Abstract
Introduction:
Microorganisms catabolize carbon-containing compounds in their environment during growth, releasing a subset of metabolic byproducts as volatile compounds. However, the relationship between growth media and the production of volatile compounds has been largely unexplored to-date.
Objectives:
To assess the core and media-specific components of the Klebsiella pneumoniae volatile metabolome via growth in four in vitro culture media.
Methods:
Headspace volatiles produced by cultures of K. pneumoniae after growth to stationary phase in four rich media (brain heart infusion broth, lysogeny broth, Mueller-Hinton broth, and tryptic soy broth) were analyzed using comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC-TOFMS). Differences in the composition of headspace volatiles as a function of growth media was assessed using hierarchical clustering analysis (HCA) and principal component analysis (PCA).
Results:
A total of 365 volatile compounds were associated with the growth of K. pneumoniae across all media, of which 36 (10 %) were common to all growth media, and 148 (41 %) were specific to a single medium. In addition, utilizing all K. pneumoniae-associated volatile compounds, strains clustered as a function of growth media, demonstrating the importance of media in determining the metabolic profile of this organism.
Conclusion:
K. pneumoniae produces a core suite of volatile compounds across all growth media studied, although the volatile metabolic signature of this organism is fundamentally media-dependent.
Keywords: Klebsiella pneumoniae, Mass spectrometry, Metabolomics, Comprehensive two-dimensional gas chromatography, Volatile compounds
1. Introduction
Klebsiella pneumoniae is an increasingly-important human pathogen capable of causing severe disease at various bodily sites, including the bloodstream, genitourinary tract, and respiratory tract (Podschun and Ullmann 1998), often in the setting of an immunocompromised host. In addition to the wide range of infections caused by this organism, K. pneumoniae has garnered increased attention recently due to the emergence and dissemination of multidrug resistant strains, which can increase mortality by upwards of 50 % (Borer et al. 2009). Current culture-based methods for the detection and antibiotic susceptibility profiling of pathogenic organisms from patient biofluids, such as sputum for respiratory infection diagnosis, can take days. One approach being investigated to decrease the time-to-diagnosis for respiratory tract infections is the use of volatile compounds, with the ultimate goal of using exhaled breath to determine infection etiology.
Exhaled breath volatile compounds are currently used in the diagnosis of Helicobacter pylori gastritis (Gisbert and Pajares 2004), and show promise for the diagnosis of both acute and chronic respiratory infections (Sethi et al. 2013). Studies have demonstrated that a subset of the volatile compounds produced during bacterial growth in vitro are either identical, or structurally similar, to those detected in ex vivo samples (e.g., breath, sputum, blood) from individuals with bacterial infections. For example, 2-aminoacetophenone has been detected in the headspace of Pseudomonas aeruginosa cultures (Bean et al. 2012) as well as the breath of cystic fibrosis patients chronically colonized with this organism (Scott-Thomas et al. 2010). The compounds α- and β-trans-bergamotene have been detected in the headspace of Aspergillus fumigatus cultures, and also in the breath of patients with invasive aspergillosis (Koo et al. 2014). Nicotinic acid has been detected in the headspace of Mycobacterium tuberculosis cultures (Syhre and Chambers 2008) and its methylated derivative, methyl nicotinate, has been detected in the breath of M. tuberculosis infected individuals (Syhre et al. 2009). Typically, only a subset of the volatile compounds from a culture headspace will translate to a respiratory infection scenario (Zhu et al. 2013b, Gao et al. 2016). Improvement in the yield from cultures might be achieved by evaluating the compounds that evolve from multiple strains, or from a single strain under multiple growth conditions.
While most prior studies measuring the volatile compounds produced by K. pneumoniae have focused on a single strain cultivated in a single media, relatively few have considered multiple strains (Julak et al. 2003, Julak et al. 2000) or the impact of different growth media (Kiviranta et al. 1998, Tait et al. 2014, Julak et al. 2003, Julak et al. 2000). Novel analytical instrumentation can provide superior resolution and sensitivity, which also aids in the characterization of volatile compounds. For example, prior studies using comprehensive two-dimensional gas chromatography coupled to time-of-flight mass spectrometry (GC×GC-TOFMS) have doubled the total number of volatile compounds associated with the growth of both P. aeruginosa (Bean et al. 2012), and K. pneumoniae (Rees et al. 2016b), in vitro, demonstrating the power of this analytical tool. This study represents the first comparative metabolomic analysis of the volatile compounds produced by K. pneumoniae in varied growth environments, using GC×GC-TOFMS, an approach that will provide us with a basis for translation to biomarkers in the context of human disease.
2. Methods
2.1. Strain Collection
Nine clinical isolates of Klebsiella pneumoniae were used in this study, a subset of which had been previously described by Hirsch and colleagues (1: 6834, 2: 6962, 3: 7097, 4: 7117, 5: 7278, 6: 7363) (Hirsch et al. 2014). All isolates were from bacteremic patients. Genetic diversity amongst isolates was determined using multilocus sequence typing (MLST), as described previously (Diancourt et al. 2005), with interpretation of results via the Institut Pasteur MLST database (http://bigsdb.pasteur.fr/). All nine isolates belonged to distinct sequence types (STs). MLST results for these strains are presented in Table 1.
Table 1.
MLST results for K. pneumoniae clinical isolates evaluated in this study
| Isolate | Allelic variants for 7 MLST genes | ST | ||||||
|---|---|---|---|---|---|---|---|---|
| gapA | infB | mdh | pgi | phoE | rpoB | tonB | ||
| 1 | 4 | 34 | 1 | 1 | 21 | 1 | 35 | 199 |
| 2 | 2 | 1 | 1 | 1 | 7 | 1 | 12 | 485 |
| 3 | 10 | 1 | 11 | 1 | 9 | 8 | 257 | 1569 |
| 4 | 1 | 6 | 1 | 1 | 248 | 124 | 1 | a |
| 5 | 2 | 1 | 2 | 1 | 7 | 1 | 25 | 834 |
| 6 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 15 |
| 7 | 2 | 1 | 2 | 1 | 9 | 4 | 26 | a |
| 8 | 4 | 1 | 2 | 52 | 1 | 1 | 7 | 307 |
| 9 | 2 | 1 | 1 | 1 | 4 | 4 | 89 | a |
Novel allelic combinations, resulting in previously unreported STs.
2.2. Culture Conditions
Culture media included Bacto™ Brain Heart Infusion (BHI), Difco™ LB Lennox (LB), Difco™ Mueller Hinton Broth (MHB), and BBL™ Trypticase™ Soy Broth (TSB) (all from Becton Dickinson (BD), Franklin Lakes, NJ). Bacteria were pre-cultured under aerobic conditions overnight (37 °C, 200 rpm shaking) in 5 mL of either BHI, LB, MHB, or TSB, and inoculated 1:1000 into 20 mL of fresh media of the same variety. Cultures were incubated aerobically (37 °C, 200 rpm shaking) in 250 mL Erlenmeyer flasks until early stationary growth phase (11 h post-inoculation). Three biological replicates were prepared per strain (n = 27 cultures per growth media), and uninoculated media controls (n = 5 per media) were prepared identically.
2.3. Sample Preparation and Concentration of Volatile Compounds
At the time of sample collection, cultures were transferred to 50 mL conical tubes and immediately submerged in ice to quench metabolism. Conical tubes were centrifuged for 5 minutes (12100 × g, 4 °C), and 4 mL of the supernatant were transferred to 20 mL air-tight headspace vials (Sigma-Aldrich, St. Louis, MO), which were immediately stored at −20 °C until analysis. All samples were analyzed within six months of collection. Headspace volatile compounds were concentrated on a 2 cm triphasic Divinylbenzene/Carboxen/Polydimethylsiloxane (DVB/CAR/PDMS) solid-phase microextraction (SPME) fiber (Supelco, Bellefonte, PA). The fiber was exposed to the culture headspace for 30 min at 50 °C, with 250 rpm agitation.
2.4. GC×GC-TOFMS Parameters
The GC×GC-TOFMS (Pegasus 4D, LECO Corporation, St. Joseph, MI) was equipped with a rail autosampler (MPS, Gerstel, Linthicum Heights, MD) and fitted with a two-dimensional column set consisting of a 60 m × 250 μm × 1.4 μm (length × internal diameter × film thickness) Rxi®−624Sil first column followed by a 1 m × 250 μm × 0.5 μm Stabilwax second column. Both columns were from Restek, Bellefonte, PA. The main oven containing column 1 was held at 35 °C for 0.5 minutes, and then ramped at 5.0 °C/min from 35 °C to 230 °C. The secondary oven containing column 2, and the quad-jet modulator (2 s modulation period, 0.5 s alternating hot and cold pulses), were heated in step with the primary oven with +5 °C and +25 °C offset relative to the primary oven, respectively. The helium carrier gas flow rate was 2 mL/min (constant flow mode). A splitless injection was used, with a 180 s desorption time. The inlet and transfer line temperatures were set to 270 °C and 250 °C, respectively. Mass spectra were acquired over the range of 30 to 500 m/z, with an acquisition rate of 200 spectra/s, and a detector voltage offset of 50 V, which serves to approximately double signal intensity relative to baseline detector voltage. Data acquisition and analysis was performed using ChromaTOF software, version 4.50 (LECO Corp.).
2.5. Determination of Retention Indices
Retention indices (RIs) were calculated using external alkane standards (C6 – C14) (Kovats 1958). The SPME fiber was exposed to a vial containing a pure retention index mixture (Sigma-Aldrich) for 10 minutes at 50 °C and desorbed at a 30:1 split. Reported RIs are between the literature values for polar and nonpolar column sets, due to the midpolarity of the Rxi®−624Sil stationary phase. Retention indices less than 600 (corresponding to C6) or greater than 1400 (corresponding to C14) were not extrapolated.
RIs were calculated using the following equation:
RI - calculated retention index, tR(analyte) - retention time of analyte, tR(n)- retention time of nearest straight-chain alkane with tR<tR(analyte), tR(N) - retention time of nearest straight-chain alkane with tR>tR(analyte), n - number of carbons in straight-chain alkane with retention time tR(n).
2.6. Data Processing and Chromatographic Alignment
Chromatographic data was processed and aligned using ChromaTOF. For peak identification, a signal-to-noise (S/N) cutoff was set at 50:1, and resulting peaks were identified by a forward search of the NIST 2011 library. A forward match score of at least 800 of 1000 and affirmation via retention indices were required for putative compound identification. For the alignment of peaks across chromatograms, maximum first and second-dimension retention time deviations were set at 6 s and 0.2 s, respectively, and the inter-chromatogram spectral match threshold between chromatograms was set at 600 (of 1000). For peaks that were detected at a S/N of 50:1 or greater in at least one chromatogram, the remaining chromatograms were searched at a minimum S/N of 10:1 for the detection of low abundance analytes.
2.7. Compound Inclusion Criteria and Statistical Analyses
All statistical analyses were performed using R v3.2.2 (R Foundation for Statistical Computing, Vienna, Austria). The relative abundance of compounds across chromatograms was first normalized using Probabilistic Quotient Normalization (Dieterle et al. 2006). Reported K. pneumoniae-associated volatile compounds were required to meet one of the following two inclusion criteria: Criteria 1) significantly more abundant in K. pneumoniae cultures relative to uninoculated media in at least one media, or Criteria 2) detection in 3 of 3 biological replicates from one or more strains in at least one media, and not detected in uninoculated media controls. A subset of the compounds that were significantly more abundant in K. pneumoniae cultures (Criteria 1) were also detected in every single sample, and we note these as a distinct sub-category (Criteria 1A), with the remaining compounds categorized as Criteria 1B. The statistical significance of compounds was tested by means of the one-sided non-parametric Mann–Whitney U test (Mann and Whitney 1947) with Benjamini-Hochberg correction (Benjamini and Hochberg 1995), and a significance level of p < 0.05 was selected.
For the comparison of compound abundance between different strains and/or growth media, the average peak intensity for each compound in a given strain was calculated as the arithmetic mean of the three biological replicates, and an arbitrarily small value of 1 was assigned for compounds that were not detected. Average peak intensities were 1) log-transformed (log10), 2) mean-centered, and 3) unit-scaled. Mean-centering and unit-scaling were performed for each compound individually.
Principal component analysis (PCA) (Hotelling 1933) was used to reduce dimensionality and visualize variance in the data. Principal component (PC) scores plots were generated using either the first two or three PCs, with each point representing either a single sample or strain. Centroid analysis was performed using PC scores, and Euclidean distance was used to determine the distance between individual samples and their corresponding centroid in three-dimensional space, utilizing average PC 1, PC 2, and PC 3 scores to calculate centroids. Hierarchical clustering analysis (HCA) was employed to assess the metabolic relatedness of isolates using all K. pneumoniae-associated volatile compounds, with Euclidean distance as the distance metric.
3. Results and Discussion
3.1. Overview of Klebsiella pneumoniae-associated volatile compounds
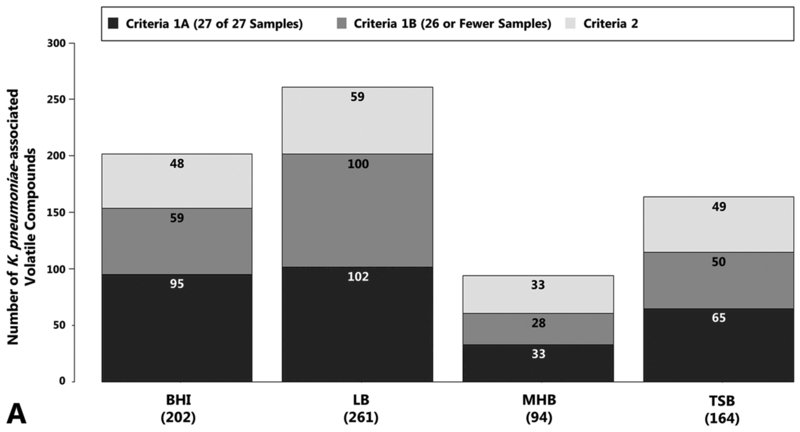
We cultured nine clinical isolates of Klebsiella pneumoniae in either brain-heart infusion (BHI) broth, lysogeny broth (LB), Mueller-Hinton broth (MHB), or tryptic soy broth (TSB) to early stationary growth phase. Bacterial cell density did not differ significantly across growth media at the time of sample collection (not shown). Headspace volatile compounds were concentrated using SPME, and analyzed using GC×GC-TOFMS. After removal of known chromatographic artifacts and environmental contaminants, a total of 1129 distinct peaks were detected across all samples after chromatographic alignment, of which 365 (32 %) were associated with the growth and metabolism of K. pneumoniae in at least one growth medium (Figure 1a), as defined by the two inclusion criteria described in our Materials and Methods (Section 2.7). The algorithm used to assign compounds to each of these categories is presented in Supplementary Figure S1.
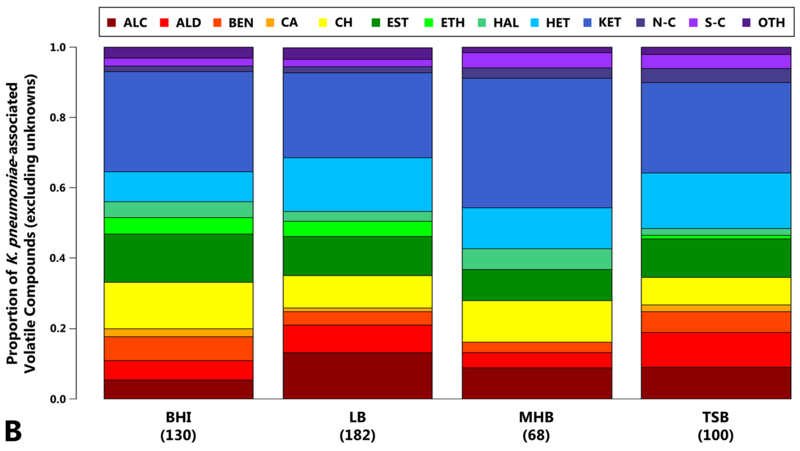
Fig. 1.
Volatile compounds produced by K. pneumoniae as a function of growth media. 1a, top: Number of compounds associated with the growth of K. pneumoniae in BHI, LB, MHB, or TSB. This includes compounds that: fulfill Criteria 1 and were detected in 27 of 27 samples (Criteria 1A, dark grey), 2) fulfill Criteria 1 and were detected in 26 or fewer samples (Criteria 1B, medium grey), or 3) fulfill Criteria 2 (light grey). Values in parentheses indicate total number of K. pneumoniae-associated volatile compounds per medium. 1b, bottom: Compound class assignments for K. pneumoniae-associated volatile compounds produced in BHI, LB, MHB, or TSB. ALC: alcohols, ALD: aldehydes, BEN: benzene derivatives, CA: carboxylic acids, CH: hydrocarbons, EST: esters, ETH: ethers, HAL: organohalides, HET: heterocycles, KET: ketones, N-C: nitrogen-containing (excluding heterocycles), S-C: sulfur-containing (excluding heterocycles), OTH: others. Only compounds for which compound class could be determined are included, and the height of each bar is scaled over the range of 0 to 1. (Supplementary Figure S2 also includes compounds for which chemical class assignments could not be determined.) Numbers in parentheses indicate total number of K. pneumoniae-associated volatile compounds for which compound class could be determined per medium.
The number and variety of K. pneumoniae-associated volatile compounds varied as a function of growth media. Growth in LB yielded the greatest total number of K. pneumoniae-associated volatile compounds (n = 261), which is more than double what was detected in MHB (n = 94), the medium that yielded the fewest number of compounds (Figure 1a). Ketones represented the single largest compound class across all growth media (ranging from 24% in LB to 37% in MHB), although alcohols, esters, heterocycles, and hydrocarbons were also highly represented (Figure 1b). The number and proportion of compounds belonging to each class is presented in Supplementary Table S1.
Our compound class results are consistent with those from the one prior study to analyze K. pneumoniae volatile compounds produced during growth in LB via GC×GC-TOFMS, which also reported on a preponderance of alcohols, esters, heterocycles, hydrocarbons, and ketones (Rees et al. 2016a). While other studies have measured the volatile compounds produced by K. pneumoniae during growth in TSB using alternative analytical techniques, the majority of these have focused on the identification of specific compound classes, such as fermentation byproducts (Lee et al. 1995, Robacker and Bartelt 1997) or alcohols (Elgaali et al. 2002), rather than a comprehensive profiling of all volatile compounds. In addition, these studies differ from our own with regards to the K. pneumoniae strains utilized and culture conditions employed (e.g., culturing time prior to concentration and analysis of volatile compounds. It is therefore not possible to directly compare our compound class results with those of prior studies. In addition, the number of K. pneumoniae-associated volatile compounds that have been previously reported in other growth media is too small for us to confidently draw conclusions regarding the relative composition of different molecular classes in these alternative growth environments (Julak et al. 2003, Julak et al. 2000, Tait et al. 2014, Junger et al. 2012, Kiviranta et al. 1998, Rees et al. 2016b, Boots et al. 2014, Storer et al. 2011).
3.2. Core K. pneumoniae-associated volatile compounds
Thirty-six of the 365 K. pneumoniae-associated volatile compounds (10 %) were common to all four growth media, and we subsequently describe these as core volatile compounds. The 17 core compounds that we could assign either: 1) a putative compound identification based on mass spectral matching and affirmation via retention indices, or 2) a compound class and molecular formula, are presented in Table 2. A complete list of core compounds is presented in Supplementary Table S3. The predominant molecular species produced by K. pneumoniae in all four growth media were 2-ketones, including hexan-2-one, heptan-2-one, nonan-2-one, and decan-2-one.
Table 2.
K. pneumoniae-associated volatile compounds common to all growth media
| CAS # | Compound | Compound Class | RI | Previously Reported |
|---|---|---|---|---|
| 75-05-8 | Acetonitrile | N-containing | - | e |
| 591-78-6 | hexan-2-one | Ketone | 831 | e, g |
| 109-08-0 | 2-methylpyrazine | Heterocycle | 862 | d, e, g |
| 100-41-4 | Ethylbenzene | Benzene derivative | 889 | e |
| 3240-09-3 | 5-methyl-5-hexen-2-one | Ketone | 922 | e |
| - | a Analyte 535 | Benzene derivative | 925 | - |
| 110-43-0 | heptan-2-one | Ketone | 933 | d-f |
| - | Analyte 558 | Heterocycle | 948 | - |
| - | Analyte 683 | Heterocycle | 1027 | - |
| 100-47-0 | Benzonitrile | N-containing | 1068 | e |
| 106-27-4 | 3-methylbutyl butanoate | Ester | 1081 | - |
| - | Analyte 772 | Ketone | 1086 | - |
| 821-55-6 | nonan-2-one | Ketone | 1138 | b-e, h |
| - | Analyte 835 | Heterocycle | 1138 | - |
| 103-79-7 | 1-phenylpropan-2-one | Ketone | 1208 | e, g |
| 693-54-9 | decan-2-one | Ketone | 1237 | e |
| 14371-10-9 | (E)-3-phenylprop-2-enal | Aldehyde | 1261 | e |
CAS#: Chemical Abstracts Service (CAS) registry number, RI: retention index (experimentally determined), -: Retention indices less than 600 not extrapolated,
Compound not produced by all strains in all media,
Note: Table 2 excludes those compounds for which putative compound identifications or compound class and molecular formula could not be determined (see Supplementary Table S3).
2-ketones have previously been associated with the growth and metabolism of K. pneumoniae in blood agar (Junger et al. 2012), human blood (Rees et al. 2016b), MHB (Boots et al. 2014), LB (Rees et al. 2016a), and TSB (Zechman et al. 1986, Lee et al. 1995, Robacker and Bartelt 1997, Elgaali et al. 2002). They have also been detected in cultures of other Enterobacteriaceae such as Escherichia coli, more distantly-related Gram-negative organisms such as Pseudomonas aeruginosa, and Gram-positive organisms such as Staphylococcus aureus (Bos et al. 2013, Schulz and Dickschat 2007). Given the detection of these compounds across distantly-related Gram-positive and Gram-negative organisms, we hypothesize that 2-ketones may represent fundamental byproducts of bacterial metabolism.
Additionally, a variety of aromatic compounds, including both benzene-derivatives (e.g., benzonitrile, cinnamaldehyde, ethylbenzene, and phenylacetone) and heterocycles (e.g., 2-methylpyrazine), are components of the core volatile metabolome. Both benzene derivatives and aromatic heterocycles have been previously detected in the headspace of K. pneumoniae cultures (Junger et al. 2012, Kiviranta et al. 1998, Lee et al. 1995, Rees et al. 2016a, Robacker and Bartelt 1997, Zechman et al. 1986), and a subset have also been detected in the headspace of other Gram-negative and Gram-positive organisms (Bos et al. 2013, Schulz and Dickschat 2007). We hypothesize that a subset of these compounds represent byproducts of essential bacterial metabolic pathways, but as the biological origins of these compounds are largely unknown, it is unclear which compounds are essential to all bacteria, and which are shared only amongst a subset.
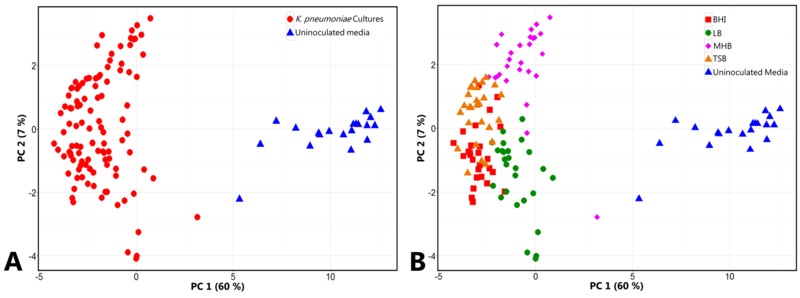
We generated a principal component (PC) scores plot using the 36 core volatile compounds for the comparison of bacterial cultures and uninoculated media controls, which reveals two distinct clusters of samples (bacterial cultures vs. uninoculated media controls) along principal component 1 (PC 1) (Figure 2a). We observe sub-structure in the core volatile compound data, with LB- and MHB-inoculated samples forming distinct clusters along PC 2 (Figure 2b), and with the incorporation of PC 4 (Supplementary Figure S3), we observe distinct clustering of all samples as a function of growth media. While the core compounds are common to all four media, these data demonstrate that the relative abundance of core compounds differs across media. Based on this observation, we hypothesize that these core volatile compounds represent metabolic byproducts of essential cellular pathways, but that the relative flux through these pathways differs based on carbon-source availability in the environment.
Fig. 2.
PC scores plots generated using the 36 core K. pneumoniae-associated volatile compounds. The core compounds result in separation between K. pneumoniae cultures (red circles, n = 108) and uninoculated media controls (blue triangles, n = 20) (2a, left), and also reveal sub-structure within the K. pneumoniae culture samples as a function of growth media (2b, right). BHI: red squares, LB: green circles, MHB: pink diamonds, TSB: orange triangles, uninoculated media: blue triangles. (PC 1: 60 %, PC 2: 7 %). A three-dimensional scores plot is presented in Supplementary Figure S3.
3.3. Media-dependent K. pneumoniae-associated volatile compounds
Of the 365 K. pneumoniae-associated volatile compounds, 329 were detected in three or fewer media, and we subsequently describe these as media-dependent volatile compounds. One hundred and forty eight (41 %) of these media-dependent compounds were specific to a single media, 114 (35 %) were common to two, and 67 (20 %) were common to three. Our observation that approximately half of the K. pneumoniae-associated volatile compounds detected in this study are specific to a single medium, while only approximately 10 % are common to all media, suggests that K. pneumoniae possesses substantial metabolic flexibility and can drastically alter its production of volatile metabolites based on growth environment. Indeed, metabolic adaptation as a function of carbon source availability has already been extensively studied in E. coli (Liu et al. 2005), a close relative of Klebsiella, revealing alterations at the transcriptomic level across a multitude of carbon sources. Therefore, while a subset of the media-specific volatile compounds that we detect may result from the catabolism of media-specific precursors into volatile byproducts, it is likely that another subset results from fundamental alterations to bacterial metabolism as a function of differential carbon (and possibly other nutrient) source availability across different growth media. This result raises important questions about in vitro experiments conducted in a single growth media that attempt to translate their results to volatile biomarkers produced under in vivo infection conditions (Zhu et al. 2013b, Gao et al. 2016). One prior study measured the differential production of volatile compounds across three growth media (BHI, TSB and enteric fermentation base [EF] broth) by K. pneumoniae and noted that of the six K. pneumoniae-associated compounds that were detected, two were specific to a single medium, two were common to two media, and two were shared across all three (Tait et al. 2014). Here, we report on 329 compounds using untargeted comprehensive 2D gas chromatography, rather than following a subset of compounds, making comparisons challenging.
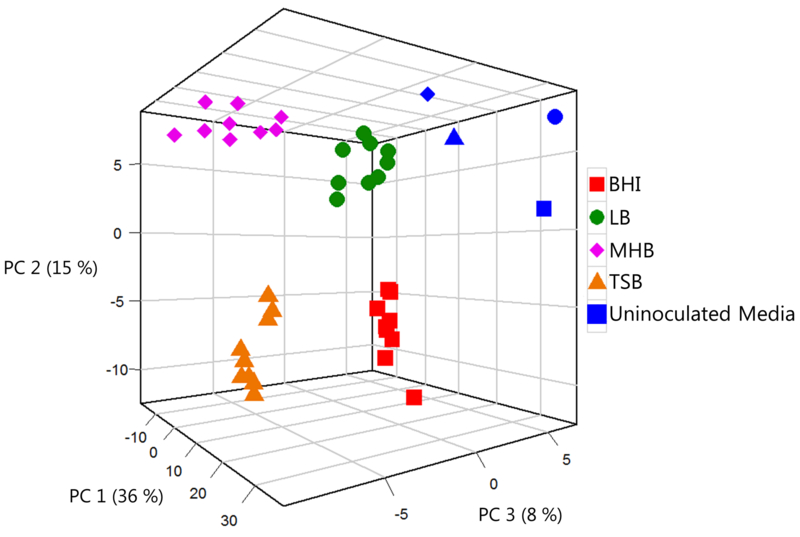
Using all 365 K. pneumoniae-associated volatile compounds (i.e., 329 media-dependent and 36 core), we generated a PC scores plot to assess broad similarities between samples (Figure 3), with each point representing the average of all biological replicates for a given strain in a single growth medium. A centroid analysis using the first three PC scores (Supplementary Table S4) demonstrates that 33 of 36 samples are closest in three-dimensional space to their own centroid, while three samples (two strains cultured in LB and one cultured in TSB) were closest to the centroids of other media. Overall, using the entire suite of 365 K. pneumoniae-associated volatile compounds resulted in clustering as a function of growth media. We therefore conclude that the volatile metabolic fingerprint of K. pneumoniae is fundamentally growth environment-dependent, and that the majority of the variation that we observe in our data is resultant from differences in growth media rather than strain-to-strain metabolic differences. This conclusion is further supported by an analysis of strain-dependent differences in the production of volatile compounds (Supplementary Figure S4), which demonstrates that there are no conserved strain-to-strain differences across media.
Fig. 3.
PC scores plot generated using all 365 K. pneumoniae-associated volatile compounds. Each point represents a single strain grown in a single growth medium, derived from the mean of three biological replicates. BHI: red squares, LB: green circles, MHB: pink diamonds, TSB: orange triangles. Uninoculated media (blue) correspond in shape to equivalent K. pneumoniae-inoculated cultures. PC 1: 36 %, PC 2: 15 %, PC 3: 8 %.
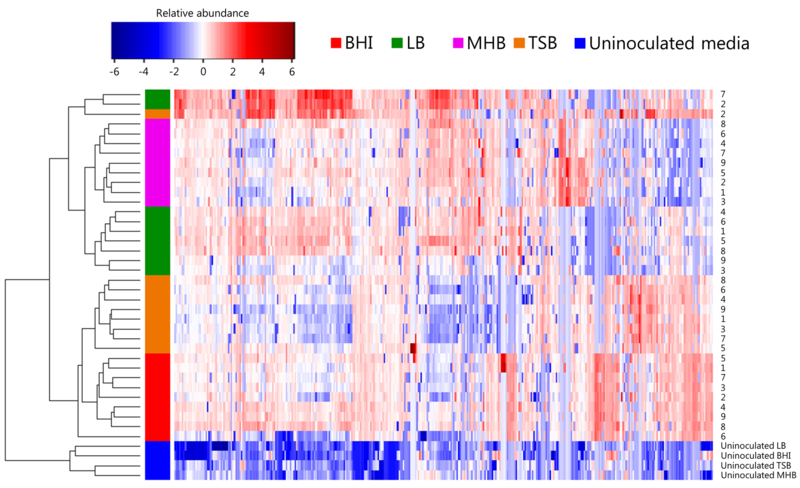
We next sought to more precisely determine the relatedness amongst samples as a function of both growth media and strain using hierarchical clustering analysis (HCA), again incorporating all K. pneumoniae-associated volatile compounds. The heat map generated using these compounds is presented in Figure 4, with each row representing a single strain grown in a single medium, and each column representing a single volatile compound. As with the PC scores plot presented in Figure 3, isolates generally clustered as a function of media, even with no feature selection algorithms employed for the separation of samples based on growth environment. Uninoculated media controls of BHI, LB, MHB, and TSB formed a single cluster distinct from K. pneumoniae-inoculated cultures, confirming that the set of K. pneumoniae-associated volatile compounds reflects differential media-dependent bacterial metabolism, and does not result from differences in the background volatile signature associated with media itself. Of interest, despite samples generally clustering by growth media, relatedness amongst strains was not conserved across media, as revealed by the dendrogram in Figure 4 (Supplementary Table S5 gives the Euclidean distance between all samples), indicating that the metabolic similarities between strains of a single species are largely conditional based on environment.
Fig. 4.
Heat map depicting the relative abundance of volatile compounds produced by K. pneumoniae during growth in BHI (red), LB (green), MHB (pink), or TSB (orange), as well as uninoculated media (blue). Each row represents a single strain derived from the mean of three biological replicates, and each column represents one K. pneumoniae-associated volatile compound. Blue = low relative abundance, red = high relative abundance. Dendogram (left) depicts the Eucleidian distance between samples, and demonstrates the relatedness of strains as a function of growth media.
3.4. Differentially-abundant volatile compounds across growth media
We utilized intraclass correlation to identify those volatile compounds whose relative abundance varied predominantly as a function of growth media. In this context, a large intraclass correlation coefficient (ICC) indicates compounds whose total variance across samples is mainly derived from between-group (e.g., comparison of samples from different growth media) rather than within-group (comparison of samples from a single growth media) differences. We identified 30 volatile compounds whose ICC was greater than 0.75, and the subset of these for which putative compound identifications or molecular formula and compound class could be determined are presented in Table 3, with the full list presented in Supplementary Table S6. A subset of the compounds presented in Table 3 are also part of the core set of K. pneumoniae-associated volatile compounds (Table 1), supporting the observation that while these core compounds are produced by K. pneumoniae across all media, their relative abundance varies as a function of growth environment.
Table 3.
K. pneumoniae-associated volatile compounds with intraclass correlation coefficient (ICC) > 0.75
| CAS # | Compound | Relative abundance (TIC, thousands) |
Between:Within Group Variance |
Previously Reported |
|||
|---|---|---|---|---|---|---|---|
| BHI | LB | MHB | TSB | ||||
| 290-37-9 | Pyrazine | 726 | 25 | 53 a | 1259 a | 36.4 | h |
| 109-08-0 | 2-methylpyrazine b | 880 | 68 | 139 | 531 | 29.0 | f-h |
| 32736-94-0 | 2,5-dimethyl-3-(2-methylpropyl)-pyrazine | 9 a | 0 a | 1 a | 3 | 19.7 | g |
| 15707-34-3 | 5-ethyl-2,3-dimethylpyrazine | 8 a | 1 | 1 a | 1 a | 14.7 | - |
| 5910-89-4 | 2,3-dimethylpyrazine | 36 a | 2 | 3 a | 7 | 13.4 | f |
| - | Analyte 835 (Pyrazine, C8H10N2) b | 68 | 3 | 3 | 23 | 12.9 | - |
| 13360-64-0 | 2-ethyl-5-methylpyrazine | 313 a | 48 | 48 a | 131 a | 6.4 | f, g |
| 13360-65-1 | 3-ethyl-2,5-dimethylpyrazine | 158 a | 19 | 26 a | 68 a | 5.8 | f, g |
| 600-14-6 | penxtane-2,3-dione | 70 a | 135 | 256 | 17 a | 5.2 | g |
| 693-54-9 | decan-2-one b | 25 | 18 | 45 | 146 | 5.1 | g |
| - | Analyte 690 (Pyrazine, C7H10N2) | 22 a | 6 | 16 | 4 | 5.0 | - |
| 13925-00-3 | 2-ethylpyrazine | 24 a | 0 a | 3 a | 10 | 4.9 | - |
| 103-79-7 | 1-phenylpropan-2-one b | 3896 | 392 | 143 | 1214 | 4.6 | d, g |
| 13925-07-0 | 2-ethyl-3,5-dimethylpyrazine | 10 a | 3 | 26 | 1 | 4.4 | g |
| - | Analyte 558 (Pyrazine, C6H8N2) b | 20311 | 8249 | 8087 | 20207 | 4.1 | - |
| - | Analyte 637 (Hydrocarbon, C10H16) | 8 | 4 | 1 | 2 a | 4.0 | - |
| 14171-88-1 | 7-phenylheptan-2-one c | 86 | 1 a | 1 a | 25 | 3.4 | - |
| 821-55-6 | nonan-2-one b | 2303 | 540 | 847 | 2187 | 3.3 | d-g, i |
| 471-12-5 | 4-methyl-1-propan-2-ylbicyclo[3.1.0]hexane | 38 | 14 | 2 a | 5 a | 3.2 | g |
TIC: total ion current,
Compound not significantly more abundant in cultures relative to uninoculated media,
Compound part of core K. pneumoniae-associated volatile compounds,
Literature retention indices for nonpolar and/or polar column sets not available,
Note: Table 3 excludes those compounds for which putative compound identifications or compound class and molecular formula could not be determined (see Supplementary Table S6).
Pyrazine derivatives represented the dominant named molecular species, accounting for 12 of 19 compounds (63 %). The one prior study to utilize GC×GC-TOFMS for the analysis of K. pneumoniae headspace volatile compounds during growth in LB identified six pyrazine-derivatives, which accounted for 7 % of detected compounds (Rees et al. 2016a), and of the 150 K. pneumoniae-associated volatile compounds reported across all studies to-date, 12 (8 %) are pyrazine-derivatives, detected with growth in either TSB (Lee et al. 1995, Robacker and Bartelt 1997) or LB (Rees et al. 2016a). We detected pyrazine-derivatives that met either Criteria 1 or Criteria 2 across all four growth media used in this study, with the widest number detected in LB (9 compounds) and TSB (8 compounds) (Table 3). Although the pyrazine-derivatives that we detected were often most abundant in BHI-inoculated cultures (10 of 12 compounds), they were often not significantly different in abundance relative to uninoculated media in this growth environment. We hypothesize that pyrazine-derivatives represent compounds associated with the growth and metabolism of K. pneumoniae in a subset of growth environments, but which are also components of the background volatile molecular signature in certain media. This demonstrates the importance of carefully selecting growth media for the purpose of identifying bacterially-derived volatile biomarkers. Future studies to determine whether these compounds exhibit a similar pattern of production in other closely-related species such as E. coli would be informative.
4. Conclusions
To our knowledge, this study is the first to assess the media-dependent production of volatile compounds by K. pneumoniae using GC×GC-TOFMS. Of the 365 compounds that we attribute to the growth and metabolism of K. pneumoniae, only 10 % were detected across all growth media used in this study; the largest subset (41 %) were specific to a single medium. We observed that the K. pneumoniae volatile metabolic signature is fundamentally media-dependent, although we hypothesize that a subset of the compounds detected under in vitro conditions, mostly likely those of the core metabolome, will also be produced under in vivo conditions. These compounds may represent novel biomarkers for the diagnosis of K. pneumoniae infections. Indeed, prior studies have suggested that while individual volatile compounds may be produced by numerous bacterial species, the volatile molecular signatures (i.e., the aggregated profile of many volatile compounds) produced by bacteria can robustly discriminate at the species level (Lechner et al. 2005, Shnayderman et al. 2005, Bruins et al. 2009, Zhu et al. 2013a, Boots et al. 2014). Based on our present findings, we hypothesize that growth conditions can be manipulated to further accentuate species-level differences between organisms.
Finally, our observation that pyrazine-derivatives are consistently detected across different media demonstrates a key, seemingly contradictory, finding associated with this study. On the one-hand, a subset of volatile compounds (e.g., pyrazine-derivatives) are detected in K. pneumoniae cultures across all growth media, while on the other, the background volatile molecular signature of certain growth media also contains these same molecules. This phenomenon may obscure our ability to detect bacterially-derived compounds in a subset of environments, demonstrating the importance of carefully selecting growth media for the purpose of identifying bacterially-derived volatile compounds. Future studies could utilize radiolabeled substrates to distinguish bacterially-derived metabolites from the background volatile metabolic signature of the growth media itself.
Supplementary Material
Acknowledgments
Funding: This study was funded by National Institutes of Health (grant number R21 AI121076) awarded to JH. CR was supported by the Burroughs Wellcome Fund (grant number 1014106). KN was supported by the Paul K. Richter and Evalyn E. Cook Richter Memorial Fund (via Dartmouth College). AL was supported by the Presidential Scholarship (via Dartmouth College).
Compliance with Ethical Standards
Footnotes
Conflict of Interest: All authors report no potential conficts of interest.
Ethical Approval: This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Bean HD, Dimandja JM & Hill JE (2012). Bacterial volatile discovery using solid phase microextraction and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci, 901, 41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y & Hochberg Y (1995). Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol, 57(1), 289–300. [Google Scholar]
- Boots AW, Smolinska A, Van Berkel JJ, Fijten RR, Stobberingh EE, Boumans ML, Moonen EJ, Wouters EF, Dallinga JW & Van Schooten FJ (2014). Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography-mass spectrometry. J Breath Res, 8(2), 027106. [DOI] [PubMed] [Google Scholar]
- Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F & Sherf M (2009). Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol, 30(10), 972–6. [DOI] [PubMed] [Google Scholar]
- Bos LD, Sterk PJ & Schultz MJ (2013). Volatile metabolites of pathogens: a systematic review. PLoS Pathog, 9(5), e1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins M, Bos A, Petit PL, Eadie K, Rog A, Bos R, Van Ramshorst GH & Van Belkum A (2009). Device-independent, real-time identification of bacterial pathogens with a metal oxide-based olfactory sensor. Eur J Clin Microbiol Infect Dis, 28(7), 775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diancourt L, Passet V, Verhoef J, Grimont PA & Brisse S (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol, 43(8), 4178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G & Senn H (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem, 78(13), 4281–90. [DOI] [PubMed] [Google Scholar]
- Elgaali H, Hamilton-Kemp TR, Newman MC, Collins RW, Yu K & Archbold DD (2002). Comparison of long-chain alcohols and other volatile compounds emitted from food-borne and related Gram positive and Gram negative bacteria. J Basic Microbiol, 42(6), 373–80. [DOI] [PubMed] [Google Scholar]
- Gao J, Zou Y, Wang Y, Wang F, Lang L, Wang P, Zhou Y & Ying K (2016). Breath analysis for noninvasively differentiating Acinetobacter baumannii ventilator-associated pneumonia from its respiratory tract colonization of ventilated patients. J Breath Res, 10(2), 027102. [DOI] [PubMed] [Google Scholar]
- Gisbert JP & Pajares JM (2004). Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection -- a critical review. Aliment Pharmacol Ther, 20(10), 1001–17. [DOI] [PubMed] [Google Scholar]
- Hirsch EB, Chang KT, Zucchi PC, Francoeur DN, Ledesma KR, Tam VH & Lasco TM (2014). An evaluation of multiple phenotypic screening methods for Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae. J Infect Chemother, 20(3), 224–7. [DOI] [PubMed] [Google Scholar]
- Hotelling H (1933). Analysis of a complex of statistical variables into principal components. J Educ Psychol, 24, 417–441. [Google Scholar]
- Julak J, Prochazkova-Francisci E, Stranska E & Rosova V (2003). Evaluation of exudates by solid phase microextraction-gas chromatography. J Microbiol Methods, 52(1), 115–22. [DOI] [PubMed] [Google Scholar]
- Julak J, Stranska E, Prochazkova-Francisci E & Rosova V (2000). Blood cultures evaluation by gas chromatography of volatile fatty acids. Med Sci Monit, 6(3), 605–10. [PubMed] [Google Scholar]
- Junger M, Vautz W, Kuhns M, Hofmann L, Ulbricht S, Baumbach JI, Quintel M & Perl T (2012). Ion mobility spectrometry for microbial volatile organic compounds: a new identification tool for human pathogenic bacteria. Appl Microbiol Biotechnol, 93(6), 2603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviranta H, Tuomainen A, Reiman M, Laitinen S, Liesivuori J & Nevalainen A (1998). Qualitative identification of volatile metabolites from two fungi and three bacteria species cultivated on two media. Cent Eur J Public Health, 6(4), 296–9. [PubMed] [Google Scholar]
- Koo S, Thomas HR, Daniels SD, Lynch RC, Fortier SM, Shea MM, Rearden P, Comolli JC, Baden LR & Marty FM (2014). A breath fungal secondary metabolite signature to diagnose invasive aspergillosis. Clin Infect Dis, 59(12), 1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovats E (1958). Gas-Chromatographische Charakterisierung Organischer Verbindungen .1. Retentionsindices Aliphatischer Halogenide, Alkohole, Aldehyde Und Ketone. Helvetica Chimica Acta, 41(7), 1915–1932. [Google Scholar]
- Lechner M, Fille M, Hausdorfer J, Dierich MP & Rieder J (2005). Diagnosis of bacteria in vitro by mass spectrometric fingerprinting:a pilot study. Curr Microbiol, 51(4), 267–9. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Demilo AB, Moreno DS & Martinez AJ (1995). Analysis of the volatile components of a bacterial fermentation that is attractive to the mexican fruit-fly, Anastrepha ludens. J Agric Food Chem, 43(5), 1348–1351. [Google Scholar]
- Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ & Blattner FR (2005). Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem, 280(16), 15921–7. [DOI] [PubMed] [Google Scholar]
- Mann HB & Whitney DR (1947). On a Test of Whether One of 2 Random Variables Is Stochastically Larger Than the Other. Ann Math Stat, 18(1), 50–60. [Google Scholar]
- Podschun R & Ullmann U (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev, 11(4), 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees CA, Franchina FA, Nordick KV, Kim PJ & Hill JE (2016a). Expanding the Klebsiella pneumoniae volatile metabolome using advanced analytical instrumentation for the detection of novel metabolites. J Appl Microbiol,. [DOI] [PubMed] [Google Scholar]
- Rees CA, Smolinska A & Hill JE (2016b). The volatile metabolome of Klebsiella pneumoniae in human blood. J Breath Res, 10(2), 027101. [DOI] [PubMed] [Google Scholar]
- Robacker DC & Bartelt RJ (1997). Chemicals attractive to Mexican fruit fly from Klebsiella pneumoniae and Citrobacter freundii cultures sampled by solid-phase microextraction. J Chem Ecol, 23(12), 2897–2915. [Google Scholar]
- Schulz S & Dickschat JS (2007). Bacterial volatiles: the smell of small organisms. Nat Prod Rep, 24(4), 814–42. [DOI] [PubMed] [Google Scholar]
- Scott-Thomas AJ, Syhre M, Pattemore PK, Epton M, Laing R, Pearson J & Chambers ST (2010). 2-Aminoacetophenone as a potential breath biomarker for Pseudomonas aeruginosa in the cystic fibrosis lung. BMC Pulm Med, 10, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Nanda R & Chakraborty T (2013). Clinical application of volatile organic compound analysis for detecting infectious diseases. Clin Microbiol Rev, 26(3), 462–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnayderman M, Mansfield B, Yip P, Clark HA, Krebs MD, Cohen SJ, Zeskind JE, Ryan ET, Dorkin HL, Callahan MV, Stair TO, Gelfand JA, Gill CJ, Hitt B & Davis CE (2005). Species-specific bacteria identification using differential mobility spectrometry and bioinformatics pattern recognition. Anal Chem, 77(18), 5930–7. [DOI] [PubMed] [Google Scholar]
- Storer MK, Hibbard-Melles K, Davis B & Scotter J (2011). Detection of volatile compounds produced by microbial growth in urine by selected ion flow tube mass spectrometry (SIFT-MS). J Microbiol Methods, 87(1), 111–3. [DOI] [PubMed] [Google Scholar]
- Syhre M & Chambers ST (2008). The scent of Mycobacterium tuberculosis. Tuberculosis (Edinb), 88(4), 317–23. [DOI] [PubMed] [Google Scholar]
- Syhre M, Manning L, Phuanukoonnon S, Harino P & Chambers ST (2009). The scent of Mycobacterium tuberculosis--part II breath. Tuberculosis (Edinb), 89(4), 263–6. [DOI] [PubMed] [Google Scholar]
- Tait E, Perry JD, Stanforth SP & Dean JR (2014). Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. J Chromatogr Sci, 52(4), 363–73. [DOI] [PubMed] [Google Scholar]
- Zechman JM, Aldinger S & Labows JN Jr. (1986). Characterization of pathogenic bacteria by automated headspace concentration-gas chromatography. J Chromatogr, 377, 49–57. [DOI] [PubMed] [Google Scholar]
- Zhu J, Bean HD, Jimenez-Diaz J & Hill JE (2013a). Secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting of multiple bacterial lung pathogens, a mouse model study. J Appl Physiol (1985), 114(11), 1544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Bean HD, Wargo MJ, Leclair LW & Hill JE (2013b). Detecting bacterial lung infections: in vivo evaluation of in vitro volatile fingerprints. J Breath Res, 7(1), 016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.