TOR inhibition leads to an immediate and substantial accumulation of amino acids, which is driven by an increase in nitrogen assimilation and amino acid synthesis.
Abstract
The Target of Rapamycin (TOR) kinase is a central regulator of growth and metabolism in all eukaryotic organisms, including animals, fungi, and plants. Even though the inputs and outputs of TOR signaling are well characterized for animals and fungi, our understanding of the upstream regulators of TOR and its downstream targets is still fragmentary in photosynthetic organisms. In this study, we employed the rapamycin-sensitive green alga Chlamydomonas reinhardtii to elucidate the molecular cause of the amino acid accumulation that occurs after rapamycin-induced inhibition of TOR. Using different growth conditions and stable 13C- and 15N-isotope labeling, we show that this phenotype is accompanied by increased nitrogen (N) uptake, which is induced within minutes of TOR inhibition. Interestingly, this increased N influx is accompanied by increased activities of glutamine synthetase and glutamine oxoglutarate aminotransferase, the main N-assimilating enzymes, which are responsible for the rise in levels of several amino acids, which occurs within a few minutes. Accordingly, we conclude that even though translation initiation and autophagy have been reported to be the main downstream targets of TOR, the upregulation of de novo amino acid synthesis seems to be one of the earliest responses induced after the inhibition of TOR in Chlamydomonas.
INTRODUCTION
The decision to allocate the available nutrient resources to selected metabolic pathways is of major importance for the successful development and survival of an organism (Yuan et al., 2013). Accordingly, several parameters, including nutrient availability and quality, environmental conditions, and energy status, have to be tightly monitored and integrated into all molecular decisions controlling organismal growth (Efeyan et al., 2015). Target of Rapamycin (TOR) is one of the essential regulatory proteins controlling growth and development in all eukaryotes (Soulard et al., 2009; Chantranupong et al., 2015). TOR, a phosphatidylinositol kinase-related protein kinase, controls the activity, localization, and stability of its target proteins by phosphorylation of their serine and/or threonine residues (Hay and Sonenberg, 2004).
The TOR protein was found as the active kinase in at least two major protein complexes (TORC1 and TORC2), which integrate a wide range of extracellular and intracellular signals, including growth hormones, energy, and nutrient availabilities, to regulate cellular growth (Cornu et al., 2013; González and Hall, 2017; Saxton and Sabatini, 2017). Interestingly, only the nutrient- and energy-controlled TORC1 is sensitive to the TOR-specific inhibitor rapamycin, while TORC2 remains largely unaffected by this drug (Loewith et al., 2002; Sarbassov et al., 2006). Only homologs of the heterotrimeric TORC1, namely TOR, Regulatory Associated Protein of TOR (RAPTOR), and lethal with SEC thirteen 8 (LST8), have been detected in genomes of photoautotrophic organisms (Dobrenel et al., 2016; Pérez-Pérez et al., 2017), implying that the TORC2 might not be present in plant or algal species.
Several of the conserved molecular processes that are controlled by TORC1 in mammals and yeast, including regulation of protein translation, autophagy, and the sensing of nutrients (González and Hall, 2017), are partially characterized in photoautotrophic model species (Dobrenel et al., 2016; Pérez-Pérez et al., 2017). Unfortunately, progress in understanding these regulatory circuits in plants and algae has been relatively slow in comparison to other model species like yeast or human cell cultures. The main reason for this delay in plants was the long-standing problem of inefficient pharmacological inhibition of TOR by rapamycin (Menand et al., 2002). This problem was partially overcome by the development of novel active site inhibitors of TOR (Benjamin et al., 2011), which are functional in plants (Montané and Menand, 2013) and algae (Imamura et al., 2016). Still, the use of rapamycin is highly desirable, due to its high specificity and little to no off-target effects (Crespo et al., 2005; Benjamin et al., 2011).
Fortunately, the green alga Chlamydomonas reinhardtii (Harris, 2001) is sensitive to rapamycin (Crespo et al., 2005). Therefore, as a well-established and fully sequenced photoautotrophic model organism, Chlamydomonas can be regarded as an excellent model system to study the TOR pathway (Merchant et al., 2007; Pérez-Pérez et al., 2017).
In recent years, a few studies attempted to elucidate the impact of TOR inhibition on primary (Lee and Fiehn, 2013) and lipid metabolism (Imamura et al., 2015; Couso et al., 2016), using long-term (24 h) treatments under photoheterotrophic growth conditions. A more recent study from our group (Jüppner et al., 2018) provided a detailed metabolic analysis of primary and lipid metabolism using synchronized Chlamydomonas CC-1690 cultures grown under photoautotrophic conditions. The analysis, which monitored the metabolic changes throughout a diel time course, clearly demonstrated that a handful of key metabolic signatures can be observed immediately after TOR inhibition. The main observed outcome of this metabolic remodeling is a shift from anabolic to catabolic metabolite utilization. This shift was characterized by decreased protein synthesis (Díaz-Troya et al., 2011) and induction of autophagy (Pérez-Pérez et al., 2010), which further led to a massive accumulation of the storage compounds starch and triacylglycerols and an increase in free amino acids (Imamura et al., 2015; Jüppner et al., 2018). While the coordinated and significant induction of starch and triacylglycerols occurred ∼1 to 2 h after TOR inhibition, the induction of amino acids could be detected as early as 15 min after rapamycin was administered (Jüppner et al., 2018). Interestingly, while the accumulation of starch/glycogen and triacylglycerols is a typical nitrogen starvation phenotype, previously described in yeast, plants, and algae (Beck and Hall, 1999; Merchant et al., 2012; Caldana et al., 2013), the accumulation of amino acids is unusual and was tentatively explained by the repression of translation and the induction of autophagy (Caldana et al., 2013; González and Hall, 2017).
In this study, we specifically targeted this question by evaluating the immediate amino acid response of Chlamydomonas cultures after TOR inhibition. For this purpose, targeted metabolic profiling, using different growth conditions and minute-scale sampling, in combination with translation and proteasomal inhibitors, was performed to elucidate the molecular mechanism behind the fast accumulation of almost all amino acids after TOR inhibition. The results of these metabolic analyses demonstrated that contrary to previous assumptions the main process leading to the rapid accumulation of almost all amino acids is mainly derived from the significant and immediate increase of de novo amino acid synthesis. We confirmed this hypothesis by performing stable isotope labeling experiments using 15N ammonium in combination with activity assays of central nitrogen assimilating enzymes. These results, in combination with measurements of the total ammonium pools in the cells demonstrate that not only the ammonium concentration within the cells is significantly increasing, but also that the accumulating amino acids are incorporating labeled nitrogen. Taken together, these results imply that nitrogen uptake, assimilation, and amino acid synthesis are coordinately upregulated immediately after the rapamycin-induced inhibition of TOR.
RESULTS
TOR Inhibition in Photoautotrophically Grown Batch Cultures of Chlamydomonas Leads to Elevated Amino Acid Levels within 5 min of TOR Inhibition
In previous studies using plants and algae, it was shown that one of the most reproducible metabolic responses after the inhibition of TORC1 leads to substantially elevated levels of amino acids (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Kleessen et al., 2015; Salem et al., 2017, 2018; Jüppner et al., 2018). Especially in the green model alga Chlamydomonas, where TOR can be pharmacologically inhibited by rapamycin, this response was observed as early as 15 to 30 min after TOR inhibition (Kleessen et al., 2015; Jüppner et al., 2018). This amino acid increase persisted for several hours (Kleessen et al., 2015) or even throughout the entire diel cell cycle (Jüppner et al., 2018) if the cultures were growing under sufficient nutritional supply.
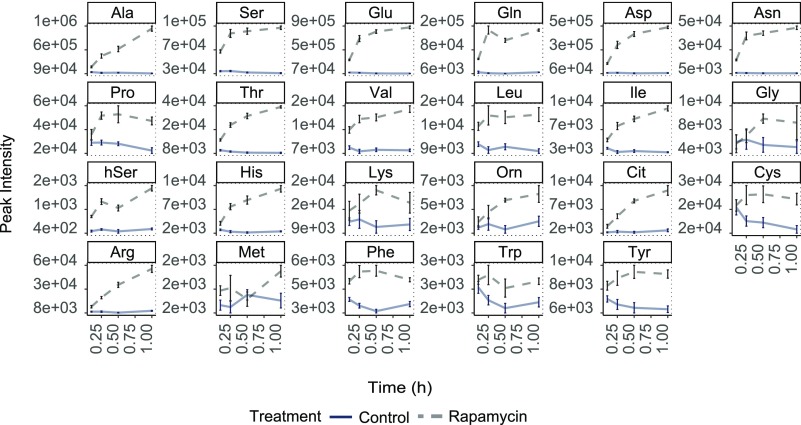
However, to better understand this metabolic response to TOR inhibition, we used a photoautotrophically growing Chlamydomonas batch culture system (Kleessen et al., 2015) and harvested the samples after 0.1, 0.25, 0.5, and 1 h of rapamycin treatment. In accordance with previous studies (Kleessen et al., 2015; Jüppner et al., 2018), we found that under full nutritional supply, all detected amino acids were significantly increased as soon as 0.1 h of TOR inhibition (Figure 1; Supplemental Data Set 1). As can be seen from the plots in Figure 1, the increase of some amino acids continued during the 1-h period, while some amino acids reached their maximum within 0.25 h but still stayed significantly elevated for the remaining time, compared with the control samples.
Figure 1.
Metabolic Changes of Indicated Amino Acid Levels upon TORC1 Inhibition (5 µM Rapamycin) Using Batch Cultures of Chlamydomonas Grown under Continuous Light.
The sampling time is given on the x axes. The y axis indicates absolute peak intensities. Samples are represented as the mean of five replicates ± se. Significance testing was performed by ANOVA2. All presented metabolite levels were significantly (P < 0.05) different between control and rapamycin treatment.
To validate this rapamycin-induced response, we included an additional experiment using an alternative ATP competitive inhibitor of TOR, namely, AZD-8055 (Chresta et al., 2010), which was shown to be functional in Chlamydomonas and plants (Montané and Menand, 2013; Imamura et al., 2016). As shown in Supplemental Figure 1 (Supplemental Data Set 2), the 0.25-h treatment of Chlamydomonas with AZD-8055 lead to an immediate and significant increase of all analyzed amino acids, confirming the phenotype obtained from the rapamycin experiment (Figure 1; Supplemental Data Set 1).
Interestingly, contrary to the fast and consistent amino acid response after rapamycin and AZD-8055 treatment, the response of metabolites from the central carbon metabolism, namely, glycolysis and the TCA cycle, was more variable. For these compounds, as described previously for synchronously growing Chlamydomonas cells (Jüppner et al., 2018), a significant decrease in the rapamycin-treated cultures was observed. Here especially phosphoenolpyruvate, malate, and succinate showed an immediate and consistent decrease after TOR inhibition (Supplemental Figure 2 and Supplemental Data Set 1). Surprisingly, these data also show that fructose-6-phosphate and glucose-6-phosphate, which are products of glycolysis, were differentially regulated after TOR inhibition. While glucose-6-phosphate was significantly elevated in the TOR-inhibited cultures, fructose-6-phosphate was continuously decreased (Supplemental Figure 2 and Supplemental Data Set 1).
Rapamycin Treatment Increases Amino Acid Pool Sizes in Translation- and Proteasome-Inhibited Chlamydomonas Cultures
The timing of the accumulation of amino acids after TOR inhibition indicated that a reduction in translation rate or an increase in nutrient recycling might not be directly responsible for the observed molecular phenotype. While we observed amino acid increases as soon as 5 min after start of the rapamycin treatment (Figure 1), the inhibition of translation or the activation of autophagy was described to occur only hours after TORC1 inhibition (Pérez-Pérez et al., 2010; Díaz-Troya et al., 2011; Crespo, 2012). Moreover, neither of these processes could explain the immediately increased levels of non-proteinogenic amino acids such as ornithine, citrulline, or β-alanine (Figure 1; Supplemental Data Set 1).
To elucidate the impact of translation inhibition and protein degradation on the amino acid phenotype in Chlamydomonas, we conducted a series of experiments utilizing pharmacological inhibition of translation and/or protein degradation in the presence and absence of rapamycin.
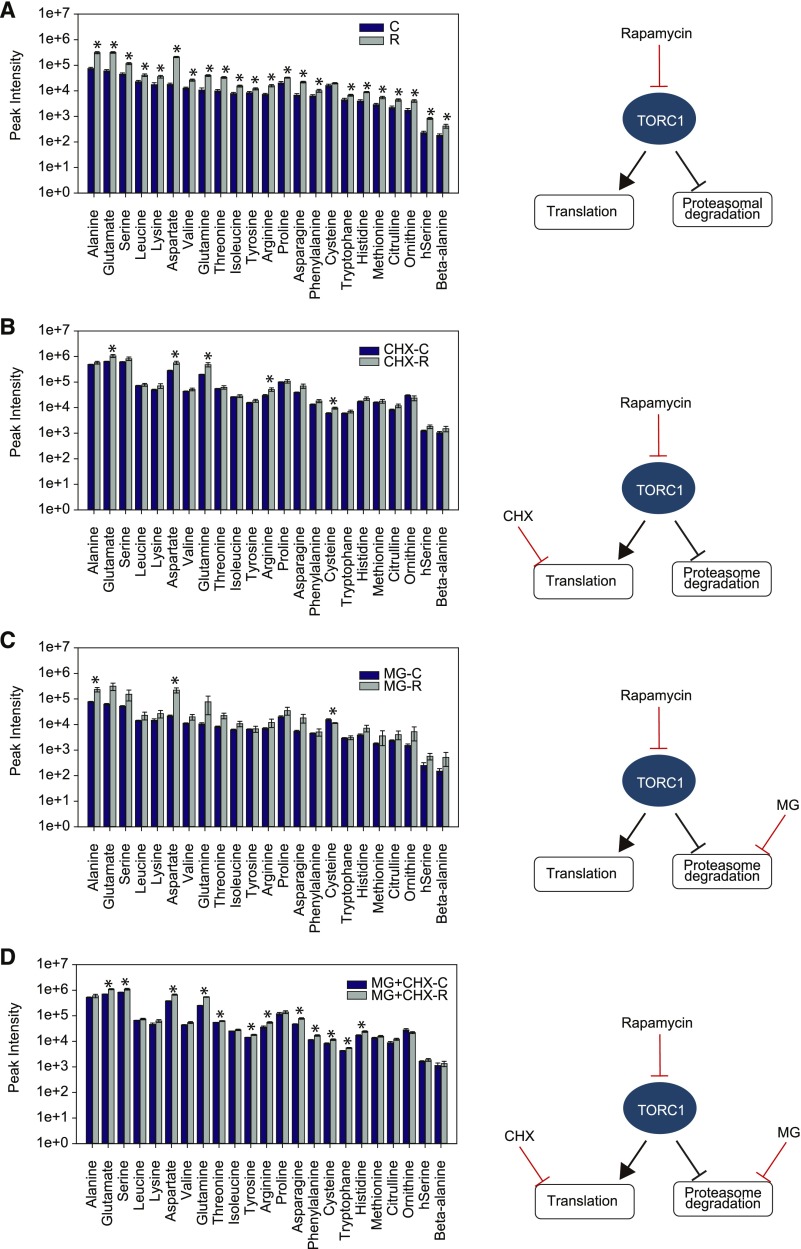
To directly test the combined impact of translation inhibition and TOR inhibition on the amino acid response, Chlamydomonas cultures were treated for 2 h with sublethal doses of the translation elongation inhibitor cycloheximide (CHX; Díaz-Troya et al., 2011) or drug vehicle, before applying rapamycin for an additional 0.25 h. As can be seen from Figure 2A (Supplemental Data Set 3) the addition of rapamycin to the control cultures led to the initially shown (Figure 1) elevated amino acid levels. Notably, the 2-h incubation of the control Chlamydomonas cultures with 35 µM CHX alone led already to an ∼5-fold increase in free amino acid levels (Figure 2B, gray bars), compared with the intensities of the untreated control cultures (no CHX and no rapamycin; Figure 2A, blue bars). This result confirms previous findings from mammalian systems, where translation inhibition led to a substantial increase in amino acid content and was therefore able to reactivate the TORC1 kinase activity (Price et al., 1989; Beugnet et al., 2003). Accordingly, the observed amino acid increase after 2 h of CHX treatment was on average higher than the increase obtained after 0.25 h treatment using rapamycin alone. Nevertheless, the results of the combined CHX and rapamycin treatment demonstrated that the two processes leading to the amino acid increase after CHX and rapamycin treatment must be partially uncoupled, since the combined rapamycin/CHX treatment led to a further increase in the concentrations of almost all detected amino acids (Figure 2B; Supplemental Data Set 3).
Figure 2.
Changes in Amino Acid Levels after 0.25 h of TORC1 Inhibition in Batch Cultures Pretreated with Different Translation (CHX) and/or Proteasome (MG) Inhibitors.
(A) Amino acid response of cultures treated with 5 µM rapamycin.
(B) Cultures pretreated for 2 h with 35 µM CHX, followed by 0.25 h of 5 µM rapamycin treatment.
(C) Cultures pretreated for 2 h with 10 µM MG, followed by 0.25 h of 5 µM rapamycin treatment.
(D) Cultures pretreated for 2 h with a combination of 10 µM MG and 35 µM CHX, followed by 0.25 h of 5 µM rapamycin. Histograms represent the mean of five replicates ± se.
Significant (P < 0.05) changes are marked with an asterisk calculated by Student’s t test.
To investigate whether the observed increase in amino acid content of CHX- and rapamycin-treated cultures was due to TOR-induced protein catabolism, a process that was described in plants and algae after TORC1 inhibition (Pérez-Pérez et al., 2010; Marshall et al., 2015), we analyzed the impact of TOR inhibition on amino acid content in combination with proteasome inhibition. For this purpose, we preincubated Chlamydomonas cultures for 2 h with 10 µM of the proteasome inhibitor MG-132 (MG; Vallentine et al., 2014), before adding either rapamycin or drug vehicle for an additional 0.25 h. Most amino acid concentrations showed an increase after the combined MG/rapamycin treatment, indicating that also under these conditions rapamycin led to a further amino acid accumulation (Figure 2C).
To complete our experimental setup, a combination of CHX and MG was used for a 2-h preincubation, before applying rapamycin. Samples were again harvested 0.25 h after rapamycin or drug vehicle treatment and amino acid content was determined. As can be seen from Figure 2D, most of the free amino acids showed substantially increased content after the combined CHX/MG/rapamycin treatment (Figure 2D), confirming that the elevated amino acid levels were not only driven by translation inhibition or proteasome-driven protein degradation.
Taken together, these results indicate that the immediate accumulation of amino acids after TOR inhibition is most likely not only contributed by translation and protein degradation (autophagy) but also by an additional, anabolic process.
Carbon-Limited Growth Conditions Restrict the Immediate Rapamycin-Induced Amino Acid Response
A possible mechanism contributing to increased amino acid accumulation after TOR inhibition might be the immediate induction of amino acid synthesis. Since de novo amino acid synthesis relies on the availability of carbon and nitrogen substrates, we conducted an experiment to test this hypothesis. Synchronized Chlamydomonas cultures (Jüppner et al., 2017) were grown and treated with rapamycin at the 11th hour of dark phase. By treating the cultures at the end of the dark phase, we ensured that neither photosynthetic carbon was fixed nor larger amounts of storage carbon were available (Jüppner et al., 2018). Samples for this experiment were harvested after 0.1, 0.25, 0.5, and 1 h of rapamycin treatment (Supplemental Figure 3A).
As hypothesized, the metabolic analysis revealed that even though all measured amino acids exhibited an immediate increase after 0.1 h of rapamycin application, most of these initially elevated amino acids did not maintain the increased concentrations, as observed under full nutrition conditions (Figure 1). As can be seen in Figure 3A, some amino acids, including Pro, Leu, Ile, Gly, Phe, Met, Lys, and Tyr, already started to decrease after 0.25 h of rapamycin treatment. Other amino acids, such as Thr, Val, hSer, His, Cit, Cys, Arg, and Trp, started to decline after 0.5 h of TOR inhibition (Figure 3A; Supplemental Data Set 4).
Figure 3.
Metabolic Changes of Indicated Amino Acid Levels upon TORC1 Inhibition.
The sampling time is given on the x axes. The y axis indicates absolute peak intensities. (A) Synchronized Chlamydomonas cultures at the end of the dark phase.
(B) Nitrogen-deprived synchronized Chlamydomonas cultures at the end of the dark phase. Change in the amino acid levels after 1 h of TORC1 inhibition (5 µM rapamycin treatment).
These results support the hypothesis that the immediate and sustained amino acid response to TOR inhibition is strictly depending on sufficient carbon availability.
Nitrogen-Deprived Growth Conditions Lead to Reduced Amino Acid Accumulation after TOR Inhibition
Since amino acid synthesis relies on carbon and other factors for its production, we tested the impact of combined carbon and nitrogen limitation on the amino acid response after TOR inhibition. For this purpose, synchronized Chlamydomonas cultures were harvested by centrifugation after 8 h of the dark phase and residual nitrogen was cleared from the cell pellets by washing them three times in nitrogen-free growth medium. The cell pellets were then resuspended in nitrogen-free medium and incubated for two additional hours in the dark under these conditions to ensure that the cells were adapted to the new nutrient regime and had consumed all residual nitrogen. At the 11th hour of the dark phase, the cultures were treated with rapamycin and samples were harvested after 0.1, 0.25, 0.5, and 1 h (Supplemental Figure 3B).
The results of this experiment showed that even though the majority of the detected amino acids still exhibited a significant initial increase during the first 0.1 h of TOR inhibition, almost all of them showed reduced levels after 0.5 h of treatment (Figure 3B; Supplemental Data Set 5). In accordance with our prediction, the double nutrient depletion experiments (low carbon, N-depletion) led to a very restricted amino acid response after the rapamycin treatment, as compared with carbon depletion alone. As illustrated in Supplemental Figure 4, the log2 fold change in the content of almost all detected amino acids between control and TOR-inhibited cultures was lower in the double starvation experiment compared with carbon starvation alone (gray versus red bars). Contrary to the low carbon experiment, some amino acids from the carbon/nitrogen starvation experiment (Met, hSer, Trp, Tyr, and Phe) not only declined to the levels obtained in the control samples after 1 h of rapamycin treatment, but were reduced even further (Figure 3B; Supplemental Figure 4).
However, 8 of the 20 detected amino acids (Glu, Gln, Asp, Asn, Ala, Ser, Thr, and Pro) did not follow this trend and stayed significantly elevated after 1 h of rapamycin treatment (Figure 3B), indicating that there might be a cellular mechanism which keeps these amino acids high under TOR inhibited conditions.
Taken together, these results illustrate that under combined carbon and nitrogen deprivation most amino acids were depleted to or even below their levels under control conditions, possibly to funnel carbon and nitrogen precursors to specific amino acids, which could be kept at elevated levels under full nitrogen supply.
TOR Inhibition Leads to Increased Ammonium Import and Assimilation
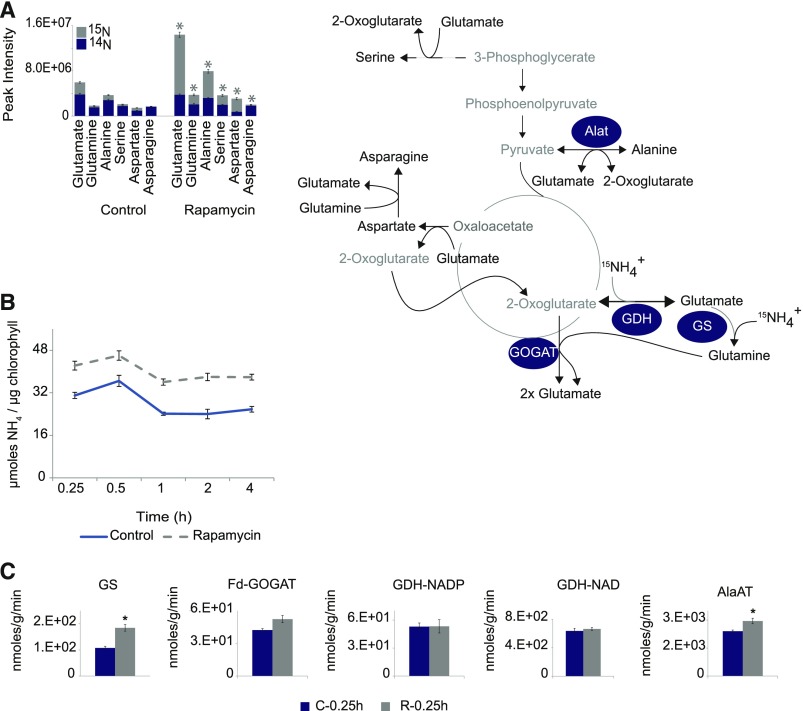
Based on the results from the nutrient depletion experiments, we wondered whether TOR inhibition might lead to changes in nitrogen uptake and/or assimilation. To answer the question whether N assimilation was affected, cells were grown under low carbon conditions (Supplemental Figure 3A), but instead of adding only rapamycin or drug vehicle at the end of the dark phase, cultures were additionally fed with 15NH4+ (Supplemental Figure 3C). To determine the 15N enrichment, samples were harvested 0.25 h after the combined rapamycin/15NH4 treatment and the most rapamycin-responsive amino acids, namely, Glu, Gln, Asp, Asn, Ala, and Ser, were analyzed for their 15N enrichment in comparison to control cells.
As expected from the previous data (Figures 1 and 3), the rapamycin-treated cultures showed a significantly higher incorporation of 15N-labeled nitrogen within 0.25 h of rapamycin treatment (Figure 4A; Supplemental Table 1). Even though all six amino acids showed significantly elevated (P < 0.05, Student’s t test) 15N-enrichment, the flux of labeled nitrogen into Glu was most striking, indicating that N assimilation was indeed stimulated by the repression of TOR.
Figure 4.
TORC1 Inhibition Alters the Nitrogen Flux into Chlamydomonas and Increased the Activities of Enzymes Involved in Nitrogen Assimilation.
(A) 15N enrichment in amino acids after 0.25 h of TORC1 inhibition (5 µM rapamycin). Blue area of the histogram represents the fraction of 14N in the measured amino acids, while the gray area represents the proportion of 15N-labeled amino acids.
(B) Time-resolved analysis of ammonium concentrations (µmol/µg chlorophyll) of Chlamydomonas cells grown as synchronized cultures. Rapamycin (5 µM) or drug vehicle (control) treatment were applied at the 11th hour of the dark phase. Five replicates of each condition and time point were harvested after indicated time on the x axis. Each measurement point of each condition represents the mean of the five replicates, and the error bars represent ±se of the mean.
(C) Changes in enzyme activities were measured after 0.25 h of rapamycin (5 µM) or drug vehicle treatment in Chlamydomonas cultures. Histograms represent the mean of five replicates ± se. Significant differences (P < 0.05) derived from Student’s t test are indicated by an asterisk.
To validate whether the increased flux of nitrogen was accompanied by an increased uptake of ammonium into the cells, we quantified the total ammonium concentration in the Chlamydomonas cultures after rapamycin treatment. As expected from the higher incorporation of 15N label, the absolute amount of ammonium also was significantly increased after 0.25 h of TOR inhibition (Figure 4B; Supplemental Table 2). Similar to the continuously elevated amino acid levels, the cellular ammonium level remained elevated across the monitored period of 4 h of rapamycin treatment (Figure 4B), indicating that the ammonium is not only more efficiently assimilated after rapamycin treatment, but also more ammonium is imported into the cells.
To investigate whether the increased nitrogen uptake is determined by the nitrogen source, we additionally performed a TOR inhibition experiment in the presence of nitrate as the sole nitrogen source. Supplemental Figure 5 shows that also under these conditions several amino acids showed substantial (P < 0.05, Student’s t test) increase after rapamycin treatment (Supplemental Data Set 6). Still, contrary to the ammonium fed cultures (Figure 1), several of the increased amino acids, including alanine and glutamic acid, showed a decline in their concentrations between 30 and 60 min of TOR inhibition. This result implies that ammonium might be the preferred nitrogen source and that its uptake is actively promoted after TORC1 inhibition.
To further elucidate whether the observed increase in 15N labeling of amino acids after TOR inhibition was only driven by the higher ammonium uptake or if the N-flux was accompanied by increased activity of the nitrogen assimilating enzymes (Fernández et al., 2009), we performed enzyme activity measurements of glutamine synthetase (GS), glutamine-2-oxoglutarate-aminotransferase (GOGAT), and GDH in the rapamycin-treated and control cells. While the activities of the two main ammonium-assimilating enzymes (GS and GOGAT) were increased after rapamycin treatment, the activity of GDH was unchanged (Figure 4B; Supplemental Table 2). Importantly, only the activity increase of GS was significant (P < 0.05 Student’s t test), while the activity of GOGAT did not fully reach the significance threshold (P value 0.07; Figure 4C). Beyond the primary assimilation process, we further determined the activity of enzymes responsible for the postassimilation distribution of amino groups in the cell. Accordingly, we selected the alanine aminotransferase (Alat), which can be responsible for the increase in nitrogen flux into alanine (Figure 4A). Similar to the activities of GS and GOGAT, we observed a strong and significant (P < 0.05, Student’s t test) increase in the Alat activity after TOR inhibition (Figure 4C; Supplemental Table 3).
Taken together, these data show that the elevated amino acid levels after TOR inhibition are driven by an increased flux of 15NH4 into the cell in combination with an increased assimilation rate of the relevant enzymes.
TOR Inhibition Drives Carbon Substrates into Amino Acid Synthesis
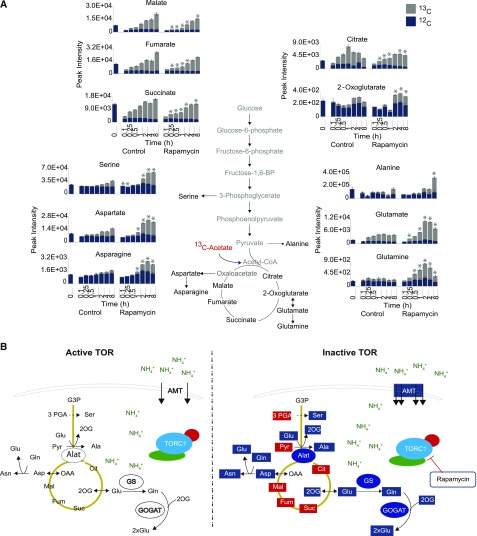
After having shown that the immediate activation of nitrogen assimilation into amino acids is occurring after or parallel to an increased uptake of 15N ammonium, we designed an additional experiment to elucidate whether active nitrogen import and increased amino acid synthesis were accompanied by higher carbon flux into the central carbon metabolism (Johnson and Alric, 2013). For this purpose, synchronized Chlamydomonas cultures were fed with fully 13C-labeled acetate at the end of the dark phase (Supplemental Figure 3D). Under these growth conditions, the 13C-labeled acetate will be used either via the direct assimilation into acetyl CoA by the acetyl-CoA synthetase or by a two-step process, which relies on the sequential action of the enzymes acetate kinase and phosphate acetyltransferase (Spalding, 2009). The produced cytosolic acetyl-CoA can then be directly employed either by the cytosolic glyoxylate cycle or by the mitochondrial TCA cycle (Johnson and Alric, 2013).
The 13C acetate was supplied together with rapamycin and for the 13C flux analysis, samples were harvested at 0.1, 0.25, 0.5, 1, 2, 4, and 8 h in the dark and selected carbon substrates from the TCA cycle next to the six consistently elevated amino acids (Glu, Gln, Asp, Asn, Ala, and Ser) were analyzed. Interestingly, even though the labeled acetate was rapidly enriched in TCA cycle intermediates (citrate, 2-oxoglutarate, succinate, fumarate, and malate), indicating that it was immediately taken up by the cells (Figure 5A; Supplemental Data Set 7), the labeling dynamics of the six amino acids were clearly changed under these experimental conditions. The cells showed an initial increase in the accumulations of Gln between 0.25 and 0.5 h, while the other five amino acids showed significantly enhanced concentrations only after 1 and 2 h of TOR inhibition (Figure 4B; Supplemental Data Set 7). Similar to the enhanced 13C incorporation into amino acids, flux into 2-oxoglutarate, the direct carbon substrate for the synthesis of Gln and Glu, was increased after rapamycin treatment. Contrary to these, the 13C flux into all other TCA cycle intermediates was significantly (P value < 0.05, Student’s t test) decreased in TOR-inhibited cells, especially at the early time points (Figure 5A; Supplemental Data Set 7).
Figure 5.
13C Enrichment in Organic and Amino Acids.
(A) The gray section of the histogram represents the fraction of 13C intensities, while the blue section represents the proportion of 12C in the indicated metabolites. Total height of the histograms shows the absolute peak intensity of the metabolite. Histograms are displayed as the mean of five replicates ± se. Asterisks indicate significant changes (Student’s t test, P < 0.05) of the enrichment of 13C in control or rapamycin-treated samples.
(B) Graphical model illustrating the proposed mechanism of increased de novo amino acid synthesis due to upregulated ammonium assimilation after TORC1 inhibition in Chlamydomonas cells. Metabolites and enzymes highlighted in red indicate reduced levels/activities, whereas the ones highlighted in blue indicate increased levels/activities upon inactivation of TORC1.
These data confirm that even though the de novo synthesis of amino acids was significantly increased by rapamycin treatment, this process was driven by an increased draining of carbon substrates out of the central metabolism but not by increased carbon flux into the TCA cycle.
DISCUSSION
In the more than 25 years since the elucidation of the TOR kinase (Heitman et al., 1991), several mechanisms explaining the upstream and downstream targets of TOR have been described in mammals and yeast (González and Hall, 2017; Saxton and Sabatini, 2017). However, even though the understanding in these well-studied heterotrophic systems is getting more detailed and mechanistic, the understanding of the function of TOR in photoautotrophic organisms is still quite fragmentary.
The focus of this study was to improve our understanding of the frequently observed amino acid accumulation phenotype in plants and algae after genetic or pharmacological inhibition of TOR (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Kleessen et al., 2015; Salem et al., 2017, 2018; Jüppner et al., 2018). This metabolic phenotype was commonly explained by the fact that TORC1 is a positive regulator of protein translation (Ma and Blenis, 2009) and a negative regulator of autophagy, thereby controlling nutrient recycling through catabolic processes directed against cellular structures (Russell et al., 2014). Accordingly, repression of protein translation (Deprost et al., 2007; Sormani et al., 2007; Díaz-Troya et al., 2011; Ahn et al., 2015), as well as the activation of autophagy (Liu and Bassham, 2010; Pérez-Pérez et al., 2010; Soto-Burgos and Bassham, 2017; Salem et al., 2018), were described in plants and algae as a result of reduced TOR activity. Even though these processes are controlled by TOR in the green lineage, it was shown in previous studies that the kinetics of TOR repression-induced amino acid accumulation was not consistent with the kinetics observed for the inhibition of protein synthesis or the induction of autophagy (Kleessen et al., 2015; Jüppner et al., 2018). While the amino acid levels increase as soon as 5 min after TOR inhibition (Figure 1), changes in the incorporation of 14C labeled amino acids into newly synthesized proteins were only observed after 4 to 8 h in TOR-inhibited Chlamydomonas cultures (Díaz-Troya et al., 2011). Similarly, induction of autophagy was only detectable 16 h after rapamycin was administered (Pérez-Pérez et al., 2010).
To elucidate this discrepancy, we performed a number of experiments where TOR inhibition through the application of rapamycin was combined with pharmacological inhibition of translation and/or proteasomal degradation (Figure 2). These data confirmed that even though translation inhibition can significantly increase amounts of free amino acids, the combined administration of CHX, MG, and rapamycin still leads to a significant increase of amino acid levels (Figure 2D), indicating that the source of this immediate molecular response should be derived from a mechanism other than translation inhibition or protein degradation.
The best assumption to explain the commonly observed amino acid phenotype was therefore that TOR inhibition leads to an increase in amino acid synthesis. Since our initial experiments were performed under full nutrient supply (Figure 1), we speculated that the observed increased metabolic levels of amino acids were only possible when sufficient carbon and nitrogen substrates were available. This assumption was additionally supported by the observation that several metabolites from central carbon metabolism were significantly depleted in TOR-inhibited cultures (Supplemental Figure 2).
In a number of nutrient-depletion experiments, we were able to confirm this hypothesis by showing that the observed amino acid increase, after the inhibition of TOR, indeed relied on the availability of sufficient carbon (Figure 3A) and nitrogen (Figure 3B) substrates. The deprivation of these essential nutrients therefore led to an altered molecular phenotype, where the elevated levels were still rapidly induced after the inhibition of TOR, but this increase could not be maintained over a period of more than 1 h (Figure 3B). These results allowed us to draw two essential conclusions: On the one hand, TOR inhibition immediately diverts carbon and nitrogen nutrients into amino acids, while on the other hand, the maintenance of this response cannot be matched by intracellular stores and relies on sufficient extracellular supply. A relevant question that results from these observations was where the carbon and nitrogen for the immediate amino acid burst, even under nutrient depletion conditions, was derived from, especially since translation and protein degradation might not be the direct sources (Figure 2). Accordingly, either a temporal redirection of cellular stocks is taking place, where less essential metabolites are sacrificed in order to synthesize the relevant amino acids or an additional carbon/nitrogen store is tapped to provide nitrogen and carbon for the increased synthesis of amino acids. One possible nutrient source could from the degradation of nucleic acids. A previous study on metabolic responses to TOR inhibition during a 24-h cell cycle indicated increased levels of intermediates from RNA degradation, namely, guanine, uracil, hypoxanthine, and β-alanine in Chlamydomonas (Jüppner et al., 2018), which is also in agreement with data from Arabidopsis, where the inhibition of TOR led to a substantial decrease in rRNA levels (Ren et al., 2011). This hypothesis requires further validation, especially concerning the kinetic parameters of this response.
Nevertheless, to more precisely answer the question of how TOR manages to trigger the increased synthesis of amino acids, stable isotope labeling experiments were performed using either 15N-labeled ammonium or 13C-labeled acetate. Using these two labeling regimes, we show that the flux of 15N from the labeled ammonium into amino acids was significantly increased after TOR inhibition (Figure 4A). Contrarily, even though carbon was drained from the central carbon metabolism, the flux of 13C into the amino acids was significantly decreased after rapamycin treatment (Figure 5A). This result is in full agreement with the significant depletion of these compounds after rapamycin treatment (Supplemental Figure 1) and indicated that TOR might specifically affect nitrogen flux into amino acid synthesis, without stimulating the carbon flux.
Beyond the increased flux of 15N into amino acids in TOR-inhibited cells, we also observed increased ammonium accumulation in the rapamycin-treated cultures (Figure 4B). This result further indicated that not only nitrogen assimilation is controlled by TOR but possibly also nitrogen uptake. This hypothesis is in full agreement with data obtained from yeast cells treated with rapamycin. In a number of experiments, it was shown that the nitrogen permease reactivator 1 (Npr1) directly upregulates the activity of the ammonium transporter Mep2 by phosphorylating and inactivating the cytosolic autoinhibitory domain of the transmembrane protein if TOR is pharmacologically repressed (Boeckstaens et al., 2014). Additionally, it was shown in yeast that TOR inhibition leads to the activation of two other ammonium transporters, namely, Mep1 and Mep3, which are negatively controlled by Par32 (protein phosphorylated after rapamycin 32). Both Npr1-centered mechanisms provide excellent blueprints for possible regulatory mechanism controlling nitrogen uptake after TOR inhibition in Chlamydomonas (Figure 5). Unfortunately, even though several Mep-type ammonium transporters can be found in Chlamydomonas (Sonnhammer and Östlund, 2015), no direct homolog of the TOR effector kinase Npr1 or the Par32 protein could as yet be identified in the green lineage.
Nevertheless, a direct, posttranslational connection between TOR and the ammonium transporter is postulated, since the response is too fast to be mediated via transcriptional/translational regulation. Interestingly, transcriptional regulation might still be an additional response layer, since rapamycin-treated Chlamydomonas but also Cyanidioschyzon merolae (a red alga) induce most of the genes encoding enzymes responsible for efficient nitrogen assimilation (nitrate reductase, nitrite reductase, and glutamine synthetase) within 24 h of TOR inkibition (Imamura et al., 2015). Rapamycin induced genes also include those encoding an ammonium and a nitrogen transporter, which were in addition induced under nitrogen-deplete conditions (Imamura et al., 2009), connecting the TOR repression to a nitrogen starvation phenotype in red and green algae.
To further elucidate whether the increased flux of nitrogen into the synthesis of amino acids is driven by the increased ammonium uptake or if the increased ammonium uptake is driven by an increased demand triggered by increased amino acid synthesis, we additionally analyzed the activities of central nitrogen assimilating enzymes such as GS/GOGAT and GDH (Figure 4C). The activity of these enzymes was always substantially elevated, even if the differences were not always statistically different. In addition to the increased nitrogen assimilation capacity of the Chlamydomonas cultures after rapamycin treatment, alanine aminotransferase activity was also significantly increased in the TOR-repressed cells (Figure 4C), indicating that the rapamycin treatment not only leads to an increased uptake of ammonium (Figure 4B) and to an increased nitrogen assimilation rate, but also to an increased distribution of the newly assimilated nitrogen in the amino acid biosynthetic pathway (Figure 4C).
Based on these data, it would be interesting to understand not only how but also why this immediate amino acid response is induced after TOR inhibition. While the answer to this question is obvious in the heterotrophic mammalian and yeast systems, it is not as clear in the photoautotrophic algae and plants. In mammals, sophisticated amino acid sensing and signaling pathways have been described for the activation or reactivation of TOR (González and Hall, 2017). At the center of these mechanisms, a number of GTPases, their guanine nucleotide exchange factors, and their GTPase-activating proteins were described (Bar-Peled and Sabatini, 2014). A family of small GTPases is responsible to tether the TOR complex to the lysosomal surface, where its activator Ras homolog enriched in brain (RHEB) is located (Menon et al., 2014). The prediction of such a positive feedback mechanism that could reactivate the inhibited TOR would be quite tempting in plants, but unfortunately, all of the essential components of this pathway are absent in the green lineage (Roustan et al., 2016). Alternatively, a more ancient amino acid sensing mechanism could be present in plants and algae, which is centered on the GCN2 (general control non-derepressible2) protein kinase (Hinnebusch, 2005). GCN2 senses amino acid concentrations based on the loading status of tRNAs. Accordingly, uncharged tRNAs activate the GCN2 kinase, which activates the translation initiations factor 2α (eIF2α), thus reducing general translation rates (Dever et al., 1992) and inducing the translation of transcripts from amino acid synthesis genes in a GCN4-dependent manner (Hinnebusch, 2005). Homologs of GCN2 and eIF2α are present in photosynthetic organisms, and in Arabidopsis thaliana, GCN2 binds tRNAs and phosphorylates eIF2α (Li et al., 2013). However, the GCN4 transcription factor downstream of the amino acid dependent GCN2 signaling cascade is still missing.
Taken together, our data elucidate a TOR-dependent, molecular phenotype in the green lineage, where the inhibition of TOR leads to the immediate and massive increase of ammonium import into the cell, followed by its assimilation into most amino acids (Figure 5B). Even though the connection between TOR activity and nitrogen uptake and assimilation seems to be similar between distantly related model species, the mechanistic implementation appears quite different. While yeast directly regulates the ammonium transporter in two Npr1-dependent pathways (Boeckstaens et al., 2014, 2015), mammalian cell cultures have a more indirect Npr1-dependent nitrogen-recycling mechanism, which relies on endocytosis and ubiquitin-based protein degradation (MacGurn et al., 2011). The direct mechanistic elucidation of this signaling cascade starting from TOR inhibition to ammonium uptake and assimilation followed by amino acid sensing in a tractable green model species like Chlamydomonas will therefore allow us to close the gap between yeast, mammals, and photosynthetic organisms, providing further dimensions to the field of growth signaling in various eukaryotes.
METHODS
Chlamydomonas Strains and Culture Conditions
Chlamydomonas reinhardtii wild-type strain CC1690 (mt+; Sagr 21 gr) obtained from the Chlamydomonas Resource Center (University of Minnesota) was used for all experiments.
Batch culture experiments were established by inoculating preculture to fresh HSM medium grown under continuous light 200 µmol m−2 s−1 (Lumilux Cool White, 30W/840; Osram), and 2 to 3% CO2 at room temperature. To assess the response of Chlamydomonas to TOR inhibition in medium containing nitrate, batch culture experiments were established as described above but the preculture was inoculated to modified HSM medium in which ammonium chloride was substituted with the same concentration of sodium nitrate as nitrogen source.
To execute TOR inhibition in absence of light, synchronized cultures of Chlamydomonas were grown, as described previously (Jüppner et al., 2017). Treatment was applied before the end of 3rd cycle (11th hour in dark). To attain TORC1 inhibition both in absence of light and nitrogen, parent cultures were harvested at the end of 3rd cycle (8th hour of the dark phase) and washed three times using Tris-HCl (pH 7) buffer to remove residual traces of ammonium. After washing, the cell pellets were gently resuspended in nitrogen-deprived medium and this culture was transferred to the fermenters already set to the same growth conditions as parent cultures (i.e., 34°C, 2–3% CO2, stirring) and left for 2 h to adapt to nitrogen-deprived condition. The treatment was applied at 11th hour of dark phase. The cultures were treated with DMSO (control) or rapamycin to the final concentration of 5 µM followed by sampling at 0.1, 0.25, 0.5, and 1 h in five replicates for all three experiments. A graphical overview of the experimental setup is displayed in Supplemental Figures 2A to 2D.
TORC1, Translation, and Proteasome Inhibition Experiments
Batch cultures of Chlamydomonas were grown under continuous light as described above. To inhibit translation activity the cultures were treated with 35 µM CHX (Sigma-Aldrich) in water (Díaz-Troya et al., 2011). The proteasome activity was inhibited by the addition of 10 µM MG (Calibochem) in DMSO (Vallentine et al., 2014). TORC1 was inhibited by the addition of 5 µM rapamycin in DMSO (Jüppner et al., 2017), while 1 µM of the ATP competitive TORC1 inhibitor AZD8055 was used as described by Imamura et al. (2016). All treatments were controlled by the addition of equal volumes of drug vehicle alone.
15N Ammonium Labeling Experiment and Amino Acid Analysis Using Hydrophilic Liquid Chromatography MS
Synchronized cultures of Chlamydomonas were grown as described in detail previously (Jüppner et al., 2018). Before the end of 3rd cycle, the parent cultures were sampled as unlabeled reference for 0 h; thereafter, they were fed with fresh medium containing 15N-labeled ammonium chloride (Sigma-Aldrich), together with the treatment with DMSO or rapamycin to a final concentration of 5 µM. Samples were harvested after 0.25 h in five replicates for each condition.
Amino acids from 15N labeling experiments were analyzed from the polar fraction of the previously published methyl tert-butyl ether extraction method (Jüppner et al., 2017). Samples extracted from 10 million cells and reference amino acid standards (1 µg/mL) were resuspended in 100 μL 80% acetonitrile in water (Biosolve). For the analysis, 2 μL of each sample or standard was injected onto a BEH amide column (100 × 2.1 mm using 1.8-µm particles; Waters) using an Acquity UPLC system (Waters). The UPLC separation was performed using buffer A (0.1% formic acid in acetonitrile) and buffer B (0.1% formic acid in water). The gradient started with 95% buffer A, which was linearly decreased to 50% A within the first 6 min. Within the next minute, the concentration of A was further decreased linearly to 30%, which was held for 0.1 min. The column was then reequilibrated for 5 min with 95% A, before the next sample was injected.
The samples from the UPLC were measured using an Exactive high-resolution mass spectrometer, which was operated as described previously (Giavalisco et al., 2011), monitoring a mass range between m/z 50 and 500. To annotate the measured amino acids, mass spectra from samples were compared with spectra obtained from reference amino acids. For this purpose, accurate m/z (mass precision below 5 ppm) and retention time values (within 0.15 min) of all amino acids were matched.
For the analysis of isotope enrichment, the ratios of monoisotopic (only 14N-containing peaks) and the 15N-labeled mass peaks were determined using Xcalibur (version 2.2; Thermo Fisher Scientific). The extracted m/z and RT pair for monoisotopic peaks and the isotopologs of each amino acid were transferred into a ToxID template file, according to the vendor’s description, which was then used for the automated peak extraction using ToxID (version 1.2; Thermo Fisher Scientific). The ToxID extraction was performed using the accurate mass scans mode, setting the retention time tolerance to 0.15 min and the m/z tolerance to 5 ppm. The resulting output was normalized by the internal standard ampicillin (Giavalisco et al., 2011), which was added to the initial extraction mixture (Jüppner et al., 2017). Relative isotopolog ratios and 15N enrichment were calculated according to the absolute intensities of the detected peaks.
13C Acetate Labeling Experiment and Data Analysis
The 13C acetate labeling was performed on synchronized cultures of Chlamydomonas CC-1690 (Jüppner et al., 2017). To ensure that the cells were depleted of intracellular carbon, the experiment was conducted during the extended dark phase when starch reserves have been shown at their minimal level (Jüppner et al., 2017). In brief, the parent culture was harvested at the end of 3rd cycle for unlabeled reference samples, described as 0-h time point samples. Thereafter, fully labeled 13C acetate stock solution was added to both fermenters to a final concentration of 10 mM, together with the treatment with DMSO or rapamycin (final concentration of 5 µM), five replicates for each condition were harvested after 0.1, 0.25, 0.5, 1, 2, 4, and 8 h.
The 13C enrichment analysis was performed as described by Huege et al. (2014) using the Corrector isotopolog calculator software.
Gas Chromatography-Based Metabolite Profiling
Primary polar metabolites, including the amino and organic acids, were extracted and analyzed by gas chromatography-mass spectrometry, as described in detail previously (Jüppner et al., 2017).
Enzyme Activity Measurements
For each performed enzyme assay, an aliquot of ∼10 million cells was harvested after 0.25 h of rapamycin (5 µM) or control (drug vehicle) treatment from synchronized cultures of Chlamydomonas during (late) light phase. Enzyme activity measurements were performed in triplicates for GS, Alat, GDH-NAD, GDH-NADP, and GOGAT as described by Gibon et al. (2004). The measured enzymatic activities were normalized to the total chlorophyll content of the cultures.
Fluorometric Measurement of Cellular Ammonium Content
Synchronized cultures of Chlamydomonas were grown as described previously (Jüppner et al., 2017). The treatment was applied at 8th hour of dark phase. The cultures were treated with DMSO (control) or rapamycin to the final concentration of 5 µM followed by sampling at 0.25, 0.5, 1, 2, and 4 h using five replicates. The harvested cultures (∼40 mio cells) were washed twice using nitrogen-free medium (HSM medium without nitrogen source) to remove any residual traces of ammonium. After the second wash, the supernatant was discarded and pellets were snap frozen in liquid nitrogen and stored at −80C until further analysis. The extraction of ammonium was performed by resuspending the pellets in 100 μL of UPLC water, followed by exposure to two freeze thaw cycles. The lysate was centrifuged for 45 min at 20,000g at 4°C and the supernatant was aliquoted for ammonium assay. The ammonium content was measured using ammonium assay kit (catalog number MAK310; Sigma-Aldrich) as per the vendor’s instructions.
The remaining pellet was resuspended in 90% methanol for chlorophyll extraction. The chlorophyll content was used to normalize the measured ammonium content in control and rapamycin-treated cultures.
Normalization of Metabolite Data
To compare the concentration of different compounds under treatment conditions, it is necessary to normalize the data to the same cell mass. Chlorophyll content, which was shown previously to be proportional to cell number, even under TOR-inhibited conditions, was used to normalize the metabolite data as a proxy for cell number and as a measure of extraction efficiency (Jüppner et al., 2017).
Statistical Data Analysis
To assess the statistically significant changes in metabolite data in response to treatment and time, ANOVA2 module of MetaboAnalyst 3.0 was employed (Xia et al., 2012). The module allows two-factor univariate ANOVA, where one factor is time and the other factor is treatment. The P values were adjusted at a cutoff of <0.05 after false discovery rate correction.
For pairwise comparison of control and treated cultures, Student’s t test statistics were applied using Microsoft Excel. The applied tests and significance levels are indicated in each figure legend.
Supplemental Data
Supplemental Figure 1. Changes in amino acid levels after 0.25 h treatment of the ATP completive inhibitor AZD-8055.
Supplemental Figure 2. Metabolic changes after 1 h of 5 µM rapamycin treatment of Chlamydomonas cultures grown in the light.
Supplemental Figure 3. Graphical representation of the experimental setup used for the different TORC1 inhibition experiments.
Supplemental Figure 4. Change in the amino acid levels of batch grown cultures under different nutritional regimes after 1 h of TOR inhibition (5 µM rapamycin treatment).
Supplemental Figure 5. Batch cultures grown on nitrate as the sole nitrogen source.
Supplemental Table 1. 15N enrichment analysis of synchronized data harvested at the end of the light phase.
Supplemental Table 2. Ammonium concentration measurement of synchronized Chlamydomonas cultures at the end of the dark phase.
Supplemental Table 3. Enzyme activity measurements of synchronized cultures.
Supplemental Data Set 1. Metabolic data of batch cultures under full nutrition inhibited with rapamycin.
Supplemental Data Set 2. Metabolic data of batch cultures inhibited with AZD-8055.
Supplemental Data Set 3. Metabolic data of cultures inhibited with cycloheximide (translation inhibitor CHX), MG-132 (proteasome inhibitor MG), and/or rapamycin.
Supplemental Data Set 4. Metabolic data of synchronized cultures harvested at the end of the light phase inhibited with rapamycin.
Supplemental Data Set 5. Metabolic data of synchronized cultures harvested at the end of the light phase and starved on nitrogen.
Supplemental Data Set 6. Metabolic data of synchronized cultures grown on nitrate as the only nitrogen source.
Supplemental Data Set 7. 13C enrichment analysis of synchronized data harvested at the end of the light phase.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Andrea Leisse for excellent technical assistance and support with the Chlamydomonas cultures. We thank Änne Michaelis and Gudrun Wolter for excellent help with the gas chromatography- and liquid chromatography-mass spectrometry measurements. We also thank Leonardo Perez de Souza and Isabel Orf for helpful discussions regarding enrichment experiments. We thank Andrew Wiszniewski for proofreading the manuscript and for many supportive scientific discussions. Furthermore, we thank the Max Planck Society for continuous support and funding that allowed us to conduct these larger scale experiments.
AUTHOR CONTRIBUTIONS
U.M., J.J., and P.G. designed the experiment. J.A. performed and optimized the enzyme activity measurements and analyzed the data together with D.K.H. U.M. performed all other experiments. U.M. and P.G analyzed the data and wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Ahn C.S., Ahn H.K., Pai H.S. (2015). Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. J. Exp. Bot. 66: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Sabatini D.M. (2014). Regulation of mTORC1 by amino acids. Trends Cell Biol. 24: 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Hall M.N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Benjamin D., Colombi M., Moroni C., Hall M.N. (2011). Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10: 868–880. [DOI] [PubMed] [Google Scholar]
- Beugnet A., Tee A.R., Taylor P.M., Proud C.G. (2003). Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 372: 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckstaens M., Llinares E., Van Vooren P., Marini A.M. (2014). The TORC1 effector kinase Npr1 fine tunes the inherent activity of the Mep2 ammonium transport protein. Nat. Commun. 5: 3101. [DOI] [PubMed] [Google Scholar]
- Boeckstaens M., Merhi A., Llinares E., Van Vooren P., Springael J.Y., Wintjens R., Marini A.M. (2015). Identification of a novel regulatory mechanism of nutrient transport controlled by TORC1-Npr1-Amu1/Par32. PLoS Genet. 11: e1005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C., Li Y., Leisse A., Zhang Y., Bartholomaeus L., Fernie A.R., Willmitzer L., Giavalisco P. (2013). Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 73: 897–909. [DOI] [PubMed] [Google Scholar]
- Chantranupong L., Wolfson R.L., Sabatini D.M. (2015). Nutrient-sensing mechanisms across evolution. Cell 161: 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chresta C.M., et al. (2010). AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70: 288–298. [DOI] [PubMed] [Google Scholar]
- Cornu M., Albert V., Hall M.N. (2013). mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 23: 53–62. [DOI] [PubMed] [Google Scholar]
- Couso I., Evans B.S., Li J., Liu Y., Ma F., Diamond S., Allen D.K., Umen J.G. (2016). Synergism between inositol polyphosphates and TOR kinase signaling in nutrient sensing, growth control, and lipid metabolism in Chlamydomonas. Plant Cell 28: 2026–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J.L. (2012). BiP links TOR signaling to ER stress in Chlamydomonas. Plant Signal. Behav. 7: 273–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J.L., Díaz-Troya S., Florencio F.J. (2005). Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 139: 1736–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolaï M., Bedu M., Robaglia C., Meyer C. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8: 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E., Feng L., Wek R.C., Cigan A.M., Donahue T.F., Hinnebusch A.G. (1992). Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596. [DOI] [PubMed] [Google Scholar]
- Díaz-Troya S., Pérez-Pérez M.E., Pérez-Martín M., Moes S., Jeno P., Florencio F.J., Crespo J.L. (2011). Inhibition of protein synthesis by TOR inactivation revealed a conserved regulatory mechanism of the BiP chaperone in Chlamydomonas. Plant Physiol. 157: 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T., Caldana C., Hanson J., Robaglia C., Vincentz M., Veit B., Meyer C. (2016). TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 67: 261–285. [DOI] [PubMed] [Google Scholar]
- Efeyan A., Comb W.C., Sabatini D.M. (2015). Nutrient-sensing mechanisms and pathways. Nature 517: 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E., Llamas Á., Galván A. (2009). Nitrogen Assimilation and its Regulation. In The Chlamydomonas Sourcebook, Stern D.B., ed (London: Academic Press; ), pp. 69–113. [Google Scholar]
- Giavalisco P., Li Y., Matthes A., Eckhardt A., Hubberten H.M., Hesse H., Segu S., Hummel J., Köhl K., Willmitzer L. (2011). Elemental formula annotation of polar and lipophilic metabolites using (13) C, (15) N and (34) S isotope labelling, in combination with high-resolution mass spectrometry. Plant J. 68: 364–376. [DOI] [PubMed] [Google Scholar]
- Gibon Y., Blaesing O.E., Hannemann J., Carillo P., Höhne M., Hendriks J.H., Palacios N., Cross J., Selbig J., Stitt M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Hall M.N. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (2001). Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 363–406. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. (2004). Upstream and downstream of mTOR. Genes Dev. 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- Heitman J., Movva N.R., Hall M.N. (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Huege J., Goetze J., Dethloff F., Junker B., Kopka J. (2014). Quantification of stable isotope label in metabolites via mass spectrometry. Methods Mol. Biol. 1056: 213–223. [DOI] [PubMed] [Google Scholar]
- Imamura S., Kanesaki Y., Ohnuma M., Inouye T., Sekine Y., Fujiwara T., Kuroiwa T., Tanaka K. (2009). R2R3-type MYB transcription factor, CmMYB1, is a central nitrogen assimilation regulator in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA 106: 12548–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S., Kawase Y., Kobayashi I., Sone T., Era A., Miyagishima S.Y., Shimojima M., Ohta H., Tanaka K. (2015). Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol. Biol. 89: 309–318. [DOI] [PubMed] [Google Scholar]
- Imamura S., Kawase Y., Kobayashi I., Shimojima M., Ohta H., Tanaka K. (2016). TOR (target of rapamycin) is a key regulator of triacylglycerol accumulation in microalgae. Plant Signal. Behav. 11: e1149285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Alric J. (2013). Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot. Cell 12: 776–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüppner J., Mubeen U., Leisse A., Caldana C., Brust H., Steup M., Herrmann M., Steinhauser D., Giavalisco P. (2017). Dynamics of lipids and metabolites during the cell cycle of Chlamydomonas reinhardtii. Plant J. 92: 331–343. [DOI] [PubMed] [Google Scholar]
- Jüppner J., Mubeen U., Leisse A., Caldana C., Wiszniewski A., Steinhauser D., Giavalisco P. (2018). The target of rapamycin kinase affects biomass accumulation and cell cycle progression by altering carbon/nitrogen balance in synchronized Chlamydomonas reinhardtii cells. Plant J. 93: 355–376. [DOI] [PubMed] [Google Scholar]
- Kleessen S., Irgang S., Klie S., Giavalisco P., Nikoloski Z. (2015). Integration of transcriptomics and metabolomics data specifies the metabolic response of Chlamydomonas to rapamycin treatment. Plant J. 81: 822–835. [DOI] [PubMed] [Google Scholar]
- Lee D.Y., Fiehn O. (2013). Metabolomic response of Chlamydomonas reinhardtii to the inhibition of target of rapamycin (TOR) by rapamycin. J. Microbiol. Biotechnol. 23: 923–931. [DOI] [PubMed] [Google Scholar]
- Li M.W., AuYeung W.K., Lam H.M. (2013). The GCN2 homologue in Arabidopsis thaliana interacts with uncharged tRNA and uses Arabidopsis eIF2α molecules as direct substrates. Plant Biol. (Stuttg.) 15: 13–18. [DOI] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., Hall M.N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Ma X.M., Blenis J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10: 307–318. [DOI] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., Smolka M.B., Emr S.D. (2011). TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147: 1104–1117. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Desnos T., Nussaume L., Berger F., Bouchez D., Meyer C., Robaglia C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99: 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S., Dibble C.C., Talbott G., Hoxhaj G., Valvezan A.J., Takahashi H., Cantley L.C., Manning B.D. (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156: 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., Kropat J., Liu B., Shaw J., Warakanont J. (2012). TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23: 352–363. [DOI] [PubMed] [Google Scholar]
- Montané M.H., Menand B. (2013). ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64: 4361–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M., Azzopardi M., Clément G., Dobrenel T., Marchive C., Renne C., Martin-Magniette M.L., Taconnat L., Renou J.P., Robaglia C., Meyer C. (2012). Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24: 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Florencio F.J., Crespo J.L. (2010). Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152: 1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Couso I., Crespo J.L. (2017). The TOR signaling network in the model unicellular green alga Chlamydomonas reinhardtii. Biomolecules 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.J., Nemenoff R.A., Avruch J. (1989). Purification of a hepatic S6 kinase from cycloheximide-treated rats. J. Biol. Chem. 264: 13825–13833. [PubMed] [Google Scholar]
- Ren M., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24: 4850–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Qiu S., Venglat P., Xiang D., Feng L., Selvaraj G., Datla R. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155: 1367–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roustan V., Jain A., Teige M., Ebersberger I., Weckwerth W. (2016). An evolutionary perspective of AMPK-TOR signaling in the three domains of life. J. Exp. Bot. 67: 3897–3907. [DOI] [PubMed] [Google Scholar]
- Russell R.C., Yuan H.X., Guan K.L. (2014). Autophagy regulation by nutrient signaling. Cell Res. 24: 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M.A., Li Y., Wiszniewski A., Giavalisco P. (2017). Regulatory-associated protein of TOR (RAPTOR) alters the hormonal and metabolic composition of Arabidopsis seeds, controlling seed morphology, viability and germination potential. Plant J. 92: 525–545. [DOI] [PubMed] [Google Scholar]
- Salem M.A., Li Y., Bajdzienko K., Fisahn J., Watanabe M., Hoefgen R., Schöttler M.A., Giavalisco P. (2018). RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiol. 177: 565–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. (2006). Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22: 159–168. [DOI] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. (2017). mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., Östlund G. (2015). InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43: D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani R., Yao L., Menand B., Ennar N., Lecampion C., Meyer C., Robaglia C. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Burgos J., Bassham D.C. (2017). SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One 12: e0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A., Cohen A., Hall M.N. (2009). TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21: 825–836. [DOI] [PubMed] [Google Scholar]
- Spalding M.H. (2009). The CO2-Concentrating Mechanism and Carbon Assimilation. In The Chlamydomonas Sourcebook D, Stern B., ed (London: Academic Press; ), pp. 257–301. [Google Scholar]
- Vallentine P., Hung C.Y., Xie J., Van Hoewyk D. (2014). The ubiquitin-proteasome pathway protects Chlamydomonas reinhardtii against selenite toxicity, but is impaired as reactive oxygen species accumulate. AoB Plants pii: plu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Mandal R., Sinelnikov I.V., Broadhurst D., Wishart D.S. (2012). MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 40: W127–W133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.X., Xiong Y., Guan K.L. (2013). Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49: 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]