Abstract
Background
Androgen deprivation therapy is a first-line treatment for disseminated prostate cancer (PCa). However, virtually all tumors become resistant and recur as castration-resistant PCa, which has no durable cure. One major hurdle in the development of more effective therapies is the lack of preclinical models that adequately recapitulate the heterogeneity of PCa, significantly hindering the ability to accurately predict therapeutic response.
Objective
To leverage the ex vivo culture method termed patient-derived explant (PDE) to examine the impact of PCa therapeutics on a patient-by-patient basis.
Design, setting, and participants
Fresh PCa tissue from patients who underwent radical prostatectomy was cultured as PDEs to examine therapeutic response.
Outcome measurements and statistical analysis
The impact of genomic and chemical perturbations in PDEs was assessed using various parameters (eg, AR levels, Ki67 staining, and desmoplastic indices).
Results and limitations
PDE maintained the integrity of the native tumor microenvironment (TME), tumor tissue morphology, viability, and endogenous hormone signaling. Tumor cells in this model system exhibited de novo proliferative capacity. Examination of the native TME in the PDE revealed a first-in-field insight into patient-specific desmoplastic stromal indices and predicted responsiveness to AR-directed therapeutics.
Conclusions
The PDE model allows for a comprehensive evaluation of individual tumors in their native TME to ultimately develop more effective therapeutic regimens tailored to individuals. Discernment of novel stromal markers may provide a basis for applying precision medicine in treating advanced PCa, which would have a transformative effect on patient outcomes.
Patient summary
In this study, an innovative model system was used to more effectively mimic human disease. The patient-derived explant (PDE) system can be used to predict therapeutic response and identify novel targets in advanced disease. Thus, the PDE will be an asset for the development of novel metrics for the implementation of precision medicine in prostate cancer.
The patient-derived explant (PDE) model allows for a comprehensive evaluation of individual human tumors in their native tumor microenvironment (TME). TME analysis revealed first-in-field insight into predicted tumor responsiveness to AR-directed therapeutics through evaluation of patient-specific desmoplastic stromal indices.
Keywords: Androgen receptor, Ex vivo, Prostate cancer, Tumor microenvironment
1. Introduction
Prostatic adenocarcinoma (PCa) is the most frequently diagnosed noncutaneous malignancy and the second leading cause of cancer-related death in American men [1,2]. As prostate tumor cells are reliant on the androgen receptor (AR), first-line therapy for disseminated disease involves targeting the AR signaling axis via androgen deprivation therapy (ADT), often coupled with antiandrogens [3,4]. Although initially effective, these patients develop resistance and experience relapse within a median of 3–4 yr on the development of castration-resistant PCa (CRPC) [5]. Intriguingly, CRPC remains largely AR-dependent due to aberrant reactivation of AR through multiple distinct mechanisms that promote cell survival and proliferation [1,6]. Currently, metastatic CRPC remains universally fatal with no effective cure, highlighting the need to further define mechanisms that control AR activity and thus develop novel means to target recurrent AR activity.
The lack of adequate preclinical models is a major hurdle in discerning innovative methods to target AR and effectively combat PCa. Many promising new therapies that perform well in preclinical evaluations ultimately fail in the clinic, which may reflect the fact that conventional cell lines lack predictive value for drug treatments in clinical disease [7–11]. Studies suggest that the generation of cell lines results in major genetic alterations, loss of tumor heterogeneity, and alterations in growth and invasion properties. Furthermore, culturing cell lines alone deprives them of microenvironmental signal reciprocity. With such key limitations, cell lines cultured as two-dimensional monolayers are not the ideal preclinical platform for studying personalized medicine. To overcome these limitations, patient-derived organoids have recently been developed as three-dimensional culture methods to better recapitulate the biological characteristics of the original tumor [12–14]. However, this technique still fails to effectively mimic the intricate microenvironmental influences contributing to human disease. Thus, there remains a need for improved systems that retain patient-specific genetic alterations and tumor microenvironment (TME) signaling to more accurately nominate effective therapies.
One exciting advance for personalized medicine is the use of patient-derived xenograft (PDX) models, which are increasingly used in translational cancer approaches for drug screening, biomarker development, and preclinical evaluation of therapies through the use of murine “avatars” in co-clinical trials [15]. However, the time to engraftment is a major limitation for the PDX model since it can require between 4 and 8 mo to develop “avatars”. Moreover, although PDXs are effective for many types of cancer, they typically have poor engraftment rates for genitourinary cancers, including PCa, and fail to recapitulate cancer heterogeneity and TME characteristics [16–19]. The TME regulates cellular responses to hormones, growth factors, and therapeutic agents, and is thus a major determinant of carcinogenesis and response to therapeutic intervention. Thus, models that preserve the native human TME are vital for effective evaluation of drug responses.
Building on models previously described [9], we used an ex vivo explant model termed patient-derived explant (PDE) to assess drug responses in clinical PCa specimens. The PDE system involves a more simplified and rapid approach to culturing of patient tissue immediately after radical prostatectomy than has been used in current patient-derived models. Importantly, the PDE maintains native TME integrity, tumor tissue morphology, viability, and endogenous AR signaling. Tumor cells in this model system exhibited de novo proliferative capacity, which was an asset when investigating the impact of PDE use for predicting the efficacy of selected therapies. Lastly, examination of the native TME in the PDE revealed first-in-field insight into patient-specific desmoplastic stromal indices that predicted responsiveness to AR-directed therapeutics. In summary, the PDE model more closely mimics TME-inclusive human disease and can facilitate the development of novel metrics for the implementation of precision medicine in PCa.
2. Patients and methods
2.1. Patient selection
We used material from patients undergoing radical prostatectomy at Thomas Jefferson University (TJU; Philadelphia, PA, USA) and the Royal Adelaide Hospital (Adelaide, Australia). Matched non-neoplastic and tumor tissues were obtained from each patient’s prostate after radical prostatectomy. Patient demographics are described in Supplementary Table 1.
2.2. Explant establishment
Patient de-identified prostate tissues were established as ex vivo explant cultures as previously described [9,20,21]. The institutional review board of TJU and the human research ethics committee of the University of Adelaide reviewed the study protocol and deemed the research to be in compliance with federal regulations [45 CFR 46.102(f)].
2.3. Immunohistochemistry
For histological analysis, formalin-fixed, paraffin-embedded (FFPE) sections were stained with hematoxylin and eosin (H&E) using standard techniques. For Ki67 analysis, tissues were stained as previously described [22] and scored by a board-certified clinical pathologist using an Aperio microscope and software. The following antibodies were used in the Ki67 protocol at 1:50 dilution for immunohistochemistry (IHC) staining of tissue: HIF1α (Novus Biologicals, Littleton, CO, USA), AR (in house), prostate-specific antigen (PSA; Abcam, Cambridge, UK) and 5-bromo-2-deoxyuridine (BrdU; BD Biosciences, Franklin Lakes, NJ, USA). Clinical grade IHC staining was performed by the clinical pathology department at TJU using a cocktail of antibodies to α-methyl-coA racemase (AMCAR) and the basal cell markers p63 and HMW keratin. Alkaline phosphatase was used to detect racemase expression, while 3,3′-diaminobenzidine was used to detect basal cells.
2.4. Enzyme-linked immunosorbent assay
PSA in PDE conditioned culture media diluted 1:50 was detected using a total PSA enzyme-linked immunosrbent assay (ELISA) kit (ALPCO, Salem, NH, USA).
2.5. Gene expression analysis
RNA was isolated from tissue using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). cDNA was generated using SuperScript VILO (ThermoFisher, Waltham, MA, USA). Quantitative PCR was conducted using primers as previously described [23] with PowerSybr and an ABI StepOne system (ThermoFisher) according to the manufacturer’s specifications.
2.6. shRNA lentiviral infection
Lenti-X HT (open Biosystems, Huntsville, AL, USA) was used to package the shCON and shAR lentiviral constructs according to the manufacturer’s instructions and as previously described [24]. In brief, once generated, the lentiviral constructs were added to the explant tissue culture medium for 48 h. The lentiviral medium was removed and replaced with fresh medium containing no lentivirus for an additional 48 and/or 96 h. Tissues were harvested and fixed as described above.
2.7. TME analysis
Simultaneous multiplex immunofluorescence (SMI) labeling of FFPE tissue sections, image acquisition, and quantification of pFAK and active α5-integrin in the TME were performed as previously described in detail [25]. High-throughput analysis of the multispectral monochromatic images acquired was performed using SMIA-CUKIE software (https://github.com/cukie/SMIA) as previously described [25].
2.7. Statistical analysis
Data are presented as mean ± standard error of the mean. Statistical significance (p < 0.05) was determined using Student’s t test on GraphPad Prism software (www.graphpad.com/scientific-software/prism).
3. Results
3.1. The PDE PCa model maintains tumor morphology, viability, and endocrine signaling
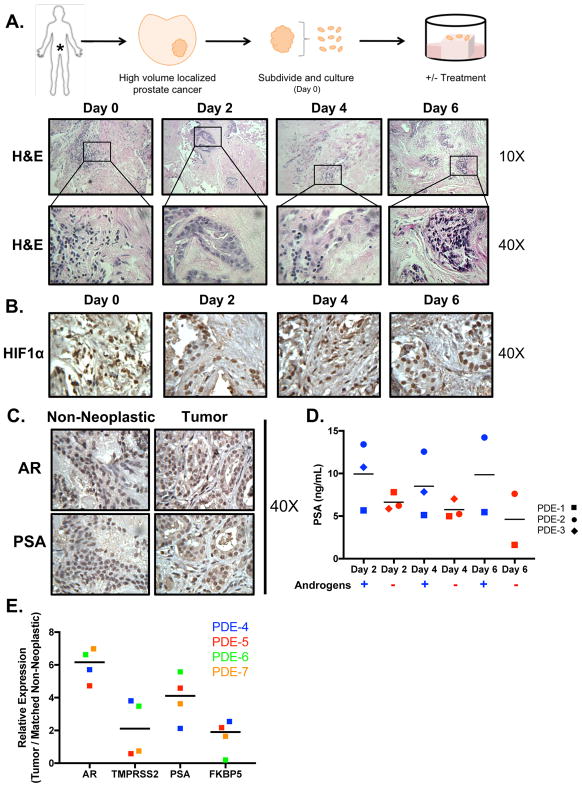
A major barrier in the PCa field is the paucity of preclinical models that effectively recapitulate human disease, thereby limiting mechanistic understanding and the development of novel therapies to combat the reactivated AR signaling observed in CRPC. Current preclinical models have limitations that include lack of complete genetic fidelity and loss of cell-to-cell cross-talk with the TME. The PDE model addresses these shortcomings through maintenance of the complex tumor heterogeneity and native local TME. To evaluate PDE utility for assessing therapeutic response in the context of an intact TME, tissue from 24 patients (Supplementary Table 1) was obtained via radical prostatectomy and analyzed. Tissues were obtained from patients with high-volume PCa, designated as Gleason grade ≥7. Tumor and matched non-neoplastic tissue from each patient were subdivided on the day of resection (day 0) and grafted onto wound-healing sponges as previously described [9,20,21] before administration of therapeutic agents or appropriate controls (Fig. 1A top). Histological analysis by a board-certified clinical pathologist revealed that PCa tissue could be maintained in culture for at least 6 d, retaining tumor and stroma morphology similar to that seen at the time of resection (Fig. 1A bottom). Moreover, the PDE sustained similar levels of HIF1α [26], indicating that changes in oxygen supply did not affect tumor biology throughout the time of ex vivo culture (Fig. 1B).
Figure 1. PCa PDE model sustains tumor morphology, viability, and endogenous endocrine signaling.
A. Top: Depiction of the method used to culture patient tumors ex vivo. Drug treatment can be added to the media to investigate the effect on tumor growth. Media (and appropriate treatment) were replaced every other day. Tissue was harvested and fixed in 4% formalin. The formalin-fixed tissue was then embedded into paraffin blocks and cut into sections with a microtome. Bottom: Slides were stained with hematoxylin and eosin (H&E). The representative images shown (10× and 40×) indicate maintenance of gross tumor morphology after culturing ex vivo for up to 6 days. B. PDE were stained for HIF1α showing that the tumors received sufficient oxygen supply to maintain viability. C. AR and PSA immunostaining of (patient #5) tissue demonstrated sustained endogenous and endocrine signaling in the explants after 6 days of ex vivo culture. All IHC images are shown as 40× magnification D. PSA secreted into media of PDE was analyzed at Days 2, 4, and 6 via ELISA in hormone proficient media (‘Androgen Proficient’ – square shape) and hormone-deficient media (‘Androgen Deprived’ – diamond shape). E. Expression of AR and AR target genes (TMPRSS2, PSA, and FKBP5) in explants from four different PDE shown as relative expression normalized to 18S. Tumor tissue was matched to their respective non-neoplastic tissue control.
As the AR is a critical driver of disease initiation and progression, alterations in AR signaling between original and cultured tissues were assessed by examining the expression of AR and a well-defined target gene, PSA. As shown in Figure 1C, the PDE model maintained endogenous AR expression and activity, as determined by IHC for AR and PSA, after 6 d (Supplementary Fig. 1A, B). Cell type–specific keratin and AMACR staining was used to validate cancer and non-neoplastic tissue histology [27] and no antibody staining served as a negative control for IHC (Supplementary Fig. 1C, D). Next, androgen-regulated PSA secretion was evaluated over the course of the PDE culture via an ELISA of the culture medium (Fig. 1D). Tissue from PDE-1 exhibited a delayed response to androgen deprivation, as PSA levels did not decrease with androgen deprivation until day 6, when there was a 29.6% reduction in PSA levels. PDE-2 and PDE-3 showed rapid responses, with 46.3% and 54.6% reductions in PSA levels, respectively, after only 2 d of androgen deprivation. PDE-2 maintained this reduction through day 6. No information for PDE-3 on day 6 was available; however, observations on days 2 and 4 showed that PSA levels consistently decreased under androgen deprivation, suggesting that this trend would probably continue to day 6. Overall, the three PDEs displayed a heterogeneous response to androgen deprivation in culture, closely recapitulating the patient variability observed in the clinic (Fig. 1D). Lastly, AR and expression of direct AR target genes assessed by quantitation of TMPRSS2, KLK3 (PSA), and FKBP5 mRNA levels were determined in the PDE after 6 d of ex vivo culture in androgen-proficient media (Fig. 1E). On average, tumor tissue showed higher endogenous AR signaling compared to the matched non-neoplastic tissue. Combined, these findings indicate that PDEs preserve pathological features of the original tumors after resection, retain AR signaling, and illustrate patient-specific variances in response to androgen deprivation.
3.2. Tumor cells in the PDE model exhibit de novo proliferative capacity
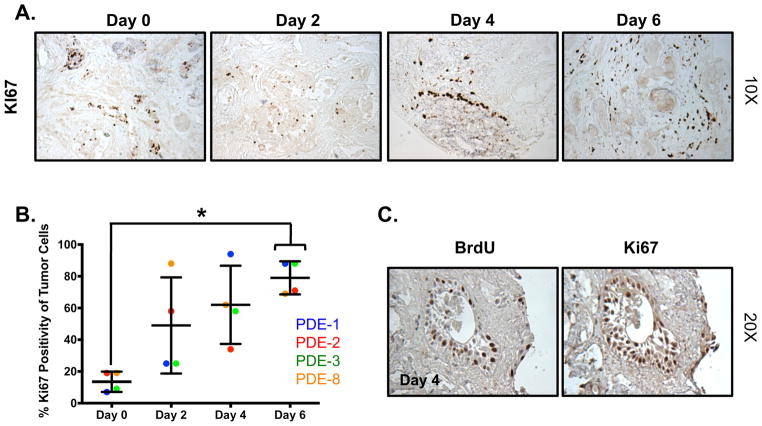
To further characterize PDE tumor biology in response to prolonged culture, the cellular proliferative index was assessed by determining the percentage of Ki67-positive epithelial tumor cell nuclei in each specimen. As shown in Figure 2A, B, tumor cells in the PDE showed an increase in proliferation from 7–19% on day 0 to 34–94% on day 4 and 69–88% on day 6. Importantly, enhanced proliferation was limited to the tumor cells, as the surrounding stroma and non-neoplastic tissue showed no change in Ki67 over the day 0 control. Next, to confirm that the increase in proliferative index as assessed by Ki67 was associated with active DNA replication, cellular uptake of BrdU was analyzed. Nuclear BrdU staining was concordant with Ki67 positivity, indicating that tumor cells were actively replicating (de novo proliferation; Fig. 2C). In sum, these findings demonstrate a time-dependent increase in proliferation of tumor cells, suggesting that the PDE model unmasks cells with proliferative potential during the culture process. These results highlight one of the strengths of the PDE model—de novo proliferation—which can be used to evaluate preclinical impact in response to hormonal stimuli and/or therapeutic agents.
Figure 2. Tumor cells in PDE exhibit de novo proliferative capacity.
A. Tissue was cultured in complete media and harvested every 48 hours for up to 6 days. Ki67 staining was performed to determine the amount of proliferation in the explants. Images are shown at 10X magnification. B. Quantification of Ki67 immunostaining showed a time-dependent increase in proliferation. PDE 3 is shown in panel A. *p<0.05. C. BrdU uptake is similar to Ki67 staining. Images are shown at 20X magnification, n=4.
3.3. The PDE model is amenable to both genetic and pharmacological perturbations
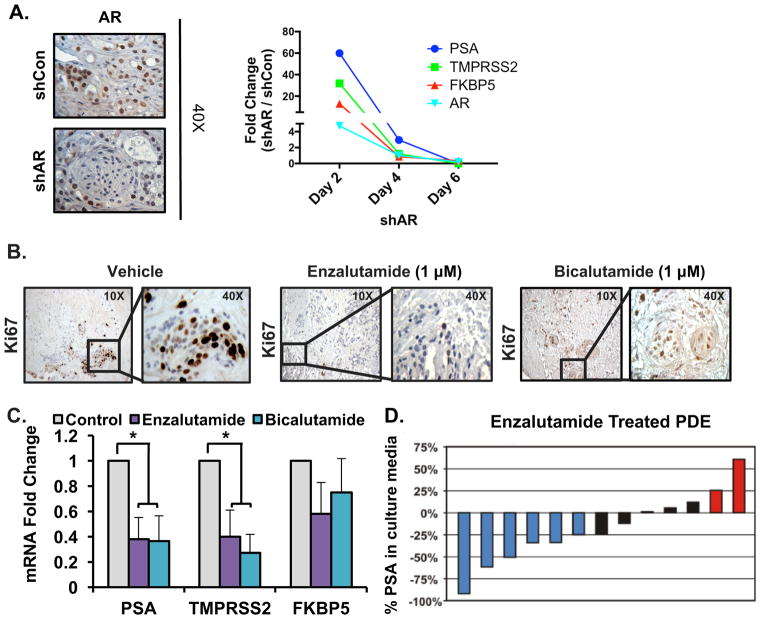
As PCa is primarily dependent on AR signaling for cell survival and proliferation, and CRPC is characterized by aberrant activation of the AR axis, AR remains a primary therapeutic target in all stages of the disease [1]. First-line therapy to combat disseminated PCa involves androgen deprivation and/or AR antagonists. To demonstrate the capacity of the PDE model to evaluate therapeutic response, genetic (shAR) and pharmacological (AR antagonist) targeting strategies were used. Genetic manipulation of AR was performed by transducing PDE with a lentiviral-delivered shRNA directed at the AR gene for isogenic suppression or a GFP-expressing lentivirus to show the efficiency of infection (Fig. 3A and Supplementary Fig. 2). PDE transduced with shAR lentivirus for 6 d exhibited a notable decrease in AR protein expression by approximately 50% (Fig. 3A). AR mRNA correspondingly decreased to less than 0.2-fold (comparing shAR and shCon in tumor tissue) on day 6, with shAR indicating efficient knockdown of AR. Furthermore, the expression of AR target genes (PSA, TMPRSS2, and FKBP5) significantly decreased to less than 0.4-fold compared to shCon in tumor tissue by day 6. Thus, PDEs are amenable to genetic manipulation of AR impacting its expression and activity ex vivo. In addition, the proliferative capacity after pharmacological inhibition was determined by scoring Ki67. PDEs exhibited varying responses to two distinct AR antagonists, enzalutamide and bicalutamide. While enzalutamide markedly decreased Ki67 staining (~50% reduction), bicalutamide treatment did not effectively impact Ki67 staining compared to control treated tissue (Fig. 3B). Nonetheless, both antagonists significantly decreased AR signaling (Fig. 3C). PSA and TMPRSS2 expression decreased by approximately 60%, while FKBP5 trended towards significance after treatment. Furthermore, PCa tissue from 13 different PDEs demonstrated a broad range of PSA secretory responses to 10 μM enzalutamide treatment. Of 13 tissue samples, six (46.1%) responded with a ≥25% decrease in secreted PSA, five (38.5%) showed no significant change in PSA, and two (15.4%) exhibited a ≥25% increase in secreted PSA (Fig. 3D). Similar to the difference in proliferative response between different AR antagonists in individual samples (Fig. 3B), the broad range of responses to enzalutamide treatment across multiple patient samples underscores the heterogeneity of clinical samples, and probably reflects variations in responsiveness across the patient population as a whole. Together, these data illustrate that AR signaling can be perturbed both genetically and pharmacologically in the PDE model and indicate the potential of this system in predicting treatment outcomes and informing personalized medical decisions.
Figure 3. AR signaling can be perturbed both genetically and pharmacologically in the PDE model.
A. Knockdown of AR was achieved through lentirviral transduction of PDE with an AR directed shRNA for 6 days. Representative images show AR and Ki67 IHC for shCon and shAR after 6 days. Right: Expression of AR and AR target genes (PSA, FKBP5, and TMPRSS2) mRNA expression from tumor samples treated with shCon and shAR for 2, 4, and 6 days, n=3. B. Representative images (10X and 40X magnification) are shown for vehicle, AR antagonist (1 μM Enzalutamide), and AR antagonist (1 μM Bicalutamide) treated tumors after 6 days of treatment, n=3. C. Expression of AR target genes (PSA, TMPRSS2, and FKBP5) mRNA expression from vehicle, 1 μM Enzalutamide, and 1 μM Bicalutamide treated PDE. Average fold change of three PDE shown. n=3, *p< 0.05. D. Waterfall plot depicting the percentage change of PSA in the culture medium with 1 μM Enzalutamide treatment. Blue bars represent patients with ≥ 25% decrease in PSA, black bars represent no change in PSA, and red bars represent ≥ 25% increase in PSA.
3.4. Impact of clinically approved and experimental PCa therapeutics on PDE models
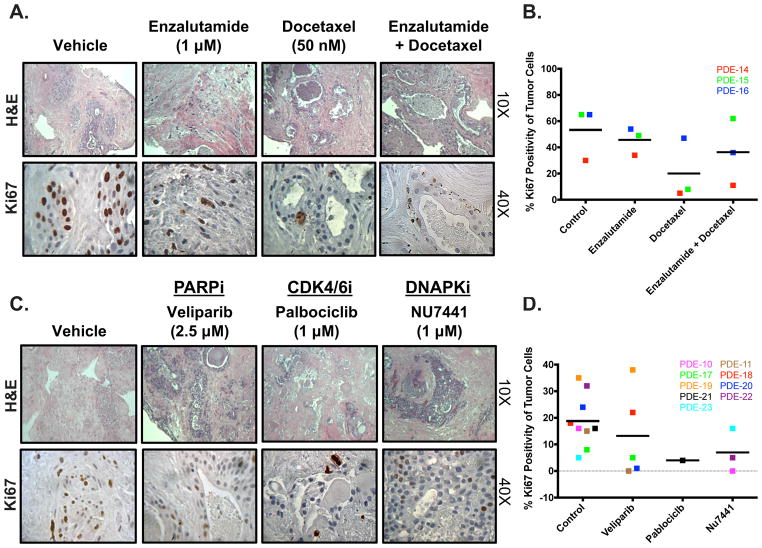
Current therapies for advanced PCa involve targeting the AR signaling axis in combination with various treatments, including taxane-based chemotherapy [28]. To evaluate the impact of such clinically approved regimens in the PDE system, PCa tissue from individual patients was treated with 1 μM enzalutamide, 50 nM docetaxel, or the combination for 6 d, at which point the proliferative index was measured (Fig. 4A, B). Figure 4A and Supplementary Figure 3 show H&E staining and Ki67 IHC for PDE-14, which did not respond to enzalutamide treatment alone. However, docetaxel alone or in combination with enzalutamide effectively reduced proliferation by 83.7% and 63.3%, respectively, compared to controls. PDE-16 showed minor decreases with single agents (reductions of 16.9% with enzalutamide and 27.7% with docetaxel) and a greater decrease with the combination (45.6% reduction in Ki67 positivity). Conversely, PDE-15 showed a more potent response to docetaxel alone (87.7% reduction) than to enzalutamide alone (24.6% reduction), while the combination did not influence proliferation. The heterogeneous nature of responses in the PDE model reflect those observed in the clinical setting, suggesting that PDEs may be able to predict the efficacy of drug treatment on an individual basis.
Figure 4. PDE model is responsive to clinically approved and experimental PCa therapeutics.
Tissue was cultured in complete media, treated with different drugs, and harvested after 6 days. Drug treatment was changed out every 48 hours. Ki67 staining was performed to determine the amount of proliferation that occurred in order to evaluate the effect of drug treatment on PDE growth. A. Representative H&E image (10X magnification) and Ki67 image (40X magnification) are shown for vehicle, AR antagonist (1 μM Enzalutamide), Taxane (50 nM Docetaxel), and AR antagonist (Enzalutamide) + Taxane (Docetaxel) treated tumors (PDE-14). B. Quantification of KI67 immunostaining from 3 separate patient samples. Sample depicted in red is the PDE used for images in A. C. Representative H&E image (10X magnification) and Ki67 image (40X magnification) are shown for vehicle, PARP Inhibitor (2.5 μM Veliparib) (PDE-17), CDK4/6 Inhibitor (1 μM Palbociclib) (PDE-21), and DNAPK Inhibitor (1 μM NU7441) (PDE-22). D. Quantification of KI67 immunostaining from 3 separate patient samples. Sample depicted in green (Veliparib), black (Palbociclib), and yellow (NU7441) are the PDE used for images in C.
Since current preclinical models lack efficacy in predicting therapeutic response, novel techniques are crucial for the development of clinically relevant targeted therapies. Previous preclinical studies showed that targeting of DNA repair factors (PARP or DNAPK) or cell cycle factors (CDK4/6) could be effective in combating a subset of advanced PCa [20,29]. Clinical trials are under way to test the efficacy and impact of PARP inhibitors and CDK4/6 inhibitors (including palbociclib) [30–32]. To assess the capacity of the PDE system to evaluate experimental PCa therapeutics, tissues were treated with 2.5 μM veliparib (PARP inhibitor), 1 μM palbociclib (CDK4/6 inhibitor), or 1 μM NU7441 (DNA-PK inhibitor) (Fig. 4C, D). Overall, five of the nine tissues responded to targeted therapy, as evidenced by a decrease in the proliferative index compared to the vehicle controls. Veliparib decreased Ki67 positivity by 99.3% (PDE-20) and 95.9% (PDE-11). DNA-PK inhibition via NU7441 decreased Ki67 positivity by 99.4% (PDE-22) and 84.4% (PDE-10). Lastly, palbociclib treatment in PDE-21 decreased the proliferative index by 75% (Fig. 4D). Four of the nine PDEs used (44.4%) did not respond to any one specific treatment tested, which could suggest clinical therapeutic resistance. In sum, these data further emphasize the capacity of the PDE system to model patient heterogeneity and its potential to fill the immediate need for better models to personalize therapeutic regimens in the clinic.
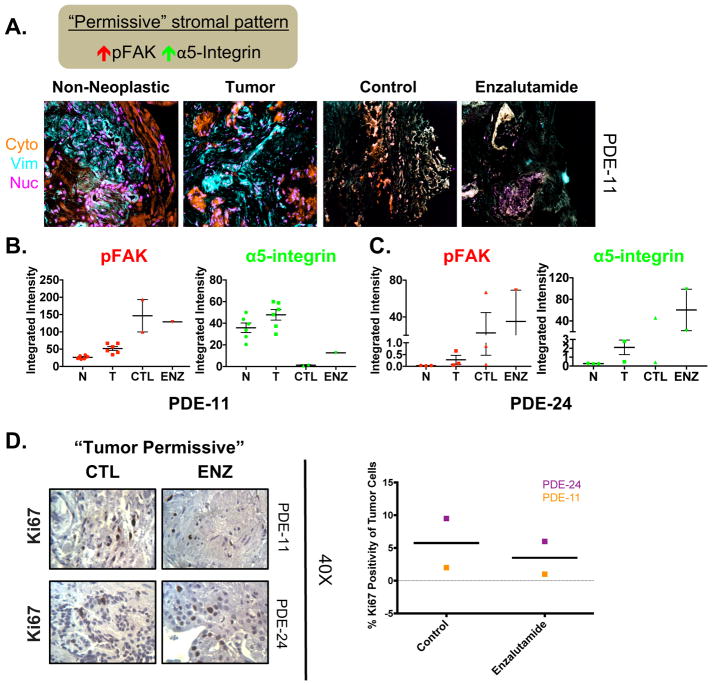
3.5. Initial tumor-permissive stromal pattern in the PDE model indicates possible resistance to enzalutamide
A normal stroma, encompassing naive fibroblastic cells and self-derived extracellular matrix, enforces cellular homeostasis and naturally suppresses tumor development [33–36]. However, it has been reported that desmoplasia, a chronic fibrosis-like state that typically accompanies epithelial carcinomas, plays critical roles in both promoting and preventing tumor progression [19,25,37]. Two stromal signatures based on combined levels of constitutively active pFAK and the active conformation of α5β1-integrin (active α5-integrin) were recently developed that are indicative of the two types of desmoplasia: tumor-permissive, with high pFAK plus active α5-integrin; and tumor-restrictive, with low pFAK plus active α5-integrin [19,25,33]. An example stroma characterized as tumor-permissive on the basis of constitutively high pFAK and active α5-integrin is shown in Fig. 5A, B. Tumor areas were recognized as cytokeratin-positive, while areas negative for cytokeratin and positive for vimentin identified areas (or masks) indicative of stroma (Fig. 5A). With tumor and stroma boundaries identified, SMI was implemented and levels of stromal pFAK and active α5-integrin were measured. Elevated pFAK and active α5-integrin levels relative to matched non-neoplastic tissue were observed, so the stroma of the tumor tissue was assessed as tumor-permissive (Fig. 5B). Sample PDE-11 exhibited a higher level of active stroma (tumor-permissive stromal pattern, with high pFAK and active α5-integrin) than PDE-24 in both its normal and tumor samples on day 0 (Fig. 5B, C). Interestingly, when treated with enzalutamide, PDE-24 exhibited a further increase in the levels of pFAK and active α5-integrin (Fig. 5C), perpetuating the tumor-permissive stromal traits despite the presence of a potent antitumor agent. To evaluate the biological impact of enzalutamide treatment in these tissues with a permissive stromal pattern, the proliferative index was assessed via Ki67 IHC. Both PDE-24 and PDE-11 maintained similar levels of Ki67 positivity with no significant decrease in proliferative index on AR antagonist treatment (Fig. 5D). Taken together, these results suggest that the initial desmoplasia state, rather than changes during treatment, could be predictive of de novo resistance to enzalutamide treatment.
Figure 5. Initial “tumor permissive” stromal pattern in the PDE model indicates possible resistance to enzalutamide.
A. Top: Schematic explaining TME characteristics indicative of “permissive” stromal type. Bottom: Representative Simultaneous Multiplex Immunofluorescence (SMI) image of masks (i.e. cytokeratin, vimentin, and nucleus) in PDE of non-neoplastic at Day 0 (N), tumor at Day 0 (T), and tumor tissue treated with either vehicle (CTL) or Enzalutamide (ENZ) for 6 days. Example of a “tumor permissive” TME with desmoplastic traits displaying high pFAK and high active α-5-integrin in tumor tissue. B. Quantification of the markers assessed in stromal areas: phospho-focal adhesion kinase (pFAK) and active α5-integrin. C. Additional patient with a “tumor permissive” stromal pattern. D. Ki67 staining was performed to determine the amount of proliferation that occurred in order to evaluate the effect of drug treatment on PDE growth. Representative Ki67 images (40X magnification) and quantification are shown for vehicle and AR antagonist (Enzalutamide) treated PDE. n=2.
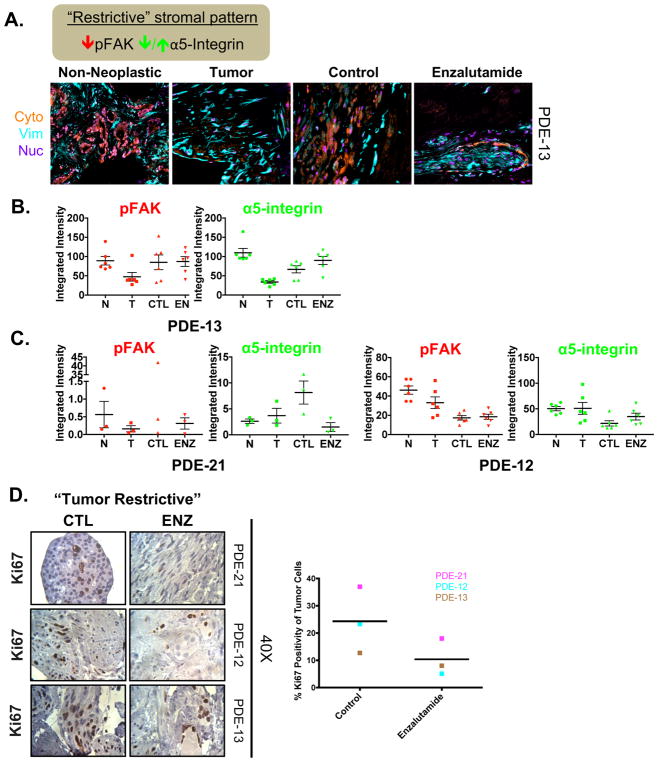
3.6. Enzalutamide reinforces the tumor-restrictive desmoplastic stromal pattern in the PCa PDE model
In contrast to the tumor-permissive pattern, low pFAK plus active α5-integrin represents a desmoplastic tumor-restrictive stromal pattern. As before, stromal areas were recognized via cytokeratin-negative and vimentin-positive masks, while the tumor region was identified via increased cytokeratin-positive staining (Fig. 6A). SMI and quantification of pFAK and α5-integrin of PDE-13 showed low pFAK in the stroma of the tumor tissue compared to the matched non-neoplastic tissue on day 0 (Fig. 6B). PDE-21 and PDE-12 also exhibited tumor-restrictive stromal patterns, with low pFAK and low or unchanged active α5-integrin in the stroma of tumor tissue compared to non-neoplastic tissue on day 0 (Fig. 6C). Intriguingly, AR antagonist treatment caused a significant reduction in pFAK and α5-integrin in PDE-21, thereby enhancing the tumor-restrictive traits (Fig. 6C). These desmoplastic traits indicate that the stroma was inactive at the initiation of PDE generation, and therefore incapable of protecting the tumor against treatment. Thus, to evaluate the biological impact of enzalutamide treatment in these samples with a tumor-restrictive stromal pattern, the proliferative index was assessed via Ki67 IHC. All three PDE samples (21, 12, and 13) showed a decrease in Ki67 positivity of 51.3%, 78.1%, and 37.0%, respectively (Fig. 6D). Thus, low pFAK in the stroma of PCa tumor tissue compared to non-neoplastic tissue before therapeutic intervention reflects an inactive tumor-associated stroma that does not interfere with AR antagonist treatment, resulting in significantly lower proliferation on treatment. Combined, these data further suggest that the initial desmoplasia state could predict responsiveness to AR antagonist treatment and provide evidence that the TME influences and/or informs PCa behavior.
Figure 6. Enzalutamide reinforces the “tumor restrictive” desmoplastic stromal pattern in the PDE model for PCa tissue.
A. Top: Schematic explaining TME characteristics indicative of “tumor restrictive” stromal type. A. Representative immunofluorescence (IF) image of masks (i.e. cytokeratin, vimentin, and nucleus) in PDE of non-neoplastic at Day 0 (N), tumor at Day 0 (T), and tumor tissue treated with either vehicle (CTL) or Enzalutamide (ENZ) for 6 days. Example of a tumor “tumor restrictive” TME with desmoplastic traits displaying low pFAK in tumor tissue. B. Quantification of the markers assessed in stromal areas: phospho-focal adhesion kinase (pFAK) and α5-integrin. C. Additional patients with “tumor restrictive” stromal patterns. D. Ki67 staining was performed to determine the amount of proliferation that occurred in order to evaluate the effect of drug treatment on PDE growth. Representative Ki67 images (40X magnification) and quantification are shown for vehicle and AR antagonist (Enzalutamide) treated PDE. n=3.
4. Discussion
One significant barrier in developing more effective therapies for PCa is the lack of adequate preclinical models to effectively mimic human disease in the laboratory. This significantly hinders the ability to accurately predict individual therapeutic responses. Recent reports characterized and used PDE as a method to culture patient tumors ex vivo and effectively recapitulate the heterogeneity and microenvironment of human cancers [20,22,29,38,39]. Building on those initial studies, the present study showed that the PDE model has several advantages: (1) maintenance of endogenous tumor morphology, viability, and AR signaling ex vivo; (2) a capacity for de novo proliferation in tumor cells that is an asset when examining the impact of therapeutic interventions; (3) amenability to both genetic and pharmacological perturbations impacting individual tumor growth; and (4) retention of the native, intact TME architecture that could potentially predict response to therapy. In short, the PDE model allows comprehensive evaluation of individual tumors in their native microenvironment; understanding the relationship between this microenvironment and tumor progression could ultimately be leveraged to develop more effective therapies.
In order to understand the role of patient-specific cancer alterations in tumorigenesis that impact therapeutic sensitivity, in vitro and in vivo model systems that accurately reflect genetic diversity are crucial. The PDE model complements current techniques and overcomes selected limitations of existing tissue-derived systems. Preclinical models such as cell lines, mouse models, organoids, and xenografts have been extensively used to evaluate the effectiveness of various therapies, leading to key discoveries. Although many of these techniques have yielded promising preclinical results, the translational impact has been limited [7–10]. One explanation for the disappointing clinical results from existing preclinical models is their inability to recapitulate the heterogeneity between individual tumors; moreover, they represent a limited spectrum of disease phenotypes. Importantly, primary patient-derived xenografts (PDXs) and organoids have been used to overcome that limitation and have had success for several cancer types [40–42]. However, PDXs have poor engraftment rates for genitourinary cancers such as PCa. Thus, mechanistic studies using PDX have proved challenging in the PCa field. Patient-derived organoids have been used as a means to develop multicellular entities capable of self-renewal and self-organization [43–45]. Similar to the PDE model, the organoid system allows for examination of the TME and its impact on tumorigenesis [46,47]. However, a limitation of this technique is that organoids are purely epithelial cultures lacking intact stromal, vascular endothelial, and/or immune cells. Importantly, the strength of the PDE model is that it maintains an intact TME that includes several stromal cell types, allowing in-depth investigation into stromal effects during tumorigenesis and/or drug treatments (Figs. 5 and 6). In addition, establishing organoids from primary PCa tissue material is technically challenging and has low efficiency rates [8], whereas the PDE model described here is rapid and technically simple.
Although the PDE model overcomes several limitations of existing tissue-derived systems, it does have some limitations. The PDE technique is ex vivo and thus is not a model in which to study de novo recruitment of immune cells. With that caveat in mind, ongoing studies have been designed to evaluate the impact of targeted therapies on existing immune cell function within the TME [48,49]. An additional limitation of PDEs is that they can only be cultured for a finite amount of time. Furthermore, the amount of tissue yielded by the PDE system is limited by the amount of starting material received following surgery. Nonetheless, the PDE model allows for discernment and manipulation of patient-specific characteristics (Figs. 3–6) and thus elegantly complements existing model systems.
Since the PDE model recapitulates the heterogeneity of responses observed in the clinical setting, this technique is currently being used in parallel with ongoing clinical trials assessing taxane-based chemotherapeutics (NCT02218606), DNA damage, and cell cycle inhibitors (NCT02555189) in metastatic PCa. These “co-clinical” trials use material obtained from needle biopsies before, during, and after progression on therapy. The goal is to query if the PDE system could serve as a rapid readout for therapeutic response at a patient-specific level. In sum, the PDE model complements existing tools aimed at discerning the molecular underpinnings of cancer biology, while adding the advantage of examining intact TME to potentially predict treatment efficacy and/or inform treatment selection.
Several studies have shown that the extracellular matrix and stroma facilitate tumor progression [34,50]; therefore, characterization of each individual TME using the PDE method could provide a novel criterion to assess responsiveness to treatment. Previous studies have characterized the TME using FAK and α5-integrin to describe tumor-permissive and tumor-restrictive phenotypes [19,25,33]. To date, no such study has been conducted in PCa; hence, this is the first study to examine these stromal parameters in relation to therapeutic sensitivity in PCa. A permissive stromal pattern before therapeutic intervention predicted resistance to enzalutamide treatment in the PDE model (Fig. 5), while the restrictive stromal pattern represented an inactive stroma that was incapable of protection against treatment (Fig. 6). While these studies are limited to a small cohort of patient tissues, they are the first in field to uncover a novel paradigm in which the initial desmoplastic characteristics of tumor-associated stroma have the potential to predict therapeutic response to AR-directed treatments. Future studies have been designed to effectively assess these stromal parameters on a large scale in clinical specimens. Furthermore, our studies suggest that the levels of stromal pFAK, in comparison to normal mesenchymal pFAK levels, may constitute a useful tool for predicting therapeutic efficacy. In addition, these desmoplastic characteristics are in accordance with previous studies in pancreatic and kidney cancer that revealed that activated stroma (ie, high pFAK and high α5-integrin) are associated with poor outcome, shorter overall survival, and rapid recurrence [25,51]. Although the specific mechanisms underpinning desmoplastic influence on drug responsiveness remain unknown, examining pathways (kinases and growth factors) impacting carcinoma-associated fibroblasts in future studies might uncover novel therapeutic targets. At present, use of the PDE model to examine native stromal localization and levels of pFAK and α5-integrin helps to distinguish patient-protective (tumor restrictive) from patient-detrimental (tumor permissive) desmoplasia in the pancreas, ovary, and kidney [19,25,33]. Retrospective studies will be needed to determine the strength of this system for TME-based prediction of drug responsiveness on an individual basis. Subsequent prospective studies could be designed using the TME desmoplastic characteristics as a novel criterion, which, alongside genomic analyses, could help unravel a patient’s unique cancer complexities and specify individual treatment regimens. In sum, the PDE is a novel and rapid model system that could define different PCa subtypes; the TME probably modulates response to AR-targeting agents, which informs individualized treatment regimens for different PCa subtypes.
5. Conclusions
In summary, this study highlights a unique model system that can be used to examine drug response in PCa clinical specimens maintained in an intact, native TME. The PDE system is amenable to genomic and pharmacological perturbations that can help in elucidating the molecular underpinnings of response to AR-directed therapeutics at a patient-specific level. Importantly, the novel criterion of TME analysis provides new insights into the complexities and heterogeneity of cancer. The TME adds an innovative measure to inform on response to AR-directed therapeutics and potentially discriminate between responders and nonresponders. Thus, PDEs are a powerful tool accounting for patient variability and can be incorporated into preclinical drug development assays to facilitate identification of new therapies and provide biological insight into the molecular pathways that lead to PCa progression. Lastly, since this model can be used to potentially identify clinically relevant tumor subpopulations, subsequent molecular profiling of cultured tissue could uncover new pathways for therapeutic intervention and influence precision medicine for PCa patients.
Supplementary Material
Acknowledgments
Funding/Support and role of the sponsor: This work was supported by Prostate Cancer Foundation Young Investigator Awards (Ayesha A. Shafi 2017, Matthew J. Schiewer 2013, Renée de Leeuw 2016); NIH/NCI grants to Karen E. Knudsen (R01 CA159945, R01 CA176401, R01 CA182569, R01 CA217329); an NIH/NCI grant to Edna Cukierman (R01 CA113451); core grant CA06927 in support of the Fox Chase Core Facilities (Talbot Library, Bio-Sample Repository, and Cell Imaging); funds from the Commonwealth of Pennsylvania and Mrs. Concetta Greenberg on behalf of Dr. Martin Greenberg; an Australian Research Council Future Fellowship to Lisa M. Butler (ST130101004); a Prostate Cancer Foundation of Australia grant to Margaret M. Centenera (YIG0412); a Movember Foundation grant to Lisa M. Butler and Wayne D. Tilley (MRTA3); and a Cancer Australia grant to Margaret M. Centenera, Lisa M. Butler, and Wayne D. Tilley (1085471). The sponsors played a role in the design and conduct of the study; data collection, management, analysis, and interpretation; and preparation, review, and approval of the manuscript.
We gratefully thank all the members of the Knudsen laboratory for their invaluable support and input.
Footnotes
Author contributions: Karen E. Knudsen had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Shafi, Schiewer, Gomella, Raj, Tilley, Cukierman, Knudsen.
Acquisition of data: Shafi, Schiewer, de Leeuw, Dylgjeri, McCue, Shah, Gomella, Lallas, Trabulsi, Centenera, Hickey, Butler, Raj, Tilley, Cukierman, Knudsen.
Analysis and interpretation of data: Shafi, Schiewer, Shah, Centenera.
Drafting of the manuscript: Shafi, Knudsen.
Critical revision of the manuscript for important intellectual content: Shafi, Schiewer, de Leeuw, Dylgjeri, McCue, Shah, Gomella, Lallas, Trabulsi, Centenera, Hickey, Butler, Raj, Tilley, Cukierman, Knudsen.
Statistical analysis: Shafi, Schiewer, de Leeuw, Dylgjeri.
Obtaining funding: Shafi, Schiewer, de Leeuw, Centenera, Butler, Tilley, Cukierman, Knudsen.
Administrative, technical, or material support: Shafi, Schiewer, de Leeuw, Dylgjeri, McCue, Shah, Gomella, Lallas, Trabulsi, Cukierman, Knudsen.
Supervision: Knudsen.
Other: None.
Financial disclosures: Karen E. Knudsen certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–38. doi: 10.1016/j.pharmthera.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Lilja H, Ulmert D, Vickers A. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–78. doi: 10.1038/nrc2351. corrigendum 403. [DOI] [PubMed] [Google Scholar]
- 4.Sharifi N, Gulley J, Dahut W. Androgen deprivation therapy for prostate cancer. JAMA. 2005;294:238–44. doi: 10.1001/jama.294.2.238. [DOI] [PubMed] [Google Scholar]
- 5.Loblaw A, Virgo K, Nam R, et al. Initial hormonal management of androgen-sensitive metastatic, recurrent, or progressive prostate cancer: 2007 update of an American Society of Clinical Oncology practice guideline. J Clin Oncol. 2007;25:1596–1605. doi: 10.1200/JCO.2006.10.1949. [DOI] [PubMed] [Google Scholar]
- 6.Knudsen K, Scher H. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–8. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drost J, Karthaus WR, Gao D, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11:347–58. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10:483–7. doi: 10.1038/nrurol.2013.126. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JI, Decker S, Zaharevitz D, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424–31. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maund SL, Nolley R, Peehl DM. Optimization and comprehensive characterization of a faithful tissue culture model of the benign and malignant human prostate. Lab Invest. 2014;94:208–21. doi: 10.1038/labinvest.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishiguro T, Ohata H, Sato A, Yamawaki K, Enomoto T, Okamoto K. Tumor-derived spheroids: relevance to cancer stem cells and clinical applications. Cancer Sci. 2017;108:283–9. doi: 10.1111/cas.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartucci M, Ferrari AC, Kim IY, Ploss A, Yarmush M, Sabaawy HE. Personalized medicine approaches in prostate cancer employing patient derived 3D organoids and humanized mice. Front Cell Dev Biol. 2016;4:64. doi: 10.3389/fcell.2016.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaira V, Fedele G, Pyne S, et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci U S A. 2010;107:8352–6. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissell M, Hines W. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–9. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeanes A, Maya-Mendoza A, Streuli C. Cellular microenvironment influences the ability of mammary epithelia to undergo cell cycle. PLoS One. 2011;6:e18144. doi: 10.1371/journal.pone.0018144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrix M, Seftor E, Seftor R, Kasemeier-Kulesa J, Kulesa P, Postovit L-M. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer. 2007;7:246–55. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 19.Rybinski B, Franco-Barraza J, Cukierman E. The wound healing, chronic fibrosis, and cancer progression triad. Physiol Genomics. 2014;46:223–44. doi: 10.1152/physiolgenomics.00158.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiewer MJ, Goodwin JF, Han S, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–49. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin JF, Schiewer MJ, Dean JL, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3:1254–71. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin JF, Kothari V, Drake JM, et al. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell. 2015;28:97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shafi AA, Cox MB, Weigel NL. Androgen receptor splice variants are resistant to inhibitors of Hsp90 and FKBP52, which alter androgen receptor activity and expression. Steroids. 2013;78:548–54. doi: 10.1016/j.steroids.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClurg UL, Nabbi A, Ricordel C, et al. Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br J Cancer. 2018;118:713–26. doi: 10.1038/bjc.2017.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco-Barraza J, Francescone R, Luong T, et al. Matrix-regulated integrin αvβ5 maintains α5β1-dependent desmoplastic traits prognostic of neoplastic recurrence. Elife. 2017;6:e20600. doi: 10.7554/eLife.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroka DM, Burkhardt T, Desbaillets I, et al. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 2001;15:2445–53. doi: 10.1096/fj.01-0125com. [DOI] [PubMed] [Google Scholar]
- 27.Herawi M, Epstein JI. Immunohistochemical antibody cocktail staining (p63/HMWCK/AMACR) of ductal adenocarcinoma and Gleason pattern 4 cribriform and noncribriform acinar adenocarcinomas of the prostate. Am J Surg Pathol. 2007;31:889–94. doi: 10.1097/01.pas.0000213447.16526.7f. [DOI] [PubMed] [Google Scholar]
- 28.Kapoor A, Wu C, Shayegan B, Rybak AP. Contemporary agents in the management of metastatic castration-resistant prostate cancer. Can Urol Assoc J. 2016;10:E414–23. doi: 10.5489/cuaj.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comstock CE, Augello MA, Goodwin JF, et al. Targeting cell cycle and hormone receptor pathways in cancer. Oncogene. 2013;32:5481–91. doi: 10.1038/onc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss KA, Herman JM, Armstrong D, et al. A final report of a phase I study of veliparib (ABT-888) in combination with low-dose fractionated whole abdominal radiation therapy (LDFWAR) in patients with advanced solid malignancies and peritoneal carcinomatosis with a dose escalation in ovarian and fallopian tube cancers. Gynecol Oncol. 2017;144:486–90. doi: 10.1016/j.ygyno.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loibl S, Turner NC, Ro J, et al. Palbociclib combined with fulvestrant in premenopausal women with advanced breast cancer and prior progression on endocrine therapy: PALOMA-3 results. Oncologist. 2017;22:1028–38. doi: 10.1634/theoncologist.2017-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis AM, Alexander A, Liu Y, et al. CDK4/6 inhibitors sensitize Rb-positive sarcoma cells to Wee1 kinase inhibition through reversible cell cycle arrest. Mol Cancer Ther. 2017;16:1751–64. doi: 10.1158/1535-7163.MCT-17-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serebriiskii I, Castelló-Cros R, Lamb A, Golemis EA, Cukierman E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biol. 2008;27:573–85. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik R, Lelkes PI, Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 2015;33:230–6. doi: 10.1016/j.tibtech.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Illmensee K, Mintz B. Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proc Natl Acad Sci U S A. 1976;73:549–53. doi: 10.1073/pnas.73.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci U S A. 1975;72:3585–9. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander J, Cukierman E. Stromal dynamic reciprocity in cancer: intricacies of fibroblastic-ECM interactions. Curr Opin Cell Biol. 2016;42:80–93. doi: 10.1016/j.ceb.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centenera MM, Gillis JL, Hanson AR, et al. Evidence for efficacy of new Hsp90 inhibitors revealed by ex vivo culture of human prostate tumors. Clin Cancer Res. 2012;18:3562–70. doi: 10.1158/1078-0432.CCR-12-0782. [DOI] [PubMed] [Google Scholar]
- 39.de Leeuw R, Berman-Booty LD, Schiewer MJ, et al. Novel actions of next-generation taxanes benefit advanced stages of prostate cancer. Clin Cancer Res. 2015;21:795–807. doi: 10.1158/1078-0432.CCR-14-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheon D-JJ, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95–119. doi: 10.1146/annurev.pathol.3.121806.154244. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Gao D, Chen Y. The potential of organoids in urological cancer research. Nat Rev Urol. 2017;14:401–14. doi: 10.1038/nrurol.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauli C, Hopkins BD, Prandi D, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–77. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 2014;1840:2506–19. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15:647–64. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rookmaaker MB, Schutgens F, Verhaar MC, Clevers H. Development and application of human adult stem or progenitor cell organoids. Nat Rev Nephrol. 2015;11:546–54. doi: 10.1038/nrneph.2015.118. [DOI] [PubMed] [Google Scholar]
- 46.Fong EL, Wan X, Yang J, et al. A 3D in vitro model of patient-derived prostate cancer xenograft for controlled interrogation of in vivo tumor-stromal interactions. Biomaterials. 2016;77:164–72. doi: 10.1016/j.biomaterials.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Åkerfelt M, Bayramoglu N, Robinson S, et al. Automated tracking of tumor-stroma morphology in microtissues identifies functional targets within the tumor microenvironment for therapeutic intervention. Oncotarget. 2015;6:30035–56. doi: 10.18632/oncotarget.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taube JM, Galon J, Sholl LM, et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol. 2018;31:214–34. doi: 10.1038/modpathol.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearce OMTM, Delaine-Smith R, Maniati E, et al. Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. 2018;8:3–4. 19. doi: 10.1158/2159-8290.CD-17-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beacham DA, Cukierman E. Stromagenesis: the changing face of fibroblastic microenvironments during tumor progression. Semin Cancer Biol. 2005;15:329–41. doi: 10.1016/j.semcancer.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Quiros RM, Valianou M, Kwon Y, Brown KM, Godwin AK, Cukierman E. Ovarian normal and tumor-associated fibroblasts retain in vivo stromal characteristics in a 3-D matrix-dependent manner. Gynecol Oncol. 2008;110:99–109. doi: 10.1016/j.ygyno.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.