Abstract
Purpose: To determine the expression and function of Delta/Notch-like EGF-related receptor (DNER) in hepatocellular carcinoma (HCC).
Methods: The expression of DNER in 84 HCC tissue samples and matched adjacent noncancerous specimens, as well as HCC cells, were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting. Survival analysis was evaluated using Kaplan–Meier method. For experiments in vitro, cell viability was measured by Cell Counting Kit-8 Assay and Colony Formation Assay. Furthermore, cell invasion and migration assays were performed with Transwell Assay.
Results: The results showed that DNER was overexpressed in the tissues and cell lines of HCC (all, p < 0.05), and the upregulated expression of DNER was significantly correlated with advanced pathologic stage (p = 0.013) and pathologic-M1 (p = 0.012) in HCC patients. Survival analysis revealed that patients with high DNER levels had worse overall survival (OS) than those with low DNER levels (p = 0.004). More importantly, DNER could be an independent predictor of prognosis for OS (HR = 2.582, 95% CI 1.239–5.380, p = 0.011). In vitro, knockdown of DNER significantly suppressed cell proliferation, colony formation, cell invasion, and cell migration in HepG2 cells. Moreover, inhibition of DNER inactivated PI3K/AKT signaling pathway by downregulating the expression of p-PI3K, p-AKT, and p-70s6k.
Conclusions: Taken together, DNER could promote proliferation, migration, and invasion of HCC cells by regulating the activation of PI3K/AKT pathway, and it might act as a potential prognostic biomarker for HCC.
Keywords: : cell growth, DNER, hepatocellular carcinoma, PI3K/AKT pathway, prognosis
Introduction
Hepatocellular carcinoma (HCC), the most common type of primary liver cancers, is the third leading cause of cancer mortality causing over 700,000 deaths annually.1,2 HCC is characterized by the high incidence of recurrence and invasive ability,3 and its pathogenesis is the combination of genetic and environmental factors, including genetic modification, infection with hepatitis B/C virus, and hepatic toxins.4 To date, despite the significant advances in HCC diagnosis and treatment, the prognosis of patients still remains unsatisfactory due to the high probability of tumor metastasis and recurrence.5 Thus, to improve the life quality of HCC patients, there is an urgent need to understand the molecular mechanisms of HCC and develop effective prognostic biomarkers.
Delta/Notch-like EGF-related receptor (DNER), also named BET and HE60, is a transmembrane protein found in the central nervous system, hair cells in inner ear, and olfactory bulb.6,7 DNER has been reported to be involved in the neural proliferation, differentiation, and development,8,9 and its deletion could lead to abnormalities and ataxia of cerebellum.10 Actually, DNER is identified as the target of anti-Tr.11 More importantly, it has also been reported that DNER could serve as the oncogenic or antioncogenic factors by regulating cellular proliferation, invasion, and metastasis.7,12 However, whether DNER plays a role in the development of HCC still remains unclear.
In this study, the authors investigated the expression of DNER and its prognostic effect in HCC. Furthermore, the authors studied the function of DNER on cell proliferation, colony formation, cell invasion, and cell migration.
Materials and Methods
Patients and specimens
A total of 84 paired cancerous and matched adjacent normal tissue specimens were collected from patients who were diagnosed with HCC by histopathology in Renmin Hospital of Wuhan University from 2008 to 2012. Tissue samples were collected immediately in liquid nitrogen and then stored in −80°C for further test. This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University, and informed consent was obtained from all patients. Clinicopathological features of all patients are summarized in Table 1.
Table 1.
The Relationship Between DNER Expression and the Clinicopathological Characteristics in HCC
| Expression of DNER | ||||
|---|---|---|---|---|
| Characteristics | n | Low | High | p |
| Age | 0.275 | |||
| <60 | 41 | 23 | 18 | |
| ≥60 | 43 | 19 | 24 | |
| Gender | 0.649 | |||
| Female | 30 | 14 | 16 | |
| Male | 54 | 28 | 26 | |
| Grade | 0.272 | |||
| G1+G2 | 47 | 21 | 26 | |
| G3+G4 | 37 | 21 | 16 | |
| Pathologic stage | 0.013* | |||
| I+II | 53 | 32 | 21 | |
| III+IV | 31 | 10 | 21 | |
| Pathologic-T | 0.059 | |||
| T1+T2 | 58 | 33 | 25 | |
| T3+T4 | 26 | 9 | 17 | |
| Pathologic-N | 0.202 | |||
| N0 | 78 | 41 | 37 | |
| N1 | 6 | 1 | 5 | |
| Pathologic-M | 0.012* | |||
| M0 | 77 | 42 | 35 | |
| M1 | 7 | 0 | 7 | |
p < 0.05.
DNER, Delta/Notch-like EGF-related receptor; HCC, hepatocellular carcinoma.
Cell culture and transfection
Human HCC cell lines, HepG2, SMMC-7721, HHCC, and Hep3B, and the normal liver cell line HL7702 were obtained from the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM-1640) at 37°C in a humidified atmosphere containing 5% CO2. The medium was supplemented with 10% fetal bovine serum, 0.1 mg/mL streptomycin, and 100 U/mL penicillin. After entering the logarithmic period, HepG2 cells were plated in six-well plates and then transfected with two DNER-siRNAs (siRNA1: 5′-AACAGGGCAGAAAGUUGUA-3′; siRNA2: 5′-AUGAAAUGGGAUCAAGUGG-3′; siRNA3: 5′-ACUUUCUGCCCUGUUUUCGGC-3′) using Lipofectamine 2000 according to the manufacturer's instructions.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and the quality of RNA was assessed with NanoDrop 2000. Complementary DNAs were synthesized by Reverse Transcriptase (Invitrogen). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the ABI 7900 (Life Technologies). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference, and primers were as follows: DNER: forward, 5′-CAGGGACCTCGTTAATGGCT-3′, reverse, 5′-CCGTTCAGACAGCTGACGTT-3′; GAPDH: forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′. Relative expression level of DNER was calculated by the 2−ΔΔCt method. Each experiment was performed in triplicate.
Western blot
After 48 h of transfection, the cell lines were lysed using RIPA lysis buffer for protein extraction, and the concentration of protein was evaluated with the BCA assay. For western detection, protein lysates were separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto the polyvinylidene fluoride (PVDF) membranes. Following blocking in 5% nonfat milk for 1 h, the membranes were incubated with primary antibodies at 4°C overnight. Next, the membranes were incubated with secondary antibody at room temperature for 1 h. Signal was visualized using the ECL blotting analysis system, and GAPDH was used as the internal control.
Cell Counting Kit-8 Assay
After 24 h of transfection, HepG2 cells were plated in 96-well plates at a density of 1 × 103 cells. To test the proliferation rate, the Cell Counting Kit-8 (CCK-8) was performed per 24 h according to the manufacturer's instructions. The absorbance was detected at the wavelength of 450 nm using the Microplate Reader.
Wound-healing assay
Scratch-healing assay was used to assess the cell migration. Briefly, HepG2 cells transfected with siRNA were cultured in six-well plates. Scratch wounds across each well were made when the cells grew to 90% confluence, and photographs were taken at 0 and 24 h, respectively.
Transwell assay
Cell invasion and cell migration were assessed using a Transwell Assay. Briefly, for cell invasion assay, 100 μL Matrigel was added to the top chamber of transwell with 24-well plates, and the lower chamber was filled with 500 μL of serum-free medium. After 24 h of transfection, the cells (1 × 105) in 100 μL were placed onto the top chamber. After incubation overnight, the cells on the upper chamber of the filter were removed with a cotton swab, while the invading cells adhered to the underside of the membrane were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet solution for 20 min. The cells were counted using a microscope in five random fields. Similar to invasive assay, the migration assay was performed without the treatment of glue, and the suspension contained 5 × 103 cells.
Colony formation assay
The cells after transfection were seeded at a density of 1500 cells/dish and further cultured at 37°C in a humidified atmosphere containing 5% CO2 for 1–2 weeks. When there were visible colonies in culture dish, the media was removed, and the colonies were fixed with 5 mL methanol for 30 min, stained with 0.1% crystal violet for 30 min, and then counted.
Statistical analyses
All statistical analyses were performed with SPSS 22.0 software (SPSS, Inc., Chicago, IL) and GraphPad Prism 5 (GraphPad Software, Inc.). The association between DNER expression and clinicopathologic parameters was evaluated by chi-square test, and comparisons between two groups were made by Student's t-test. Survival analysis was performed by Kaplan–Meier method with log-rank test. Univariate and multivariate analyses were performed using the Cox proportional hazards regression model. A p-value <0.05 was considered statistically significant.
Results
Upregulation of DNER and its correlation with progression of human HCC
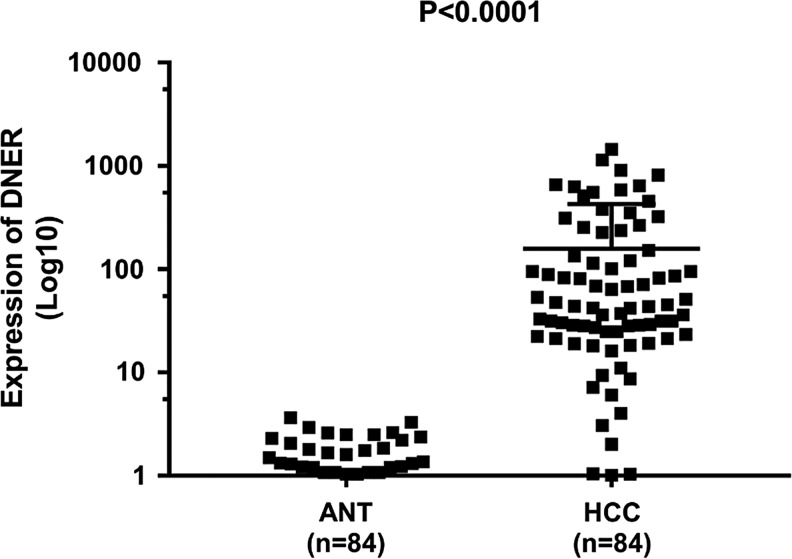
To examine the effect of DNER in the development of HCC, the expression of DNER was determined by qRT-PCR in 84 HCC and matched adjacent normal tissue samples. As shown in (Fig. 1), DNER expression in HCC at the level of mRNA was significantly higher than that in normal controls (p < 0.0001).
FIG. 1.
Relative DNER expression detected by qRT-PCR. DNER expression was significantly upregulated in HCC tissues compared to that in adjacent normal tissues (p < 0.0001). DNER, Delta/Notch-like EGF-related receptor; HCC, hepatocellular carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction.
In addition, to evaluate the clinical significance of DNER in HCC patients, the authors assessed the relationship between DNER level and clinicopathological parameters. Primarily, the authors divided the patients into two groups according to low or high levels of DNER in HCC. The results showed that high DNER expression levels significantly correlated with advanced pathologic stage (p = 0.013) and pathologic-M1 (p = 0.012). However, there was no significant correlation between DNER expression levels and other clinical characteristics such as age, gender, grade, pathologic-T, and pathologic-N (all, p > 0.05; Table 1). Taken together, these results indicate that aberrant expression of DNER might play a role in the progression of HCC although serving as the carcinogen.
Prognostic significance of DNER in patients with HCC
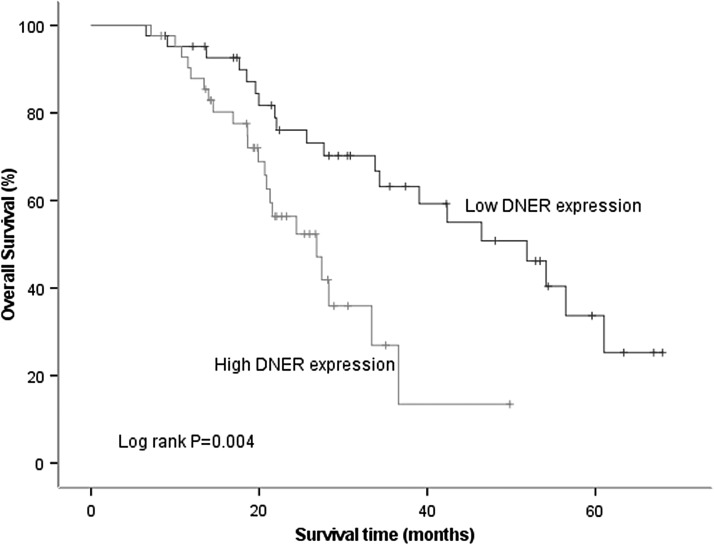
To demonstrate the prognostic role of DNER in HCC, the authors examined the correlation between DNER level and overall survival (OS) of HCC patients. As shown in (Fig. 2), patients with high expression of DNER had shorter OS compared to those with low level of DNER (p = 0.004). Univariate analysis revealed that DNER expression (p = 0.005), gender (p = 0.030), pathologic stage (p = 0.021), and pathologic-T (p = 0.031) were predictors for OS. Moreover, multivariate Cox regression analysis suggested that DNER could be an independent predictor of prognosis for OS (HR = 2.582, 95% CI 1.239–5.380, p = 0.011; Table 2).
FIG. 2.
Survival analysis. Patients with high DNER expression had worse OS compared to those with low expression in HCC (p = 0.004). OS, overall survival.
Table 2.
Univariate and Multivariate Analysis of Variables Associated with OS in Patients with HCC
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | p | HR | 95% CI | p | HR | 95% CI |
| DNER expression (high/low) | 0.005* | 2.701 | 1.352–5.398 | 0.011* | 2.582 | 1.239–5.380 |
| Age (<60/≥60) | 0.456 | 1.264 | 0.683–2.340 | |||
| Gender (female/male) | 0.030* | 0.509 | 0.276–0.937 | 0.023* | 0.480 | 0.254–0.906 |
| Grade (G1+G2/G3+G4) | 0.841 | 1.065 | 0.577–1.964 | |||
| Pathologic stage (I + II/III+IV) | 0.021* | 2.054 | 1.113–3.789 | 0.701 | 0.739 | 0.157–3.472 |
| Pathologic-T (T1+T2/T3+T4) | 0.031* | 1.981 | 1.066–3.680 | 0.296 | 2.266 | 0.489–10.506 |
| Pathologic-M (M0/M1) | 0.298 | 1.739 | 0.613–4.937 | |||
| Pathologic-N (N0/N1) | 0.076 | 2.996 | 0.891–10.067 | |||
p < 0.05.
HR, hazard ratio; OS, overall survival.
Reduced expression of DNER suppressed cell proliferation, colony formation, invasion, and migration
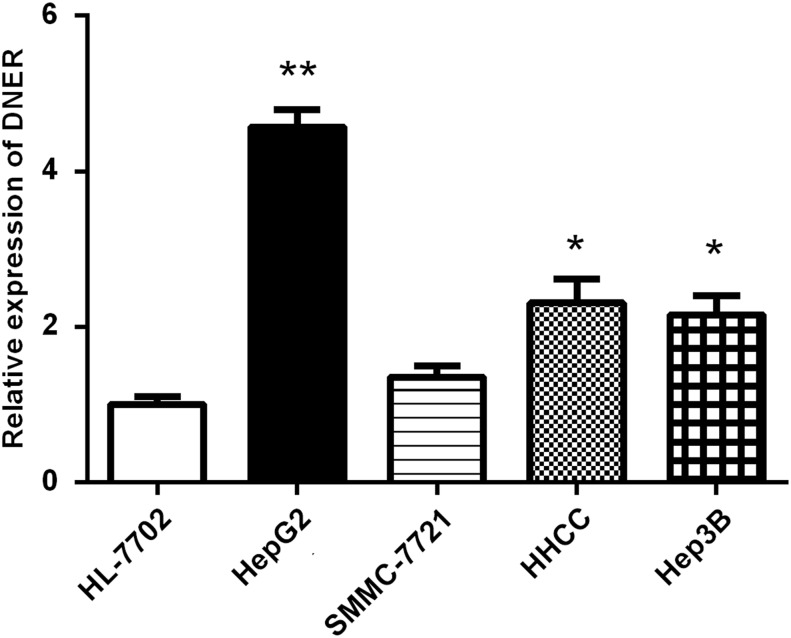
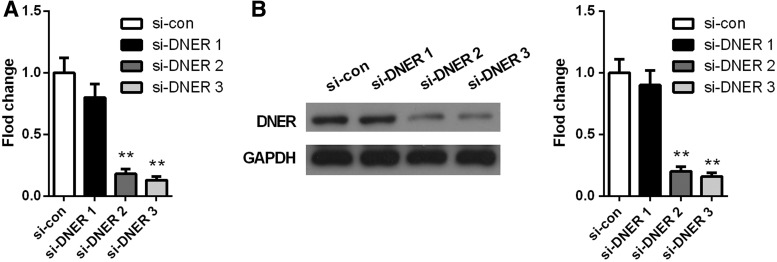
To further investigate the molecular function of DNER in tumorigenesis of HCC, the authors detected its expression level in human liver cell line HL-7702 and HCC cell lines. In accordance with the results of HCC tissues, DNER expression in HCC cell lines (HepG2, HHCC, and Hep3B) was significantly increased compared with HL-7702 (p < 0.05; Fig. 3). Among HCC cell lines, DNER performed the highest expression in HepG2 cells, and thus, HepG2 cell line was chosen for further functional investigation in vitro. Next, the authors transfected HepG2 cells with siRNA1, siRNA2, and siRNA3 and found that siRNA2 and siRNA3 significantly decreased the expression of DNER at mRNA and protein levels compared with siRNA1 (p < 0.05; Fig. 4), and therefore, siRNA2 and siRNA3 were selected for subsequent studies.
FIG. 3.
DNER mRNA expression in HCC cell lines. DNER levels in HepG2, HHCC, and Hep3B cell lines were significantly increased compared with HL-7702 (*p < 0.05, **p < 0.01).
FIG. 4.
The expression of DNER in HCC cells transfected with siRNA1, siRNA2, and siRNA3. DNER levels detected by qRT-PCR (A) and western blotting (B). **p < 0.01.
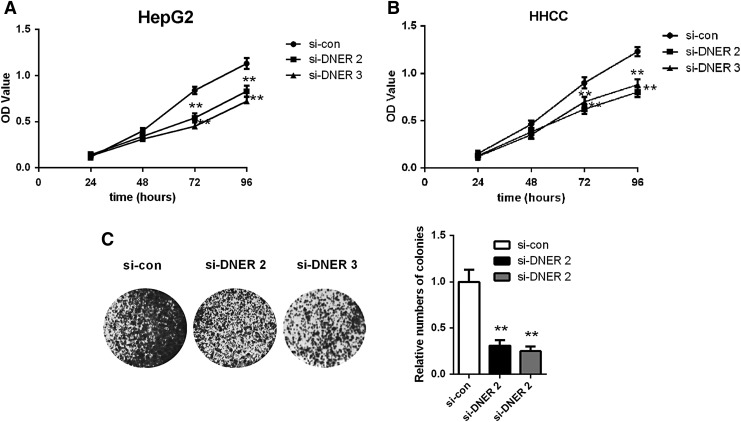
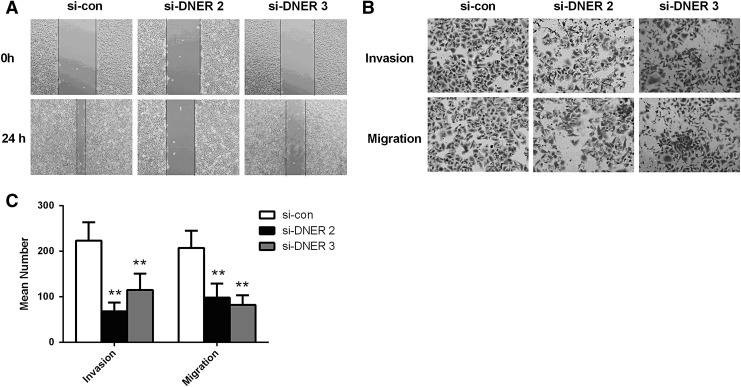
To understand the functional mechanism of DNER in the carcinogenesis of HCC, the authors detected cell proliferation, colony forming ability, cell invasion, and cell migration. By CCK-8 assays, the authors found that the proliferation rate of HCC cells was significantly downregulated after the knockdown of DNER at 72 or 96 h in both HepG2 cell (p < 0.05; Fig. 5A) and HHCC cell (p < 0.05; Fig. 5B). Similarly, colony formation assay showed that the number of colonies in HepG2 cells transfected with siDNER was lower than that in controls, and the difference was significant (p < 0.05; Fig. 5C). Consequently, the wound healing assay showed that there was a significant reduction in wound closure in cells with decreased DNER expression compared with control cells, and transwell assay demonstrated that the number of invaded and migrated cells in HepG2 transfected with siDNER was significantly reduced compared to the controls (p < 0.05; Fig. 6), suggesting that DNER could increase the migration and invasive ability of HCC cells significantly.
FIG. 5.
Knockdown of DNER inhibits proliferation ability of HCC cells. Cell proliferation detected by CCK-8 assay (A, B) in HepG2 and HHCC cell lines and colony formation assay (C) in HepG2 line as indicated, **p < 0.01. CCK, Cell Counting Kit.
FIG. 6.
Knockdown of DNER inhibits migration and invasion ability of HCC cells. (A) Cell migration detected by wound healing assay. (B, C) Cell invasion and cell migration were assessed using a transwell assay in HepG2 line as indicated. **p < 0.01.
Downregulation of DNER could suppress the activation of PI3K/AKT signaling pathway
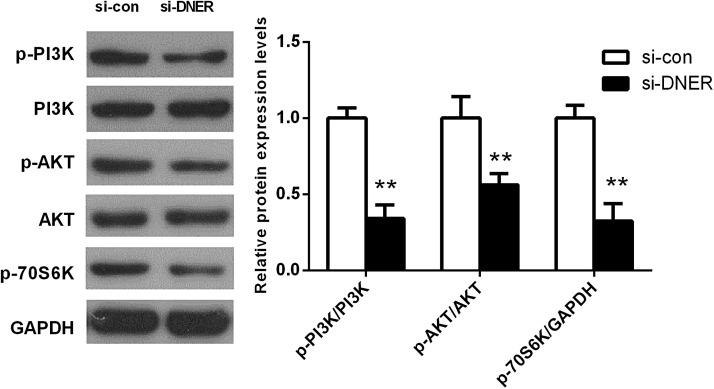
PI3K signaling pathway is involved in tumor progression with essential protein of PI3K and AKT, which has played important roles in proliferation and metastasis of tumor cells. In this study, western blot analysis demonstrated that the phosphorylated PI3K, AKT, and 70s6k in cells transfected with DNER-siRNA were significantly lower than in cells transfected with negative control (all, p < 0.05; Fig. 7), indicating that downregulated DNER expression could significantly suppress the activation of PI3K/AKT pathway.
FIG. 7.
Reduced DNER inhibits PI3K/AKT signaling pathway in HepG2 cells. Western blot analysis showed that reduced DNER expression significantly decreased the expression of phosphorylated PI3K, AKT, and 70s6k. **p < 0.01.
Discussion
Today, it has made HCC prognosis and treatment difficult because of the lack of reliable potential prognostic biomarkers and therapy target. In the present study, the authors found that DNER was a potential prognostic biomarker for HCC, and it altered the cell proliferation, migration, and invasion ability in HCC by interacting with PI3K/AKT signaling pathway.
As a novel EGF-like repeat-containing single-pass transmembrane protein, DNER, on chromosome 2, was identified to be linked to carcinogenesis.13 Wang et al.7 demonstrated that DNER expression was upregulated in prostate cancer, and the knockdown of DNER inhibited tumorigenesis in PC-3 xenografts. In accordance with the previous research findings, the authors found that the expression of DNER in HCC tissues and HCC cell lines was significantly increased compared with those in normal controls in the present study. Moreover, their results suggested that patients with high DNER expression had worse OS than those with low DNER expression, and DNER could be a potential prognostic biomarker for HCC, which has not been reported in other studies till now. Previous studies revealed that the clinicopathological features of HCC, such as TNM stage, tumor differentiation, and tumor size,14–16 were in connection with the outcome of HCC patients. In the present study, high DNER expression was significantly associated with the advanced pathologic stage and pathologic-M1. All these studies indicated that DNER might play a role as a tumor promoter in HCC.
DNER has been identified as the central nervous system-specific ligand for the Notch receptors that are important for cellular differentiation, growth, and cancer through mediating plenty of cell signaling.17,18 For glioblastoma, DNER inhibited the growth of neurospheres and was involved in the inhibition of their engraftment and growth as tumor xenografts.13 In addition, knockdown of DNER was also found to play an important role in prostate cancer by inhibiting cell proliferation, migration, and invasion.7 In the present study, the authors demonstrated that knockdown expression of DNER in HCC cells could significantly inhibit cell proliferation, colony formation, invasion, and migration.
DNER mediates notch signaling by affecting expression of several notch pathway genes in pancreatic β-cells.19 Accumulating studies had found that PI3K/AKT signaling pathway is closely involved in the carcinogenesis of cancers.20,21 Between notch and PI3K signaling pathway, Song et al.22 revealed that inhibition of Notch signaling could promote the Adipogenic Differentiation of Mesenchymal Stem Cells by autophagy involving PTEN-PI3K/Akt/mTOR pathway. Furthermore, the interplay between the Notch and PI3K/Akt pathways was involved in high glucose–induced podocyte apoptosis.23 At present, the enhancers of PI3K, such as LAMP3,24 EDG2,25 and RAB27B,26 had been reported to strengthen the PI3K/AKT signaling and improve the growth of HCC. In the present study, the knockdown of DNER resulted in the decreased level of p-PI3K, p-AKT, and p-70S6K, indicating that siDNER-mediated suppression of HCC cell proliferation, migration, and invasion might be caused by the inactivated PI3K/AKT signaling.
Taken together, this study first reported that the expression of DNER was upregulated in HCC tissues and a risk factor for poor prognosis of HCC. Moreover, the decrease of DNER expression could inhibit cell proliferation, colony formation, cell invasion, and cell migration through interacting with PI3K/AKT signaling pathway. However, further investigation with larger samples is still required in the future.
Acknowledgments
This work was supported by the “Foundation of Hubei Provincial Department of Education” (No. B2014059). The authors thank all the members.
Disclosure Statement
No competing financial interests exist.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87. [DOI] [PubMed] [Google Scholar]
- 2.Guo S, Chen W, Luo Y, et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol 2015;8:5395. [PMC free article] [PubMed] [Google Scholar]
- 3.Bruix J, Boix L, Sala M, et al. Focus on hepatocellular carcinoma. Cancer Cell 2004;5:215. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Song C, Duan B, et al. LncRNA-SVUGP2 suppresses progression of hepatocellular carcinoma. Oncotarget 2017;8:97835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu AX, Dan GD, Sahani DV, et al. HCC and angiogenesis: Possible targets and future directions. Nat Rev Clin Oncol 2011;8:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J, Wang X, Zhang X, et al. DNER modulates the length, polarity and synaptogenesis of spiral ganglion neurons via the Notch signaling pathway. Mol Med Rep 2018;17:2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Wu Q, Zhu S, et al. Delta/notch-like epidermal growth factor-related receptor (DNER) orchestrates stemness and cancer progression in prostate cancer. Am J Transl Res 2017;9:5031. [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh FY, Ma TL, Shih HY, et al. Dner inhibits neural progenitor proliferation and induces neuronal and glial differentiation in zebrafish. Dev Biol 2013;375:1. [DOI] [PubMed] [Google Scholar]
- 9.Richardson L, Venkataraman S, Stevenson P, et al. EMAGE mouse embryo spatial gene expression database: 2014 update. Nucleic Acids Res 2014;42(Database issue):D835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tohgo A, Eiraku M, Miyazaki T, et al. Impaired cerebellar functions in mutant mice lacking DNER. Mol Cell Neurosci 2006;31:326. [DOI] [PubMed] [Google Scholar]
- 11.de Graaff E, Maat P, Hulsenboom E, et al. Identification of delta/notch-like epidermal growth factor-related receptor as the Tr antigen in paraneoplastic cerebellar degeneration. Ann Neurol 2012;71:815. [DOI] [PubMed] [Google Scholar]
- 12.Sun P, Xia S, Lal B, et al. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem Cells 2009;27:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hein R, Flesch-Janys D, Dahmen N, et al. A genome-wide association study to identify genetic susceptibility loci that modify ductal and lobular postmenopausal breast cancer risk associated with menopausal hormone therapy use: A two-stage design with replication. Breast Cancer Res Treat 2013;138:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu WB, Jia WD, Ma JL, et al. Knockdown of GTPBP4 inhibits cell growth and survival in human hepatocellular carcinoma and its prognostic significance. Oncotarget 2017;8:93984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu SL, Wu JC, Liu PF, et al. Up-regulation of RNF187 induces hepatocellular carcinoma cell epithelial to mesenchymal transitions. Oncotarget 2017;8:101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WG, Hu B, Sun HX, et al. Long non-coding RNA00364 represses hepatocellular carcinoma cell proliferation via modulating p-STAT3-IFIT2 signaling axis. Oncotarget 2017;8:102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eiraku M, Tohgo A, Ono K, et al. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci 2005;8:873. [DOI] [PubMed] [Google Scholar]
- 18.Chillakuri CR, Sheppard D, Lea SM, et al. Notch receptor-ligand binding and activation: Insights from molecular studies. Semin Cell Dev Biol 2012;23:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson RL, Muller YL, Kobes S, et al. A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes 2014;63:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thapa N, Choi S, Tan X, et al. Phosphatidylinositol phosphate 5-kinase Iγ and phosphoinositide 3-kinase/Akt signaling couple to promote oncogenic growth. J Biol Chem 2015;290:18843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol 2010;28:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song B, Chi Y, Li X, et al. Inhibition of notch signaling promotes the adipogenic differentiation of mesenchymal stem cells through autophagy activation and PTEN-PI3K/AKT/mTOR pathway. Cell Physiol Biochem 2015;36:1991. [DOI] [PubMed] [Google Scholar]
- 23.Wang XM, Yao M, Liu SX, et al. Interplay between the Notch and PI3K/Akt pathways in high glucose-induced podocyte apoptosis. Am J Physiol Renal Physiol 2014;306:205. [DOI] [PubMed] [Google Scholar]
- 24.Liao X, Song L, Zhang L, et al. LAMP3 regulates hepatic lipid metabolism through activating PI3K/Akt pathway. Mol Cell Endocrinol 2017;470:160. [DOI] [PubMed] [Google Scholar]
- 25.Xu M, Liu Z, Wang C, et al. EDG2 enhanced the progression of hepatocellular carcinoma by LPA/PI3K/AKT/mTOR signaling. Oncotarget 2017;8:66154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y, Ye X, Le S, et al. Downregulation of serum RAB27B confers improved prognosis and is associated with hepatocellular carcinoma progression through PI3K-AKT-P21 signaling. Oncotarget 2017;8:61118. [DOI] [PMC free article] [PubMed] [Google Scholar]