Abstract
Radix Astragali (RA) herb with warm property and significant “tonifying qi” effects is indicated for the syndrome of internal cold due to Yang deficiency. The purpose of this research was to explore effects of Radix Astragali (RA) through PPAR signaling pathway on gene expression profiles related to energy metabolism in rats with the Yang-deficiency cold (YDC) syndrome, for identifying the pathological mechanism of Yang-deficiency cold (YDC) syndrome and the effects mechanism of RA. The results indicated that RA could significantly increase body weight (BM), cold and heat tendency (CT), overall temperature (OT), rectum temperature (RT), toe temperature (TT), energy intake (EI), and V(O2)/V(CO2) ratio (which indicates basal metabolism, BM) (P<0.05), enhancing the depressed metabolic function in YDC syndrome model rat. Our data also indicated differentially expressed genes (DEGs) related to energy metabolism involving lipids, carbohydrates, and amino acids metabolic process; the expression of CPT-1 and FABP4 (ap2) was improved; PPAR, Glycolysis, Wnt, cAMP, MAPK, AMPK, and fatty acid degradation signaling pathway may be related to energy metabolism. However, the Chinese herbal medicine RA plays a certain role in promoting the metabolism of substances and energy in rats by its warming and beneficial effect. Our results suggest that the mechanism underlying the function of RA may take effect through the regulation of PPAR signaling pathway and related gene expression. Lipids, carbohydrates, and amino acids metabolic process may be affected to adjust the reduced metabolic function in the model animals. In general, results indicate that RA could promote energy metabolism in rats with the YDC syndrome via PPAR signaling pathway regulating the expression of CPT-1 and FABP4 (ap2), which reflected the warm and qi tonifying properties of RA.
1. Introduction
Yang-deficiency cold syndrome (YDCS) has long been a common and frequently occurring clinical disease. In the modern period, YDCS is even more commonly seen in multiple clinical disorders. Most acute diseases (such as craniocerebral injury combined with lung infection, hepatic encephalopathy, etc.) show this syndrome. YDCS also frequently appears in gynecological and digestive diseases and even more often in chronic diseases (such as type II diabetes) and advanced cancer [1–3]. Currently, there are many studies focusing on YDCS. However, the pathological mechanism is still not clear [4, 5]. “973” project is the most important fund of Chinese government in basic theory research. Our research group luckily applied for this project “Traditional Chinese Medicine Drug Properties Theory Related Basis Problem Study” (2007CB512601), the chills and fever of Chinese traditional medicine in the process of pharmacodynamics research. Under the guidance of traditional Chinese medicine theory, we established a mature animal model that mimics the characteristics and symptoms of YDCS. According to the previous results, the test on gene expression profile of the liver demonstrated that the cytokine network dysregulation is represented by carbohydrate and lipid metabolism imbalance in YDCS model rats [6, 7]. According to the preliminary experimental results of our research group combined with literature research and clinical observation, it is speculated that the occurrence of YDCS may be closely related to the abnormal changes of energy metabolism. PPAR is one of the main pathways for energy metabolism and plays a crucial role in changing energy metabolism. Therefore, this experiment will use both macroscopic observation and microscopic detection to verify the mechanism of YDCS and the role of PPAR signaling pathway along with its target genes CPT-1 and FABP4.
Promotion or inhibition of carbohydrate and lipid metabolism is mainly affected by the target genes CPT-1 and FABP4 in the PPAR signaling pathway. Peroxisome proliferator-activated receptor (PPAR) is a subfamily of nuclear hormone receptors; three isoforms of PPARs have been identified, namely, PPAR-α, PPAR-β/θ, and PPAR-γ. PPAR-α and PPAR-γ are mainly involved in regulating glucose and lipid metabolism, insulin sensitivity, and immune response. Therefore, specific focus on the therapeutic potential of PPAR modulators in gene regulation and metabolic diseases may become a promising direction of developing treatment for metabolic syndrome [8–11]
CPT1 (carnitine palmitoyltransferase 1B) is an integral membrane protein [12]. It is the first element and rate-limiting step of the carnitine palmitoyltransferase system and also an essential step in the beta-oxidation of long-chain fatty acids [13]. In addition, the important role of CPT1 in fatty acid metabolism makes it a potentially useful enzyme in developing the treatment for many other metabolic disorders [14]. CPT1 is inhibited by malonyl-CoA. This inhibition is an ideal target to regulate CPT1 for the treatment of metabolic disorders [15]. FABP4 (adipocyte-fatty acid-binding protein 4, also known as aP2) is involved in the regulation of glucose and lipid metabolism. FABP4 is also capable of resisting the development of atherosclerosis and improving the sensitivity of insulin [16]. Along with many other previous publications, people showed that FABP4 has a close relationship with metabolic syndrome, inflammation, atherosclerosis, obesity, insulin resistance, diabetes mellitus, hypertension, cardiac dysfunction, cardiovascular events, etc. [17]. These two target genes, namely, CPT-1 and FABP4, are in close relation to the activation of PPAR signaling pathway.
This important multiple syndrome is often ignored in the early stages of many diseases [1–5], and the efficacy of treatment is poor. Therefore, a highly effective and mild Chinese medicine that can better suit early treatment and can be taken in long-term is urgently needed to be applied in the treatment of YDCS. It is helpful to suppress the development of diseases and improve the prognosis of diseases. Our research group studied the medicinal properties of a mild beneficial traditional Chinese herbal medicine, RA membranaceus. RA is a commonly used TCM that has sweet taste with warm property. It belongs to lung and spleen meridians and it has such functions as warming the interior and dispelling cold, strengthening and uplifting Yang, and warming and activating meridian; because of its ability to replenish Qi and Yang, RA is often used clinically to treat YDCS caused by insufficient Yang. Modern pharmacology studies show that RA can enhance energy metabolism and immunity. The mechanism of its action may be through the PPAR signal pathway, which regulates energy metabolism. Therefore, our investigation took the change in PPAR signal pathway affecting energy metabolism as a starting point.
We conducted and explained our study mainly through the following aspects: observation of the overall pharmacodynamics focused on energy metabolism; detection and real-time monitoring of rat whole body surface temperature using forward looking infrared (FLIR) cameras; monitoring of rat energy metabolism changes and trends in a 24-hour time frame; and analysis of the possible mechanism of warmth production using RNA-seq transcriptome sequencing from the perspective of gene expression profiles.
2. Materials and Methods
2.1. Animals
Thirty-six specific-pathogen-free (SPF) grade, six-week-old Wistar rats (half male and half female), weighing 200±20 g, were acquired from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China), experimental animal license number: SCXK (Beijing) 2016-000140. All experiments were done in accordance with the guideline and regulation by the Administration Office of Laboratory Animals at Jinan, China. All experimental protocols were approved by medical Ethics Committee of Shandong University of Traditional Chinese Medicine.
2.2. Animal Model
Rats were acclimated for three days in our animal facility and then randomly divided into three groups: normal control group (NCG), YDCS model group, and RAWA treatment group (12 animals in each group). Model rats and RAWA treatment group were fed with TCM mixture every afternoon for two weeks to establish YDCS model.
The animal model were established using TCM mixture (Gypsum, Radix Gentianae, Cortex Phellodendri, and Rhizoma Anemarrhenae mixed in a 2:1.2:1:1.5 ratio [7]) according to the relevant literature [18, 19]. The mixture was enriched to 4 g/ml of water abstract and stored in 4°C for later use. The abstract was administered by gavage with the dose of 64g·kg−1·d−1, 4g/ml, 16ml/kg (the model group and the treatment group). The administration was performed once a day, fourteen consecutive days. The evaluation standards for the establishment of the YDCS model were based on previous experimental studies and with reference to the literature [20]. The partial least-regression method (PLS) was used to select indicators that can determine the degree of the deficient cold model: fecal texture, hair cleanliness, water intake (in positive correlation with the model), urine color, body weight, average toe temperature, and autonomic activity (in negative correlation with the model). Therefore, the standards for successful replication of the YDCS model are as follows: Rats have loose stools. Hair is dirty and messy. Water intake is large (macroscopic representation). There is weight loss. Basal (respiratory) metabolism (RER) is reduced without interference for 24 hours (changes in metabolism index). The anal temperature, toe temperature, and overall body temperature showed a decreasing trend (temperature decreased, and metabolism was significantly reduced). These are accurately consistent with the symptoms of the deficiency cold syndrome and its evaluation indicators have reached the standard. The deficiency cold model was successfully replicated in this experiment.
The BCG was given equal amounts of normal saline at the same time.
2.3. Preparation of RAWA
RAWA was prepared using RA membranaceus. The herb was cultivated in Gansu Province, China, and identified by Prof. Feng Li from the Department of Traditional Chinese Medicine, Shandong University of Traditional Chinese Medicine. RA membranaceus was boiled into water decoction. The concentration of the crude drug was 0.54g/ml and stored at 4°C.
2.4. Experimental Protocol
After the model was successfully established, RAWA was administrated for treatment, according to People's Republic of China Pharmacopoeia (2015 edition), and RA was administered at twice the dose used in adults (60g/70kg). Based on the equivalent dose coefficient algorithm, the rat equivalent dose was 6.3 times that of adults per body weight unit. The abstract was administered by gavage with the dose of 5.40g·kg−1·d−1, 0.54g/ml, 10ml/kg [21]. The administration was performed once a day, seven consecutive days [6]. The normal control group and the model group were given equal amounts of normal saline.
After the last administration, the animals were euthanasia. The livers were rapidly separated at 4° and frozen in liquid nitrogen.
2.5. Temperature Change Detection
7 days, 10 days, and 14 days after drug administration, the overall temperature (OT), rectum temperature (RT), and toe temperature (TT) of the experimental animals were examined using forward looking infrared (FLIR) cameras and DT-880B hand-held infrared body surface temperature rapid screening instrument. Each group of animals was tested alternately in parallel experiments.
2.6. Metabolic Rate Monitoring
Basal metabolism changes were automiaticly detected in rats for 24 hours by using LabMaster. Noninterfering basal metabolism monitoring was performed after the establishment of animal model following the administration of RAWA. Oxygen consumption, carbon dioxide production, respiratory exchange rate, and energy consumption were calculated by indirect calorimetry. The frequency and time of drinking and feeding were captured by high-precision sensors. The total activity of rats in cages was measured by beam sensors. Therefore, noninterfering basal metabolism changes and their significance in YDCS model group and RA treatment group can be analyzed.
2.7. Statistical Analysis
Data analysis was performed using SPSS 24.0 (IBM, LSD, P <0.05) and presented by mean ± standard deviation. F-test was used for the comparison of consecutive variables between groups. Chi-square test was used in the categorical variable group. Single-factor variation analysis of variance was used for comparison. And LSD test was used for further comparisons between pairs. Significant p-values were as follows: ∗∗ P < 0.01,∗ P < 0.05 versus normal control; ##P <0.01, #P <0.05 versus model.
2.8. Transcriptomic Sequencing and Analysis
Sample total RNA was extracted using TRIzol reagent (Ambion-1561), and DNA was digested using DNase. RNA integrity was assessed using the mirVana miRNA Isolation Kit (Ambion) of the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Briefly speaking, a sequencing library was generated using the TruSeq chain mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. Oligo (dT) magnetic beads are used to enrich eukaryotic mRNAs. Interrupting reagents are added to break the mRNA into short fragments. The interrupted mRNA is used as a template to synthesize one-strand cDNA using six-base random primers. A two-stranded synthesis reaction system is then used to synthesize the double-stranded cDNA, after which PCR is performed to enrich the cDNA template. Purify the PCR product (AMPure XP system). Using TruSeq chain type LT Sample Prep Kit and cBot cluster generation system to cluster exponentially encoded samples (Illumina). The sequencing library was then sequenced (Illumina HiSeq X 10) on an Illumina sequencing platform by collecting 150 bp of double-ended data. The entire experiment was conducted at Shanghai Quanmai Biotechnology Co., Ltd. (Shanghai, China). Raw reads are sequences in fastq format. The NGS QC Toolkit software [22] was used for quality control and removal of the linker. Based on this, low-quality base and N base reads were filtered out, resulting in high-quality clean reads. Clean reads were then aligned to the reference genome and reference genes of this species using Bowtie2 [23] or TopHat [24]] (http://tophat.cbcb.umd.edu/). The status of the sample was assessed by the genome and the rate of gene alignment. The result of the comparison is stored as a binary file, namely, a bam file. Using HTSeq [25] software to obtain the number of reads aligned with the gene in each sample. Cufflinks [26] software was used to calculate the gene expression quantity FPKM (Fragments Per kb Per Million Reads) [27], which is the number of fragments from each kilobase of a gene per million fragments. Then the clustering method is used to calculate the distance between samples, examine the similarity between the samples, and calculate the direct correlation of the samples. Generally speaking, the same type of sample can appear in the same cluster through clustering. Genes in the same cluster may have similar biological functions.
2.9. Functional and Pathway Enrichment Analysis of DEGs
Differential expression analysis was performed to identify differentially expressed genes among different control groups, YDCS model group, and RA treatment group (using the estimateSizeFactors function of DESeq (2012) R package [28] to normalize the data and using the nbinomTest function to compute the P-value and fold change values for the difference comparison; P-value<0.05 and fold change>2 or fold change<0.5 are set as the threshold of significant differential expression). Using the negative binomial distribution test to calculate the gene differential expression level DESeq software [29]. (http://bioconductor.org/packages/release/bioc/html/DESeq.html). Using NB (negative binomial distribution test) to test the significance of reads. The base mean value was used to estimate the expression level of the gene expression. GO function significance and KEGG pathway significance analysis were used to determine the major biological function or pathway affected by differential genes. Subsequently, alternative splicing analysis and SNP analysis (Single Nucleotide Polymorphisms) were performed on differential genes. By alternative splicing of one gene, multiple proteins may be produced. This can greatly increase protein diversity. Based on the sequencing data, we used ASprofile software [30] to detect the presence of alternative splicing in the sample. Further comparisons of the differential genes were performed by comparing the assembled gene with the gene annotation information of the reference sequence. This may reveal that the assembled gene will extend the 5′ or 3′ end of the gene annotation, thereby optimizing the gene structure. The new annotation information is compared with known annotation information to obtain new transcript information. With the above comparison results, new transcript predictions can be performed simultaneously. Assembly results must satisfy the following conditions to be considered as new transcripts: 200 bp or more from the existing annotation gene and no less than 180 bp in length. Based on the TopHat-Cufflinks analysis platform, reads were assembled using Cufflinks software and new transcripts were assembled and compared with the gene annotation information of the reference sequence by Cuffcompare software. Read data from raw-sequence RNA-seq are unloaded to ftp://ftp.ensembl.org/pub/release-84/fasta/rattus_norvegicus/cdna/Rattus_norvegicus.Rnor_6.0.cdna.all.fa.gz.
2.10. Quantitative Real-Time PCR Analysis
Quantification was performed with a two-step reaction process: reverse transcription and PCR. Each RT reaction has two steps.
The first step is 0.5 μg RNA, 2 μl of 4×gDNA wiper Mix, adding Nuclease-free H2O to 8 μl. Reactions were performed in a GeneAmp® PCR System 9700 (Applied Biosystems, USA) for 2 min at 42°C. The second step is add 2μl of 5 × HiScript II Q RT SuperMix IIa. Reactions were performed in a GeneAmp® PCR System 9700 (Applied Biosystems, USA) for 10 min at 25°C; 30 min at 50°C; 5 min at 85°C. The 10 μl RT reaction mix was then diluted × 10 in nuclease-free water and held at -20°C. Real-time PCR was performed using LightCycler® 480 II Real-time PCR Instrument (Roche, Swiss) with 10 μl PCR reaction mixture that included 1 μl of cDNA, 5 μl of 2× QuantiFast® SYBR® Green PCR Master Mix (Qiagen, Germany), 0.2 μl of forward primer, 0.2 μl of reverse primer, and 3.6 μl of nuclease-free water. Reactions were incubated in a 384-well optical plate (Roche, Swiss) at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s. Each sample was run in triplicate for analysis. At the end of the PCR cycles, melting curve analysis was performed to validate the specific generation of the expected PCR product. The primer sequences were designed in the laboratory and synthesized by Generay Biotech (Generay, PRC) based on the mRNA sequences obtained from the NCBI database. The expression levels of mRNAs were normalized to input the reference gene, e.g., GAPDH, ACTB, and were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
3. Results
3.1. Model Evaluation
Based on preliminary experiments and literature studies, the standards for successful replication of the YDCS model are compared with the normal control group; the model rats mainly changed in the macroscopic symptoms, metabolic indicators, and temperature indicators.
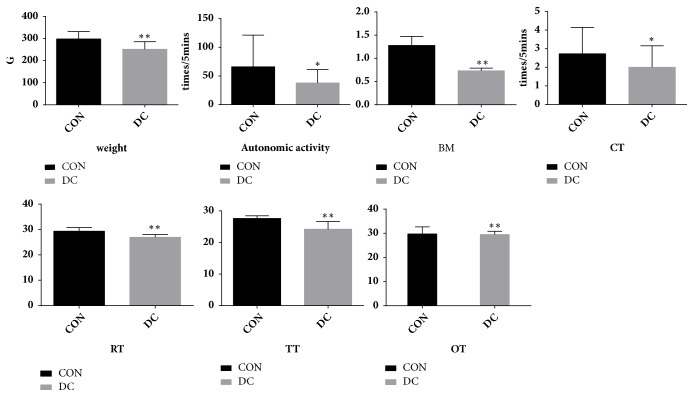
After the model was established, compared with the normal group, the YDCS model rats showed macroscopic symptoms: the stool was loose, the hair was dirty and messy, the amount of water intake was high, the spirit was wilting, and there were symptoms of deficiency cold syndrome. On the metabolic indicators, the body weight and basal (respiration) metabolism (RER) was significantly lower (P<0.01). The autonomic activity and CT were significantly lower (P<0.05). On the temperature index, RT, TT, and OT were significantly decreased (P<0.01) ( Figure 1).
Figure 1.
After the establishment of the YDCS model, compared with the blank control group, the changes shown in weight, autonomic activity, BM, CT, RT, TT, and OT in the model rats further explain the state and extent of YDCS. ∗P < 0.05, ∗∗P < 0.01 versus control group.
3.2. Test Results on Metabolism-Related Macroscopic Indicators
In this study, macroscopic observations were made on the energy metabolism of YDCS model and RAWA treatment group rats in terms of symptoms, weights, automatic monitoring of basal metabolic changes in noninterfering environments, cold and heat tendency, temperature changes, etc.
3.3. Measurement of the Symptoms
The results are shown as follows: there were no differences between the initial status of each group. Compared with the normal control group, rats in the model group appeared to have an addiction of lying, a decrease in food and water intake, loose stools, pale and partial darkness on lips and paws, temperature drop, messy and dry hair, clear urine in large amount, etc. As shown in Supplementary Figure 1, compared with clinical patients, the YDCS stage was characterized by symptoms including clear urine in large amount, loose stools, pale face, pale tongue, white coating on the tongue, weak and delayed pulse, and preference to warmth and soft touching, etc. These symptoms are consistent with YDCS model rats. After 7 days of treatment with RA water abstract, the overall condition of the animals improved significantly. The total activity, food intake, and water intake increased. Weight of the YDCS model animals increased stools became moderate. Lips and nails turned white and pink. The overall, rectal, and toe temperature all rise. Hair became smoother and shiner. There was improvement in urine color, etc. (Supplementary Figure 2). The differences between normal control group, YDCS model group, and RAWA treatment group in hair and paw color of the rats indicate the improvement in the condition of YDCS model group rats.
3.4. Measurement of the Temperature
According to the test of OT, RT, and TT, there was no difference between the initial status of three groups. Compared with the normal control group, the OT of the YDCS model rats was decreased (P<0.05). The average TT and the RT were significantly lower (P<0.01) as shown in Figure 2(a). However, in the rats treated with RAWA for 7 days, the OT and TT increased (P>0.05) and the RT increased significantly (P<0.05) as shown in Figure 2(b).
Figure 2.
(a) The infrared imaging images of the rats in the YDCS model group and measurement of the temperature (OT, TT, RT). Data are represented as mean ±SD from three experiments. ∗P < 0.05, ∗∗P < 0.01 versus control group. #P < 0.05, ##P < 0.01 versus model group. (b) The infrared imaging images of the rats in the AWA treatment group and measurement of the temperature (OT, TT, RT). Data are represented as mean ±SD from three experiments. ∗P < 0.05, ∗∗P < 0.01 versus control group. #P < 0.05, ##P < 0.01 versus model group.).
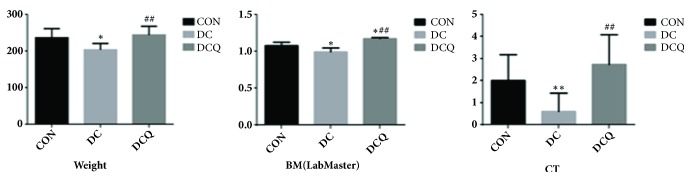
3.5. Measurement of the Body Weight, Cold and Heat Tendency, and Basal Metabolism
Effects on body weight, cold and heat tendency, and basal metabolism of rats were as follows: There was no difference in performance between the initial status of different groups. Compared with the normal control group, the body weight was significantly lower (P<0.05), the tendency of cold and heat was significantly colder (P<0.01), and the basal metabolism also dropped significantly (P<0.05) in the YDCS model group, as shown in Figure 3.
Figure 3.
Measurement of the body weight, cold and heat tendency, and basal metabolism in YDCS model group. The results present averages of three independent experiments (mean ± SD). ∗P < 0.05, ∗∗P < 0.01 versus control group. #P < 0.05, ##P < 0.01 versus model group.
After seven days of treatment with RAWA, the body weight and basal metabolism of the rats were significantly higher than those of YDCS model group (tending towards neutrality and the cold-tolerant capacity increased), which was statistically significant (P<0.01). This shows that RA can treat YDCS efficiently and improve the reduced metabolism, which may be the intrinsic mechanism of its positive role in compensating Qi and Yang, as shown in Figure 3.
3.6. A Systematic Study of Molecular Mechanisms of YDCS and RAWA Effect Focusing on Energy Metabolism Using RNA-Seq Transcriptome Sequencing GO Functional and KEGG Enrichment Analysis of DEGs
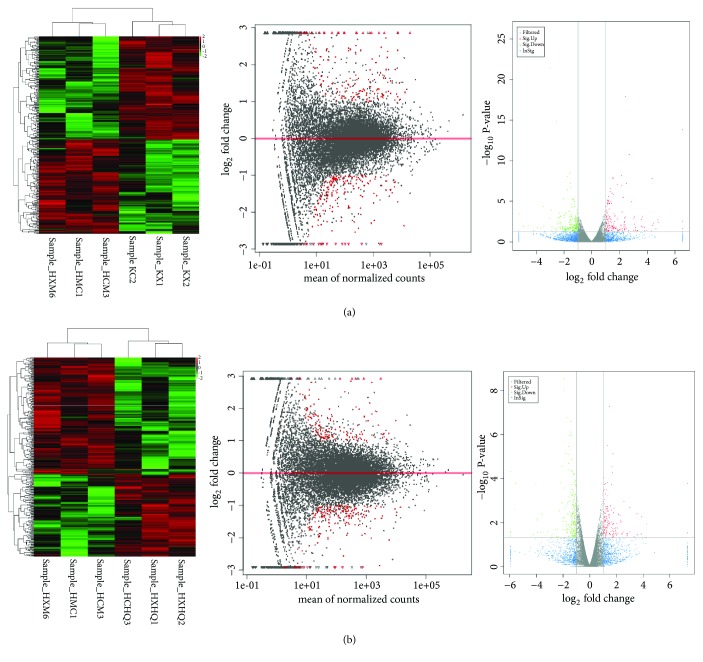
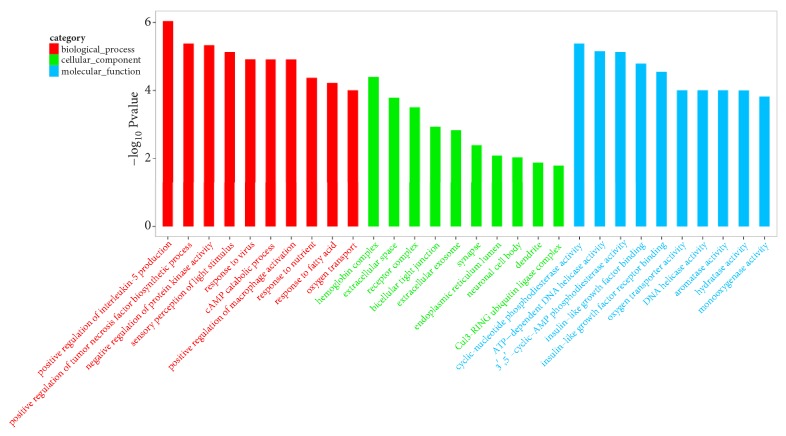
In order to investigate the mechanism of the therapeutic effect of RAWA on YDCS, we used the second-generation sequencing to detect the whole-genomic level transcriptome of the rat liver DNA. The goal is to study the changes of gene expression level in metabolic-related genes. We chose Illumina HiSeq X Ten [31, 32] platform for our sequencing experiment. The results showed that the expression of metabolism-related genes in rats in YDCS model group presented a “reverse regulation” tendency. On the other hand, treatment with RAWA could reverse the trend of reduction in the expression of most metabolism-related genes (Figure 4(a)). More specifically, 38 typical metabolism-related genes were significantly downregulated in YDCS model group. After treatment with RAWA, 49 metabolism-related genes were upregulated (Figure 4(b)). GO analysis was performed to analyze the biological processes that influence the major differences in their genes, such as RNA metabolism, tissue development, and regulation of GTPase activity (Figure 5). Interestingly, RAWA downregulated genes associated with cell death; apoptotic, ERK1, and ERK2 cascades were downregulated. Meanwhile, RAWA can upregulate ATP binding, DNA repair, glucose metabolism, and cholesterol and lipid metabolism, which is contrary to the trend of changes in the YDCS model group (Figure 6). The GO functional annotations are mainly focused on several categories including glucose metabolism, lipid metabolism, inflammation, and amino acid metabolism. We found that after RAWA treatment, the expression of most metabolic genes increased, such as genes related to coenzyme metabolism, glucose metabolism, lipid metabolism and GTPase activity, and DNA replication initiation. On the contrary, the expression of these energy metabolism processes was downregulated in the YDCS model group. KEGG enrichment focusing on glucose, lipid, and amino acid metabolism showed that YDCS group and RAWA group were both enriched in PPAR signaling pathway.
Figure 4.
(a) Hierarchical clustering of the quantitative information from significantly changed gene expression by RNA-seq. Compared with the normal control group, the rat metabolism was related to YDCS; red indicates the high expression gene, and green indicates the low expression gene. (b) Hierarchical clustering of the quantitative information from significantly changed gene expression by RNA-seq, compared with YDCS group. The rat metabolism was related to the RAWA group; red indicates the high expression gene, and green indicates the low expression gene. The difference that generated is reflected in the MA diagram. The X-axis is the mean value of all samples used for comparison after standardization, and the Y-axis is log2 fold change. Red marked the difference genes in significance (according to differential screening conditions). The differences generated by comparison were reflected in the volcanic map, the gray and blue genes were nonsignificant differences, and the red and green genes were the significant difference genes. The horizontal axis is the display of log2 fold change, and the vertical axis is the display of log10 P-value.).
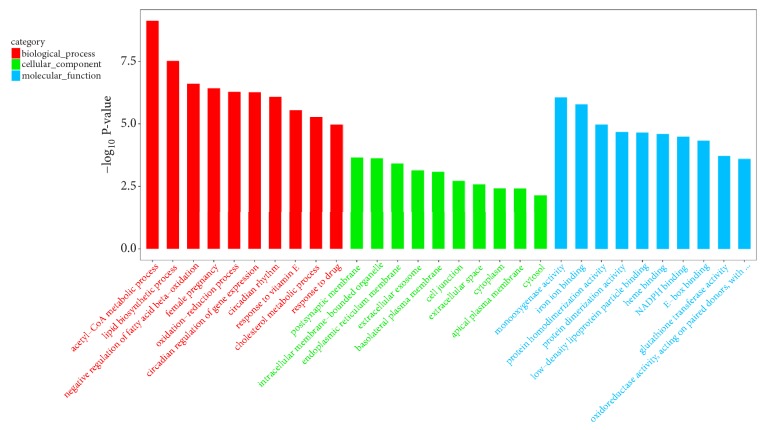
Figure 5.
Compared with the normal control group, GO analysis result of YDCS model group.
Figure 6.
Compared with the YDCS model group, GO analysis result of RAWA treatment group.
Therefore, these data indicate that the use of RAWA to treat YDCS may play a role in regulating energy metabolism by downregulating apoptosis and upregulating glucose, lipid, and amino acid metabolism-related genes.
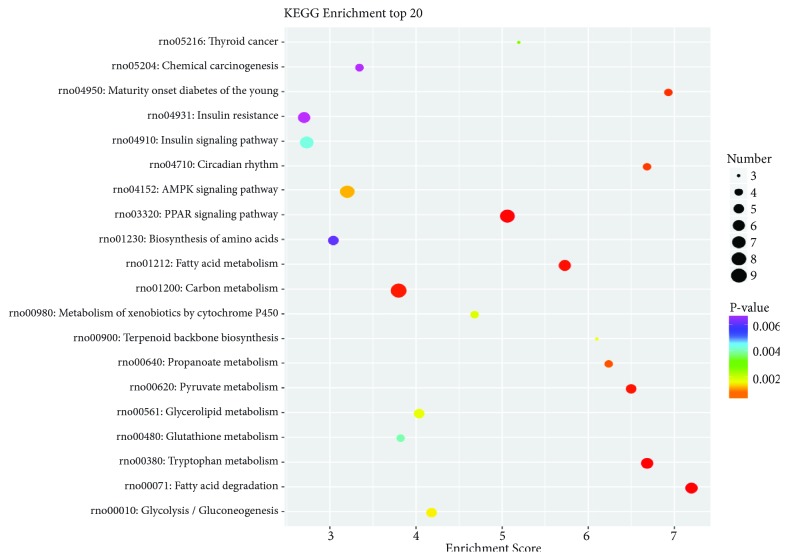
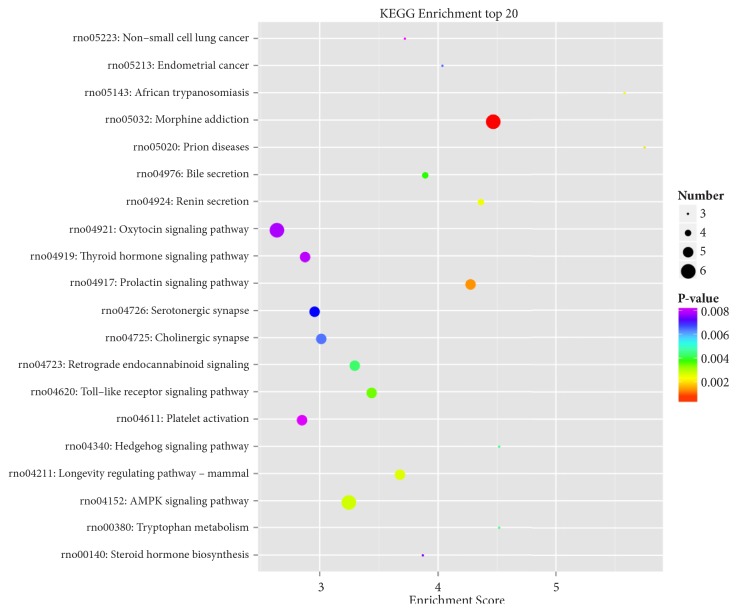
3.7. KEGG Pathway Enrichment Analysis of DEGs
Figures 7 and 8 showed significantly enriched pathways of DEGs through KEGG enriched analysis. DEGs of the model group were enriched in fatty acid degradation, PPAR signaling pathway, and AMPK signaling pathway (Table 1). In the RAWA treatment group, Wnt, cAMP, AMPK, and PPAR signaling pathway were enriched (Table 2), all related to energy metabolism.
Figure 7.
Compared with the normal control group, the rat metabolism was related to the YDCS in KEGG pathway.
Figure 8.
Compared with YDCS group, the rat metabolism was related to the RAWA in KEGG pathway. KEGG concentration analysis top 20 (the pathway entry with the corresponding number of corresponding genes greater than 2, according to the corresponding -log10 P-value of each entry). KEGG is the pathway of the main public database; using KEGG pathway analysis was carried out on the differences in gene database (in combination with the results KEGG comments), using hypergeometric inspection method to calculate gene enrichment of significant difference in each entry pathway. The result of the calculation will return a significant P-value with a small P-value indicating that the difference gene is enriched in the pathway. The X-axis in the figure represents the enrichment score, and the larger the bubbles, the more different the genes included in the entry. The color of the bubbles is changed by red-blue-green and yellow-yellow, which is rich in P-value.
Table 1.
KEGG pathway enrichment analysis DEGs of the model group related to energy metabolism.
| Id | Term | P-val | Enrichment score | Gene Symbol |
|---|---|---|---|---|
| path:rno00071 | Fatty acid degradation | 1.53E-05 | 7.194942 | LOC100911186;Acat2;Eci1;Cpt1b; |
| Cyp4a1;Aldh1b1 | ||||
| path:rno03320 | PPAR signaling pathway | 2.47E-05 | 5.055905 | Plin5;Cyp8b1;Gk;Fabp5;Cpt1b;Scd1; |
| Cyp4a1;Dbi | ||||
| path:rno00620 | Pyruvate metabolism | 9.89E-05 | 6.495434 | Acat2;Pklr;Acss2;Aldh1b1;Acacb |
| path:rno01212 | Fatty acid metabolism | 7.17E-05 | 5.726587 | LOC100911186;Acat2;Fasn;Fads1; |
| Cpt1b;Scd1 | ||||
| path:rno00061 | Fatty acid biosynthesis | 0.00234 | 7.194942 | Scd1;Acacb |
| path:rno00380 | Tryptophan metabolism | 2.55E-05 | 6.681018 | Kynu;LOC100911186;Acat2;Cyp1a1; |
| Ddc;Aldh1b1 | ||||
| path:rno04152 | AMPK signaling pathway | 0.000848 | 3.197752 | Srebf1;Creb3l1;Fasn;Ccnd1;Cpt1b; |
| Scd1;Hmgcr;Acacb | ||||
| path:rno00010 | Glycolysis / Gluconeogenesis | 0.001161 | 4.175636 | Gck;Pklr;Acss2;Aldoc;Aldh1b1 |
| path:rno00561 | Glycerolipid metabolism | 0.001397 | 4.031649 | Gpat3;Gk;Tkfc;Pnpla3;Aldh1b1 |
| path:rno00980 | Metabolism of xenobiotics by cytochrome P450 | 0.001503 | 4.676712 | LOC102550391;Cyp1a1;Gstt1;Gsta3 |
| path:rno00480 | Glutathione metabolism | 0.003734 | 3.817724 | LOC102550391;Gpx2;Gstt1;Gsta3 |
| path:rno01230 | Biosynthesis of amino acids | 0.005878 | 3.036826 | Tkt;Pklr;Pycr1;Aldoc;Gpt |
| path:rno04922 | Glucagon signaling pathway | 0.007072 | 2.922945 | Gck;Slc2a2;Creb3l1;Cpt1b;Acacb |
| path:rno00100 | Steroid biosynthesis | 0.007219 | 4.922855 | Fdft1;Tm7sf2 |
| path:rno00220 | Arginine biosynthesis | 0.007219 | 4.922855 | Gpt;Nos1 |
| path:rno00340 | Histidine metabolism | 0.010949 | 4.251557 | Ddc;Aldh1b1 |
| path:rno01040 | Biosynthesis of unsaturated fatty acids | 0.01395 | 3.89726 | Fads1;Scd1 |
| path:rno00330 | Arginine and proline metabolism | 0.016333 | 3.05003 | Pycr1;Aldh1b1;Nos1 |
| path:rno04146 | Peroxisome | 0.027864 | 2.338356 | Crot;Ephx2;Ech1;Pex11a |
Table 2.
KEGG pathway enrichment analysis DEGs of the AWA treatment group related to energy metabolism and immunity.
| Id | Term | P-val | Enrichment score | Gene Symbol |
|---|---|---|---|---|
| path:rno04152 | AMPK signaling pathway | 0.002426 | 3.242165 | Foxo3;Ccnd1;Cpt1b;Scd1;Adipor2;Pik3r1 |
| path:rno00380 | Tryptophan metabolism | 0.004174 | 4.515873 | Inmt; Cyp1a1; Ddc |
| path:rno00140 | Steroid hormone biosynthesis | 0.007262 | 3.870748 | RGD1559459;Cyp1a1; Cyp2d4 |
| path:rno04310 | Wnt signaling pathway | 0.021086 | 2.30738 | Prickle2;Prkacb; Ccnd1; Nfatc4; Lrp6 |
| path:rno04024 | cAMP signaling pathway | 0.024854 | 2.084249 | Adora2a; Prkacb; Pde4d; Adcyap1r1; Pde4c; Pik3r1 |
| path:rno00980 | Metabolism of xenobiotics by cytochrome P450 | 0.02489 | 3.161111 | RGD1559459; Cyp1a1 |
| path:rno00982 | Drug metabolism - cytochrome P450 | 0.026553 | 3.084011 | RGD1559459;Fmo2 |
| path:rno03320 | PPAR signaling pathway | 0.029248 | 2.563063 | Fabp4; Cpt1b; Scd1 |
| path:rno04922 | Glucagon signaling pathway | 0.037441 | 2.370833 | Prkacb; Gnaq; Cpt1b |
| path:rno01212 | Fatty acid metabolism | 0.041939 | 2.580499 | Cpt1b;Scd1 |
| path:rno00340 | Histidine metabolism | 0.046618 | 2.873737 | Ddc |
| path:rno00360 | Phenylalanine metabolism | 0.035579 | 3.327485 | Ddc |
| path:rno04620 | Toll-like receptor signaling pathway | 0.00323 | 3.43599 | Mapk12; Cxcl10; Lbp; Pik3r1 |
| path:rno04115 | p53 signaling pathway | 0.017262 | 3.010582 | Sesn3; Igfbp3; Ccnd1 |
| path:rno04750 | Inflammatory mediator regulation of TRP channels | 0.019943 | 2.554433 | Prkacb; Mapk12; Gnaq; Pik3r1 |
| path:rno04630 | Jak-STAT signaling pathway | 0.023181 | 2.257937 | Cish; Ccnd1; Il13ra1; Stam2; Pik3r1 |
| path:rno04670 | Leukocyte transendothelial migration | 0.030841 | 2.278278 | Cd99; Mapk12;Cldn15; Pik3r1 |
| path:rno04068 | FoxO signaling pathway | 0.041079 | 2.107407 | Mapk12; Foxo3; Ccnd1; Pik3r1 |
Others, such as Toll-like receptor, p53 and Jak-STAT signaling pathway, Bile secretion, and calcium signaling pathway, were related to immunity.
Taken together, this KEGG pathway enrichment indicates that the pathology of YDCS and the effect of RAWA both focus on energy metabolism and immune responses.
3.8. Expression of Metabolism-Related Genes in YDCS Group and RAWA Treatment Group
Compared with normal model group, 96 genes showed most significant differences in expression levels among genes related to metabolism. Compared with YDCS model group, 59 genes had most significant differences among genes related to metabolism (Figure 4), 28 of which were biological processes, such as fatty acid metabolism, nutrient and glucose transport and catabolism, cAMP decomposition, protein kinase activity, regulation of glycoproteins, regulation of lipid and triglyceride biosynthesis. 31 of the 59 genes are responsible for molecular functions, including lipid metabolism, regulatory responses to GTP, AMP, and ADP, cyclic nucleotide phosphodiesterase activity, ATP-dependent DNA helicase activity, monooxygenase and hydrolase activity (Figure 7).
3.9. GO Analysis of Significant Differential Energy Metabolism-Related Genes in YDCS Group and AWA Treatment Group
According to the GO classification criteria, the YDCS group possessed energy metabolism gene functions compared with normal control group. The AWA treatment group possessed energy metabolism (concentration) gene functions compared with YDCS model group. Among those genes, 20 genes were related to glucose metabolism, 26 genes were related to lipid metabolism, and 35 genes were related to amino acid metabolism (Tables 3 and 4).
Table 3.
GO analysis of significant differential energy metabolism related genes in YDCS group and RAWA treatment group.
| The serial number | Gene name | GOid | Fold change | P-val | GO function |
|---|---|---|---|---|---|
| ENSRNOG00000003259 | C1qtnf1 | GO:0051897 | 0.414337 | 0.000326 | Sugar and protein metabolism |
| ENSRNOG00000010438 | Cpt1b | GO:0006635 | 0.435098 | 0.016545 | Lipid metabolism |
| ENSRNOG00000014532 | Lbp | GO:0001889 | 0.46867 | 0.000321 | Lipid metabolism |
| ENSRNOG00000003028 | Dnah17 | GO:0005524 | 0.417407 | 0.02197 | Energy metabolism |
| ENSRNOG00000020369 | Igf2 | GO:0045953 | 0.34108 | 0.035391 | Sugar acid metabolism |
| ENSRNOG00000030154 | Cyp4a2 | GO:0005504 | 0.084145 | 0.035647 | Lipid metabolism |
| ENSRNOG00000010438 | Cpt1b | GO:0015909 | 0.435098 | 0.002426 | Lipid metabolism |
| ENSRNOG00000031233 | Mapk12 | GO:000552 | 0.443239 | 40.009524 | Amino acid metabolism |
| ENSRNOG00000014524 | S1pr3 | GO:0005515 | 0.392174 | 0.009524 | Amino acid metabolism |
| ENSRNOG00000001302 | Adora2a | GO:003081 | 0.482049 | 0.013993 | Sugar metabolism |
| ENSRNOG00000020585 | Tbxa2r | GO:000718 | 0.46712 | 0.013993 | Amino acid metabolism |
| ENSRNOG00000003033 | Plcd3 | GO:000487 | 0.401252 | 0.013993 | Lipid metabolism |
| ENSRNOG00000036828 | Pde1b | GO:0043025 | 0.123816 | 0.013993 | Amino acid metabolism |
| ENSRNOG00000012098 | Adcyap1r1 | GO:0030819 | 0.462201 | 0.041939 | Amino acid metabolism |
| ENSRNOG00000019518 | Pde4c | GO:0006198 | 0.305552 | 0.041939 | Amino acid metabolism |
| ENSRNOG00000010805 | Ap2 | GO:0008289 | 3.398621 | 0.040759 | Lipid metabolism |
| ENSRNOG00000010438 | CPT-1 | GO:0015909 | 2.298333 | 0.016545 | Lipid metabolism |
| ENSRNOG00000023546 | Hspb1 | GO:2001028 | 8.084827 | 0.00208 | Heat shock protein |
| ENSRNOG00000014882 | Fgf11 | GO:0008083 | 2.089544 | 0.025097 | growth factor activity |
Table 4.
The genes of cytochrome P450 family were significantly downregulated, suggesting that the YDCS model can reduce lipid metabolism and biotransformation function in rats. On the contrary, RAWA can increase lipid metabolism and biotransformation function.
| The serial number | Gene name | Fold change | P-val | GOid | GO function |
|---|---|---|---|---|---|
| ENSRNOG00000055078 | Cyp4b1 | 0.42533 | 0.03924 | GO:0055114 | Energy metabolism |
| ENSRNOG00000019500 | Cyp1a1 | 0.253766 | 0.0269 | GO:0032496 | Lipid metabolism |
| ENSRNOG00000002134 | Cyp4a2 | 2.497622 | 0.00689 | GO:0003924 | Amino acid metabolism |
| ENSRNOG00000032261 | Cyp2d4 | 0.463307 | 0.00951 | GO:0019369 | Lipid metabolism |
| ENSRNOG00000003510 | Fmo2 | 0.285916 | 0.02557 | GO:0006739 | NADP metabolism |
| ENSRNOG00000029478 | Cyp4f39 | 2.243402 | 0.0016 | GO:0016705 | Energy metabolism |
| ENSRNOG00000042714 | RGD1559459 | 2.439187 | 0.03076 | GO:0052696 | NADP metabolism |
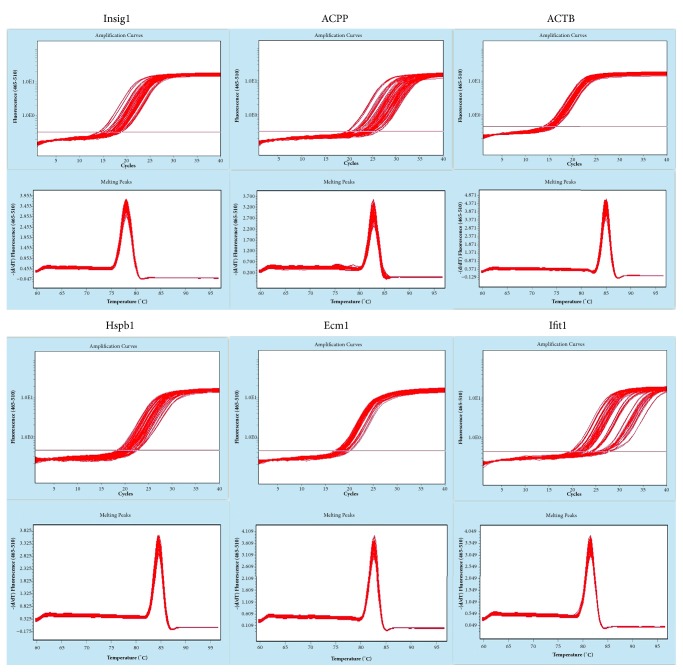
3.10. Results of Quantitative Real-Time PCR
To confirm RNA-seq results, qRT-PCR was performed on Hspb1, Ecm1, Ifit1, Insig1, and Acpp being 0.1884, 0.8315, 2.3439, 5.0865, 0.1174, respectively, in the model group. And in RAWA group, the 2−ΔΔCt values were 6.4745, 2.2208, 0.0521, 0.3910, and 0.3234, respectively (Table 5). The values, being consistent with the study of transcriptome sequencing technique, indicated the reliability of the results (Figure 9).
Table 5.
The primers of selected genes for qRT-PCR
| GenBank accession number | Gene symbol | Forward primer (5 -3 ) | Reverse primer (5 -3 ) |
|---|---|---|---|
| ENSRNOG00000023546 | Hspb1 | GGAGATCACCATTCCGGTC | GCAAGCTGAAGGCTTCTAC |
| ENSRNOG00000021166 | Ecm1 | TCTTGACCGTTGACCTTAGC | GCCAGGGATCTGTTTATGC |
| ENSRNOG00000019050 | Ifit1 | CCTTTGCCTGGAGGAAACTA | CACAAGCCGGACATTCTG |
| ENSRNOG00000006859 | Insig1 | GTCTCATCAGTGTGGGAACTA | CTGGGCTGTCAGTAAGATTG |
| ENSRNOG00000011820 | Acpp | CAAGCCAAGGAGTTGAAGT | GGCCACGAGGATTCCTTA |
| NM_031144 | ACTB | CCACCATGTACCCAGGCATT | CGGACTCATCGTACTCCTGC |
Figure 9.
4. Discussion
The common feature of YDCS is the change of energy metabolism and weakened immunity, resulting in significant loss of body weight, drop in food/water intake and temperature, and reduced cold resistance. The pathogenic causes of YDCS are in accordance with the record of the ancient Chinese medicine classic Nei jing, which said that too much cold herb intake is harmful to Yang. The basic process of YDCS development is as follows: take too much cold herb, hurt the body righteousness, and result in damage of lung, spleen, kidney, liver, and other organs, poor circulation of blood, fatigue, chills, laziness, long-time lying, lack of movement, reduced water intake, loose stools, and other symptoms [2]. It can be divided into two major categories: Yang-deficiency (YD) constitution and the YDCS stage due to the development of the disease. In YD constitution, due to congenital deficiency, deficiency of kidney Yang, impaired function of lung, spleen, liver, and kidney, and other causes of deficiency of Yang, the body develops glucose-6-phosphate dehydrogenase [5], G protein (by controlling ATP, ADP, AMP, and GTP), and T cell immune functions, etc. reduction. According to this theory, in this experiment, we used a traditional cold compound formula of Chinese medicine to replicate the model of YDCS under the guidance of the comprehensive diagnosis and treatment of “Nature-Humanity Unity” proposed by Nei jing. Based on the difference in infrared thermal sensitivity of various parts of the human body, we utilized infrared imaging to detect the temperature changes in the YDCS model group and the RAWA treatment group. The principle of detection agrees with the traditional Chinese medicine diagnosis principle “inference of the inside comes from the outside”. As a result, we found that the YDCS model showed a reduction in energy metabolism represented by a drop of temperature, cold-tolerance, and basal metabolism on a macroscopic scale. After treatment with RAWA, the reduced energy metabolism increased significantly. Because these changes are closely related to the change of metabolic function in the human body, the transcriptome sequencing was used to sequence the whole genome of the liver, which is the main organ of metabolism. The sequencing results became more specific after the “noise” is excluded.
The emergence of YDCS because of the body's energy metabolism dysregulation was represented by glucose metabolism, lipid metabolism, and amino acid metabolism. Reduction by different degrees occurred in these metabolic functions causing further pathological changes in the body's endocrine and immunity. YDCS is a chronic pathological process, and it is closely related to PPAR signaling pathway and downregulation of the target genes CPT-1, FABP4 (Supplementary Figure 3) [33]. Therefore, as a typical mild Chinese medicine with few side effects, RAWA is suitable for long-term use for prevention and treatment of YDCS by focusing on various pathogenic and influencing factors in the early stage.
RA belongs to a kind of classical Chinese medicine with mild properties and warming effects [29]. Modern pharmacology shows that RA contains RA polysaccharides, astragaloside, flavonoids, alkaloids, vitamins, and other active ingredients [34]. RA membranaceus is widely used in clinical treatment of digestive system, respiratory system, cardiovascular diseases, endocrine diseases, and metabolic diseases. It has an enhancing effect on regulating energy metabolism and immune and metabolic functions [35]. RA also showed advantages including significant treatment effect, no toxic side effects, and less adverse reactions [36–40]. Previous tests showed that making RA into water abstract form can better unleash its function. This study found that RA can significantly increase the body weight, temperature, basal metabolism, and cold resistance of YDCS model rats. Through pharmacodynamic studies, it has been found that RA has a beneficial effect on the generation, storage, and use of energy in the body. Combined with the transcriptome results, the effect of RA membranaceus on energy metabolism may be due to the action on PPAR signaling pathway, resulting in the upregulation of the target gene CPT-1 and FABP4 (Supplementary Figure 4). RA can also enhance production of heat and promote the reduced energy metabolism in the YDCS model rats. These benefits are the specific manifestation of its warming effect [32, 33].
In the transcriptome sequencing, the major differential genes and pathway enrichment have shown that energy metabolism represented by carbohydrates, lipids, and amino acid metabolism is the core of the analysis of YDCS pathogenesis and RAWA treatment mechanism. RAWA upregulates lipid metabolism represented by ap2 (Fabp4) under the action of cytochrome P450 family member Cyp1a1 in vivo. Activation of PPAR signaling pathway (path: rno03320) and MAPK signaling pathway (path: rno04010) through activation of FABP resulted in the activation of PPARα and PPARγ pathways and significant downregulation of Scd1. This leads to increased lipid metabolism and upregulation of CPT-1 and ap2 (FABP4), which further affect AMPK signaling pathway (path: rno04152) in lipolysis. Also, the oxidation of fatty acids enhances lipid metabolism. All these effects will further influence the enhancement of the immune and endocrine system.
Among the molecules mentioned above, SCD-1 (stearoyl-CoA desaturase) is a key enzyme in lipid metabolism. It can catalyze the formation of monounsaturated fatty acids from saturated fatty acids, activate adenylate-activated protein kinases, and affect the synthesis of cholesterol and triglycerides, increasing fatty acid oxidation mostly in the liver, heart, and skeletal muscles [41]. It can affect the heat production of skeletal muscles as well [42, 43]. Furthermore, inflammation and cell stress response are regulated by the activation of MAPK signaling pathway and NF-κB signaling pathway [44, 45]. The decrease of SCD-1 after the treatment with RA water decoction can reduce fat accumulation, increase heat production and lipid metabolism [46], accelerate fatty acid decomposition, and promote energy metabolism of the body.
CPT1 is a key enzyme in the fatty acid oxidation process. It is located in the mitochondrial outer membrane and is the rate-limiting enzyme in the fatty acid β oxidation process [47]. CPT1 can be divided into three subtypes, increase FFA (nonesterified fatty acid), promote lipolysis.
Among which CPT1A has the strongest activity in the liver [48]. Therefore, CPT1A can regulate the oxidation of fatty acids in the body to a greater extent and increase the rate of fatty acid oxidation, which plays an important role in the changes of energy metabolism in the body.
Ap2 (Fabp4) belongs to the FABP family, which are a family of fatty acid-binding proteins that bind to hydrophobic lipids and carry out intracellular transportation. It binds reversibly to saturated and unsaturated long-chain fatty acids. Fabp4 executes its function in regulating the metabolism and transport of fatty acids through the promoter-binding elements PPRE1 and PPRE2 presented in PPARγ pathway, leading to the important role in the body's energy metabolism [47, 48]. More literature has shown that Fabp4 has an essential effect on the metabolic syndrome. The increased Fabp4 expression can induce the upregulation of acetyl-CoA cholesterol acyltransferase 1 (ACAT1) gene expression [49], but it can also regulate the gene expression of ATP binding cassette transporter A1 (ABCA1) and hormone-sensitive lipolytic enzyme (HSL) in the opposite direction, resulting in decreased degradation of triglycerides and cholesterol. It is worth noting that literature studies have pointed out that overexpression of FABP4 can cause increased metabolism and inflammatory enhancement. The latter can lead to atherosclerosis and other related diseases. However, our experiment is comparing with the metabolically deficient model rats of YDCS. The focus is on the recovery to normal level, so it does not involve the overexpression of FABP4.
Based on symptoms and the macroscopic/microscopic observations of temperature and metabolism, combined with modern research, we discussed the effects of RA and YDCS on energy metabolism. From the genetic level, changes in key pathway PPARs are the main entry point. PPAR signaling pathway is the key metabolic pathway and CPT-1, FABP4 are the key target genes that regulate lipid metabolism. CPT-1, FABP4 are also key targets for the treatment of metabolic and immune diseases. According to transcriptome sequencing results, in the YDCS model group, PPAR pathway was activated, in which CPT-1 and FABP4 were significantly reduced, while after RAWA treatment, CPT-1 and FABP4 in the PPAR pathway were significantly upregulated, indicating that RAWA acts on these targets. RAWA showed a clear adjusting role in metabolic system. This role may take effect through the role of PPAR signaling pathway and the target genes CPT-1 and FABP4 by improving metabolism reduced by YDCS. In addition, FABP4 upregulation can enhance basal metabolism and improve the metabolic dysfunction caused by reduced metabolism in YDCS. The purpose of enhancing immunity can be achieved by enhancing the expression of inflammatory factors such as TNF-α, IL-1, and IL-6. All the effects mentioned above will achieve the ultimate purpose of treatment of cold syndrome [50].
5. Conclusion
In summary, the mechanism of YDCS pathology is energy metabolism deficiency caused by PPAR signaling pathway and target genes CPT-1, FABP4. RAWA had well effects that could upregulate CPT-1, FABP4 via multiple pathways to promote energy metabolism of rats with the YDCS syndrome, especially through the PPAR signaling pathway. RAWA can also further upregulate immunity. These effects indicate the tonic and warm properties of RAWA. Further study is needed for the details of the regulative mechanism.
Acknowledgments
This research was supported by the National Basic Research Program of China (973 Program, 2013CB531803), Natural Science Foundation of China (81673852, 81703838, and 81603419), the Shandong Provincial Natural Science Foundation of China (ZR2016HM66), and Key Technological Innovation Project of Key Industries in Shandong Province (2016CYJS08A01-3 and 2016CYJS08A01-4).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
Wanchen Yu contributed to this research and should be considered as first author.
Conflicts of Interest
All the authors declare that they have no financial or commercial conflicts of interest.
Authors' Contributions
Wanchen Yu wrote the first draft of the manuscript. Shijun Wang designed the study. Wanchen Yu conducted the experiments, wrote the manuscript, performed the experiments, and analyzed the data. Wanchen Yu, Xin Zong, and Shijun Wang contributed to the writing of the manuscript. Wanchen Yu, Xin Zong, and Shijun Wang agree with manuscript results and conclusions. Wanchen Yu, Haijun Zhao, Xin Zong, and Shijun Wang jointly developed the structure and arguments for the paper. Wanchen Yu, Haijun Zhao, and Shijun Wang made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
Supplementary Materials
Supplementary Figure 1: Morphological observation of rats: Compared with the normal control group, rats in YDCS model group appeared to have an addiction of lying, messy and dry hair, squinting, listless, pale and partial darkness on lips and paws, clear urine in large amount, loose stools, etc. Supplementary Figure 2: Morphological observation of rats: After 7 days of the treatment with RA water abstract, the overall condition of the animals improved significantly. Hair became smoother and shiner; rat activity increased. Lips, stools forming became moderate; nails turned white and pink; urine became of normal color. Supplementary Figure 3: The change of PPAR signal pathway in YDCS model: The PPAR signal pathway is an important pathway for metabolism and immunity. In YDCS pathological process, rat glucose metabolism, lipid metabolism, and amino acid metabolism reduction were disordered. The body's energy metabolism dysregulation of rats reflects the abnormal PPAR signal pathway. It is closely related to PPAR signaling pathway and downregulation of the target genes CPT-1, FABP4. Supplementary Figure 4: The change of PPAR signaling pathway in RAWA treatment group: Combined with the transcriptome results, the effect of RA membranaceus on energy metabolism may be due to the action on PPAR signaling pathway, resulting in the upregulation of the target gene CPT-1 and FABP4. RA can also enhance production of heat and promote the reduced energy metabolism in the YDCS model rats. These benefits are the specific manifestation of its warming effect. RAWA upregulates lipid metabolism represented by ap2 (Fabp4) under the action of cytochrome P450 family member Cyp1a1 in vivo. Activation of PPAR signaling pathway (path: rno03320).
References
- 1.Yu R., Yang Y., Han Y., et al. Expression profiling of transcriptome and its associated disease risk in yang deficiency constitution of healthy subjects. Evidence-Based Complementary and Alternative Medicine. 2016;2016:9. doi: 10.1155/2016/1493098.1493098 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Huang X., Chen Q., Yang G., et al. Metabolic profiling study of yang deficiency syndrome in hepatocellular carcinoma by h1 NMR and pattern recognition. Evidence-Based Complementary and Alternative Medicine. 2012;2012:6. doi: 10.1155/2012/843048.843048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong Y., Liu L., He X., et al. The Th17/Treg immune balance in ulcerative colitis patients with two different Chinese syndromes: Dampness-heat in large intestine and spleen and kidney yang deficiency syndrome. Evidence-Based Complementary and Alternative Medicine. 2015;2015:10. doi: 10.1155/2015/264317.264317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J., Kim K. H. Protocol for the evaluation and validation of Qi Blood Yin Yang deficiency pattern questionnaire: prospective observational study. Integrative Medicine Research. 2015;4(4):249–255. doi: 10.1016/j.imr.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C.-H., Li T.-C., Tsai C.-I., et al. Yang deficiency body constitution acts as a predictor of diabetic retinopathy in patients with Type 2 Diabetes: Taichung diabetic body constitution study. Evidence-Based Complementary and Alternative Medicine. 2015;2015 doi: 10.1155/2015/940898.940898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H.-Y., Wang S.-J., Teng J.-L., et al. Effects of Radix aconiti lateralis preparata and Rhizoma zingiberis on energy metabolism and expression of the genes related to metabolism in rats. Chinese Journal of Integrative Medicine. 2012;18(1):23–29. doi: 10.1007/s11655-012-0964-7. [DOI] [PubMed] [Google Scholar]
- 7.Han B., Wang S., Li L., Wang Y., Zhao H. Gene expression profiling of rat livers with Yin-deficiency-heat syndrome. Journal of Traditional Chinese Medicine. 2013;33(3):378–383. doi: 10.1016/s0254-6272(13)60182-x. [DOI] [PubMed] [Google Scholar]
- 8.Monsalve F. A., Pyarasani R. D., Delgado-Lopez F., Moore-Carrasco R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediators of Inflammation. 2013;2013:8. doi: 10.1155/2013/549627.549627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S., Sun X., Rafikov R., et al. PPAR-γ regulates carnitine homeostasis and mitochondrial function in a lamb model of increased pulmonary blood flow. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0041555.e41555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feige J. N., Gelman L., Michalik L., Desvergne B., Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Progress in Lipid Research. 2006;45(2):120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Guan Y., Breyer M. D. Peroxisome proliferator-activated receptors (PPARs): Novel therapeutic targets in renal disease. Kidney International. 2001;60(1):14–30. doi: 10.1046/j.1523-1755.2001.00766.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C., Zhu C., Ling Y., et al. The clinical value of huangqi injection in the treatment of leucopenia: A meta-analysis of clinical controlled trials. PLoS ONE. 2013;8(12) doi: 10.1371/journal.pone.0083123.e83123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S., Keung W., Serra D., et al. Malonyl-CoA mediates leptin hypothalamic control of feeding independent of inhibition of CPT-1a. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2011;301(1):R209–R217. doi: 10.1152/ajpregu.00092.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreurs M., Kuipers F., van der Leij F. R. Regulatory enzymes of mitochondrial β-oxidation as targets for treatment of the metabolic syndrome. Obesity Reviews : an official journal of the International Association for the Study of Obesity. 2010;11(5):380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 15.Shi J., Zhu H., Arvidson D. N., Woldegiorgis G. The first 28 N-terminal amino acid residues of human heart muscle carnitine palmitoyltransferase I are essential for malonyl CoA sensitivity and high-affinity binding. Biochemistry. 2000;39(4):712–717. doi: 10.1021/bi9918700. [DOI] [PubMed] [Google Scholar]
- 16.Coe N. R., Simpson M. A., Bernlohr D. A. Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. Journal of Lipid Research. 1999;40(5):967–972. [PubMed] [Google Scholar]
- 17.Furuhashi M., Saitoh S., Shimamoto K., Miura T. Fatty acid-binding protein 4 (FABP4): Pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clinical Medicine Insights: Cardiology. 2014;8(suppl 3):23–33. doi: 10.4137/CMC.S17067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bing-bing H., Shi-jun W., Hua-yun Y., Hai-jun Z., Yuan W. Study of gene chip technology on whole genome expression of liver of rats with asthenia cold syndrome. Liaoning Journal of Traditional Chinese Medicine. 2011;38(12):2464–2466+2513. [Google Scholar]
- 19.Chenggang W., Bingbing H., Yuan W., Shijun W. Evaluation on animal model of deficiency cold syndrome and deficiency heat syndrome by using partial least squares-discriminant analysis. Liaoning Journal of Traditional Chinese Medicine. 2014;41(06):1275–1278. [Google Scholar]
- 20.Bing-bing H., Shi-jin W., Hai-jun Z. A study based on PLS the characterization and evaluation system of asthenia cold syndrome models in rats. Shaanxi Zhongyi. 2011;32(12):1676–1678. [Google Scholar]
- 21.Zhao W. X., Cui N., Jiang H., et al. Effects of radix astragali and its split components on gene expression profiles related to water metabolism in rats with the dampness stagnancy due to spleen deficiency syndrome. Evidence-Based Complementary and Alternative Medicine. 2017;2017:10. doi: 10.1155/2017/4946031.4946031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue S.-J., Liu J., Feng W.-W., et al. System pharmacology-based dissection of the synergistic mechanism of huangqi and huanglian for diabetes mellitus. Frontiers in Pharmacology. 2017;8 doi: 10.3389/fphar.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel R. K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030619.e30619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4, article R36) doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C., Roberts A., Goff L., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S., Pyl P. T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., Araki M., Goto S., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36(1):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang E.-W., Girke T., Jiang T. Differential gene expression analysis using coexpression and RNA-Seq data. Bioinformatics. 2013;29(17):2153–2161. doi: 10.1093/bioinformatics/btt363. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C., Wang G., Wang J., et al. Analysis of differential gene expression and novel transcript units of ovine muscle transcriptomes. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089817.e89817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu P., Zhao H., Luo Y. Anti-aging implications of Astragalus Membranaceus (Huangqi): A well-known Chinese tonic. Aging and Disease (A&D) 2017;8(6):868–886. doi: 10.14336/AD.2017.0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenk S., Saberi M., Olefsky J. M. Insulin sensitivity: Modulation by nutrients and inflammation. The Journal of Clinical Investigation. 2008;118(9):2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng H.-S., Wang J., Zhang H.-T., et al. Rapid identification of growth years and profiling of bioactive ingredients in Astragalus membranaceus var. mongholicus (Huangqi) roots from Hunyuan, Shanxi. Chinese Medicine. 2017;12(1):p. 14. doi: 10.1186/s13020-017-0135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao H., Banbury L. Different Proportions of Huangqi (Radix Astragali Mongolici) and Honghua (Flos Carthami) Injection on alpha-Glucosidase and alpha-Amylase Activities. Evidence-Based Complementary and Alternative Medicine. 2015;2015:7. doi: 10.1155/2015/785193.785193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C., Song H., Wang X., et al. Urinary metabolomics reveals the therapeutic effect of HuangQi Injections in cisplatin-induced nephrotoxic rats. Scientific Reports. 2017;7(1, article no. 3619) doi: 10.1038/s41598-017-03249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong-li X. Advances in clinical application of Astragalus membranceus and drug adverse reaction. Pharmaceutical Care and Research. 2002;(03):180–182. [Google Scholar]
- 37.Jin-hua L., Jing-ran C. Advances in clinical application of Astragalus membranceus and drug adverse reaction. Li Shi Zhen Medicine and Materia Medica Research. 2008;(09):94–95. [Google Scholar]
- 38.Yu S.-Y., OuYang H.-T., Yang J.-Y., et al. Subchronic toxicity studies of Radix Astragali extract in rats and dogs. Journal of Ethnopharmacology. 2007;110(2):352–355. doi: 10.1016/j.jep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W. D., Zhang C., Liu R.-H., et al. Preclinical pharmacokinetics and tissue distribution of a natural cardioprotective agent astragaloside IV in rats and dogs. Life Sciences. 2006;79(8):808–815. doi: 10.1016/j.lfs.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 40.Qi Z., Zhu L. L., Chen G. G., et al. Pharmacokinetics of astragaloside iv, in beagle dogs. European Journal of Drug Metabolism and Pharmacokinetics. 2007;32(2):75–79. doi: 10.1007/bf03190995. [DOI] [PubMed] [Google Scholar]
- 41.McGarry J. D., Brown N. F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. European Journal of Biochemistry. 1997;244(1):1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 42.Doh K.-O., Kim Y.-W., Park S.-Y., Lee S.-K., Park J. S., Kim J.-Y. Interrelation between long-chain fatty acid oxidation rate and carnitine palmitoyltransferase 1 activity with different isoforms in rat tissues. Life Sciences. 2005;77(4):435–443. doi: 10.1016/j.lfs.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 43.Alečković-Halilović M., Mešić E., Sinanović O., Zukić S., Mustedanagić J. Carnitine palmitoyl transferase deficiency-unrecognized cause of recurrent acute kidney injury. Renal Failure. 2013;35(5):732–734. doi: 10.3109/0886022X.2013.780979. [DOI] [PubMed] [Google Scholar]
- 44.Ruvolo P. P. Intracellular signal transduction pathways activated by ceramide and its metabolites. Pharmacological Research. 2003;47(5):383–392. doi: 10.1016/S1043-6618(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 45.Jiang M., Zhang L., Ma X., et al. Tamoxifen inhibits macrophage FABP4 expression through the combined effects of the GR and PPARγ pathways. Biochemical Journal. 2013;454(3):467–477. doi: 10.1042/BJ20130580. [DOI] [PubMed] [Google Scholar]
- 46.Tertov V. V., Orekhov A. N., Kacharava A. G., Sobenin I. A., Perova N. V., Smirnov V. N. Low density lipoprotein-containing circulating immune complexes and coronary atherosclerosis. Experimental and Molecular Pathology. 1990;52(3):300–308. doi: 10.1016/0014-4800(90)90071-k. [DOI] [PubMed] [Google Scholar]
- 47.Guo L., Li X., Tang Q.-Q. Transcriptional regulation of adipocyte differentiation: a central role for CCAAT/enhancer-binding protein (C/EBP) beta. The Journal of Biological Chemistry. 2015;290(2):755–761. doi: 10.1074/jbc.r114.619957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Y., Luo L., Luo N., Timothy Garvey W. Lipid metabolism mediated by adipocyte lipid binding protein (ALBP/aP2) gene expression in human THP-1 macrophages. Atherosclerosis. 2006;188(1):102–111. doi: 10.1016/j.atherosclerosis.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 49.Hamm J. K., El Jack A. K., Pilch P. F., Farmer S. R. Role of PPARγ in regulating adipocyte differentiation and insulin-responsive glucose uptake. Annals of the New York Academy of Sciences. 1999;892:134–145. doi: 10.1111/j.1749-6632.1999.tb07792.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhu J., Zhang Y., Fan F., Wu G., Xiao Z., Zhou H. Tumor necrosis factor-α-induced protein 8-like-2 is involved in the activation of macrophages by Astragalus polysaccharides in vitro. Molecular Medicine Reports. 2018;17(5):7428–7434. doi: 10.3892/mmr.2018.8730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Morphological observation of rats: Compared with the normal control group, rats in YDCS model group appeared to have an addiction of lying, messy and dry hair, squinting, listless, pale and partial darkness on lips and paws, clear urine in large amount, loose stools, etc. Supplementary Figure 2: Morphological observation of rats: After 7 days of the treatment with RA water abstract, the overall condition of the animals improved significantly. Hair became smoother and shiner; rat activity increased. Lips, stools forming became moderate; nails turned white and pink; urine became of normal color. Supplementary Figure 3: The change of PPAR signal pathway in YDCS model: The PPAR signal pathway is an important pathway for metabolism and immunity. In YDCS pathological process, rat glucose metabolism, lipid metabolism, and amino acid metabolism reduction were disordered. The body's energy metabolism dysregulation of rats reflects the abnormal PPAR signal pathway. It is closely related to PPAR signaling pathway and downregulation of the target genes CPT-1, FABP4. Supplementary Figure 4: The change of PPAR signaling pathway in RAWA treatment group: Combined with the transcriptome results, the effect of RA membranaceus on energy metabolism may be due to the action on PPAR signaling pathway, resulting in the upregulation of the target gene CPT-1 and FABP4. RA can also enhance production of heat and promote the reduced energy metabolism in the YDCS model rats. These benefits are the specific manifestation of its warming effect. RAWA upregulates lipid metabolism represented by ap2 (Fabp4) under the action of cytochrome P450 family member Cyp1a1 in vivo. Activation of PPAR signaling pathway (path: rno03320).
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.