Abstract
Long considered inert fat storage depots, it has become clear that lipid droplets (LDs) are bone fide organelles. Like other organelles, they have a characteristic complement of proteins and lipids, and undergo a lifecycle that includes biogenesis, maturation, interactions with other organelles, and turnover. I will discuss recent insights into mechanisms governing the lifecycle of LDs, and compare and contrast the LD lifecycle with that of other metabolic organelles such as mitochondria, peroxisomes, and autophagosomes, highlighting open questions in the field.
Keywords: cell biology, organelles, organelle biogenesis, lipid droplets, membrane contact sites, lipolysis, autophagy
1. Introduction
Lipid droplets (LDs) are cellular structures that store fat in the form of neutral lipids. Nearly all eukaryotic cells can make LDs, which are composed of a core of triglycerides and sterol esters. Because the core of the LD is hydrophobic, LDs are surrounded by a phospholipid monolayer, with the hydrophobic phospholipid tails oriented toward the neutral lipid core (Thiam et al., 2013). This is different from most other cellular organelles, which have aqueous lumens and are surrounded by phospholipid bilayers. Despite their unique structure, it has become clear that LDs are bone fide organelles. Like other organelles, LDs have a characteristic complement of proteins and lipids. Over 100 LD-associated proteins have been identified (Bersuker and Olzmann, 2017; Krahmer et al., 2013). These include proteins involved in lipid synthesis and metabolism, as well as proteins involved in membrane trafficking and organelle transport (Bersuker and Olzmann, 2017). LDs undergo a lifecycle that includes biogenesis, maturation, and turnover (Hashemi and Goodman, 2015; Figure 1). Under certain conditions LDs can undergo fusion and fission (Thiam et al., 2013), and LDs also make close contacts with many other organelles, presumably to exchange lipids (Barbosa et al., 2015; Schuldiner and Bohnert, 2017; Figure 1). In this review, I will discuss these aspects of the LD lifecycle, highlighting similarities and differences with other cellular organelles, as well as open questions in this exciting field.
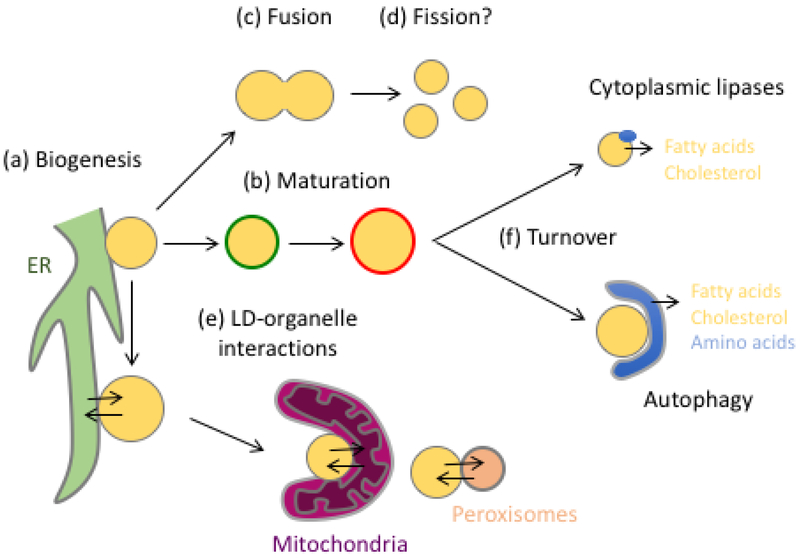
Figure 1: The LD Lifecycle.
LDs undergo a complex lifecycle that begins with biogenesis from the ER (a). LDs undergo a maturation process involving the sequential recruitment of different proteins, such as perilipins (b); the change from green to red indicates the sequential recruitment of proteins. Under certain conditions LDs may undergo fusion (c) or fission (d), and LDs may make contacts with a number of other organelles, including the ER, mitochondria, and peroxisomes (e). Arrows indicate lipid and/or protein transfer between organelles, although in many cases the types of lipid being transferred and direction of transfer are as yet unknown. Finally, LDs can be turned over via lipolysis involving cytoplasmic lipases, or via macroautophagy, referred to as lipophagy (f).
2. LD biogenesis
2.1. LD biogenesis
LDs can form in response to various stimuli, including excess lipids or a variety of stresses (Gubern et al., 2009). There is evidence that LD biogenesis occurs in the endoplasmic reticulum (ER), which is the site of many lipid synthesis enzymes (Pol et al., 2014; Walther et al., 2017). In the prevailing models of LD biogenesis, neutral lipid synthesis occurs within the ER membrane, at ER microdomains enriched in acyl-CoA synthetase 3, an enzyme involved in the first step of triacylglycerol synthesis (Kassan et al., 2013). Once the neutral lipid concentration exceeds that which can be supported within the membrane, phase separation occurs, and triglyceride molecules dispersed in the phospholipid bilayer coalesce to form lipid lenses between the leaflets of the ER membrane bilayer (Pol et al., 2014; Thiam and Foret, 2016; Wilfling et al., 2014a). FIT2 is an ER protein that binds diacylglycerol and triacylglycerol, and may play a role in partitioning neutral lipids within the ER to support lens formation (Gross et al., 2011; Kadereit et al., 2008). The accumulation of diacylglycerol in the ER membrane is likely also one of the factors mediating recruitment of cytoplasmic LD proteins, such as Plin3, to the sites of LD formation (Bulankina et al., 2009; Skinner et al., 2009).
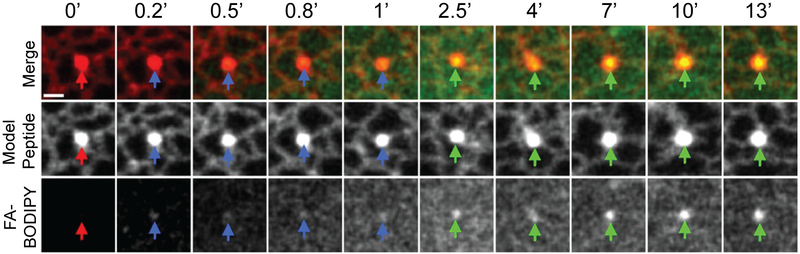
Figure 2 shows the biogenesis of a LD after the addition of oleic acid to COS-1 cells (Kassan et al., 2013). Cells were transfected to express a model peptide, which accumulates in the ER microdomains that will give rise to LDs. Upon the addition of oleic acid, a potent inducer of LD biogenesis, a fluorescent fatty acid analogue (FA-BODIPY) was observed to accumulate within these microdomains, giving rise to new LDs. The addition of oleic acid also induced new punctae of the model peptide, indicating that LDs can form either at pre-existing ER microdomains, or at domains that form in response to a stimulus such as excess fatty acid (Kassan et al., 2013).
Figure 2: LD Biogenesis.
COS-1 cells were transfected to express a model peptide consisting of the hydrophobic domain of the methyl transferase ALDI, fused to the LD-targeting signal of caveolin. This model peptide is initially targeted to the ER, and accumulates in microdomains where LD biogenesis will occur. Transfected cells were treated with oleic acid and a fluorescent fatty acid analogue (FA-BODIPY). During LD biogenesis, the FA-BODIPY accumulated in an ER microdomain, forming a nascent LD. Numbers indicate time in minutes after the addition of oleic acid and FA-BODIPY. Modified with permission from Kassan et al., 2013.
The final step of LD biogenesis involves budding (i.e., separating) from the ER. In yeast, LDs may never completely dissociate from ER, remaining attached by thin stalks (Jacquier et al., 2011). In mammalian cells, at least some LDs do seem to separate from the ER completely (Soni et al., 2009; Wilfling et al., 2013). LD budding may occur spontaneously, via a de-wetting mechanism (Thiam et al., 2013; Thiam and Foret, 2016; Walther et al., 2017). In emulsion physics, wetting describes the spreading of a liquid on a substrate, while dewetting describes the rupture of a thin liquid film on a substrate (Thiam and Foret, 2016). In the case of LD formation, the contact angle between the nascent LD and the ER bilayer could gradually increase as the LD grows, leading to spontaneous fission when a sufficient angle is achieved (Thiam et al., 2013; Thiam and Foret, 2016; Walther et al., 2017). Nevertheless, proteins are also likely involved in mediating or regulating this step. LD budding requires the generation of negative curvature at the base of the LD, which has been proposed to occur via the generation of phosphatidic acid by AGPAT, DGK1, or PLD at the ER-LD interface (Pol et al., 2014). Finally, FIT2, in addition to partitioning neutral lipids within the ER, also seems to regulate the directionality of budding of LDs in yeast, promoting budding towards the cytoplasm and away from the ER lumen (Choudhary et al., 2015).
Seipin is another protein that has been implicated in LD biogenesis. In yeast, when LD formation was initiated by inducing expression of the triglycerol synthesis enzyme DGA1 in the absence of seipin, LD biogenesis was impaired and triglycerides accumulated within ER membranes (Cartwright et al., 2015). In the same study, intranuclear LDs were frequently observed in seipin-deficient cells, suggesting that seipin also plays a role in the directionality of LD budding (Cartwright et al., 2015). Finally, in both yeast and mammalian cells, seipin localizes to ER-LD contact sites and plays additional roles in the trafficking of lipids and proteins to LDs at a stage after budding (Salo et al., 2016; Szymanski et al., 2007; Wang et al., 2016; see “interactions between LDs and other organelles“ below).
2.2. Crosstalk between LD, autophagosome, and peroxisome biogenesis
Several other organelles originate from the ER (Joshi et al., 2017). Autophagosomes are double membrane structures that engulf cytoplasm and organelles, and deliver material to the lysosome to be catabolized and recycled. Autophagy is initiated in response to a variety of stresses, including starvation, ER stress, and infection (Galluzzi et al., 2017). The source of the membrane for autophagosome biogenesis depends on the type of stress experienced by the cell. Depending on the cell type and source of stress, autophagosomes can originate from the mitochondria (Hailey et al., 2010), ER (Axe et al., 2008), ER-mitochondria contact sites (Hamasaki et al., 2013), plasma membrane (Ravikumar et al., 2010), or ER-Golgi intermediate compartment (Ge et al., 2013). Intriguingly, both autophagosomes and LDs have been observed cradled within “cups” of ER membrane (Hayashi-Nishino et al., 2009; Robenek et al., 2006). In addition, LC3 and Atg2, which are involved in autophagosome biogenesis, also localize to LDs (Shibata et al., 2009; Shibata et al., 2010; Velikkakath et al., 2012). These observations suggest the possibility that autophagosome and LD biogenesis may share some common machinery.
Peroxisomes are metabolic organelles involved in bile acid synthesis, as well as the metabolism of glyoxylate, dicarboxylate, amino acids and lipids (Tripathi and Walker, 2016). Peroxisomes can multiply via division of existing peroxisomes, or can form de novo from the ER (Kim, 2017; Smith and Aitchison, 2013). It has very recently been shown that mitochondria-derived vesicles also play a role in the biogenesis of peroxisomes (Sugiura et al., 2017). This suggests that multiple membrane sources may actually be a common theme in organelle biogenesis. It remains to be determined whether organelles other than the ER can contribute to LD biogenesis. The lipid synthesis enzyme acyl-CoA:diacylglycerolO-acyltransferase 2 (DGAT2) can localize to the ER, LDs, and mitochondria (Stone et al., 2009). In addition, LDs have been observed apparently entirely enclosed within mitochondria by electron microscopy (Aon et al., 2014). Therefore, it will be interesting to see whether organelles other than the ER can give rise to LDs, or contribute to LD biogenesis in some way.
Intriguingly, targeting of the LD protein UBXD8 from the ER to LDs depends on Pex19 and Pex13, proteins previously implicated in peroxisome biogenesis (Schrul and Kopito, 2016). Like for LDs and autophagosomes, this suggests the possibility of shared biogenesis machinery for LDs and peroxisome. The possibility that LD, autophagosome and peroxisome biogenesis may share machinery raises a number of intriguing questions. How are proteins destined for LDs sorted from those meant for other organelles? Are the pathways co-regulated? Is the shared machinery limiting? If so, the implication is that LD biogenesis competes with autophagosome or peroxisome biogenesis. At first glance competition between LD and autophagosome biogenesis appears to make sense, since LD biogenesis occurs under conditions of excess lipids, while autophagy is triggered in response to nutrient deprivation. However, nutrient deprivation that triggers macroautophagy can actually lead to the recycling of large quantities of membrane, causing the release of excessive fatty acids to the cytoplasm (see “crosstalk between lipolysis and autophagy” below). These cytoplasmic fatty acids can be extremely toxic, acting as detergents that damage cellular membranes (Aon et al., 2014). An alternative implication of shared machinery is that it could allow for a coordinated increase in the biogenesis of both autophagosomes and LDs. In this case, conditions that trigger autophagy would concomitantly up-regulate the machinery for LD biogenesis, which would serve to package fatty acids released through autophagy into inert neutral lipids, and protect the cell from lipotoxicity. Likewise, LD and peroxisome biogenesis could both be upregulated in response to conditions where both increased lipid storage and metabolism are required.
3. LD maturation and heterogeneity
LDs show remarkable heterogeneity in protein and lipid composition. Some of this heterogeneity may reflect subpopulations of LDs with different functions. For example, when cells are loaded with excess fatty acids and sterols, they form distinct populations of LDs enriched either in sterol esters or triglycerides (Hsieh et al., 2012). However, it has also been proposed that LDs undergo a maturation process over time. Organelle maturation and heterogeneity is an emerging theme in cell biology, and has been observed for organelles including endocytic structures, mitochondria, and peroxisomes.
3.1. Protein targeting to LDs
To discuss LD maturation and heterogeneity, it is first necessary to consider how proteins target LDs. Although LDs have a unique architecture, being surrounded by a phospholipid monolayer, there are proteins that can move between the ER and LD surface. It has recently been proposed that LD proteins fall into two categories, class I and class II proteins, which are targeted to LDs by two general mechanisms (Kory et al., 2016).
Class I proteins target the LD surface from the ER bilayer using a hydrophobic hairpin structure. These targeting regions consist of α-helical domains that form a v-shaped sequence that embeds into the membrane, often with proline residues at the midpoint of the hydrophobic sequence to cause a kink (Kory et al., 2016). This hairpin topology has been best characterized for the plant oleosins (Abell et al., 2002; Abell et al., 1997). Other proteins that may form similar hairpin structures include the lipid synthesis enzymes glycerol-3-phosphate acyltransferase 4 (GPAT4) and DGAT2 (Kory et al., 2016). Class I proteins appear to lack ER luminal domains, which allows them to insert either into the ER bilayer or LD monolayer (Kory et al., 2016). These proteins may move from the ER to LDs either during initial LD formation, or after LD budding, via membrane bridges (see below).
Class II proteins are translated in the cytoplasm, and target the LD surface using amphipathic helices or short, hydrophobic-rich sequences (Kory et al., 2016). Current models suggest that amphipathic sequences are unfolded in solution, but fold into a helix when binding a membrane surface (Seelig, 2004). Examples of class II LD proteins include the phosphatidyl choline synthesis enzyme CTP:phosphocholine cytidyltransferase α (CCTα), and the perilipin family of proteins (Kory et al., 2016; Krahmer et al., 2011). The perilipins are an ancient family consisting of five genes (Plin1–5) that regulate lipid metabolism (Kimmel and Sztalryd, 2016). The perilipins share two conserved protein motifs: a hydrophobic perilipin/ADRP/TIP47 (PAT) domain at the N-terminus, followed by a repeating 11-mer helical repeat of varying lengths (Kimmel and Sztalryd, 2016). The C-termini of the perilipins are more divergent, and the family members are expressed to different degrees in different tissues, and have unique but overlapping functions (Kimmel and Sztalryd, 2016). The localization of class II proteins to LDs can be prevented by protein crowding at the LD surface, or by competition with other class II proteins (Kory et al., 2016; Kory et al., 2015).
LD maturation
The idea that LDs undergo a maturation process began with the observation that in adipocytes, LDs of different sizes were coated with different proteins; small LDs stained positive for Plin3, while medium LDs had Plin2 and the largest LDs were enriched with Plin1 (Wolins et al., 2005). In adipocytes, small LDs merge during differentiation, resulting in one very large LD in mature adipocytes (Gao et al., 2017). Thus, it was proposed that a unique complement of proteins is recruited and subsequently lost as LDs mature, fine-tuning lipid metabolism as cells differentiate (Wolins et al., 2006). The observation that proteins are recruited to LDs at different rates as they expand has been corroborated by videomicroscopy of LD biogenesis (Kassan et al., 2013), and by a proteomics study that identified the proteins present on purified LDs of different sizes (Zhang et al., 2016).
Plin1 is predominantly expressed in adipocytes, so this particular progression of protein recruitment to LDs is likely tissue-specific. However, LD maturation has been observed in non-adipose cell types as well. After biogenesis and detachment from the ER, a subpopulation of LDs later undergoes remodeling by the COPI machinery, which allows for the formation of membrane bridges to the ER (Wilfling et al., 2014b). In fly and mammalian cells, these bridges mediate the transfer of lipid synthesis proteins, such as GPAT4 and DGAT2, from the ER to LDs, resulting in the expansion of the subset of LDs positive for these proteins (Wilfling et al., 2013). It has been proposed that small, GPAT4-negative LDs function in temporary storage of “overflow” lipids, protecting the ER from lipotoxicity, while large, GPAT4-positive LDs are dedicated to long term lipid storage (Wilfling et al., 2013).
3.2. LD maturation vs. heterogeneity
It is currently unclear whether different types of LDs are completely interchangeable, through a maturation process, or whether there are fundamentally distinct classes of LDs. The transfer of GPAT4 and other lipid synthesis enzymes to existing LDs and their subsequent expansion is an example of LD maturation. In adrenal cells loaded with excess oleic acid and cholesterol, it was shown that lipids can segregate into separate populations of LDs, which are enriched in different LD proteins (Hsieh et al., 2012). Cholesterol ester-rich LDs have more Plin1c and Plin4, while triglyceride-rich LDs have more Plin1a, Plin 1b, and Plin5 (Hsieh et al., 2012). Proteomic studies have identified additional proteins that differ between cholesterol ester- versus triglyceride-rich LDs (Khor et al., 2014). These two populations of LDs may serve different functions. Triglyceride-rich LDs must clearly play a role in energy storage. Consistent with this, Plin5 is highly expressed in oxidative tissues such as heart and muscle, and plays a role in mediating LD-mitochondria contacts, which are important for fatty acid metabolism (Wang and Sztalryd, 2011; see “interactions between LDs and other organelles“). In contrast, Plin1c is highly expressed in steroidogenic cells (Kimmel and Sztalryd, 2016). Thus, cholesterol-rich LDs are likely to play a role in the synthesis of various steroid hormones. It is possible that cholesterol ester-rich LDs can convert to triglyceride-rich LDs, and vice versa, through remodeling of proteins on the LD surface. It is also possible that cholesterol-rich and triglyceride-rich LDs represent completely separate populations, which may even originate through different mechanisms. In fission yeast, sterol-rich and triglyceride-rich LDs were observed to form in different regions of the cell (Meyers et al., 2016), favoring the latter possibility.
In yeast, an additional subpopulation of LDs with a unique protein composition has also been identified (Eisenberg-Bord et al., 2017; Moldavski et al., 2015). This subpopulation is localized to the nucleus-vacuole junction by Ldo16, while another protein derived from an overlapping gene, Ldo45, is responsible for recruiting additional proteins to the LD subpopulation (Eisenberg-Bord et al., 2017). The function of this LD subpopulation is not entirely clear, but cells lacking Ldo16 and Ldo45 showed impaired autophagy of LDs during stationary phase (Teixeira et al., 2017).
Finally, another distinct population of LDs exists within the nucleus of certain cell types (Farese and Walther, 2016; Layerenza et al., 2013; Uzbekov and Roingeard, 2013). These nuclear LDs are especially prominent in hepatocytes, and again have a characteristic complement of proteins (Ohsaki et al., 2016). Nuclear LDs are closely associated with the inner nuclear membrane and with nuclear structures called promyelocytic leukemia (PML) bodies (Ohsaki et al., 2016). Knockdown of the PML body protein PML-II decreased the number of nuclear LDs, while knockdown of SUN proteins, which link the outer and inner nuclear membranes, increased the number of nuclear LDs (Ohsaki et al., 2016). The function of nuclear LDs remains mysterious, but the authors of the PML-II study speculate that they may play a role in regulating nuclear receptors such as peroxisome proliferator-activated receptors, which in turn regulate cell metabolism (Ohsaki et al., 2016).
3.3. Heterogeneity of other organelles
Organelle heterogeneity is an emerging theme in cell biology. Mitochondria have many cellular functions. There is emerging evidence for different populations of mitochondria that are ultrastructurally, biochemically, and functionally distinct (Woods, 2017). The endocytic system is probably the best-studied example of organelle maturation, with fission/fusion events and sequential protein recruitment resulting in the maturation of early endosomes to late endosomes to lysosomes (Maxfield and McGraw, 2004). However, even within a “mature” class of organelle such as lysosomes, heterogeneity has been observed. For example, it was recently shown that lysosomes positioned in the cell periphery are less acidic and have less protease activity than those in the juxtanuclear region (Johnson et al., 2016). This may reflect different functions: if peripheral lysosomes function in plasma membrane repair, for example, reduced luminal pH and protease/lipase activity could be advantageous (Johnson et al., 2016). These observations highlight the need to develop methods that allow specific populations of organelles, including LDs, to be specifically studied. Caution must be used when interpreting results that are an average value for a whole class of organelle, including LDs, within a cell or population of cells. Such measurements may obscure subtle differences between different populations of LDs, or between LDs that are at different stages of maturation.
4. LD fusion and fission
4.1. LD fusion
LDs can undergo fusion via two separate mechanisms. During the differentiation of adipocytes, many small LDs combine to form one large LD (Gao et al., 2017). The resulting LD occupies much of the volume of the adipocyte, and allows for efficient lipid storage by minimizing the surface area to volume ratio of the hydrophobic neutral lipids. This type of fusion occurs by Ostwald ripening, in which lipids are transferred from the smaller to larger LD by diffusion (Thiam et al., 2013). The close apposition of LDs for lipid transfer is mediated by the CIDE family of proteins, including CIDEC/Fsp27 (Gong et al., 2011; Jambunathan et al., 2011; Sun et al., 2013). It is also possible for two LDs to merge via coalescence. Coalescence involves the fusion of two LDs without directional transfer from the smaller to larger LD (Thiam et al., 2013). Usually, the surfactant properties of the phospholipids on the LD surface prevent LD fusion. In cases where the phospholipid composition is altered and surface tension reduced, LDs spontaneously merge (Guo et al., 2008; Krahmer et al., 2011). LDs can also coalesce in response to a number of pharmacological agents (Murphy et al., 2010). Thus, LD coalescence is likely to be important for various pathophysiological conditions, but it is not clear whether this type of LD fusion occurs during normal physiological processes.
4.2. LD fission
LD fission has also been reported. In the yeast Schizosaccharomyces pombe, LD fission was observed by videomicroscopy using stringent criteria to avoid mistakenly scoring the dispersion of two pre-existing LDs as a fission event (Long et al., 2012). In addition, electron microscopy showed “dumbbell”-shaped LDs, which may represent a transition state during LD fission (Long et al., 2012). LD fission may also occur in mammalian cells. LD fragmentation was initially reported in response to PKA activation/lipolysis in 3T3-L1 adipocytes (Marcinkiewicz et al., 2006). However, subsequent studies have suggested that the appearance of “micro-LDs” during lipolysis represents release of fatty acids from large LDs, followed by biogenesis of new small LDs, rather than LD fission (Hashimoto et al., 2012; Paar et al., 2012). These micro-LDs aid in lipolysis by increasing the total surface area to volume ratio of LDs in the cell, and therefore allowing cytoplasmic lipases greater access to triglycerides stored within LDs (Hashimoto et al., 2012). A recent study showed that the formation of micro-LDs in adipocytes depends on DGAT1, and also plays a role in protecting the ER from lipotoxic stress (Chitraju et al., 2017). LD remodeling clearly has important consequences for lipid metabolism, however it remains unclear whether true LD fission occurs in mammalian cells.
4.3. LD fusion-fission compared to mitochondrial fusion-fission
Mitochondria are dynamic organelles that constantly undergo fusion and fission. The machinery for these dynamics is relatively well characterized. Mitochondrial fusion is mediated by the mitofusins Mfn1 and Mfn2 on the outer mitochondrial membrane and Opa1 on the inner mitochondrial membrane, while mitochondrial fission is mediated by Drp1 and other members of the dynamin family of large GTPases (Lee et al., 2016; Pernas and Scorrano, 2016). Mitochondrial fission involves the ER wrapping around mitochondria at sites of constriction (Friedman et al., 2011). Inverted formin 2 on the ER and Spire1C on mitochondria interact at ER-mitochondria contact sites to nucleate actin assembly, which in turn plays a role in the recruitment of Drp1 to the outer mitochondrial membrane (Korobova et al., 2013; Manor et al., 2015). Actin polymerization at ER-mitochondria contact sites also stimulates a rise in mitochondrial matrix calcium, activating inner mitochondrial membrane constriction (Chakrabarti et al., 2017).
It is not yet clear whether LD fusion and/or fission use similar mechanisms to those used by mitochondria. Mitochondria are surrounded by two membrane bilayers, while LDs are surrounded by a phospholipid monolayer. Thus, there will certainly be differences between how mitochondrial fusion/fission and LD fusion/fission are achieved. For example, mitofusins are transmembrane proteins that have been proposed to span the mitochondrial outer membrane, with the C-terminus residing in the mitochondrial intermembrane space (Mattie et al., 2017). Any proteins involved in LD fusion must necessarily have a different topology from mitofusins, since they reside in a phospholipid monolayer. However, mitochondrial and LD fusion/fission may share some similar features. Recently, it was reported that non-muscle myosin IIa (NMIIA) and formin-like 1 nucleate actin assembly at LD-LD contact sites (Pfisterer et al., 2017). Interestingly, NMIIA depletion resulted in larger LDs (Pfisterer et al., 2017). The authors propose that actin at LD-LD contact sites prevents LD coalescence rather than actively promoting LD fission, yet the parallel with mitochondrial fission is intriguing.
LD fusion/fission may also share functions with mitochondrial fusion/fission. Mitochondrial fusion tends to occur under conditions of high energy demand, including during starvation, cell division, and differentiation (Pernas and Scorrano, 2016). Likewise, LD fusion occurs during cell differentiation and in response to changing metabolic conditions (Gao et al., 2017). Mitochondrial fission is crucial for mitochondrial quality control, and is involved in removing damaged proteins and mitochondrial DNA (Ni et al., 2015), and in remodeling the protein composition of mitochondria in response to metabolic cues (Hughes et al., 2016). Under these conditions, fission allows part of a mitochondrion to be degraded by autophagy, while sparing the rest of the mitochondrion. Similarly, it is conceivable that LD fission could play a role in the removal of oxidized proteins and lipids from a LD, or in remodeling of LDs to remove a subset of LD proteins to alter LD function.
5. Interactions between LDs and other organelles
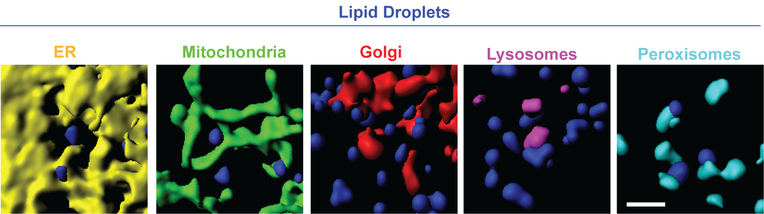
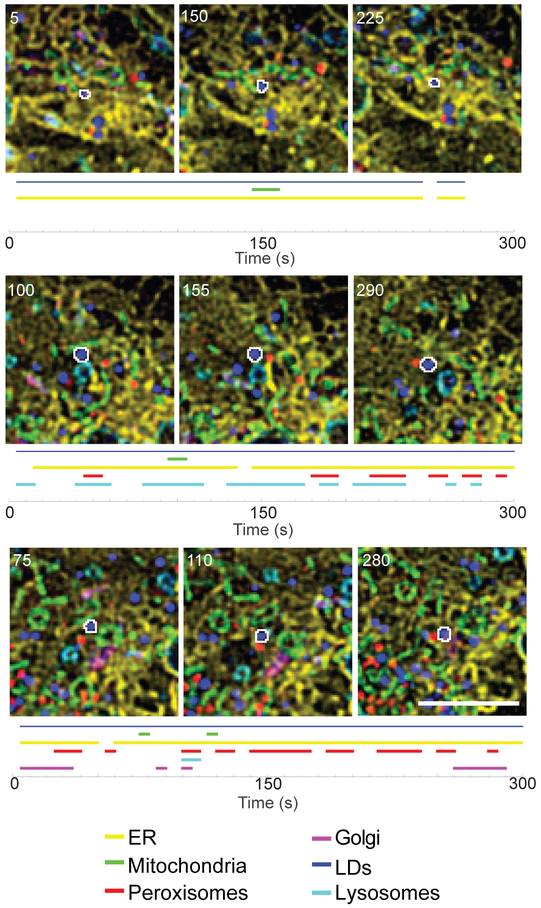
LDs can make close contacts with a variety of other organelles (Barbosa et al., 2015; Schuldiner and Bohnert, 2017). Membrane contact sites are increasingly understood to be key sites of communication and metabolite exchange between organelles. A recent study using multispectral imaging to simultaneously image six organelles found that LDs frequently moved within 1 pixel (~100 nm) of every other labeled organelle, namely the ER, mitochondria, peroxisomes, lysosomes, and the Golgi (Valm et al., 2017). Some of these organelle contact sites had elaborate morphologies, with the ER or mitochondria wrapping around LDs (Figure 3). Surprisingly, when LDs were tracked and their organelle contacts mapped over time, some LDs touched every other labeled organelle over the course of only five minutes (Valm et al., 2017; Figure 4). In contrast, some LDs made contact with only one or two other organelles during a five-minute imaging period. It is unclear whether these differences reflect subpopulations of LDs with different functions, or LDs that are at different stages of maturation. For many LD-organelle contact sites it is not yet clear what types of proteins or lipids are being exchanged, in which direction they are moving, or how the contacts are formed or regulated.
Figure 3: LD-organelle contacts in space.
Examples of LD-organelle contacts in segmented multispectral lattice light-sheet images. Shown are contacts between LDs (blue) and ER (yellow), mitochondria (green), Golgi (red), lysosomes (magenta), or peroxisomes (cyan). Scale bar, 2 μm. Image credit: Sarah Cohen, Alex Valm, Wesley Legant, Eric Betzig, and Jennifer Lippincott-Schwartz. Images acquired as described in (Valm et al., 2017).
Figure 4: LD-organelle contacts over time.
Micrographs of a COS-7 cell expressing fusion proteins targeted to the ER (yellow), mitochondria (green), Golgi (red), lysosomes (magenta), and peroxisomes (cyan), and labeled with a lipophilic dye to label LDs (blue). The LDs outlined in white were tracked, and their interorganelle contacts mapped over time. A blue line indicates that the LD was successfully tracked, while colored lines indicate that the LD was within 1 pixel (97 nm) of the indicated organelle at the specified time point. Numbers on the micrographs represent time (s). Scale bar, 5 μm. Modified with permission from Valm et al., 2017.
LDs share an especially intimate connection with the ER, their site of biogenesis. In yeast, LDs may never really detach from the ER (Jacquier et al., 2011). In mammalian cells, at least some of the LDs do seem to pinch off and separate from the ER (Soni et al., 2009; Wilfling et al., 2013). As described above, lipid synthesis enzymes can transfer from the ER to LDs via membrane bridges (Wilfling et al., 2013). Membrane contact sites also function in the exchange of lipids between LDs and the ER. While at least some proteins seem to transfer unidirectionally from the ER to LDs (Wilfling et al., 2013), lipids can transfer bi-directionally between LDs and the ER. A protein complex involving the acyl-CoA synthetase FATP1 on the ER, and the diacylglycerol acyltransferase DGAT2 on LDs, was shown to mediate LD expansion by facilitating lipid synthesis at the ER-LD interface in worm and mammalian cells (Xu et al., 2012). In yeast, Ice2p mediates ER-LD contacts that facilitate the transfer of lipids in the opposite direction, from LDs to the ER (Markgraf et al., 2014). This occurs under conditions where the ER needs to increase phospholipid synthesis in order to undergo expansion, for example during early exponential phase (Markgraf et al., 2014).
In addition to being involved in LD biogenesis, seipin plays a role at ER-LD contact sites (Salo et al., 2016; Szymanski et al., 2007; Wang et al., 2016). Seipin is an oligomeric ER transmembrane protein (Binns et al., 2010). Yeast lacking seipin have LDs with aberrant morphology, either small and clustered or supersized (Boutet et al., 2009; Fei et al., 2008; Szymanski et al., 2007). Lack of seipin also correlates with altered protein composition of LDs in yeast (Cartwright et al., 2015; Fei et al., 2008; Grippa et al., 2015; Wang et al., 2014a). Recently, Ldo16 and Ldo45 were identified as LD proteins that interact with seipin at ER-LD contact sites (Eisenberg-Bord et al., 2017; Teixeira et al., 2017). In fly and human cells, seipin forms foci in the ER that move along ER tubules, become colocalized with sites of triglyceride accumulation, and enable the growth and maturation of nascent LDs (Wang et al., 2016). Using pulse-chase and fluorescence recovery after photobleaching experiments, it was shown that in human cells the initial incorporation of proteins and lipids into LDs during LD biogenesis is normal (Salo et al., 2016). However, the transfer of class I proteins and fatty acids from the ER to pre-existing LDs was impaired in seipin knockout cells (Salo et al., 2016). These data suggest that in mammalian cells, the primary function of seipin is to facilitate transfer of proteins and lipids between the ER and LDs at membrane contacts sites after LDs form and detach from the ER.
LDs also make close contacts with mitochondria and peroxisomes. In mammalian cells, mitochondria are the primary site of β-oxidation of fatty acids. Contacts between LDs and mitochondria occur on detyrosinated microtubules, and increase in response to starvation (Herms et al., 2015). Pulse-chase experiments using a fluorescent fatty acid suggested that in response to starvation, fatty acids are transferred from LDs to mitochondria at sites of close apposition (Rambold et al., 2015). It has also been proposed that transfer of lipids from LDs to mitochondria at contact sites may protect mitochondria from lipotoxicity (Wang and Sztalryd, 2011). Overexpression of the LD protein Plin5 increases LD-mitochondria contacts (Wang et al., 2011). However, it is not clear whether Plin5 binds directly to a protein or lipid on mitochondria, or affects contact sites more indirectly. In some yeasts and in plants, peroxisomes are the only site of β-oxidation, and accordingly in these organisms LD-peroxisome contacts are very prominent (Binns et al., 2006; Hayashi et al., 2001). LD-peroxisome contacts have also been observed in mammalian cells (Schrader, 2001). In addition to transfer of fatty acids from LDs to peroxisomes, these contact sites may play a role in the transfer of ether linked lipids from peroxisomes to LDs (Barbosa et al., 2015).
Finally, LDs have been observed in close proximity to autophagosomes and lysosomes. This may be because LDs can be digested by autophagosomes in a process termed lipophagy (see below). It is also possible that lipids from lipoprotein particles, taken up via endocytosis, transfer from lysosomes to LDs, either directly or via the ER, at membrane contact sites. Clearly, there is much work to be done to further elucidate the mechanism, function, and regulation of LD-organelle contact sites.
6. LD turnover
Like other organelles, LDs have a life cycle that involves LD turnover and breakdown. As the organelle responsible for lipid storage, LDs must be able to respond rapidly to changes in the energetic needs of the cell. The best characterized mechanism for the release of fatty acids from triglycerides within LDs is via cytoplasmic lipases, called lipolysis (Zechner et al., 2017). However, pieces of LDs and even whole LDs can also be degraded via autophagy, in a process termed lipophagy (Singh et al., 2009; Zechner et al., 2017). It is not entirely clear when cells rely on lipolysis versus lipophagy to release lipids from LDs. This likely depends on the cell and tissue type, the stimulus, and whether there is a need to release fatty acids or cholesterol, or to remodel the proteins on the LD surface.
6.1. Release of lipids from LDs by lipases
The best-characterized mechanism by which fatty acids are released from LDs involves cytoplasmic lipases. In response to various cellular signals, lipolysis is initiated by the enzyme adipose triglyceride lipase (ATGL), which is the primary triglyceride lipase in most cell types (Smirnova et al., 2006; Zechner et al., 2017). Two proteins directly interact with the enzymatic domain of ATGL: CGI-58 activates ATGL (Lass et al., 2006), while G0/G1 switch protein inhibits ATGL (Yang et al., 2010). Many signaling pathways involved in nutrient sensing and differentiation directly or indirectly affect ATGL-mediated lipolysis (Zechner et al., 2017). The action of ATGL is followed by hormone sensitive lipase (HSL), the main diacylglycerol lipase, while the final step in the lipolytic cascade is performed by monoacylglycerol lipase (MGL) (Zechner et al., 2017). Together, these enzymes break down triglycerides, releasing fatty acids and glycerol. ATGL, CGI-58, G0/G1 switch gene 2, and HSL have all been validated as LD-associated proteins (Bersuker and Olzmann, 2017). However, open questions remain about the precise trafficking, localization and regulation of these enzymes.
6.2. LD turnover by autophagy
Autophagy can be divided into macroautophagy, microautophagy, and chaperone-mediated autophagy. In macroautophagy, a double-membraned autophagosome engulphs cytoplasm, protein aggregates, and/or organelles. These autophagosomes then fuse with lysosomes, resulting in the delivery of proteases and lipases, which break down the contents of the autophagosome (Galluzzi et al., 2017). In microautophagy, organelles or pieces of organelles are directly internalized into the vacuole of plants or yeast via direct membrane invaginations (Galluzzi et al., 2017). Finally, in chaperone-mediated autophagy, cytoplasmic proteins are transported into the lysosome for degradation through a protein translocation complex at the lysosomal membrane (Kaushik and Cuervo, 2012). All three forms of autophagy have been implicated in lipid metabolism.
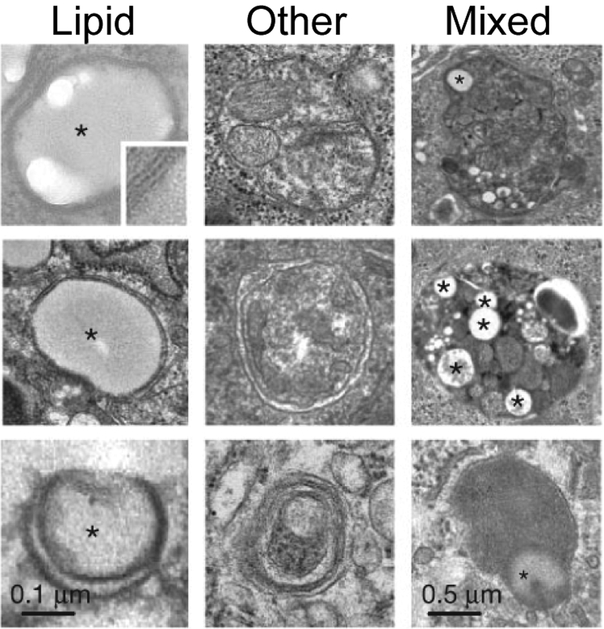
Macroautophagy of LDs, termed lipophagy, was first observed in hepatocytes treated with excess oleic acid or grown in methionine/choline-deficient medium (Singh et al., 2009). Under these conditions, LDs associated with components of the autophagy machinery, including LC3 (Singh et al., 2009). In addition, pharmacological inhibition of autophagy or depletion of Atg5 resulted in increased triglyceride storage within LDs (Singh et al., 2009). In hepatocytes, dynamin 2 and Rab10 also play roles in autophagy of LDs (Schroeder et al., 2015; Schulze et al., 2013). Figure 5 shows autophagosomes containing lipids only, other cargo, or a combination of lipids and other cargo. These images imply that either whole LDs or portions of LDs can be digested through autophagy. Interestingly, lipids were also observed within autophagic compartments in vivo in the livers of mice after 6 hours of fasting (Singh et al., 2009). Although some of these experiments involve nutrient deprivation, they actually all result in lipid loading in the cells where autophagy was observed. While methionine/choline-deficient medium is a form of nutrient deprivation, it is used to induce hepatic steatosis (Anstee and Goldin, 2006), and is therefore more appropriately interpreted as a model of lipid loading. Likewise, fasting mice results in lipolysis and the release of fatty acids from adipose tissue, and therefore actually causes lipid loading in the liver, where lipophagy was observed. Taken together, these results suggest that the primary role of lipophagy is to deal with excess lipids. Consistent with these observations, lipophagy was also found to be important for the hydrolysis of cholesterol esters in macrophage foam cells in response to atherogenic lipoprotein particles (Ouimet et al., 2011; Ouimet and Marcel, 2012).
Figure 5: LD turnover by lipophagy.
Electron micrographs of cultured hepatocytes treated with excess oleic acid or methionine/choline-deficient medium. Shown are autophagic vacuoles containing only lipids, other cargo, or mixed (lipids plus other cargo). Lipids are indicated by stars. Modified with permission from Singh et al., 2009.
Microlipophagy of LDs has been observed in yeast. In response to glucose restriction, LDs are internalized directly into the vacuole using a form of microlipophagy that requires components of the core autophagy machinery as well as endosomal sorting complexes required for transport (ESCRTs) (Seo et al., 2017; Vevea et al., 2015; Wang et al., 2014b). This form of lipophagy occurs on specialized sterol-enriched vacuolar microdomains, which are triggered by AMPK activation (Seo et al., 2017; Wang et al., 2014b).
Finally, chaperone-mediated autophagy plays a role in lipid metabolism as well. The LD proteins Plin2 and Plin3 can be degraded by chaperone-mediated autophagy, which increases the access of lipases to neutral lipids and facilitates lipolysis (Kaushik and Cuervo, 2015, 2016). This hints at the complex interplay between lipolysis and the various forms of autophagy, which is further discussed below.
6.3. Crosstalk between lipolysis and autophagy
It is not well understood what dictates whether LDs are digested via cytoplasmic lipases (lipolysis) or autophagy (lipophagy). This likely depends on the cell type and the stimulus triggering changes in metabolism. Lipophagy has been most robustly observed in hepatocytes and macrophages (Ouimet et al., 2011; Schulze et al., 2013; Singh et al., 2009). In addition, the stimulus inducing changes in metabolism is likely a key factor. Lipophagy was initially observed under conditions of lipid loading (Singh et al., 2009). Whether lipophagy occurs in response to starvation is more controversial. When cells in culture are deprived only of serum, LD number is reduced, and LDs colocalize with autophagy machinery (Rambold et al., 2015; Singh et al., 2009). This may be because serum is the main source of exogenous lipids for cultured cells, so serum starvation is a form of lipid-deprivation. However, whether the colocalization of autophagy machinery with LDs in response to serum starvation represents functional lipophagy remains to be formally shown.
In contrast to serum starvation, more general nutrient deprivation does not induce lipophagy. When mouse fibroblast cells were starved in HBSS (no serum, no amino acids, low glucose), it was observed that ATGL-mediated lipolysis, and not lipophagy, contributed to the transfer of fatty acids from LDs to mitochondria (Rambold et al., 2015). Under these conditions, autophagy machinery did not colocalize with LDs (Rambold et al., 2015). This is likely because amino acid deprivation triggers bulk autophagy of organelle membranes, which actually results in the release of fatty acids from phospholipids; thus, amino acid deprivation results in elevated fatty acid levels and increased LD biogenesis, rather than lipid deprivation and lipophagy (Rambold et al., 2015). It was recently shown that the release of fatty acids in response to amino acid deprivation depends on mTORC1, and that the fatty acids released by autophagy are packaged into LDs by DGAT1 (Nguyen et al., 2017). This prevents mitochondrial dysfunction due to lipotoxicity (Nguyen et al., 2017), and is consistent with a previously proposed role for DGAT1 in preventing ER stress (Wilfling et al., 2013).
In addition to autophagy contributing to LD biogenesis, the reverse has also been proposed. Because deletion of enzymes involved in triglyceride or sterol ester synthesis impaired autophagy, it was proposed that LDs supply lipids for autophagosome biogenesis (Shpilka et al., 2015). However, a subsequent study found that LDs were dispensable as a membrane source for autophagosome biogenesis in yeast (Velazquez et al., 2016). Instead, LDs played an important role in maintaining lipid homeostasis in the ER: the absence of LDs caused changes in the phospholipid composition of the ER, which in turn compromised the regulation of autophagy (Velazquez et al., 2016). This conclusion was also supported in mammalian cells by experiments that showed that inhibition of DGAT1-dependent LD biogenesis did not affect autophagic flux (Nguyen and Olzmann, 2017). Thus, there is clearly crosstalk between lipid synthesis, lipolysis, and autophagy, at many levels.
8. Conclusions and future perspectives
LDs are bone fide organelles that undergo a lifecycle including biogenesis, maturation, and turnover. They may also undergo fusion and fission, and they interact with and exchange materials with other organelles. Many aspects of the LD lifecycle remain incompletely understood. LDs have unique biophysical properties, due to their hydrophobic core and phospholipid monolayer membrane (Thiam et al., 2013; Thiam and Foret, 2016). Therefore, certain aspects of the LD lifecycle are unique to LDs. For example, LD regulation by cytoplasmic lipases does not apply to other organelles, and the fusion/fission of LDs may be different from that of other organelles due to their phospholipid monolayer. However, certain aspects of the LD lifecycle share similarities with other organelles. LDs arise from the ER (Pol et al., 2014; Walther et al., 2017). This is similar to peroxisomes and autophagosomes, for which the ER can also be a source of membrane (Axe et al., 2008; Hamasaki et al., 2013; Joshi et al., 2017). Like other organelles, LDs are heterogeneous, undergo a maturation process, and can be turned over by autophagy (macro-, micro- and chaperone-mediated). It will be interesting to investigate how LD biogenesis, function, and turnover are coordinated with that of other organelles, and whether any of these processes share common machinery. Because LDs have been traditionally less studied than other organelles, it will also be useful to apply tools developed for the study of other organelles to LD biology. Due to the heterogeneity of LDs, I am particularly excited about the use of imaging-based approaches that allow for the observation of individual LDs. The last ten years have seen an explosion in interest in the cell biology of LDs, and I am confident that the next ten years will lead to even further insights into the lifecycle of this metabolically important organelle.
Acknowledgements:
This work was supported by the University of North Carolina at Chapel Hill, and by the National Institute on Aging of the National Institutes of Health, under award number K99AG052570.
References:
- Abell BM, High S, Moloney MM, 2002. Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J Biol Chem 277, 8602–8610. [DOI] [PubMed] [Google Scholar]
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM, 1997. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9, 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstee QM, Goldin RD, 2006. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol 87, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aon MA, Bhatt N, Cortassa SC, 2014. Mitochondrial and cellular mechanisms for managing lipid excess. Front Physiol 5, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT, 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Savage DB, Siniossoglou S, 2015. Lipid droplet-organelle interactions: emerging roles in lipid metabolism. Curr Opin Cell Biol 35, 91–97. [DOI] [PubMed] [Google Scholar]
- Bersuker K, Olzmann JA, 2017. Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation. Biochim Biophys Acta 1862, 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM, 2006. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 173, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Lee S, Hilton CL, Jiang QX, Goodman JM, 2010. Seipin is a discrete homooligomer. Biochemistry 49, 10747–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E, El Mourabit H, Prot M, Nemani M, Khallouf E, Colard O, Maurice M, Durand-Schneider AM, Chretien Y, Gres S, Wolf C, Saulnier-Blache JS, Capeau J, Magre J, 2009. Seipin deficiency alters fatty acid Delta9 desaturation and lipid droplet formation in Berardinelli-Seip congenital lipodystrophy. Biochimie 91, 796–803. [DOI] [PubMed] [Google Scholar]
- Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KN, Honing S, 2009. TIP47 functions in the biogenesis of lipid droplets. J Cell Biol 185, 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM, 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol Biol Cell 26, 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Ji WK, Stan RV, de Juan Sanz J, Ryan TA, Higgs HN, 2017. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitraju C, Mejhert N, Haas JT, Diaz-Ramirez LG, Grueter CA, Imbriglio JE, Pinto S, Koliwad SK, Walther TC, Farese RV Jr., 2017. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab 26, 407–418 e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V, Ojha N, Golden A, Prinz WA, 2015. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol 211, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Mari M, Weill U, Rosenfeld-Gur E, Moldavski O, Castro IG, Soni KG, Harpaz N, Levine TP, Futerman AH, Reggiori F, Bankaitis VA, Schuldiner M, Bohnert M, 2017. Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV Jr., Walther TC, 2016. Lipid droplets go nuclear. J Cell Biol 212, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H, 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK, 2011. ER tubules mark sites of mitochondrial division. Science 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jaattela M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Munz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G, 2017. Molecular definitions of autophagy and related processes. EMBO J 36, 1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Chen FJ, Zhou L, Su L, Xu D, Xu L, Li P, 2017. Control of lipid droplet fusion and growth by CIDE family proteins. Biochim Biophys Acta 1862, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R, 2013. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2, e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P, 2011. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol 195, 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A, Buxo L, Mora G, Funaya C, Idrissi FZ, Mancuso F, Gomez R, Muntanya J, Sabido E, Carvalho P, 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J Cell Biol 211, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DA, Zhan C, Silver DL, 2011. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc Natl Acad Sci U S A 108, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubern A, Barcelo-Torns M, Casas J, Barneda D, Masgrau R, Picatoste F, Balsinde J, Balboa MA, Claro E, 2009. Lipid droplet biogenesis induced by stress involves triacylglycerol synthesis that depends on group VIA phospholipase A2. J Biol Chem 284, 5697–5708. [DOI] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV, 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453, 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J, 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T, 2013. Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. [DOI] [PubMed] [Google Scholar]
- Hashemi HF, Goodman JM, 2015. The life cycle of lipid droplets. Curr Opin Cell Biol 33, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Segawa H, Okuno M, Kano H, Hamaguchi HO, Haraguchi T, Hiraoka Y, Hasui S, Yamaguchi T, Hirose F, Osumi T, 2012. Active involvement of micro-lipid droplets and lipid-droplet-associated proteins in hormone-stimulated lipolysis in adipocytes. J Cell Sci 125, 6127–6136. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Hayashi H, Hara-Nishimura I, Nishimura M, 2001. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218, 83–94. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A, 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature cell biology 11, 1433–1437. [DOI] [PubMed] [Google Scholar]
- Herms A, Bosch M, Reddy BJ, Schieber NL, Fajardo A, Ruperez C, Fernandez-Vidal A, Ferguson C, Rentero C, Tebar F, Enrich C, Parton RG, Gross SP, Pol A, 2015. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat Commun 6, 7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR, 2012. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci 125, 4067–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Hughes CE, Henderson KA, Yazvenko N, Gottschling DE, 2016. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R, 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci 124, 2424–2437. [DOI] [PubMed] [Google Scholar]
- Jambunathan S, Yin J, Khan W, Tamori Y, Puri V, 2011. FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One 6, e28614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Ostrowski P, Jaumouille V, Grinstein S, 2016. The position of lysosomes within the cell determines their luminal pH. J Cell Biol 212, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Zhang H, Prinz WA, 2017. Organelle biogenesis in the endoplasmic reticulum. Nat Cell Biol 19, 876–882. [DOI] [PubMed] [Google Scholar]
- Kadereit B, Kumar P, Wang WJ, Miranda D, Snapp EL, Severina N, Torregroza I, Evans T, Silver DL, 2008. Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci U S A 105, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan A, Herms A, Fernandez-Vidal A, Bosch M, Schieber NL, Reddy BJ, Fajardo A, Gelabert-Baldrich M, Tebar F, Enrich C, Gross SP, Parton RG, Pol A, 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol 203, 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2012. Chaperones in autophagy. Pharmacol Res 66, 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2015. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol 17, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2016. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 12, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor VK, Ahrends R, Lin Y, Shen WJ, Adams CM, Roseman AN, Cortez Y, Teruel MN, Azhar S, Kraemer FB, 2014. The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One 9, e105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P, 2017. Peroxisome Biogenesis: A Union between Two Organelles. Curr Biol 27, R271–R274. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, Sztalryd C, 2016. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu Rev Nutr 36, 471–509. [DOI] [PubMed] [Google Scholar]
- Korobova F, Ramabhadran V, Higgs HN, 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Farese RV Jr., Walther TC, 2016. Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol 26, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Thiam AR, Farese RV Jr., Walther TC, 2015. Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Dev Cell 34, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, Farese RV Jr., Walther TC, 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab 14, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV Jr., Walther TC, 2013. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics 12, 1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R, 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab 3, 309–319. [DOI] [PubMed] [Google Scholar]
- Layerenza JP, Gonzalez P, Garcia de Bravo MM, Polo MP, Sisti MS, Ves-Losada A, 2013. Nuclear lipid droplets: a novel nuclear domain. Biochim Biophys Acta 1831, 327–340. [DOI] [PubMed] [Google Scholar]
- Lee JE, Westrate LM, Wu H, Page C, Voeltz GK, 2016. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540, 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AP, Manneschmidt AK, VerBrugge B, Dortch MR, Minkin SC, Prater KE, Biggerstaff JP, Dunlap JR, Dalhaimer P, 2012. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic 13, 705–714. [DOI] [PubMed] [Google Scholar]
- Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J, 2015. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL, 2006. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J Biol Chem 281, 11901–11909. [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Klemm RW, Junker M, Hannibal-Bach HK, Ejsing CS, Rapoport TA, 2014. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep 6, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie S, Riemer J, Wideman JG, McBride HM, 2017. A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE, 2004. Endocytic recycling. Nat Rev Mol Cell Biol 5, 121–132. [DOI] [PubMed] [Google Scholar]
- Meyers A, Del Rio ZP, Beaver RA, Morris RM, Weiskittel TM, Alshibli AK, Mannik J, Morrell-Falvey J, Dalhaimer P, 2016. Lipid Droplets Form from Distinct Regions of the Cell in the Fission Yeast Schizosaccharomyces pombe. Traffic 17, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldavski O, Amen T, Levin-Zaidman S, Eisenstein M, Rogachev I, Brandis A, Kaganovich D, Schuldiner M, 2015. Lipid Droplets Are Essential for Efficient Clearance of Cytosolic Inclusion Bodies. Dev Cell 33, 603–610. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG, 2010. Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens. PLoS One 5, e15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, Olzmann JA, 2017. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev Cell 42, 9–21 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TB, Olzmann JA, 2017. Lipid droplets and lipotoxicity during autophagy. Autophagy, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, Williams JA, Ding WX, 2015. Mitochondrial dynamics and mitochondrial quality control. Redox Biol 4, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Kawai T, Yoshikawa Y, Cheng J, Jokitalo E, Fujimoto T, 2016. PML isoform II plays a critical role in nuclear lipid droplet formation. J Cell Biol 212, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL, 2011. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab 13, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, Marcel YL, 2012. Regulation of lipid droplet cholesterol efflux from macrophage foam cells. Arterioscler Thromb Vasc Biol 32, 575–581. [DOI] [PubMed] [Google Scholar]
- Paar M, Jungst C, Steiner NA, Magnes C, Sinner F, Kolb D, Lass A, Zimmermann R, Zumbusch A, Kohlwein SD, Wolinski H, 2012. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem 287, 11164–11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L, Scorrano L, 2016. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu Rev Physiol 78, 505–531. [DOI] [PubMed] [Google Scholar]
- Pfisterer SG, Gateva G, Horvath P, Pirhonen J, Salo VT, Karhinen L, Varjosalo M, Ryhanen SJ, Lappalainen P, Ikonen E, 2017. Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat Commun 8, 14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Gross SP, Parton RG, 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol 204, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott-Schwartz J, 2015. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC, 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature cell biology 12, 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ, 2006. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci 119, 4215–4224. [DOI] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magre J, Thiele C, Holtta-Vuori M, Jokitalo E, Ikonen E, 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J 35, 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, 2001. Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J Histochem Cytochem 49, 1421–1429. [DOI] [PubMed] [Google Scholar]
- Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA, 2015. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrul B, Kopito RR, 2016. Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat Cell Biol 18, 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Bohnert M, 2017. A different kind of love - lipid droplet contact sites. Biochim Biophys Acta 1862, 1188–1196. [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA, McNiven MA, 2013. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol 203, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J, 2004. Thermodynamics of lipid-peptide interactions. Biochim Biophys Acta 1666, 40–50. [DOI] [PubMed] [Google Scholar]
- Seo AY, Lau PW, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, Lippincott-Schwartz J, 2017. AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, Uchiyama Y, 2009. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochemical and biophysical research communications 382, 419–423. [DOI] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Tamura H, Ueno T, Nishimura T, Inoue T, Sasaki M, Koike M, Arai H, Kominami E, Uchiyama Y, 2010. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochemical and biophysical research communications 393, 274–279. [DOI] [PubMed] [Google Scholar]
- Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z, 2015. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J 34, 2117–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ, 2009. Autophagy regulates lipid metabolism. Nature 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE, 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. The Journal of biological chemistry 284, 30941–30948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Goldberg EB, Makarova KS, Lin L, Brown WJ, Jackson CL, 2006. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep 7, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD, 2013. Peroxisomes take shape. Nat Rev Mol Cell Biol 14, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS, 2009. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci 122, 1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV Jr., 2009. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem 284, 5352–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, Mattie S, Prudent J, McBride HM, 2017. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542, 251–254. [DOI] [PubMed] [Google Scholar]
- Sun Z, Gong J, Wu H, Xu W, Wu L, Xu D, Gao J, Wu JW, Yang H, Yang M, Li P, 2013. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat Commun 4, 1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM, 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira V, Johnsen L, Martinez-Montanes F, Grippa A, Buxo L, Idrissi FZ, Ejsing CS, Carvalho P, 2017. Regulation of lipid droplets by metabolically controlled Ldo isoforms. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Farese RV Jr., Walther TC, 2013. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14, 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Foret L, 2016. The physics of lipid droplet nucleation, growth and budding. Biochim Biophys Acta 1861, 715–722. [DOI] [PubMed] [Google Scholar]
- Tripathi DN, Walker CL, 2016. The peroxisome as a cell signaling organelle. Curr Opin Cell Biol 39, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R, Roingeard P, 2013. Nuclear lipid droplets identified by electron microscopy of serial sections. BMC Res Notes 6, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J, 2017. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M, 2016. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol 212, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N, 2012. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Molecular biology of the cell 23, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, Pon LA, 2015. Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev Cell 35, 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV Jr., 2017. Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol 33, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Miao YH, Chang YS, 2014a. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci 127, 1214–1228. [DOI] [PubMed] [Google Scholar]
- Wang CW, Miao YH, Chang YS, 2014b. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 206, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, Garg A, Olarte MJ, Lin Q, Frohlich F, Hannibal-Bach HK, Upadhyayula S, Perrimon N, Kirchhausen T, Ejsing CS, Walther TC, Farese RV, 2016. Seipin is required for converting nascent to mature lipid droplets. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sreenivasan U, Hu H, Saladino A, Polster BM, Lund LM, Gong DW, Stanley WC, Sztalryd C, 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res 52, 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sztalryd C, 2011. Oxidative tissue: perilipin 5 links storage with the furnace. Trends Endocrinol Metab 22, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Haas JT, Walther TC, Farese RV Jr., 2014a. Lipid droplet biogenesis. Curr Opin Cell Biol 29, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV Jr., Walther TC, 2014b. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3, e01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV Jr., Walther TC, 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24, 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE, 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett 580, 5484–5491. [DOI] [PubMed] [Google Scholar]
- Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE, 2005. S3–12, Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem 280, 19146–19155. [DOI] [PubMed] [Google Scholar]
- Woods DC, 2017. Mitochondrial Heterogeneity: Evaluating Mitochondrial Subpopulation Dynamics in Stem Cells. Stem Cells Int 2017, 7068567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV Jr., Mak HY, 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol 198, 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lu X, Lombes M, Rha GB, Chi YI, Guerin TM, Smart EJ, Liu J, 2010. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab 11, 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Madeo F, Kratky D, 2017. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang Y, Cui L, Deng Y, Xu S, Yu J, Cichello S, Serrero G, Ying Y, Liu P, 2016. Morphologically and Functionally Distinct Lipid Droplet Subpopulations. Sci Rep 6, 29539. [DOI] [PMC free article] [PubMed] [Google Scholar]