LIKE PROTEIN KINASE2 (RPK2), two groups of leucine-rich repeat receptor-like protein kinases, are required for early anther cell specification. However, little is known about the molecular mechanisms underlying these two RLK-mediated signaling pathways. Here, we show that CLAVATA3 INSENSITIVE RECEPTOR KINASEs (CIKs), a group of novel coreceptor protein kinase-controlling stem cell homeostasis, play essential roles in BAM1/2- and RPK2-regulated early anther development in Arabidopsis thaliana. The archesporial cells of cik1/2/3 triple and cik1/2/3/4 quadruple mutant anthers perform anticlinal division instead of periclinal division. Defective cell division and specification of the primary and inner secondary parietal cells occur in these mutant anthers. The disordered divisions and specifications of anther wall cells finally result in excess microsporocytes and a lack of one to three parietal cell layers in mutant anthers, resembling rpk2 or bam1/2 mutant anthers. Genetic and biochemical analyses indicate that CIKs function as coreceptors of BAM1/2 and RPK2 to regulate archesporial cell division and determine the specification of anther parietal cells.
INTRODUCTION
Anthers, the male part of flowering plants, bear pollen grains for producing sperm cells and thus play important roles in plant sexual reproduction. In Arabidopsis thaliana, anther development is artificially divided into fourteen stages according to the morphological and cellular features of developing anthers under light microscopy (Sanders et al., 1999). At stage 1, anther primordia, containing L1, L2, and L3, three distinct cell layers, originate from the floral meristem. At stage 2, some L2 cells develop into the archesporial cells at four corners of the anther where four anther lobes will be formed later. Then, at stage 3, the archesporial cells divide periclinally and asymmetrically to produce the primary sporogenous cells and primary parietal cells. Subsequently, cell divisions of the primary parietal cells generate the inner and outer secondary parietal cells at stage 4. The inner secondary parietal cells periclinally divide again to produce the middle layer and the tapetum at stage 5 (Albrecht et al., 2005; Walbot and Egger, 2016). In the meantime, the L1 layer cells develop into the epidermis and the L3 layer cells divide and differentiate to establish the connective and vascular tissues. Thus, a four-lobed anther is established with each lobe consisting of four somatic cell layers from outside to inside: the epidermis, the endothecium, the middle layer, and the tapetum, surrounding the microspore mother cells (MMCs). At stage 6 and stage 7, MMCs undergo meiosis to give rise to tetrads; the tapetal cells become vacuolated, and the middle layer cells begin to degenerate. Then, at stage 8, microspores are released from the tetrads following the degradation of the surrounding callose wall. During stages 9 to 14, microspores further develop to form mature pollen grains that will be finally released for pollination when anther dehiscence occurs.
Coordinated cell division and differentiation of the L2 cells during the early process of anther development have great importance for establishing a normal anther structure. Diverse factors regulating cell fate determination during early anther development have been identified in Arabidopsis, rice (Oryza sativa), and maize (Zea mays) (Zhang and Yang, 2014). For example, SPOROCYTELESS/NOZZLE (SPL/NZZ), an adaptor-like transcription repressor, is expressed during microsporogenesis and megasporogenesis. Normal archesporial cells are produced in spl anthers and divide successfully to form the primary parietal cells and primary sporogenous cells. However, the primary sporogenous cells degenerate directly and are unable to form microsporocytes (Schiefthaler et al., 1999; Yang et al., 1999; Wei et al., 2015). A recent study showed that MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3) and MPK6 phosphorylate and stabilize SPL to regulate archesporial cell division but only in the adaxial anther lobes (Zhao et al., 2017). It has been found that ROXY1 and ROXY2, two glutaredoxins, function downstream of SPL to control the periclinal division of the archesporial cells also in the adaxial anther lobes (Xing and Zachgo, 2008), suggesting an essential role of redox state in specifying archesporial cell fate (Zhang and Yang, 2014).
BARELY ANY MERISTEM1 (BAM1) and BAM2, two leucine-rich repeat receptor-like protein kinases (LRR-RLKs), play key roles in regulating the correct formation of the L2-derived cells (Hord et al., 2006). Double mutation of these two homologous genes leads to disordered division and differentiation of L2 cells, generating anthers with more MMC-like cells while lacking the endothecium, the middle layer, and the tapetum. Consistently, the expression of SPL is expanded to all the L2-derived cells in bam1/2 anthers. RECEPTOR-LIKE PROTEIN KINASE2 (RPK2), another LRR-RLK, is also required for early anther development (Mizuno et al., 2007). rpk2 mutants generate anthers lacking the middle layer, with abnormally hypertrophic tapetal cells and inadequately thickened and lignified endothecium cells, that finally fail to produce and release functional pollen grains. BAM1 was shown to physically interact with RPK2 to regulate cell proliferation in the root meristem (Shimizu et al., 2015). Whether BAM1/2 and RPK2 function in the same pathway to modulate early anther development is an open question. EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS), an LRR-RLK with a long extracellular domain, and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) and SERK2, two LRR-RLKs with only five LRRs in their short extracellular domains, were found to form a receptor/coreceptor complex for perceiving the TAPETUM DETERMINANT1 (TPD1) peptide signal to control the specification of tapetal cells and microsporocytes during early anther development (Canales et al., 2002; Zhao et al., 2002; Yang et al., 2003; Albrecht et al., 2005; Colcombet et al., 2005; Jia et al., 2008; Huang et al., 2016; Li et al., 2017). Mutation of these genes leads to anthers with excess microsporocytes but lacking tapetal cells and abnormally maintaining the middle layer even at stage 9. Recently, β-carbonic anhydrases were identified as the direct downstream targets of EMS1 (Huang et al., 2017). TPD1-like 1A (OsTDL1A)/MICROSPORELESS2 (MIL2) and MULTIPLE SPOROCYTE1 (MSP1), homologs of Arabidopsis TPD1 and EMS1, respectively, interact to specify anther cell fate possibly by affecting redox status in rice (Yang et al., 2016). ERECTA (ER) and its close homologs ERECTA-LIKE1 (ERL1) and ERL2 are also involved in regulating both anther lobe formation and anther cell differentiation in Arabidopsis (Hord et al., 2008).
LRR-RLK, consisting of at least 223 members in Arabidopsis, is one of the major categories of plant transmembrane RLKs (Shiu and Bleecker, 2001; Torii, 2004; Gou et al., 2010). Some LRR-RLKs function as receptors to regulate a variety of biological processes. For example, BRASSINOSTEROID INSENSITIVE1 (BRI1) perceives brassinosteroids (Clouse et al., 1996; Li and Chory, 1997). ER and its homologs are involved in controlling inflorescence architecture, stomata patterning, and ovule development (Shpak et al., 2003, 2005; Pillitteri et al., 2007). FLAGELLIN-SENSITIVE2 (FLS2) mediates the plant pathogen response (Gómez-Gómez and Boller, 2000). CLAVATA1 (CLV1) regulates stem cell fate (Clark et al., 1993). Unlike these LRR-RLKs with a long extracellular domain for direct binding of ligands, SERKs possess a short extracellular domain that cannot perceive ligands alone. However, SERKs can bind the new surfaces generated by the ligand-binding receptors and their corresponding ligands (Santiago et al., 2013; Sun et al., 2013; Wang et al., 2015; Song et al., 2016) to function as vital coreceptors in multiple RLK-mediated signaling pathways, including brassinosteroid perception, plant innate immune response, cell proliferation, stomata patterning, floral organ abscission, root meristem maintenance, vascular development, and anther somatic cell differentiation (Li et al., 2002, 2017; Nam and Li, 2002; Chinchilla et al., 2007; Roux et al., 2011; Ladwig et al., 2015; Meng et al., 2015, 2016; Ou et al., 2016; Zhang et al., 2016).
However, it is unknown whether BAM1, BAM2, and RPK2 need coreceptors to determine cell fate specification during early anther development. The bam1/2 mutant anthers show defects during the first periclinal division of the archesporial cell (Hord et al., 2006), while the serk1/2 mutant anthers show defects until the division of the inner secondary parietal cell (Albrecht et al., 2005). On the other hand, RPK2 has already been shown to be required for specification of the middle layer (Mizuno et al., 2007), whereas SERKs are involved in formation of the tapetum but not the middle layer (Albrecht et al., 2005; Colcombet et al., 2005). These phenotypic differences indicate that SERKs are not involved in BAM1/2- and RPK2-regulated anther cell differentiation. Besides five SERK members, the nine remaining LRR II-RLKs in Arabidopsis hold similar short extracellular structures, suggesting that they might also act as coreceptors in different signaling pathways. A reverse genetic approach was then employed to elucidate the biological functions of these RLKs. We found that a group of four LRR II-RLKs, named CLAVATA3 INSENSITIVE RECEPTOR KINASE1 (CIK1) to CIK4, function as coreceptors of CLV1 and RPK2 to regulate stem cell homeostasis (Hu et al., 2018). In this study, we show that CIKs also associate with BAM1/2 and RPK2 to control somatic cell fate determination during early anther development in Arabidopsis.
RESULTS
Loss-of-Function Mutants of CIKs Show Reduced Fertility
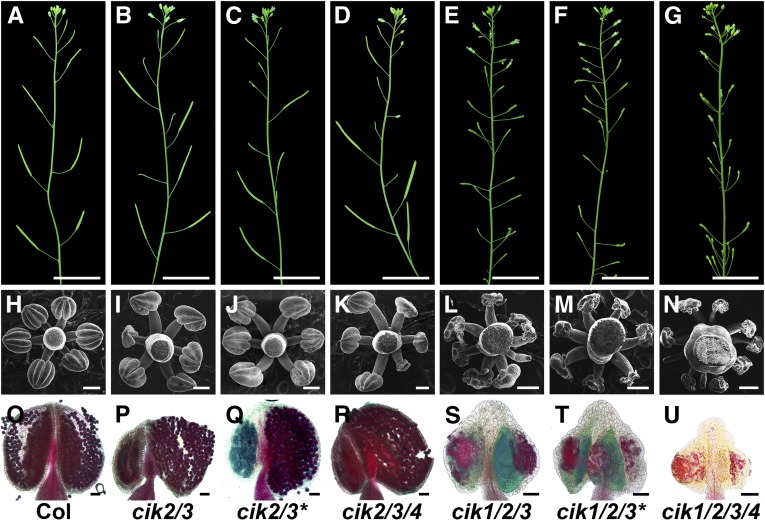
Arabidopsis LRR II-RLK family has 14 members. Among them, five SERKs have been intensively investigated in the past decade. To elucidate the functions of other LRR II-RLKs, we obtained null T-DNA insertion lines of these RLK genes and created high-order mutants (Hu et al., 2018; Supplemental Figures 1A to 1D and Supplemental Data Set 1). All of the single knockout cik mutants show phenotypes indistinguishable from the wild type (Supplemental Figures 1E to 1R). Interestingly, a double mutant, cik2/3, but not other double mutants or even the cik1/2/4 and cik1/3/4 triple mutants, produces some short and sterile siliques (Figure 1B; Supplemental Figures 1J and 1K). cik2/3/4 mutants show a reduced fertility phenotype similar to that of cik2/3 mutants (Figure 1D). Moreover, cik1/2/3 and cik1/2/3/4 mutants display more severe defects without any seeds produced (Figures 1E and 1G). Independent cik2-2/3-2 (cik2/3*) and cik1-2/2-2/3-2 (cik1/2/3*) mutants created using a different set of single T-DNA insertion alleles of cik1, cik2, and cik3 show phenotypes similar to the first cik2/3 and cik1/2/3 mutants (Figures 1C and 1F), further demonstrating that CIKs are responsible for the defective fertility phenotypes.
Figure 1.
Loss of Function of CIKs Leads to Reduced Fertility.
(A) to (G) Inflorescence stems of 6-week-old plants.
(H) to (N) Scanning electron microscopy images showing stamens at flower stage 12.
(O) to (U) Alexander’s staining of mature anthers to show pollen viability.
(A), (H), and (O), wild type (Col); (B), (I), and (P), cik2/3; (C), (J), and (Q), cik2/3* (the second cik2/3 mutant); (D), (K), and (R), cik2/3/4; (E), (L), and (S), cik1/2/3; (F), (M), and (T), cik1/2/3* (the second cik1/2/3 mutant); (G), (N), and (U), cik1/2/3/4. Bars = 2 cm in (A) to (G), 200 µm in (H) to (N), and 50 µm in (O) to (U).
Anther Development in cik Mutants Is Abnormal
When cik1/2/3/4 mutants were used as the male to cross with wild-type plants, no viable seeds were produced in any of the ovules, indicating that mutations of CIKs caused abnormal male fertility. Observations under a stereomicroscope revealed that stamens of cik1/2/3 and cik1/2/3/4 mutants cannot reach the stigma (Supplemental Figures 2A to 2F). Further scanning electron microscopy analyses showed that six anthers surround the gynoecium, and each anther develops four plump locules in the wild type (Figure 1H; Supplemental Figures 1L and 2G). However, nearly half of the anthers in cik2/3 flowers displayed abnormal morphology, as 33.3% of anthers showed an asymmetric shape with two collapsed locules and 11.7% of anthers had four collapsed locules (Figures 1I and 1J; Supplemental Figure 2H). A similar defective anther phenotype was observed in cik2/3/4 mutants (Figure 1K). Remarkably, cik1/2/3 mutants produce more stamens than the wild type but without any normal plump locules (Figures 1L and 1M). Notably, 16.6% of anthers in cik1/2/3 mutants have only two or three collapsed locules, and 8% of stamens are club-shaped without any observable locules (Supplemental Figure 2I). cik1/2/3/4 mutants produce more than nine stamens with even more severe defects compared with cik1/2/3 mutants (Figure 1N; Supplemental Figure 2J). All single knockout mutants and the other two triple knockout mutants, cik1/2/4 and cik1/3/4, showed normal anther morphology under scanning electron microscopy analyses (Supplemental Figures 1L to 1R).
Alexander’s staining analyses were performed to examine the viability of pollen grains in cik anthers. Only viable pollen grains stain red (Alexander, 1969). The results showed that only the plump locules of cik2/3 and cik2/3/4 anthers produce normal pollen grains similar to the wild type (Figures 1O to 1R). cik1/2/3 anthers almost produce no observable pollen grains or occasionally produce very few pollen grains that cannot be released (Figures 1S and 1T). Although degenerated debris without normal shape and size of pollen grains can be stained red, cik1/2/3/4 anthers cannot produce any pollen grains (Figure 1U).
As expected, the anther defects in cik1/2/3/4 mutants can be rescued by the complementation of CIKs driven by an Arabidopsis UBQ10 promoter or native CIK promoters (Supplemental Figure 3). Taken together, these results demonstrate that CIKs function redundantly in regulating anther development.
Anther Cell Differentiation Is Defective in cik Mutants
To determine when the mutant anthers exhibit defective morphology, flowers of wild-type and mutant plants after flower stage 9 were examined by scanning electron microscopy. The results showed that most of the cik2/3 anthers exhibit a similar size indistinguishable from that of the wild type at three examined stages (Supplemental Figures 2K to 2P). Anthers of cik1/2/3 and cik1/2/3/4 mutants are obviously smaller than those of the wild type even early at flower stage 9, and the anther size of these two mutants does not further increase even when the anthers develop to flower stage 12 (Supplemental Figures 2Q to 2V), suggesting that early anther development in these mutants is affected.
Semithin transverse sections of anthers were then examined to further reveal the detailed defects of early anther development in cik mutants. In the wild type, the L2 layer cells in the stamen primordium form the archesporial cells at four corners. The archesporial cells then generate MMCs surrounded by three parietal cell layers through three rounds of periclinal cell division and cell differentiation. Meanwhile, the L1 cells form the epidermis that encloses these L2-derived cells (Figures 2A, 2G, 2M, and 2S). Later, MMCs form tetrads via meiosis that will be released as microspores at anther stage 8 when the middle layer degenerates (Supplemental Figures 4A, 4G, and 4M).
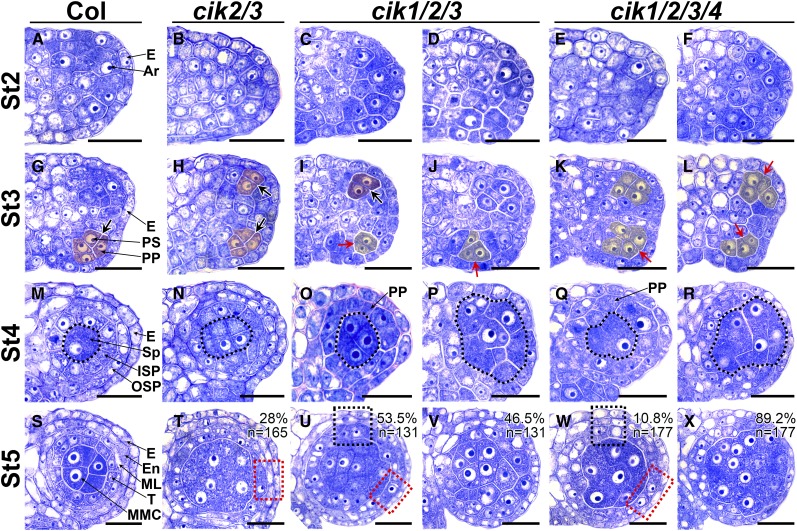
Figure 2.
Cell Division and Differentiation Are Abnormal in Anthers of cik Mutants.
(A) to (F) Semithin sections of stage 2 (St2) anthers. The archesporial cells (Ar) are produced under the epidermis (E). No difference was observed between the wild type (A) and cik mutants ([B] to [F]).
(G) to (L) Semithin sections of St3 anthers. The archesporial cell periclinally divides once to produce an outer primary parietal cell (PP) and an inner primary sporogenous cell (PS) in the wild type (G). No obvious defects can be observed in cik2/3 (H). Anticlinal cell divisions occur in cik1/2/3 ([I] and [J]) and cik1/2/3/4 ([K] and [L]). Thick black arrows indicate normal periclinal division of cells shaded in red; red arrows indicate anticlinal division of cells shaded in yellow.
(M) to (R) Semithin sections of St4 anthers. The primary parietal cell divides once to produce an outer secondary parietal cell (OSP) and an inner secondary parietal cell (ISP) in the wild type (M) and cik2/3 (N). No OSP and ISP cells can be correctly specified in cik1/2/3 and cik1/2/3/4 ([O] to [R]). Sporogenous cells (Sp) and Sp-like cells are indicated with dotted loops.
(S) to (X) Semithin sections of St5 anthers. The inner secondary parietal cell divides once to produce an outer middle layer cell (ML) and an inner tapetal cell (T), forming a locule with four cell layers that enclose the microspore mother cells (MMC) in the wild type (S). In cik2/3, 28% of locules display a discontinuous middle layer (dotted red rectangle) (T). In cik1/2/3, 53.5% of locules display one or two parietal cell layers (dotted red and black rectangles) (U), and 46.5% of locules produce MMC-like cells enclosed only by the epidermis (V). In cik1/2/3/4, 10.8% of locules have one or two parietal cell layers (dotted red and black rectangles) (W), and 89.2% of locules produce MMC-like cells enclosed only by the epidermis (X). All cik mutants produce extra MMC-like cells.
Half anther is shown in (A) to (L); one locule is shown in (M) to (X). n indicates the total number of examined locules. Bars = 20 µm.
Although no obvious defects can be found in early anthers of cik2/3 from stage 2 to stage 4, 28% of anther locules show more MMC-like cells and discontinuous middle layer at stage 5 (Figures 2B, 2H, 2N, and 2T). Later, at stage 6, the tapetum in these defective anthers becomes abnormally hypertrophic, leading to collapsed locules with no microspores at stage 8 (Supplemental Figures 4B, 4H, and 4N’). Normal pollen grains can be produced in other wild-type-like anthers (Supplemental Figure 4N). The second cik2/3 mutant shows similar defects (Supplemental Figure 4X).
Anthers of cik1/2/3 mutants can initiate archesporial cells like the wild type at stage 2 (Figures 2C and 2D). However, only some archesporial cells in cik1/2/3 anthers can perform normal asymmetric periclinal division to generate the primary parietal cells and the primary sporogenous cells (Figure 2I). The remaining archesporial cells in the same anther show disordered cell divisions, producing fewer and larger cells at stage 3 (Figure 2J). At stage 4, the larger sporogenous cell-like cells are surrounded either by a layer of primary parietal cell-like cells under the epidermis or only by the epidermis (Figures 2O and 2P). Subsequently, 53.5% of locules produce only one or two layers of parietal cells surrounding the MMC-like cells, and 46.5% of them generate many more MMC-like cells surrounded directly by the epidermis at stage 5 (Figures 2U and 2V). Later, those MMC-like cells in locules with one or two layers of parietal cells can enter meiosis to form the tetrads that cannot be released as microspores (Supplemental Figures 4C, 4I, and 4O). Very occasionally, some locules can produce a few defective pollen grains that cannot be released either (Supplemental Figure 4O’). The MMC-like cells without surrounding parietal cells cannot complete meiosis to form tetrads, finally resulting in collapsed and sterile locules (Supplemental Figures 4D, 4J, and 4P). The second cik1/2/3 mutant shows the same defective anther development (Supplemental Figures 4Y and 4Z). Another triple mutant, cik2/3/4, produces defective anthers similar to those of cik2/3 (Supplemental Figure 4A1). Consistent with the scanning electron microscopy results, the cik1/3/4 mutant produces normal anthers (Supplemental Figure 4B1). Although no obvious anther defects were revealed by scanning electron microscopy observation, semithin sectioning found that a few cik1/2/4 anthers display a discontinuous middle layer (Supplemental Figures C1 and D1).
Anthers of cik1/2/3/4 mutants show similar but more severe defects compared with cik1/2/3 mutants. Only 10.8% of locules produce one or two layers of parietal cells (Figures 2E, 2K, 2Q, and 2W), while 89.2% of locules show more MMC-like cells surrounded directly by the epidermis (Figures 2F, 2L, 2R, and 2X). Finally, all cik1/2/3/4 anthers fail to produce any pollen grains (Supplemental Figures 4E, 4F, 4K, 4L, 4Q, and 4R). All these data demonstrate that disruption of CIKs leads to disorganized L2-derived cells because of abnormal cell division and differentiation.
MMC and Tapetum Marker Genes Show Altered Expression Patterns in cik Mutants
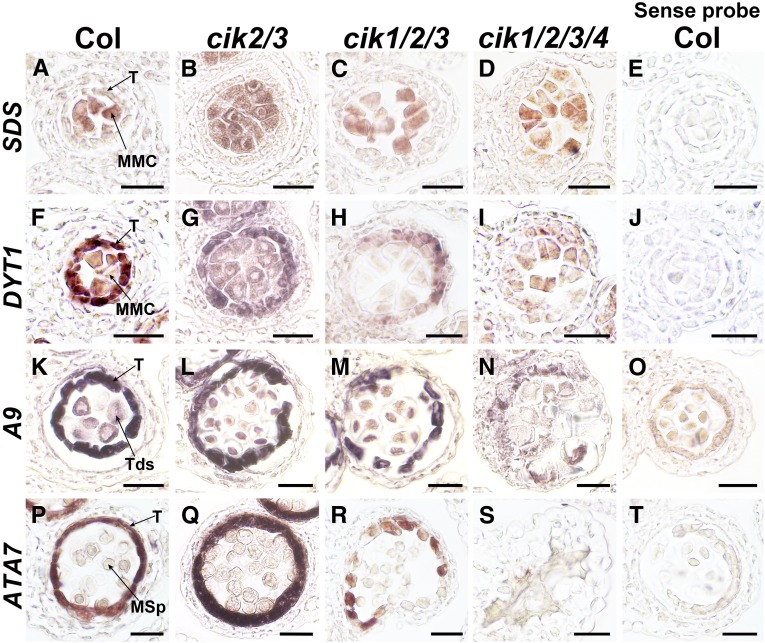
To examine the identities of the L2-derived cells in cik anthers, RNA in situ hybridization was used to investigate the expression patterns of specific cell markers. SOLO DANCERS (SDS) encoding a cyclin-like protein required for homologous recombination during meiotic prophase I is specifically expressed in meiocytes and used to indicate MMCs (Azumi et al., 2002). Consistent with the increased MMC-like cells observed by transverse sectioning (Supplemental Figure 4S), the expression of SDS was detected in all the MMC-like cells in anthers of cik mutants (Figure 3A to 3E), indicating that more cells with MMC identity are specified in cik mutants during anther development.
Figure 3.
RNA in Situ Hybridization Analyses of MMC and Tapetum Markers in Wild-Type and cik Anthers.
(A) to (D) SDS was detected in MMCs of wild-type (A), cik2/3 (B), cik1/2/3 (C), and cik1/2/3/4 (D) anthers at stage 6.
(F) to (I) DYT1 was detected in the tapetum and MMCs of wild-type (F) and cik2/3 (G) anthers at stage 6. Higher expression in the tapetum than MMCs is observed. Weak DYT1 signal was detected in MMCs and the tapetum of the cik1/2/3 mutant (H) and only in MMC-like cells of the cik1/2/3/4 mutant (I).
(K) to (N) A9 was detected in the tapetum and tetrads of wild-type (K), cik2/3 (L), and cik1/2/3 (M) anthers at stage 7. No typical A9 expression was detected in cik1/2/3/4 (N).
(P) to (S) ATA7 was detected in the tapetum of wild-type (P), cik2/3 (Q), and cik1/2/3 (R) anthers at stage 8. No signal was detected in cik1/2/3/4 (S).
Sense probes of SDS (E), DYT1 (J), A9 (O), and ATA7 (T) were used as negative controls. These experiments were repeated three times independently with similar results. MSp, microspore; T, tapetum; Tds, tetrads. Bars = 20 µm.
The DYSFUNCTIONAL TAPETUM1 (DYT1) gene encoding a basic helix-loop-helix transcription factor shows specific high levels of expression in the tapetum and low levels of expression in MMCs in wild-type anthers from late stage 5 to early stage 6 (Zhang et al., 2006). cik2/3 mutant anthers exhibit expression specificity of DYT1 similar to the wild type, but the expression region is greatly expanded because of the increased MMC-like cells (Figures 3F and 3G). While some cik1/2/3 anthers show much lower expression of DYT1 in the tapetum-like cells, indicating that the tapetal cell identity is at least partially impaired in these anthers (Figure 3H), the other cik1/2/3 anthers and all cik1/2/3/4 anthers show very low expression of DYT1 in MMC-like cells and no tapetum could be identified (Figure 3I).
Two more tapetum-specific markers, A9 and ATA7 (Paul et al., 1992; Rubinelli et al., 1998), were further analyzed to examine the identity of anther cell layers at later stages. In wild-type anthers at stage 7, A9 is expressed at high level in the tapetal cells, with weak expression in the tetrads (Figure 3K). In cik2/3 and some cik1/2/3 anthers, clear A9 signals were detected in a cell layer surrounding the tetrads, indicating that tapetal cells still exist in these cik anthers (Figures 3L and 3M). However, no typical A9 expression was detected in the remaining cik1/2/3 and all cik1/2/3/4 anthers, indicating that no tapetal cell layer can be properly specified in them (Figure 3N). ATA7 is expressed later in the tapetum at stage 8, showing the same expression pattern in wild-type and cik2/3 anthers (Figures 3P and 3Q). While some cik1/2/3 anthers express ATA7 at much lower level in the tapetum-like cells (Figure 3R), others show no detectable expression, similar to cik1/2/3/4 anthers, further confirming that the tapetal cell layer cannot be successfully specified in these anthers (Figure 3S). These data suggest that anther cell fate of cik mutants is disordered, consistent with the semithin sectioning results.
Expression Patterns of CIKs Support Their Functions during Early Anther Development
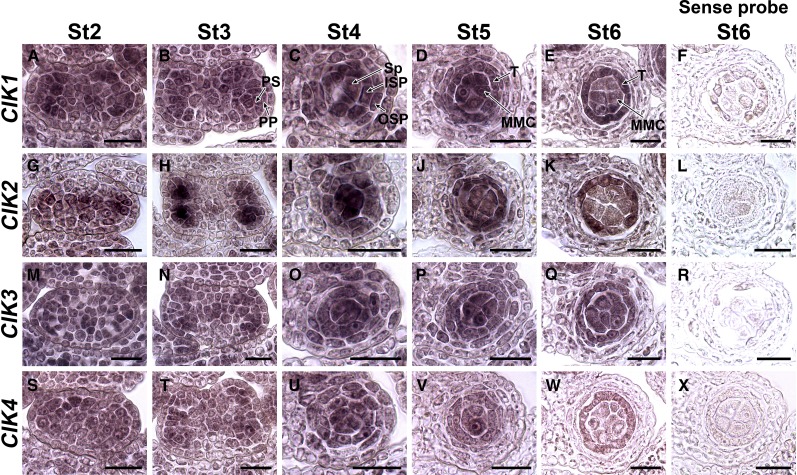
To further confirm the functions of CIKs in regulating anther cell specification, RNA in situ hybridization was performed to investigate their expression patterns during early anther development. All four CIK genes are expressed in anthers at the examined stages with similar patterns (Figure 4). At stage 2, CIKs are expressed ubiquitously in anthers (Figures 4A, 4G, 4M, and 4S). The expression is then gradually concentrated to four lobes at stage 3 and the L3-derived cells but with lower expression level (Figures 4B, 4H, 4N, and 4T). CIK2 shows a more specific pattern at this stage, with obviously higher expression in the primary sporogenous cells (Figure 4H). At stage 4, the sporogenous cells and two layers of secondary parietal cells show high expression of CIKs (Figures 4C, 4I, 4O, and 4U). Later, the expression of CIKs was detected in MMCs, the tapetum, and the middle layer (Figures 4D, 4J, 4P, and 4V). Finally, the expression is concentrated in MMCs and the tapetum (Figures 4E, 4K, 4Q, and 4W). These results support the functions of CIKs in regulating cell specification during early anther development.
Figure 4.
RNA in Situ Hybridization Showing the Expression Patterns of CIKs in Early Wild-Type Anthers.
(A) to (E) CIK1 is expressed in the whole anther at stage 2 (St2) (A) and St3 (B), with higher level in L2-derived cells. The expression is concentrated in four locules with higher expression in the sporogenous cells and two layers of secondary parietal cells at St4 (C), MMCs, the tapetum and the middle layer at St5 (D), and MMCs and the tapetum at St6 (E).
(G) to (K) CIK2 shows expression patterns similar to CIK1 except that the signal is more obvious in the primary sporogenous cells at St3 (H).
(M) to (Q) CIK3 shows expression patterns similar to CIK1.
(S) to (W) CIK4 shows expression patterns similar to CIK1.
Sense probes of CIK1 (F), CIK2 (L), CIK3 (R), and CIK4 (X) were used as negative controls. One locule is shown for anthers at St4, St5, and St6. These experiments were repeated at least three times independently with similar results. ISP, inner secondary parietal cell; OSP, outer secondary parietal cell; PP, primary parietal cell; PS, primary sporogenous cell; Sp, sporogenous cell; T, tapetum. Bars = 20 µm.
CIK-Regulated Early Anther Development Is Independent of CLV Signaling
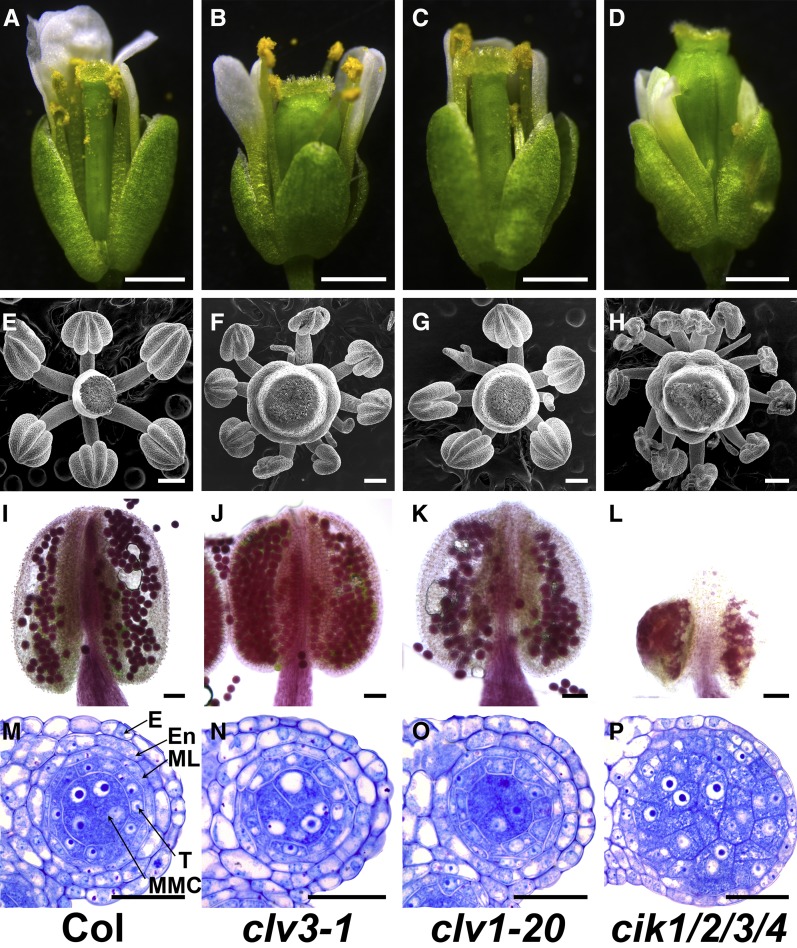
Since CIKs play vital roles in maintaining stem cell homeostasis, and cik1/2/3 and cik1/2/3/4 mutants produce extra stamen-like structures, a phenotype also observed in clv mutants (Clark et al., 1993; Kayes and Clark, 1998; Fletcher et al., 1999; Brand et al., 2000; Hu et al., 2018), flowers and anthers of clv3-1 and clv1-20 were examined to elucidate whether the anther defects in cik mutants are caused by impaired CLV signaling. The results showed that mature stamens of clv3-1 and clv1-20 can reach the stigma (Figures 5A to 5D). Besides those defective stamen-like structures, the clv mutants can produce anthers displaying normal morphology that cannot be observed in cik1/2/3/4 mutants (Figures 5E to 5H). Alexander’s staining analyses indicated that clv mutants can produce functional pollen grains (Figures 5I to 5L). Moreover, semithin sections of clv anthers do not reveal defects similar to cik mutants (Figures 5M to 5P). These data indicate that CIKs regulate anther development through a CLV-independent pathway.
Figure 5.
clv Mutants Produce Functional Anthers.
(A) to (D) Flowers of the wild type (A), clv3-1 (B), clv1-20 (C), and cik1/2/3/4 (D) at stage 13. Bars = 1 mm.
(E) to (H) Scanning electron microscopy images showing anthers of the wild type (E), clv3-1 (F), clv1-20 (G), and cik1/2/3/4 (H) at flower stage 12. Bars = 200 µm.
(I) to (L) Alexander’s staining of stage 13 anthers to show pollen viability of wild type (I), clv3-1 (J), clv1-20 (K), and cik1/2/3/4 (L). Bars = 50 µm.
(M) to (P) Semithin sections showing one locule of stage 5 anthers of wild type (M), clv3-1 (N), clv1-20 (O), and cik1/2/3/4 (P). Bars = 20 µm.
E, epidermis; En, endothecium; ML, middle layer; T, tapetum.
CIKs Genetically Interact with BAM1/2 to Regulate Archesporial Cell Division
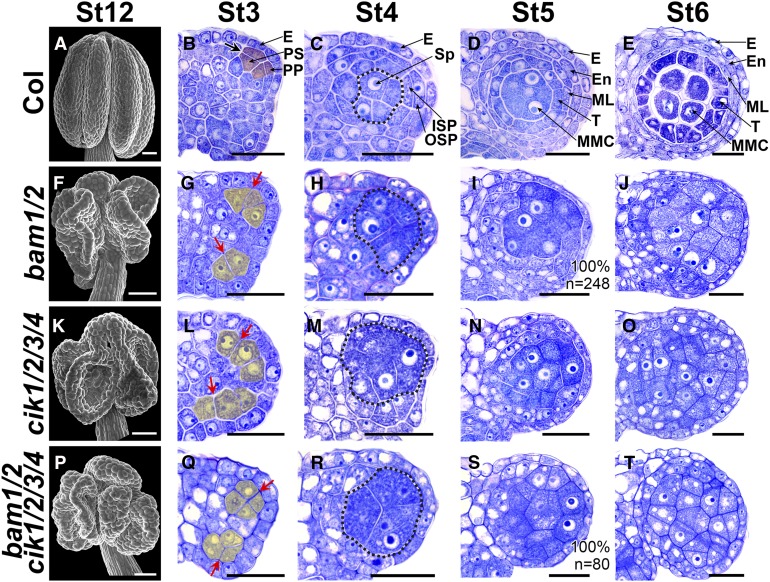
In cik1/2/3/4 anthers, no parietal cell layers can be successfully specified at stage 5, resembling those of bam1/2 mutants (Figures 2X and 6I), suggesting that CIKs may function genetically with BAM1/2 to control early anther development. Therefore, the bam1/2 cik1/2/3/4 mutant was created to investigate the genetic relationship between CIKs and BAM1/2. Scanning electron microscopy observations found that bam1/2, cik1/2/3/4, and bam1/2 cik1/2/3/4 mutants all produce smaller and collapsed anthers compared with wild-type plants (Figures 6A, 6F, 6K, and 6P). Semithin sections showed that the periclinal division of the archesporial cell in all these three mutants is changed to an anticlinal division, and the primary parietal cell and primary sporogenous cell cannot be properly produced and specified at stage 3 (Figures 6B, 6G, 6L, and 6Q). Then, at stage 4, no inner and outer secondary parietal cells can be recognized in the sections, while more sporogenous cell-like cells are produced in anthers of all three mutants (Figures 6C, 6H, 6M, and 6R). Finally, when a typical anther structure is formed in the wild type at stage 5 and stage 6, 89.2% of locules in cik1/2/3/4 and all locules of bam1/2 and bam1/2 cik1/2/3/4 generate extra MMC-like cells enclosed only by the epidermis (Figures 6D, 6E, 6I, 6J, 6N, 6O, 6S, and 6T). These data demonstrate that bam1/2, cik1/2/3/4, and bam1/2 cik1/2/3/4 mutants produce very similar defective anthers in which no functional locules can be properly formed because of disorganized division of archesporial cells and unsuccessful differentiation of anther parietal cells.
Figure 6.
CIKs Genetically Interact with BAM1/2 to Regulate Archesporial Cell Division.
(A) to (E) Wild-type anthers. Scanning electron microscopy image showing a mature anther with four plump lobes at flower stage 12 (St12) (A). Semithin sections showing normal anther development at stage 3 (St3) (B), St4 (C), St5 (D), and St6 (E).
(F) to (J) bam1/2 anthers. Scanning electron microscopy image showing a mature anther with collapsed lobes at flower St12 (F). At St3, the archesporial cell divides anticlinally, and no primary parietal cell and primary sporogenous cell can be properly specified (G). At St4, no outer secondary parietal cell and inner secondary parietal cell are produced (H). MMC-like cells are enclosed only by the epidermis in St5 (I) and St6 (J) anthers.
(K) to (O) cik1/2/3/4 anthers showing defects similar to bam1/2 mutants.
(P) to (T) bam1/2 cik1/2/3/4 anthers showing defects similar to bam1/2 and cik1/2/3/4 mutants.
Thick black arrow indicates normal periclinal division of cells shaded in red (B). Red arrows indicate abnormal anticlinal division of cells shaded in yellow ([G], [L], and [Q]). Sp-like cells are indicated with dotted loops. n indicates the total number of examined locules. E, epidermis; En, endothecium; ISP, inner secondary parietal cell; ML, middle layer; OSP, outer secondary parietal cell; PP, primary parietal cell; PS, primary sporogenous cell; Sp, sporogenous cell; T, tapetum. Bars = 50 µm in (A), (F), (K), and (P) and 20 µm in (B) to (E), (G) to (J), (L) to (O), and (Q) to (T).
A previous study proposed that BAM1/2 restrict the expression of SPL in MMCs in wild-type anthers at stage 5 (Hord et al., 2006). In bam1/2 anthers, SPL expression is expanded to all MMC-like cells. A similar SPL expression pattern was observed in cik1/2/3/4 anthers (Supplemental Figures 5A to 5C). Moreover, CIKs exhibit expression patterns similar to BAM1/2 (Figure 4; Hord et al., 2006). Taken together, these results indicate that CIKs and BAM1/2 likely function in the same genetic pathway to regulate archesporial cell division during early anther development.
CIKs Function with RPK2 to Regulate Early Anther Development
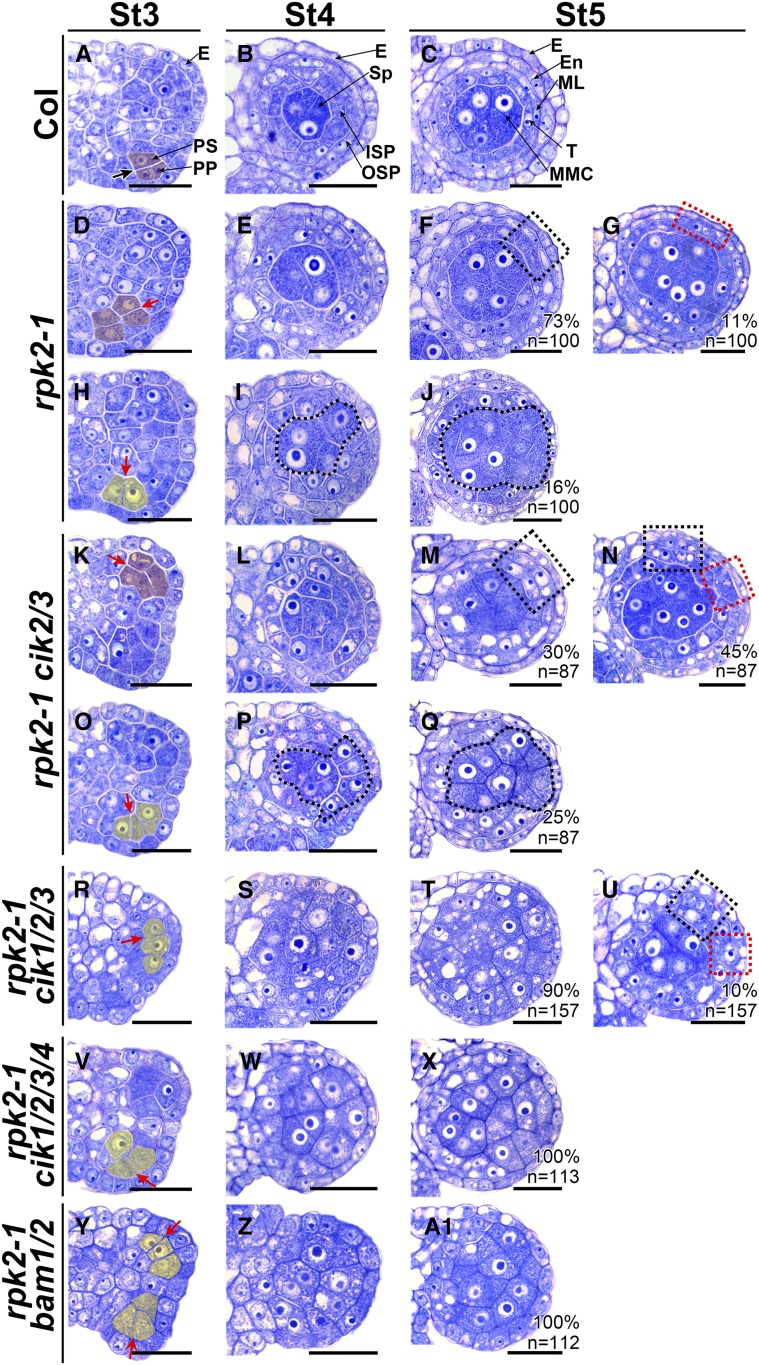
RPK2 is a vital player in regulating anther development (Mizuno et al., 2007). As previously reported, we found that most rpk2-1 locules lack only the middle layer because the inner secondary parietal cell cannot properly differentiate (Figures 7A to 7F). On the other hand, we observed that 11% of rpk2-1 locules lack two parietal cell layers in some anther wall areas (Figure 7G), suggesting that cell fate of the primary parietal cell is changed and no secondary parietal cells can be generated through a periclinal division. Moreover, we noticed that some archesporial cells carry out anticlinal instead of periclinal division at stage 3 (Figure 7H), subsequently resulting in 16% of locules lacking a parietal cell layer and with extra MMCs enclosed directly by the epidermis, reminiscent of bam1/2 and cik1/2/3/4 anthers (Figures 6I, 6J, 6N, 6O, 7I, and 7J). In summary, our observations revealed that the rpk2-1 mutation affects early anther cell fate, leading to defective anthers lacking either one, two, or three parietal cell layers.
Figure 7.
CIKs Genetically Interact with RPK2 to Coordinate Cell Specification during Early Anther Development.
(A) to (C) Wild-type anthers at stage 3 (St3) (A), St4 (B), and St5 (C).
(D) to (J) Anthers of rpk2-1. At St3, some archesporial cells periclinally and symmetrically divide once to produce primary parietal cell- and primary sporogenous cell-like cells (shaded in red) (D). The primary parietal cell-like cells can divide once to produce two layers of secondary parietal cell-like cells at St4 (E). At St5, the MMC-like cells are enclosed by the tapetum-like cells and the endothecium-like cells (dotted black rectangle) (F) or by only one parietal cell layer (dotted red rectangle) (G). The other archesporial cells divide anticlinally (shaded in yellow), and no primary parietal cells can be produced (H), resulting in sporogenous cell-like cells (I) and MMC-like cells (J) enclosed only by the epidermis.
(K) to (Q) Anthers of rpk2-1 cik2/3 showing phenotypes similar to rpk2-1. In 75% of locules, one or two parietal cell layers can be produced at St5 (dotted red or black rectangle) ([M] and [N]). In 25% of locules, the MMC-like cells (dotted loops) are enclosed only by the epidermis (Q).
(R) to (U) Anthers of rpk2-1 cik1/2/3. Ninety percent of rpk2-1 cik1/2/3 locules produce MMC-like cells enclosed only by the epidermis (T). The other locules are enclosed by one or two parietal cell layers (dotted red or black rectangle) (U).
(V) to (X) rpk2-1 cik1/2/3/4 anthers showing phenotypes similar to bam1/2.
(Y) to (A1) rpk2-1 bam1/2 anthers showing phenotypes similar to bam1/2.
Thick black arrow indicates normal periclinal division of cells shaded in red (A). Red arrows indicate abnormal periclinal division of cells shaded in red ([D] and [K]) or anticlinal division of cells shaded in yellow ([H], [O], [R], [V], and [Y]). Sp-like cells ([I] and [P]) and MMC-like cells ([J] and [Q]) are indicated with dotted loops. Percentages of each phenotype are indicated. n indicates the total number of examined locules. E, epidermis; En, endothecium; ISP, inner secondary parietal cell; ML, middle layer; OSP, outer secondary parietal cell; PP, primary parietal cell; PS, primary sporogenous cell; Sp, sporogenous cell; T, tapetum. Bars = 20 µm.
Because cik anthers show phenotypes similar to rpk2-1, lacking either the middle layer (Figures 2T, 2U, and 2W), the middle and endothecium layers (Figures 2U and 2W), or all three parietal cell layers (Figures 2V and 2X), high-order mutants were created by crossing rpk2-1 with cik mutants to elucidate the genetic relationship between RPK2 and CIKs. Interestingly, rpk2-1 cik2/3 mutants produce defective anthers exhibiting the above three phenotypes, which is similar to rpk2-1 and cik mutants, indicating that the archesporial cell division is affected or the parietal cell layers cannot be correctly specified in these mutant anthers (Figures 7K to 7Q). rpk2-1 cik1/2/3 and rpk2-1 cik1/2/3/4 anthers show more severe phenotypes compared with rpk2-1 cik2/3 mutants. Ten percent of rpk2-1 cik1/2/3 locules develop one or two parietal cell layers (Figure 7U), and the others produce extra MMC-like cells and lack all the parietal cell layers similar to rpk2-1 cik1/2/3/4 and cik1/2/3/4 anthers because of altered archesporial cell division (Figures 7R to 7X). Since no parietal cell layers can be specified in 16% of rpk2-1 locules (Figure 7J), and BAM1 can physically interact with RPK2 (Shimizu et al., 2015), the rpk2-1 bam1/2 triple mutant was then created to investigate the genetic interaction between RPK2 and BAM1/2. The results showed that, similar to the bam1/2 mutant, the archesporial cells of rpk2-1 bam1/2 triple mutants divide anticlinally at anther stage 3, and no parietal cell layers can be finally generated (Figures 7Y and 7A1). Consistently, expanded SPL expression was detected in all MMC-like cells of rpk2-1 anthers, as observed in cik1/2/3/4 and bam1/2 anthers (Supplemental Figures 5B to 5E). Taken together, these results indicate that CIKs and RPK2 likely function in the same genetic pathways to regulate not only the specification of the middle layer, but also the differentiation of the secondary parietal cell layers during early anther development. Moreover, CIKs and RPK2 play a vital role in determining the archesporial cell fate.
CIKs Physically Interact with BAM1/2 and RPK2
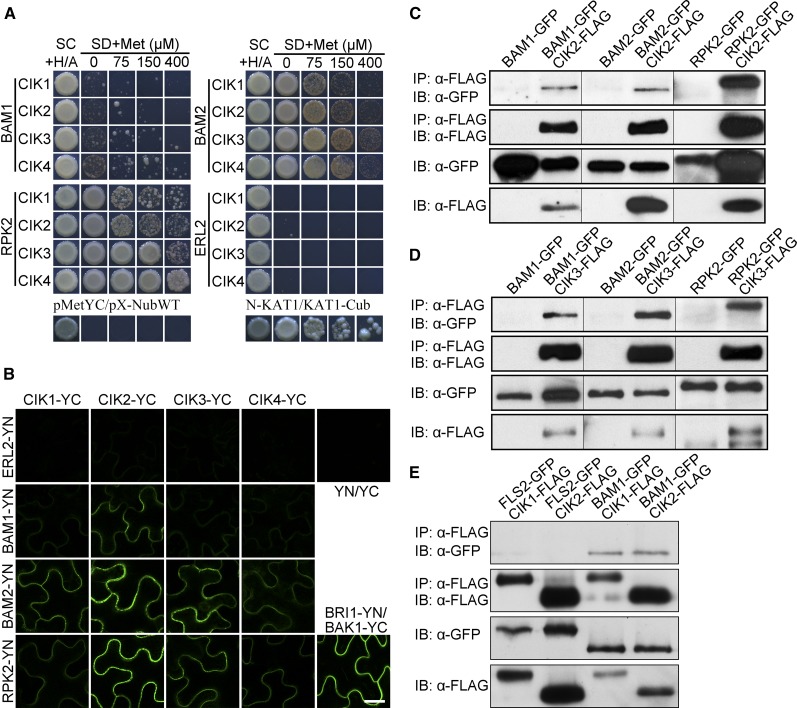
Physical interactions between CIKs and BAM1/2 or RPK2 were then detected to further investigate whether they function together to regulate early anther development. Results of yeast two-hybrid analyses using a mating-based split ubiquitin system (mbSUS) indicated that BAM1 has weak interactions with CIKs, whereas both BAM2 and RPK2 exhibit stronger interactions with CIKs (Figure 8A). Bimolecular fluorescence complementation (BiFC) analyses further demonstrated that BAM1/2 and RPK2 associate with CIKs on the plasma membrane, consistent with their property of receptor-like protein kinases (Figure 8B). Additionally, coimmunoprecipitation assays using coexpressed FLAG-tagged CIKs and GFP-tagged BAM1, BAM2, RPK2, or FLS2 in Arabidopsis indicated that CIKs are able to coimmunoprecipitate BAM1/2 and RPK2 but not FLS2 (Figures 8C to 8E). Similar results of interactions between CIKs and RPK2 were also reported in a previous study (Hu et al., 2018). In summary, these biochemical data demonstrate that CIKs interact with BAM1/2 and RPK2 to form receptor complexes for regulating early anther development.
Figure 8.
CIKs Physically Interact with BAM1/2 and RPK2.
(A) mbSUS yeast two-hybrid results showing that CIKs interact with BAM1/2 and RPK2. SC, synthetic complete medium; SD, synthetic minimal medium. pMetYC/pX-NubWT, negative control; N-KAT1/KAT1-Cub, positive control. ERL2 was used as a negative control.
(B) BiFC results showing that CIKs interact with BAM1/2 and RPK2 in epidermal cells of tobacco leaves. BRI1-YN/BAK1-YC, positive control; YN/YC, negative control. ERL2 was used as a negative control. Bar = 20 µm.
(C) to (E) Coimmunoprecipitation analyses showing that CIKs interact with BAM1/2 and RPK2 in vivo. Total proteins were extracted from leaves of 3-week-old double transgenic plants expressing GFP-tagged BAMs, RPK2, or FLS2 and CIKs-FLAG. Proteins immunoprecipitated with an α-Flag antibody were analyzed with an α-GFP antibody (IP, α-Flag; IB, α-GFP) or an α-Flag antibody (IP, α-Flag; IB, α-Flag). Protein extracts before immunoprecipitation were analyzed with an α-GFP antibody (IB, α-GFP) or an α-Flag antibody (IB, α-Flag) as input controls. FLS2-GFP CIKs-FLAG double transgenic plants and BAM1-GFP, BAM2-GFP, or RPK2-GFP single transgenic plants were used as negative controls. These experiments were repeated at least three times independently with similar results.
CIKs Can Be Phosphorylated by BAM1/2 and RPK2
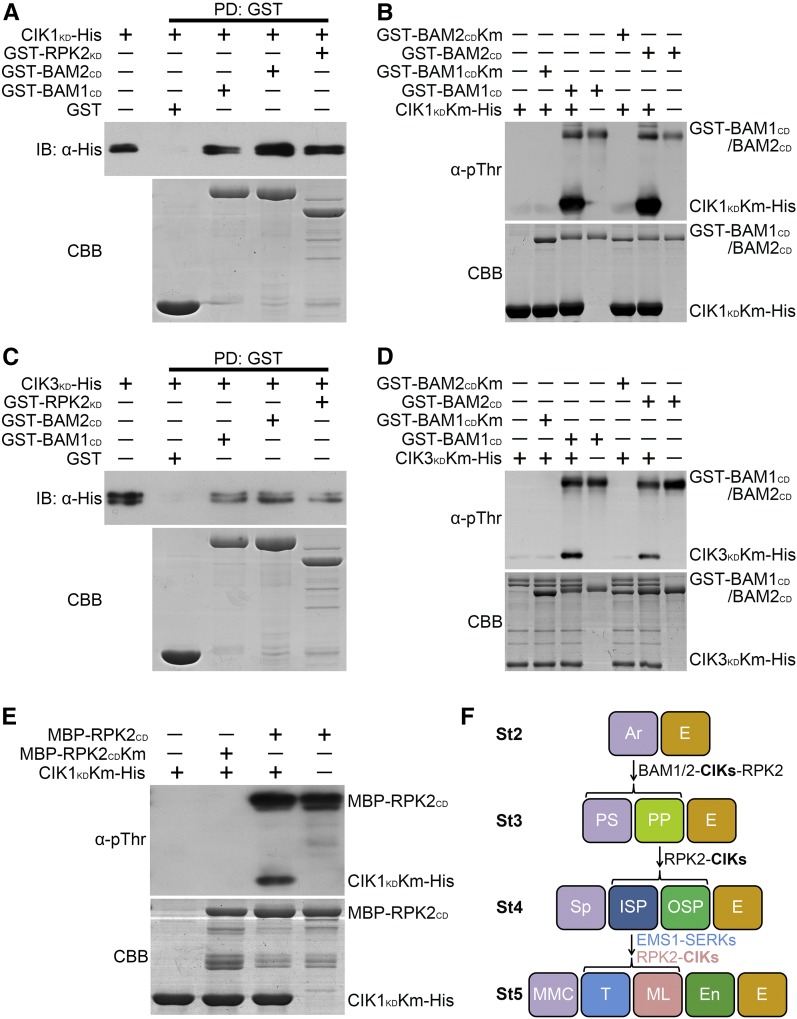
We next inspected whether BAM1/2 and RPK2 can phosphorylate CIKs to transduce signal downstream for controlling early anther development. First, His-tagged kinase domains of CIK1 and CIK3 (CIK1KD-His and CIK3KD-His), GST-fused cytoplasmic or kinase domains of BAM1, BAM2, and RPK2 (GST-BAM1CD, GST-BAM2CD, and GST-RPK2KD), and MBP-tagged cytoplasmic domain of RPK2 (MBP-RPK2CD) were expressed in Escherichia coli cells. Then the purified fusion proteins were used to test whether CIKs can interact with BAM1/2 and RPK2 in vitro. The results showed that CIK1KD-His and CIK3KD-His can be pulled down by GST-BAM1CD, GST-BAM2CD, and GST-RPK2KD, respectively, but not GST itself, indicating that CIKs can associate with BAM1/2 and RPK2 through their kinase domains in vitro (Figures 9A and 9C).
Figure 9.
CIKs Can Be Phosphorylated by BAM1/2 and RPK2.
(A) and (C) CIK1KD and CIK3KD interact with BAM1CD, BAM2CD, and RPK2KD in vitro. CIK1KD-His (A) and CIK3KD-His (C) fusion proteins were incubated with glutathione beads bound with GST, GST-BAM1CD, GST-BAM2CD, or GST-RPK2KD. The pulled-down (PD) proteins were immunoblotted with an α-His antibody (IB, α-His).
(B) and (D) In vitro kinase assays showing phosphorylation of CIK1KDKm (B) or CIK3KDKm (D) by BAM1CD and BAM2CD. BAM1CDKm and BAM2CDKm cannot phosphorylate CIK1KDKm and CIK3KDKm.
(E) In vitro kinase assays showing phosphorylation of CIK1KDKm by RPK2CD. RPK2CDKm cannot phosphorylate CIK1KDKm.
An α-pThr antibody was used to detect the phosphorylation levels. The input proteins were stained with Coomassie Brilliant Blue (CBB). These experiments were repeated three times independently with similar results.
(F) CIKs function as coreceptors of BAM1/2 and RPK2 to regulate early anther development. CIKs physically interact with BAM1/2 and RPK2 to ensure specification and maintenance of the parietal cell fate. In the quadruple cik mutant, the primary parietal cell cannot be properly specified because of the altered division of the archesporial cell, which produces MMC-like cells enclosed only by the epidermis. The inner secondary parietal cell and the outer secondary parietal cell cannot be properly specified in some areas of cik anther wall either, resulting in MMC-like cells surrounded by one or two parietal cell layers and the epidermis. RPK2-CIKs and EMS1-SERKs complexes coordinate to determine the inner secondary parietal cell fate. The inner secondary parietal cell only forms the tapetal cell without the function of CIKs or RPK2, whereas it only forms the middle layer cell in ems1 or serk1/2 mutants. Anther stages (St) 2 to 5 are represented. Ar, archesporial cell; E, epidermis; En, endothecium; ML, middle layer; PP, primary parietal cell; PS, primary sporogenous cell; ISP, inner secondary parietal cell; OSP, outer secondary parietal cell; Sp, sporogenous cell; T, tapetum.
A phosphothreonine antibody (α-pThr) was then used to detect the phosphorylation of CIKs by BAM1/2 and RPK2. The results showed that GST-BAM1CD, GST-BAM2CD, and MBP-RPK2CD exhibit strong autophosphorylation activity in a kinase assay buffer. Mutation of the highly conserved lysine residue in the ATP binding site of these RLKs led to kinase-dead BAM1 (GST-BAM1CDKm), BAM2 (GST-BAM2CDKm), RPK2 (MBP-RPK2CDKm), CIK1 (CIK1KDKm-His), and CIK3 (CIK3KDKm-His) with no detectable kinase activity when detected by the pThr antibody. When GST-BAM1CD, GST-BAM2CD, or MBP-RPK2CD was incubated with CIK1KDKm-His or CIK3KDKm-His in a kinase assay buffer, respectively, strong phosphorylation of CIK1KDKm-His or CIK3KDKm-His was detected, whereas no phosphorylation signals were detected when GST-BAM1CDKm, GST-BAM2CDKm, and MBP-RPK2CDKm were incubated with CIK1KDKm-His or CIK3KDKm-His (Figures 9B, 9D, and 9E). Together, these results indicate that BAM1/2 and RPK2 can phosphorylate CIK1/3 directly in vitro.
DISCUSSION
CIKs Play Redundant and Unequal Roles in Regulating Early Anther Development through a CLV-Independent Pathway
Our previous study revealed that CIKs play essential roles in the CLV signaling pathway for maintaining stem cell homeostasis in Arabidopsis (Hu et al., 2018). We noticed that cik1/2/3 and cik1/2/3/4 mutants show not only expanded shoot apical meristems, but also complete sterility (Figures 1E to 1G). Further phenotypic analyses indicated that cik2/3, cik1/2/3, and cik1/2/3/4 mutants show increasingly severe anther defects similar to bam1/2 mutants early at stage 3 (Figures 1 and 2; Supplemental Figure 2). However, mutations of the ligand gene CLV3 and the receptor gene CLV1 in the CLV signaling pathway do not lead to the anther defects seen in cik mutants (Figure 5). These results suggest that CIKs regulate anther development independently of the CLV signaling pathway, which is also supported by a previous report that CLV1 and BAM1/2 play different roles in meristem and organ development because of their different expression patterns (DeYoung et al., 2006).
All four CIK genes are expressed in early anthers with quite similar patterns (Figure 4). Moreover, complementation of CIKs under a UBQ10 or native CIK promoter can rescue anther defects of cik1/2/3/4 mutants (Supplemental Figure 3). On the other hand, only cik2/3 mutants, but not other double mutants, show anther defects (Figure 1), indicating a principal effect of CIK2 and CIK3 on anther development. Furthermore, cik1/2/3 mutants show much more severe anther defects than do cik2/3/4 mutants, indicating that CIK1 contributes more to anther development than does CIK4 (Figure 1). Together, our data demonstrate that CIKs play essential and redundant roles but contribute differentially to regulating early anther development in Arabidopsis.
It is notable that most cik1/2/3/4 anther lobes show abnormal division of the archesporial cell. While about half of cik1/2/3 lobes show abnormal division of the archesporial cell, the others display defective division and differentiation of the primary parietal cell. However, only 28% of cik2/3 lobes exhibit defective division and differentiation of the inner secondary parietal cell at later development stages. These results may reflect the functional diversity of CIKs. Different mutation combinations make it possible to completely reveal the functions of CIKs in the anther at different developmental stages.
CIKs Are Required for Normal Cell Fate Determination in Early Anther Development
cik mutant anthers produce extra MMC-like cells and fewer parietal cell layers than wild-type plants (Figures 2S to 2X and 3; Supplemental Figures 4S to 4V), which can be caused either by extra divisions of the sporogenous cells or by mis-specified divisions and differentiations of the archesporial cells or parietal cells. Detailed microscopy observations revealed that the first periclinal division of the archesporial cells in triple and quadruple cik mutants is abnormal. The archesporial cell usually divides periclinally and asymmetrically into the outer primary parietal cell and the inner primary sporogenous cell in wild-type anthers (Figure 2G), whereas the archesporial cell in cik mutant anthers either produces two similar daughter cells via a symmetrical periclinal division or divides anticlinally (Figures 2I to 2L). These data indicate that the abnormal division of the archesporial cells results in defective cell fate determination in those cik anthers lacking proper parietal cells early at stage 3 but producing more MMC-like cells later at stage 5.
It is notable that the MMC-like cells in cik1/2/3 and cik1/2/3/4 anthers containing two parietal cell layers can complete meiosis to form the tetrads (Supplemental Figures 4I and 4K). However, the MMC-like cells in anthers lacking all the parietal cell layers cannot undergo meiosis to produce tetrads (Supplemental Figures 4J and 4L). These data demonstrate that CIKs ensure correct division of the archesporial cell and successful specification of the parietal cells, which play essential roles for proper transformation of sporophytic cells to gametophytic cells.
CIKs Function with BAM1/2 to Regulate Archesporial Cell Fate
The importance of BAM1/2 in controlling archesporial cell fate was revealed 12 years ago (Hord et al., 2006). However, the downstream signaling events of BAM1/2 have remained as an unsettled issue until this study was performed. We believe that CIKs function with BAM1/2 in the same signaling pathway to ensure the asymmetrical periclinal division of the archesporial cell based on the following observations (Figure 9F). First, cik1/2/3 and cik1/2/3/4 locules display typical bam1/2 anther phenotypes, with significantly increased MMC-like cells surrounded only by the epidermis (Figures 2V and 2X). Second, genetic analyses demonstrated that CIKs function with BAM1/2 in the same pathway because bam1/2 cik1/2/3/4 mutant anthers exhibit the same parietal-cell-less phenotype as bam1/2 and cik1/2/3/4. The periclinal division of the archesporial cell is altered to an abnormal anticlinal division in these mutant anthers (Figures 6G, 6L, and 6Q). The resulted cells lose their identity, causing the production of excess MMC-like cells (Figures 6J, 6O, and 6T). Additionally, SPL expression is expanded into all MMC-like cells in both bam1/2 and cik1/2/3/4 mutants (Supplemental Figures 5B and 5C). Finally, biochemical analyses revealed that CIKs physically associate with BAM1/2 (Figure 8).
CIKs Cooperate with RPK2 to Control Archesporial Cell Fate and Promote Parietal Cell Specification
Besides lacking the middle layer, our results showed that mutation of RPK2 impairs archesporial cell division, producing defective anthers without three parietal cell layers (Figure 7J), as observed in bam1/2, cik1/2/3, and cik1/2/3/4 mutants (Figures 2V, 2X, and 6J), indicating that RPK2 also participates in determining archesporial cell fate. Moreover, we found that 25% of rpk2-1 cik2/3, over 90% of rpk2-1 cik1/2/3, and all rpk2-1 cik1/2/3/4 locules show the same parietal-cell-less phenotype with excess MMC-like cells enclosed only by the epidermis (Figures 7Q, 7T, and 7X). Semithin sections of anthers at stage 3 indicate that the archesporial cells carry out similar anticlinal division in cik1/2/3, cik1/2/3/4, rpk2-1, rpk2-1 cik2/3, rpk2-1 cik1/2/3, and rpk2-1 cik1/2/3/4 mutants (Figures 2I to 2L, 7H, 7O, 7R, and 7V). Further biochemical analyses showed that CIKs can physically interact with and be phosphorylated by RPK2 (Figures 8 and 9E). Together, these data support that CIKs also function together with RPK2 to regulate archesporial cell division and daughter cell fate (Figure 9F).
Some areas of the anther wall exhibit only one parietal cell layer in rpk2-1, cik1/2/3, cik1/2/3/4, rpk2-1 cik2/3, and rpk2-1 cik1/2/3 mutants (Figures 2U, 2W, 7G, 7N, and 7U), indicating that the archesporial cell may periclinally divide once to generate the primary parietal cell that cannot produce the secondary parietal cell in some mutant anthers. On the other hand, some locules of cik2/3, cik1/2/3, cik1/2/3/4, rpk2-1 cik2/3, and rpk2-1 cik1/2/3 anthers can develop into two parietal cell layers, likely without a middle layer, as in the rpk2-1 mutant (Figures 2T, 2U, 2W, 7F, 7M, and 7U) (Mizuno et al., 2007), which demonstrates that the inner secondary parietal cell cannot divide in these mutants. It was noticed that high-order mutants of rpk2-1 and cik have more locules with earlier anther defects than the background possibly because strong defects at an earlier stage disguise the later phenotype. For example, rpk2-1 cik1/2/3 mutants produce more locules without any parietal cell layers than rpk2-1 and cik1/2/3 mutants. However, none of the rpk2-1 cik mutants exhibit earlier anther defects than cik1/2/3/4 mutants, further indicating that CIKs and RPK2 function in the same genetic pathway to regulate early anther development. Together, our results support the conclusion that CIKs and RPK2 form a complex that regulates archesporial cell division and promotes parietal cell differentiation throughout the process of anther structure development (Figure 9F).
Multiple RLKs Cooperate to Determine Cell Fate during Early Anther Development
Several lines of evidence suggest that RPK2, BAM1/2, and CIKs may form a complex to control archesporial cell division. First, anthers of bam1/2, cik1/2/3/4, rpk2-1, rpk2-1 bam1/2, and rpk2-1 cik1/2/3/4 mutants exhibit similar defective division of the archesporial cell. Second, CIKs can directly interact with BAM1/2 and RPK2, and CIKs can be phosphorylated by BAM1/2 and RPK2. A previous study already showed that BAM1 can interact with RPK2 (Shimizu et al., 2015). Third, similar expression patterns of SPL have been detected in bam1/2, cik1/2/3/4, and rpk2-1 mutant anthers. We noticed that not all rpk2-1 locules display division defects of the archesporial cell and finally lack all parietal cell layers possibly because of the remaining homologous gene RPK1. The rpk1/2 double mutant may show anther phenotype similar to bam1/2 anthers. However, the rpk1/2 mutant is embryo lethal (Nodine et al., 2007), and no rpk1/2 anthers were available for this study. On the other hand, because defective archesporial cell division occurs at a very early stage in bam1/2 anthers, it is impossible to tell whether BAMs also regulate later anther cell divisions and parietal cell differentiations.
Both the tapetum and the middle layer are derived from the inner secondary parietal layer (Albrecht et al., 2005; Walbot and Egger, 2016). It is possible that two RLK complexes, EMS1-SERK1/2 and RPK2-CIKs, coordinate cell division and specification in the inner secondary parietal cells for properly generating the tapetum and the middle layer in wild-type anthers. In ems1 or serk1/2 mutants, the RPK2-CIKs receptor complex is functional in the inner secondary parietal cells, resulting in specification and maintenance of the middle layer cells. By contrast, in rpk2 or cik mutants, the EMS1-SERK complex ensures specification and maintenance of the tapetum (Figure 9F).
CIKs May Be a Group of Novel Coreceptors Functioning in Multiple Signaling Pathways
Phylogenetic analyses indicated that CIKs belong to the LRR II-RLK family that also contains well-characterized coreceptor kinases SERKs (Supplemental Figure 1A). Moreover, CIKs display structures similar to BAK1 (Supplemental Figure 1B). Our genetic results indicated that CIKs function together with BAM1/2 and RPK2 to regulate early anther development. Biochemical data showed that CIKs directly interact with and can be phosphorylated by BAM1/2 and RPK2. Moreover, we have shown that CIKs function as coreceptors of CLV1, CLV2/CORYNE (CRN), and RPK2 to regulate shoot apical meristem homeostasis (Hu et al., 2018). Furthermore, previous studies showed that CIK1 and CIK3 regulate the antivirus response and leaf senescence, respectively, and therefore they were named NSP-INTERACTING KINASE3 (Fontes et al., 2004) and SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE (Xu et al., 2011). Recently, it was reported that CIK2 is required for sensing CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptides in the root (Anne et al., 2018). Together, these results suggest that CIKs may function as a group of novel coreceptors in a variety of RLK-mediated signaling pathways to regulate plant growth, development, and immunity. Phylogenetic analyses identified homologous CIKs in representative angiosperms, suggesting that CIKs may be functionally conserved in these species (Supplemental Figure 6 and Supplemental Data Set 2).
It is remarkable that the RPK2-CIK complex regulates not only CLV3-mediated meristem maintenance (Hu et al., 2018), but also early anther development. However, a previous study showed that RPK2 cannot directly bind CLV3 (Shinohara and Matsubayashi, 2015). Furthermore, clv3 mutants develop plump and fertile anthers, resembling those produced by wild-type plants. These data indicate that the functions of RPK2 and CIKs in regulating early anther development are independent of CLV3 but possibly dependent on another CLE peptide. In addition, the ligand perceived by BAM1/2 in regulating anther development has yet to be identified. Identifying and characterizing the ligands of BAM1/2 and RPK2 during early anther development would provide important insights into cell fate determination.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col) and mutant plants in the Col background were grown in a greenhouse under long-day light conditions (16 h white light per day, ∼100 µmol m−2 s−1 light intensity; Foshan Lighting WT5-14-65/A212/B4, 22 ± 2°C). All T-DNA insertion alleles were obtained from the ABRC, including cik1-1 (at1g60800, SALK_034037), cik1-2 (at1g60800, SALK_059440), cik2-1 (at2g23950, SALK_066568), cik2-2 (at2g23950, SALK_071166), cik3-1 (at4g30520, SALK_111290), cik3-2 (at4g30520, CS106143), cik4-1 (at5g45780, SALK_049669), rpk2-1 (SALK_062412), bam1 (SALK_101542), and bam2 (SAIL_1053_E09). The first set of single knockout mutants was crossed to obtain high-order mutants cik2/3, cik2/3/4, cik1/3/4, cik1/2/4, cik1/2/3, cik1/2/3/4, rpk2-1 cik2/3, rpk2-1 cik1/2/3, rpk2-1 cik1/2/3/4, and bam1/2 cik1/2/3/4. Single knockout mutants cik1-2, cik2-2, and cik3-2 were crossed to create the second cik2/3 (cik2/3*) and cik1/2/3 (cik1/2/3*) mutants that are indicated with an asterisk. bam1/2/+, cik1/+/2/3, cik1/+/2/3*, cik1/+/2/3/4, rpk2-1/+, rpk2-1/+/bam1/+/2, rpk2-1/+/cik2/3, rpk2-1/+/cik1/+/2/3, rpk2-1/+/cik1/+/2/3/4, and bam1/2/+/cik1/+/2/3/4 mutants were self-pollinated to segregate the corresponding homozygous mutants that are sterile. Genotypes of all the mutants were determined via PCR. All primers used in this study are listed in Supplemental Table 1.
Characterization of Mutant Phenotypes
Flowers approximately at flower stages 12 and 13 were used for stereomicroscopy analyses (Leica M165C). Sepals and petals were removed to expose stamens for scanning electron microscopy observation using a Hitachi 4700 scanning electron microscope. Alexander’s staining was employed to examine the viability of pollen grains as previously described (Alexander, 1969). For semithin section analysis of anthers, inflorescences with flower buds were fixed in FAA fixer (50% [v/v] ethanol, 5% [v/v] acetate acid, and 3.7% [v/v] formaldehyde), embedded in Technovit 7100 resin, and then sectioned with a Leica microtome (RM2245). The sections were stained with 0.05% (w/v) toluidine blue O for 2 min and then washed with double-distilled water. After dried on a heating plate at 42°C and mounted in Neutral Balsam Mounting Medium (Sangon Biotech; E675007), the sections were observed using a Leica microscope (DM6000B). All the images were photographed using a DFC420C digital camera.
Phylogenetic and Statistical Analyses
Amino acid sequences of CIKs from six dicotyledons (AT, Arabidopsis thaliana; ATR, Amborella trichopoda; Cpa, Carica papaya; Gorai, Gossypium raimondii; Medtr, Medicago truncatula; Potri, Populus trichocarpa) and three monocotyledons (Bradi, Brachypodium distachyon; LOC_Os, Oryza sativa ssp japonica; Zm, Zea mays) were retrieved from the PLAZA database (https://bioinformatics.psb.ugent.be/plaza/). Arabidopsis LRR II-RLK and BAK1-ASSOCIATING RECEPTOR-LIKE KINASE1 (BARK1) protein sequences were retrieved from TAIR (https://www.arabidopsis.org). Multiple sequence alignment was performed in ClustalW. The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates in MEGA (https://www.megasoftware.net).
Anthers of wild type and cik mutants at flower stage 12 were photographed using a scanning electron microscope (Hitachi S-3700N), and the percentages of each type of anthers were calculated. Anthers of wild type and cik mutants at anther stage 5 were sectioned and photographed for counting anther wall phenotypes and cell numbers of the MMCs, tapetum, middle layer, and endothecium in examined genotypes.
RNA in Situ Hybridization
RNA in situ hybridization experiments were performed as previously described (Hord et al., 2006). Briefly, inflorescences of 5-week-old plants were fixed in 4% (w/v) paraformaldehyde for 6 h. The samples were kept on ice for subsequent fixation and dehydration. After cleared in Histo-Clear (National Diagnostics; SGHS-202), the samples were embedded into Paraplast (Leica) for sectioning. Prepared 8-μm sections on slides were dewaxed and treated with proteinase K (1 μg/mL) for 30 min. Digoxigenin-labeled RNA probes were subsequently hybridized to the slides. To synthesize RNA probes, specific cDNA fragments of target genes were PCR-amplified with primers containing T3 and T7 promoter sequences (Supplemental Table 1). In vitro RNA probe synthesis, hybridization and signal detection were performed according to the manual of the DIG RNA labeling kit (Roche; 11175025910).
Molecular Cloning and Generation of Transgenic Plants
CDS sequences of CIK1, CIK2, CIK3, CIK4, BAM1, BAM2, RPK2, FLS2, and ERL2 and genomic sequences of CIK1, CIK2, and CIK3 were PCR-amplified and cloned into a pDONR/zeo vector with the help of Gateway technology (Invitrogen). The obtained entry clones were then used to transfer target sequences into appropriate destination vectors for plant expression. The binary vector pB35GWF (Gou et al., 2010) was restricted with HindIII and KpnI to replace the CaMV35S promoter sequence with an Arabidopsis UBQ10 promoter or native promoter sequences of CIK1, CIK2, and CIK3, resulting in destination vectors pUBQ10-GWR-FLAG and pCIKs-GWR-FLAG for expressing FLAG-tagged CIK proteins in planta. Another binary vector pH35GWG was used to express BAM1-GFP, BAM2-GFP, RPK2-GFP, and FLS2-GFP fusion proteins in planta. These constructs were used either to complement cik1/2/3/4 mutants or to create double transgenic plants for coimmunoprecipitation analyses.
Protein Interaction Analyses and Phosphorylation Assays
A mbSUS yeast two-hybrid system (Obrdlik et al., 2004) was employed to detect interactions between BAM1/2, RPK2, ERL2, and CIKs according to the manual. Full-length CDS sequences of CIKs, BAM1/2, ERL2, and RPK2 were PCR-amplified and mixed with linearized pMetYCgate vector or pX-NubWTgate vector to transform yeast strain THY.AP4 or THY.AP5 through in vivo DNA recombination. Diploid yeast cells were selected by growing on a synthetic complete medium (SC) containing adenine (Ade) and histidine (His). Interactions were examined by growing diploid yeast cells on a synthetic minimal medium (SD) containing 0, 75, 150, or 400 µM Met. pMetYC, pX-NubWT, and ERL2 were used as negative controls, and KAT1 was used as a positive control.
For BiFC analyses, Agrobacterium tumefaciens strain GV3101 containing binary construct CIK-YC was respectively mixed with clones containing BAM1-YN, BAM2-YN, RPK2-YN, or ERL2-YN and infiltrated into young leaves of Nicotiana benthamiana grown for 3 to 4 weeks. YFP fluorescence signals were detected by a confocal microscope (Leica SP8) 3 d later. BRI1 and BAK1 were used as positive controls. ERL2, YN, and YC were used as negative controls.
The in vivo coimmunoprecipitation analyses were performed as described (Meng et al., 2015). Briefly, rosette leaves of 3-week-old double transgenic plants were ground to fine powder in liquid nitrogen for protein extraction by 0.5 to1 mL of cold extraction buffer (10 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% [v/v] glycerol, 0.5% [v/v] Triton X-100, 1:100 complete protease inhibitor cocktail [Roche; 04693132001]). The cleared protein extracts were then incubated with α-FLAG beads (Sigma-Aldrich; A2220) for 2 h at 4°C with gentle shaking. The immunoprecipitated proteins were analyzed by immunoblotting with an α-GFP antibody (Roche; 11814460001, lot 19958500) and an α-FLAG antibody (Abmart; M20008L, lot 293881).
For pull-down and phosphorylation assays, kinase domains (KD) or cytoplasmic domains (CD) of CIKs, BAM1/2, and RPK2 were PCR-amplified and cloned into the pDEST15 vector with a GST tag (Invitrogen; 11802014), the pMal-cRi vector with a MBP tag (New England Biolabs) or the pET-28 vector with a His tag (Novagen; 69865) to produce fusion proteins of CIK1KD-His, CIK3KD-His, GST-RPK2KD, GST-BAM1CD, GST-BAM2CD, and MBP-RPK2CD. A conserved lysine in the putative ATP binding site of CIK1KD, CIK3KD, BAM1CD, BAM2CD, and RPK2CD was mutated to an aspartic acid for making kinase-dead CIK1KDKm (K329E), CIK3KDKm (K331E), BAM1CDKm (K722E), BAM2CDKm (K718E), and RPK2CDKm (K902E). Proteins expressed in Escherichia coli were purified with the Ni-IDA Sefinose Resin (Sangon Biotech; C600029), the glutathione agarose beads (Sangon Biotech; C600031) or the amylose resin (New England Biolabs; E8021S) according to the manufacturer’s instructions. For GST pull-down, 10 µg of CIK1KD-His or CIK3KD-His fusion proteins was incubated with GST, GST-BAM1CD, GST-BAM2CD, or GST-RPK2KD in 0.5 mL pull-down buffer (10 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% [v/v] glycerol, and 1% [v/v] Triton X-100) for 2 h at 4°C with gentle shaking. The pulled-down proteins were analyzed by immunoblotting with an α-His antibody (Proteintech; 66005-1, lot 10003861). For in vitro phosphorylation assay, 10 µg of each purified fusion protein was incubated in 30 μL kinase buffer (25 mM Tris-HCl, pH 8.0, 10 mM MgCl2, and 300 µM ATP) for 1 h at 25°C with gentle shaking. The proteins were then analyzed by immunoblotting with an α-phosphothreonine antibody (α-pThr; Cell Signaling Technology; 9381, lot 08).
Accession Numbers
Sequence data from this article can be found in TAIR (http://www.arabidopsis.org/) under the following accession numbers: A9, AT5g07230; ATA7, AT4G28395; BAK1, AT4g33430; BAM1, AT5G65700; BAM2, AT3G49670; BARK1, AT3G23750; BRI1, AT4g39400; CIK1, AT1G60800; CIK2, AT2G23950; CIK3, AT4G30520; CIK4, AT5G45780; CLV1, AT1G75820; CLV3, AT2G27250; DYT1, AT4G21330; ERL2, AT5G07180; RPK2, AT3G02130; SDS, AT1G14750; and SPL, AT4G27330.
Supplemental Data
Supplemental Figure 1. Characterization of cik mutants.
Supplemental Figure 2. Defective flowers and anthers of cik mutants.
Supplemental Figure 3. Anther defects of cik1/2/3/4 can be rescued by CIKs.
Supplemental Figure 4. Semithin sections of wild-type and cik anthers.
Supplemental Figure 5. SPL expression is expanded in cik1/2/3/4, bam1/2, and rpk2-1 anthers.
Supplemental Figure 6. CIKs are conserved in angiosperms.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Alignments used to generate the phylogeny presented in Supplemental Figure 1A.
Supplemental Data Set 2. Alignments used to generate the phylogeny presented in Supplemental Figure 6.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the ABRC for providing the T-DNA insertion lines used in this study. We thank Liang Peng, Liping Guan, Yuhong Niu, Haiyan Li, and Yahu Gao (Core Facility for Life Science Research, Lanzhou University) for technical assistance. This work was supported by National Natural Science Foundation of China (31770312, 31471402, 31530005, 31720103902, 31270229, and 31070283), China Postdoctoral Science Foundation (2018T111116), the Ministry of Education (113058A and NCET-12-0249), the 111 Project (B16022), the Fundamental Research Funds for the Central Universities (lzujbky-2017-it01 and lzujbky-2017-kb05), and the Gansu Provincial Science and Technology Department (17ZD2NA015-06 and 17ZD2NA016-5).
AUTHOR CONTRIBUTIONS
X.G. and Y.C. conceived the project, designed all experiments, and analyzed the data. X.G., Z.W., and Y.C. wrote the manuscript. Y.C. performed most of the experiments. C.H. performed the complementation experiments. Y.Z. conducted the in vitro phosphorylation assays. K.C. and F. L. contributed to the high-order mutants. X.L., L.X., and H.S. contributed to the generation and analysis of mutant plants. J.Y., S.H., K.H., and J.L. helped prepare the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Albrecht C., Russinova E., Hecht V., Baaijens E., de Vries S. (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Anne P., Amiguet-Vercher A., Brandt B., Kalmbach L., Geldner N., Hothorn M., Hardtke C.S. (2018). CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 145: dev162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi Y., Liu D., Zhao D., Li W., Wang G., Hu Y., Ma H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21: 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- Canales C., Bhatt A.M., Scott R., Dickinson H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12: 1718–1727. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418. [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Boisson-Dernier A., Ros-Palau R., Vera C.E., Schroeder J.I. (2005). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B.J., Bickle K.L., Schrage K.J., Muskett P., Patel K., Clark S.E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45: 1–16. [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fontes E.P., Santos A.A., Luz D.F., Waclawovsky A.J., Chory J. (2004). The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 18: 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gou X., He K., Yang H., Yuan T., Lin H., Clouse S.D., Li J. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord C.L., Chen C., Deyoung B.J., Clark S.E., Ma H. (2006). The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord C.L., Sun Y.J., Pillitteri L.J., Torii K.U., Wang H., Zhang S., Ma H. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1: 645–658. [DOI] [PubMed] [Google Scholar]
- Hu C., et al. (2018). A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4: 205–211. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhang T., Linstroth L., Tillman Z., Otegui M.S., Owen H.A., Zhao D. (2016). Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet. 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Li Z., Biener G., Xiong E., Malik S., Eaton N., Zhao C.Z., Raicu V., Kong H., Zhao D. (2017). Carbonic anhydrases function in anther Cell differentiation downstream of the receptor-like kinase EMS1. Plant Cell 29: 1335–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Liu X., Owen H.A., Zhao D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. USA 105: 2220–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes J.M., Clark S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851. [DOI] [PubMed] [Google Scholar]
- Ladwig F., Dahlke R.I., Stührwohldt N., Hartmann J., Harter K., Sauter M. (2015). Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 27: 1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang Y., Huang J., Ahsan N., Biener G., Paprocki J., Thelen J.J., Raicu V., Zhao D. (2017). Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol. 173: 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Chen X., Mang H., Liu C., Yu X., Gao X., Torii K.U., He P., Shan L. (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25: 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhou J., Tang J., Li B., de Oliveira M.V.V., Chai J., He P., Shan L. (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Reports 14: 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S., Osakabe Y., Maruyama K., Ito T., Osakabe K., Sato T., Shinozaki K., Yamaguchi-Shinozaki K. (2007). Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50: 751–766. [DOI] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Nodine M.D., Yadegari R., Tax F.E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev. Cell 12: 943–956. [DOI] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y., et al. (2016). RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res. 26: 686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul W., Hodge R., Smartt S., Draper J., Scott R. (1992). The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol. Biol. 19: 611–622. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Bemis S.M., Shpak E.D., Torii K.U. (2007). Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134: 3099–3109. [DOI] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinelli P., Hu Y., Ma H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37: 607–619. [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Santiago J., Henzler C., Hothorn M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892. [DOI] [PubMed] [Google Scholar]
- Schiefthaler U., Balasubramanian S., Sieber P., Chevalier D., Wisman E., Schneitz K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N., Ishida T., Yamada M., Shigenobu S., Tabata R., Kinoshita A., Yamaguchi K., Hasebe M., Mitsumasu K., Sawa S. (2015). BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 208: 1104–1113. [DOI] [PubMed] [Google Scholar]
- Shinohara H., Matsubayashi Y. (2015). Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82: 328–336. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., Lakeman M.B., Torii K.U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293. [DOI] [PubMed] [Google Scholar]
- Song W., et al. (2016). Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26: 674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B., Chai J. (2013). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23: 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U. (2004). Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234: 1–46. [DOI] [PubMed] [Google Scholar]
- Walbot V., Egger R.L. (2016). Pre-meiotic anther development: cell fate specification and differentiation. Annu. Rev. Plant Biol. 67: 365–395. [DOI] [PubMed] [Google Scholar]
- Wang J., Li H., Han Z., Zhang H., Wang T., Lin G., Chang J., Yang W., Chai J. (2015). Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268. [DOI] [PubMed] [Google Scholar]
- Wei B., Zhang J., Pang C., Yu H., Guo D., Jiang H., Ding M., Chen Z., Tao Q., Gu H., Qu L.J., Qin G. (2015). The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res. 25: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Zachgo S. (2008). ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53: 790–801. [DOI] [PubMed] [Google Scholar]
- Xu F., Meng T., Li P., Yu Y., Cui Y., Wang Y., Gong Q., Wang N.N. (2011). A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 157: 2131–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Qian X., Chen M., Fei Q., Meyers B.C., Liang W., Zhang D. (2016). Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol. 171: 2085–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.L., Xie L.F., Mao H.Z., Puah C.S., Yang W.C., Jiang L., Sundaresan V., Ye D. (2003). TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.C., Ye D., Xu J., Sundaresan V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yang L. (2014). Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17: 49–55. [DOI] [PubMed] [Google Scholar]
- Zhang H., Lin X., Han Z., Wang J., Qu L.J., Chai J. (2016). SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol. Plant 9: 1406–1414. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095. [DOI] [PubMed] [Google Scholar]
- Zhao D.Z., Wang G.F., Speal B., Ma H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Zheng Y.F., Zeng T., Sun R., Yang J.Y., Li Y., Ren D.T., Ma H., Xu Z.H., Bai S.N. (2017). Phosphorylation of SPOROCYTELESS/NOZZLE by the MPK3/6 kinase is required for anther development. Plant Physiol. 173: 2265–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]