Abstract
The Accrual to Clinical Trials (ACT) network is a federated network of sites from the National Clinical and Translational Science Award (CTSA) Consortium that has been created to significantly increase participant accrual to multi-site clinical trials. The ACT network represents an unprecedented collaboration among diverse CTSA sites. The network has created governance and regulatory frameworks and a common data model to harmonize electronic health record (EHR) data, and deployed a set of Informatics for Integrating Biology and the Bedside (i2b2) data repositories that are linked by the Shared Health Research Information Network (SHRINE) platform. It provides investigators the ability to query the network in real time and to obtain aggregate counts of patients who meet clinical trial inclusion and exclusion criteria from sites across the United States. The ACT network infrastructure provides a basis for cohort discovery and for developing new informatics tools to identify and recruit participants for multi-site clinical trials.
Keywords: clinical trials, accrual, cohort discovery, clinical data research network, electronic health records
INTRODUCTION
Advancing translational research to improve human health requires that sufficient numbers of participants are available for clinical investigations. It is therefore critical to efficiently identify eligible participants, provide them with research opportunities, and enroll them in research studies. These tasks are particularly challenging for multi-site clinical trials, the majority of which are unable to recruit their proposed number of participants within their planned time frame.1–4 Failure to accrue eligible participants in trials is inefficient and wasteful, limits generalizability of results, and leads to premature closure of trials.5
Electronic health records (EHRs) are now used clinically at nearly all major medical centers and hospitals. EHR data provide an opportunity to significantly increase the efficiency of clinical trials by identifying eligible participants based on demographic, diagnostic, procedural, laboratory, medication, and other information.6,7 However, there are important privacy and regulatory concerns that need to be addressed in order to use the EHR data cooperatively.8 In addition, differences in data representation across institutional EHRs pose technical challenges in harmonizing and sharing data.9
THE ACCRUAL TO CLINICAL TRIALS NETWORK
To address these challenges, four Clinical and Translational Science Award (CTSA) sites collaborated to create the Accrual to Clinical Trials (ACT) network supported by funding from the National Center for Advancing Translational Sciences (NCATS).10 The network is expanding its reach by adding new CTSA sites in waves and is enhancing the functionality of the network in three stages. Stage I will enable rapid real-time cohort exploration across the federated network. Stage II will enable identification and contact of participants who are eligible for clinical trial recruitment. Stage III will develop approaches, tools, educational materials, and infrastructure to enable patients and care providers to identify clinical trials.
The ACT network has developed a governance structure, constructed a regulatory framework, deployed technical infrastructure, created a common data model (CDM), and linked CTSA sites to enable rapid cohort exploration. Institutional leaders at participating sites signed a data sharing agreement and the ACT protocol was approved by local Institutional Review Boards (IRBs). The network links 21 CTSA sites and has implemented Stage I functionality to enable cohort discovery across more than 40 million patients for clinical trials as well as for other cohort studies.
The ACT network complements the National Patient-Centered Clinical Research Network (PCORnet) that is funded by the Patient-Centered Outcomes Research Institute (PCORI). The goal of PCORnet is to create a network of clinical data research networks (CDRNs) that leverage EHR data for comparative effectiveness and other types of research.11 The goal of the ACT network is to enable efficient, safe and lower cost multi-site clinical trials and translational research studies for all CTSAs.
ORGANIZATION
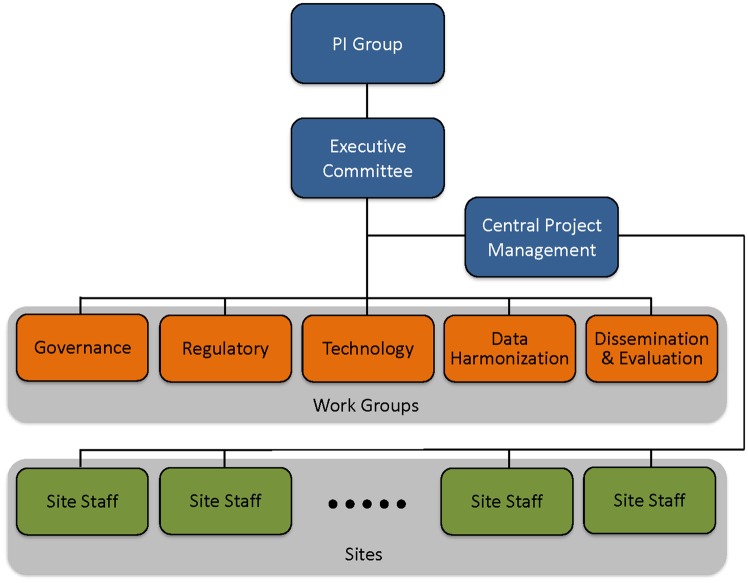
The ACT network organization consists of a group of four Principal Investigators (PIs), an executive committee, five work groups, staff at each site including a project manager, technical and dissemination personnel, and a central project management team (see Figure 1). The PI group is responsible for the strategic goals, benchmarks, metrics, and timely implementation. The executive committee is formed by the PI group, work group leads, the central project management team, and a NCATS program officer. The committee functions to optimize all operational decisions and decides on the priorities and deliverables for each work group.
Figure 1.
The Accrual to Clinical Trials network organization that includes the PI Group, the executive committee, the five work groups, staff at each site, and the central project management team.
The work group leads are responsible for scheduling work group meetings, implementation of their assigned tasks, recording the completion of milestones and facilitating face-to-face meetings to ensure integration, collaboration, and meeting benchmarks. The governance work group is responsible for defining the structure of ACT operations, including the creation of a governance process. ACT governance attempts to strike a balance between ensuring an equitable decision making process that facilitates consensus seeking while minimizing unnecessary bureaucracy. The regulatory work group is responsible for identifying a common regulatory approach to enable compliant access of EHRs from within and across the ACT sites, and develops regulatory guidance to assist sites in securing IRB approval for their participation in the ACT network. The technology work group is responsible for the development, coordination and implementation of software and processes for the network. The data harmonization work group is responsible for the specification of the CDM and distribution of ontologies to the sites. The dissemination and evaluation work group is responsible for the rollout of the network to investigators and for developing tools to evaluate network use.
Each ACT site has a local project manager who is responsible for tracking the site’s progress and status, recording milestones, and reporting risks and issues to the central project management team. The central project management team oversees overall project planning and coordination among the various work groups, serves as the single point of contact of information for the entire project, and is responsible for managing all communication and reporting across the participating sites.
PARTICIPATING SITES
At present, the ACT network brings together 21 CTSA sites from 16 U.S. states and the District of Columbia and includes several pediatric academic health centers (see Figure 2). In the future, additional CTSA sites will be invited to join the network, and by project completion, up to 64 sites will be connected. The current ACT network contains data on more than 40 million patients (see Tables 1 and 2). Each site contributed at a minimum data from January 2012 though many sites have contributed far more.
Figure 2.
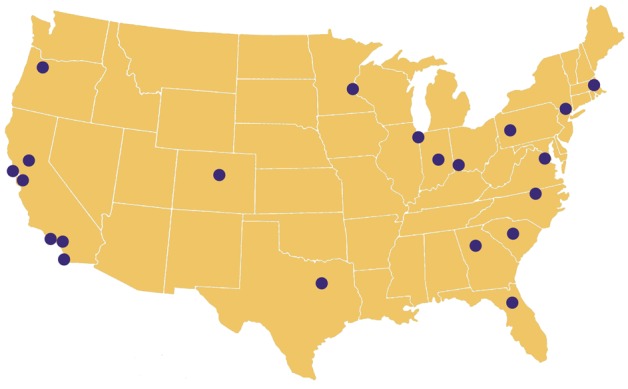
Locations of sites in the Accrual to Clinical Trials network.
Table 1.
The Accrual to Clinical Trials network patient counts
| CTSA site, state | Number of patients (%) | |
|---|---|---|
| 1 | Children’s National Medical Center, DC | 666 600 (2) |
| 2 | Columbia University, NY | 621 200 (2) |
| 3 | Duke University, NC | 1 332 900 (3) |
| 4 | Emory University, GA | 1 153 300 (3) |
| Morehouse University, GA | 105 300 (0.3) | |
| 5 | Harvard University, MA | 1 419 700 (4) |
| 6 | Indiana University, IN | 2 343 800 (6) |
| 7 | Medical University of South Carolina, SC | 1 287 500 (3) |
| 8 | Northwestern University, IL | 3 095 800 (8) |
| 9 | Oregon Health & Science University, OR | 2 875 800 (7) |
| 10 | Stanford University, CA | 579 900 (1) |
| 11 | University of California, Davis, CA | 2 372 100 (6) |
| 12 | University of California, Irvine, CA | 1 603 500 (4) |
| 13 | University of California, Los Angeles, CA | 4 562 300 (11) |
| 14 | University of California, San Diego, CA | 2 330 600 (6) |
| 15 | University of California, San Francisco, CA | 3 282 400 (8) |
| 16 | University of Cincinnati, OH | 836 700 (2) |
| 17 | University of Colorado/Children's Hospital Colorado, CO | 997 300 (2) |
| 18 | University of Florida, FL | 593 200 (1) |
| 19 | University of Minnesota, MN | 2 337 200 (6) |
| 20 | University of Pittsburgh, PA | 1 368 300 (3) |
| 21 | UT Southwestern, TX | 4 428 800 (11) |
| Total | 40 194 200 (100) |
Table 2.
The Accrual to Clinical Trials network demographics
| Number of patients (%) | |
|---|---|
| Age (years) | |
| 0–9 | 3 262 900 (8) |
| 10–17 | 3 065 800 (8) |
| 18–34 | 7 577 500 (19) |
| 35–44 | 4 866 700 (12) |
| 45–54 | 5 371 600 (14) |
| 55–64 | 5 477 200 (14) |
| 65–74 | 4 233 900 (11) |
| 75–84 | 2 456 200 (6) |
| 85–90 | 708 700 (2) |
| ≥90 | 2 224 300 (6) |
| Gender | |
| Female | 20 286 800 (54) |
| Male | 17 337 000 (46) |
| Race | |
| Asian | 941 000 (4) |
| Black or African American | 3 276 700 (16) |
| American Indian or Alaskan Native | 119 200 (0.6) |
| Native Hawaiian or Other Pacific Islander | 175 500 (0.8) |
| White | 16 533 000 (79) |
| Ethnicity | |
| Hispanic | 2 421 000 (14) |
| Not hispanic | 37 773 100 (86) |
INFORMATICS INFRASTRUCTURE
Technology
The network consists of local Informatics for Integrating Biology at the Bedside (i2b2) EHR data repositories12 that are integrated by the Shared Health Research Information Network (SHRINE) platform.13 i2b2 was chosen as the platform for local data repositories, since it is widely used by many CTSA sites; provides a core set of tools to manage projects, ontologies, data, and workflows; has additional tools for cohort exploration; and has an active group of users distributed worldwide who create and share software enhancements. SHRINE provides a federated query and response system that enables investigators to query EHR data housed in i2b2 repositories across multiple independent institutions.14 For the ACT network, new functionality in SHRINE was developed that supports a true hub and spoke network topology, eases network setup and management, enables a distributed data steward model, betters error reporting for users, and improves administrative reporting. Informatics and technical needs for individual sites, including configuration, testing and security needs are managed through weekly calls, web-based wikis and email list discussions.
For operational efficiency, there are three separate networks that include test, stage, and production networks. The test network, comprising of four sites, is used for testing software and ontology upgrades. The stage network consists of new sites that are connecting to the network for the first time and provides a mechanism to evaluate a site’s readiness and troubleshoot technical, ontology, and data issues. The production network is used by investigators for cohort exploration.
Data model and i2b2/SHRINE ontologies
The ACT CDM specifies data domains and data elements to be loaded at each site’s i2b2 repository. The domains in the current ACT CDM include demographics, diagnoses, procedures, medications, laboratory test results, and visit characteristics; these comprise a subset of the domains in the PCORnet CDM.13 For each data domain such as diagnoses or procedures, the ACT CDM (version 1.4) has fewer data elements when compared with the corresponding data domain in the PCORnet CDM (version 4.1); however, a data element that is included in the ACT CDM has the same definition as the corresponding data element in PCORnet CDM. The ACT CDM is available from the ACT website at http://www.actnetwork.us.
SHRINE, as does i2b2, employs a query language that allows querying of data across the sites in the network using predefined terms. For each data domain (such as diagnoses or medications) the predefined collection of terms that describe the data for the domain are arranged in a hierarchy for easy navigation. These ontologies are developed centrally and distributed for installation at each ACT site and are available from the ACT wiki (https://ncatswiki.dbmi.pitt.edu). The ACT ontologies are updated periodically to keep up with expansion of data domains, changes and updates in source terminologies, and to fix errors found in the deployed ontologies.
Data updates and data characterization
To ensure recency, each site updates its i2b2 repository monthly. To ensure data quality, development and deployment of data characterization processes is ongoing. Guidance for extract-transform-load process including recommendations for the types of data and sources of data and i2b2 and ontology specific formats is provided through regular conference calls. An online data characterization survey has been implemented that will be completed by each site. Data from this survey will provides site-specific information on the time period of data, amount of data along several dimensions, expected gaps in data (eg, a children’s hospital may not have patients older than 18 years of age), and results of checks on data formats in the i2b2 repository.
Test cases
Several high-priority clinical trials were employed as test cases for preliminary evaluation of the network. Examples of clinical trials that were used as test cases included identification of patients with early rheumatoid arthritis and an inadequate response to methotrexate, identification of patients with early stage fibrosis secondary to Hepatitis C infection, and identification of patients with coronary artery disease who are eligible for chelation therapy. For each test case, inclusion and exclusion criteria of the corresponding clinical trial was translated into a SHRINE query using the ACT ontologies, and the query was executed from the University of Pittsburgh site. Since ACT uses a hub and spoke network topology, the query that is issued from the University of Pittsburgh site is transmitted to the SHRINE hub that is located at the Harvard Medical School from where it is broadcast to all network sites. Counts resulting from the execution of the query at each site’s i2b2 repository are transmitted to the site that issued the query via the SHRINE hub. Queries often execute in 5 min or under and the investigator is provided with a list of sites on the network and corresponding counts. As an example, the inclusion and exclusion criteria for the chelation therapy test case and coverage for these criteria in the ACT ontologies are shown in Supplementary Appendix Tables S1 and S2 respectively. This query returned counts from 21 sites on the network in 5 min and identified a total of 8383 patients across the network.
Challenges
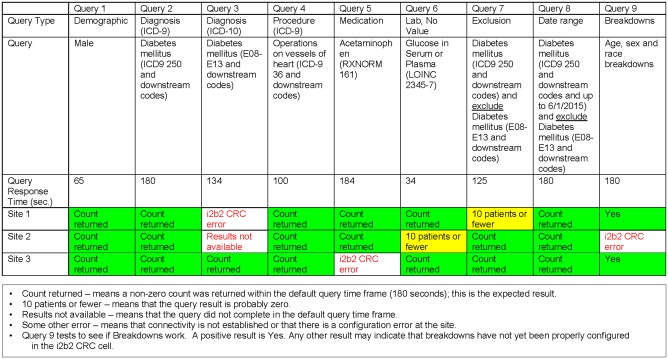
The ACT network is a large, real-time and self-serve network and these features pose several challenges. Maintaining continuous connectivity of every site to the network is exacting. Currently, network status is assessed weekly using a “smoke test” that interrogates every data domain based on which a report is circulated that provides technical and connectivity status of each site’s i2b2 repository (see Figure 3). A helpdesk and a mailing list provide guidance and assistance with troubleshooting. Update of software or ontologies have to be done in a narrow maintenance window so that the network is available for querying for the maximum length of time. In addition, in a large network, it is useful to be able to issue a series of queries without waiting for each query to complete at each site. A future version of SHRINE will include this capability. The source terminologies that are used to develop the ontologies typically retire codes. However, to maintain ability to query historical data the ontologies have to include retired codes, and the process of obtaining retired codes can be challenging depending on the terminology. Ensuring the quality of the data at each site is critical for large scale use of the network; this is particularly challenging since data characterization needs to be performed with every update of the data repository at each site. Work is ongoing in developing data characterization processes that can be efficiently deployed across the network.
Figure 3.
An example report from the “smoke test” that provides an overview of connectivity and configuration status of the data repository at each site. A complete report will contain a row of results for each site that is connected to the Accrual to Clinical Trials network.
SUMMARY
The long-term goal of the ACT network is to transform clinical and translational research by developing an efficient and extensible digital infrastructure that enables CTSA sites to collaborate in cohort exploration, to identify and contact patients who are eligible for clinical trials, and to enable patients and care providers to identify clinical trials. Currently, the ACT network enables cohort exploration by providing aggregate counts from more than 40 million patients who meet clinical trial inclusion criteria across 21 sites. In the future, the ACT network will attempt to extend to all CTSA sites, add additional data domains and data elements to the ACT CDM, and use the ACT infrastructure to develop and deploy novel recruitment tools to enable investigators to efficiently recruit participants across institutions. The ACT network represents an unprecedented collaboration among diverse CTSA sites and serves as a national resource for accelerating recruitment of research participants.
FUNDING
This work was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under grant numbers UL1 TR000005-09S1, UL1 TR001857-01S1, UL1 TR001876, UL1 TR001873, UL1 TR002553, UL1 TR002378, UL1 TR002541, UL1 TR002529, UL1 TR001450, UL1 TR001422, UL1 TR002369, UL1 TR001085, UL1 TR001860, UL1 TR001414, UL1 TR001881, UL1 TR001442, UL1 TR001872, UL1 TR001425, UL1 TR002535, UL1 TR001427, UL1 TR002494, UL1 TR001857, and UL1 TR001105.
CONTRIBUTORS
S.V. conceived and designed the study, participated in data collection, analysis, and interpretation, drafted and revised the manuscript and approved the final version for submission. M.J.B. made critical manuscript revisions and approved the final version for submission. V.S.D. participated in data collection, made critical manuscript revisions, and approved the final version for submission. E.R.S participated in data collection and approved the final version for submission. D.M. participated in data collection, drafted and revised the manuscript and approved the final version for submission. N.R.A. drafted and revised the manuscript and approved the final version for submission. K.A.A. drafted and revised the manuscript and approved the final version for submission. D.R. made critical manuscript revisions and approved the final version for submission. S.N.M. participated in data collection and approved the final version for submission. E.H.M. participated in data collection and approved the final version for submission. H.A.P. made critical manuscript revisions, and approved the final version for submission. R.T. obtained funding, made critical manuscript revisions and approved the final version for submission. G.S.F. obtained funding, made critical manuscript revisions and approved the final version for submission. L.M.N. obtained funding, made critical manuscript revisions and approved the final version for submission. S.E.R. obtained funding, made critical manuscript revisions and approved the final version for submission.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1. Kitterman DR, Cheng SK, Dilts DM, Orwoll ES.. The prevalence and economic impact of low-enrolling clinical studies at an academic medical center. Acad Med 2011; 8611: 1360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tice DG, Carroll KA, Bhatt KH, et al. Characteristics and causes for non-accrued clinical research (NACR) at an ccademic medical institution. J Clin Med Res 2013; 53: 185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Irving SY, Curley MA.. Challenges to conducting multicenter clinical research: ten points to consider. AACN Adv Crit Care 2008; 192: 164–9. [DOI] [PubMed] [Google Scholar]
- 4. Cheng SK, Dietrich MS, Dilts DM.. A sense of urgency: evaluating the link between clinical trial development time and the accrual performance of cancer therapy evaluation program (NCI-CTEP) sponsored studies. Clin Cancer Res 2010; 1622: 5557–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kost RG, Mervin-Blake S, Hallarn R, et al. Accrual and recruitment practices at Clinical and Translational Science Award (CTSA) institutions: a call for expectations, expertise, and evaluation. Acad Med 2014; 898: 1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elkhenini HF, Davis KJ, Stein ND, et al. Using an electronic medical record (EMR) to conduct clinical trials: Salford Lung Study feasibility. BMC Med Inform Decis Mak 2015; 151: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weng C, Bigger JT, Busacca L, Wilcox A, Getaneh A. Comparing the effectiveness of a clinical registry and a clinical data warehouse for supporting clinical trial recruitment: a case study. In: AMIA Annual Symposium Proceedings. 2010: 867–71. Washington, DC. [PMC free article] [PubMed]
- 8. Coorevits P, Sundgren M, Klein GO, et al. Electronic health records: new opportunities for clinical research. J Intern Med 2013; 2746: 547–60. [DOI] [PubMed] [Google Scholar]
- 9. Bayley KB, Belnap T, Savitz L, Masica AL, Shah N, Fleming NS.. Challenges in using Electronic Health Record data for CER: experience of 4 learning organizations and solutions applied. Med Care 2013; 51: S80–6. [DOI] [PubMed] [Google Scholar]
- 10. Reis SE, Berglund L, Bernard GR, Califf RM, Fitzgerald GA, Johnson PC.. Reengineering the national clinical and translational research enterprise: the strategic plan of the National Clinical and Translational Science Awards Consortium. Acad Med 2010; 853: 463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS.. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014; 214: 578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 172: 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): A prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009; 165: 624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anderson N, Abend A, Mandel A, et al. Implementation of a deidentified federated data network for population-based cohort discovery. J Am Med Inform Assoc 2012; 19 (e1): e60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.