Abstract
Background
Inflammation is a fundamental factor that contributes to the development and progression of several types of cancer including colon cancer. Toll-like receptors (TLRs) and their signaling pathways have been reported to be associated with chronic inflammation and thereby induced cancer. Our aim was to investigate the expression and polymorphisms of TLR2 and their association with colon cancer.
Methods
Real-time PCR and immunohistochemistry were used to investigate TLR2 gene expression and to evaluate the potential risk of predisposition to colon cancer caused by three tagging single-nucleotide polymorphisms (SNPs) on TLR2, including rs3804100, rs4696480, and rs3804099. TaqMan assay was conducted on samples from 115 patients with colon cancer and 102 age- and sex-matched normal individuals.
Results
We found that, TLR2 was highly expressed in epithelial colon cancer cells and both TLR2 mRNA and protein levels, and significantly decreased in tumor tissues compared to normal tissues. Two of three TLR2 SNPs increased the risk of colon cancer. However, TLR2 rs3804099 increased the risk of colon cancer development by more than 3.8- and 5-fold in female patients and patients aged less than 57 years, respectively. The T allele of TLR2 rs3804100 showed a significant association with patients less than 57 years. In silico analysis of the TLR2 nucleotide substitution in SNP rs3804100 and rs3804099 determined that 67% and 70% probability of these single nucleotide variants alter splicing phenotypes, rs3804100 more specifically result on activating an additional splice site. Genotype and allele frequencies of rs4696480 were similar between the overall study populations. Thus, TLR2 rs4696480 appear to be not involved in colon cancer in our study population.
Conclusions
There was a significant link between innate immunity deregulation through disruption of the TLRs and potential development of colon cancer. These SNPs can be used as screening markers for predicting colon cancer risk earlier in life to implement necessary prevention.
Keywords: colon cancer, gene expression, genotyping, polymorphism, Toll-like receptors, innate immunity
Introduction
Inflammation is the most important factor that contributes to colon cancer development and growth. Several studies have reported a close association between chronic inflammation and cancer involving Toll-like receptors (TLRs) and their signaling pathways.1,2 Other studies showed that TLRs and their agonists can serve as therapeutic targets in several cancer and immune diseases to induce either apoptosis or anti-tumor responses.3 These receptors are mainly expressed in normal human mature cells that play a role in innate immunity such as monocytes, neutrophils, macrophages, dendritic cells, T cells, B cells, and natural killer cells.4 TLRs are also expressed on tumor cells in a wide variety of tissues, suggesting their important roles in tumor cell evasion from immune-surveillance.1,3,5 These receptors act as innate immune sensors against endogenous and exogenous danger signals by recognizing pattern recognition molecules (damage-associated molecular pattern [DAMPs] and pathogen-associated molecular pattern [PAMPs])6–8 and driving an adaptive immune response through their signaling pathways, leading to nuclear factor-kappa B (NF-κB) and interferon regulatory factor 3 translocation and transactivation.9 This activates various inflammatory cytokines10 such as tumor necrosis factor and interleukin-6. TLR signaling consists of at least two distinct pathways: one is a myeloid differentiation primary-response protein-88 (MyD88)-dependent pathway, while the other is MYD88-independent. Most TLRs types activate MYD88 signaling pathways,11 which requires TLRs adaptor proteins including MYD88, Toll-interleukin 1 receptor domain-containing adaptor protein, Toll/interleukin-1 receptor domain-containing adaptor protein inducing interferon beta, and Toll-interleukin 1 receptor domain-containing adapter molecule 2.12 So far, more than 13 types of TLRs have been identified; 10 functional TLRs family members (TLRs 1–10) have been identified in humans,13 while the other TLRs 11–13 are found in rodent species (rats and mice).14 TLR polymorphisms influence dysregulation of the immune system and the development of different types of cancers. Recently, Slattery et al15 and Semlali et al16 reported that TLR4 single-nucleotide polymorphism (SNP) (rs11536898) is associated with colon cancer development. SNPs in the TLR1, TLR6, and TLR10 cluster increase prostate cancer susceptibility.17 In addition, recent data revealed a strong link between the risk of gastric cancer development and presence of SNPs in TLR2 and TLR4.18,19 These previous studies suggest an association between innate immunity gene polymorphism and inflammation process, which increases the susceptibility of gastric carcinogenesis development.20 Recently, the TLR4 (Asp299Gly) SNP was found to be associated with an increased risk of prostate cancer development in a north Indian population.21 A synthetic meta-analysis based on data from 22 studies also confirmed the association of the TLR4 Asp299Gly SNP with increased gastrointestinal cancer risk, but with decreased prostate cancer risk.22 Additionally, Bhide et al11 found a close association between novel mutations in TLR genes (TLR1-Ser150Gly and Val 220 Met; TLR2 – Phe 670Leu) and an increased susceptibility to Mycobacterium avium subsp. Para tuberculosis.
More than 175 SNPs have been reported in TLR2, which is located on chromosome 4q32.23,24 It has been shown that TLR2 polymorphisms increase prostate cancer risk in a north Indian population.25 Two SNPs in TLR2 (rs3804099, rs3804100) and the clinicopathologic features of Korean patients with papillary thyroid cancer and age-matched controls were analyzed using direct sequencing.26 Interestingly, Caws et al27 found that TLR2 polymorphisms are associated with disseminated tuberculosis. SNPs rs3804100 and rs4696480 were reported to be associated with several cancers, but not colon cancer.28,29 TLR2 rs3804099 has been studied in other populations.15 Because the SNP association differs between populations, we focused on these three SNPs in a Saudi population and evaluated the roles of the rs3804100 and rs4696480 SNPs in colorectal cancer. This goal of this study was to determine the association of innate immunity genes such as TLR2 SNPs (rs3804099, rs3804100, and rs4696480) and the risk of colon cancer development in a Saudi Arabia population.
Methods
Study population
A total of 115 patients with colon cancer and 102 age- and sex-matched controls were included in this study, which was conducted in the Kingdom of Saudi Arabia. Patients were recruited between 2012 and 2016 from King Khalid University Hospital in Riyadh. A group of surgical doctors and pathologists specialized in colon cancer recruited, monitored, diagnosed, and collected clinical data from all patients using standard clinical, endoscopic, radiological, and histological criteria confirming adenocarcinoma and thus the eligibility of patients to participate in this study. Healthy participants had no clinical signs or symptoms of colon cancer, based on colonoscopy screening. The Ethics Committee at King Khalid University Hospital approved this current study with IRB number CAMS 13/3536. Each participant provided written informed consent and completed a written survey. Participants were asked to complete a self-administered questionnaire regarding their socio-demographic characteristics (eg, age, family history of cancer, personal medical history, and social behavior [eg, smoking habits and alcohol intake]). The clinical information and genetic materials were obtained from each participant after obtaining informed consent in accordance with the Ethics Committee guidelines at King Khalid University Hospital. The tissue samples were used for RNA/DNA extraction and immunohistochemistry to evaluate TLR2 expression. Blood samples were used for genomic DNA extraction for genotyping assays to investigate the possible association between TLR2 polymorphisms and colon cancer susceptibility.
Cell culture
HCT-116, Lovo, and SW-480 were cultured according to the recommendations of the provider ATCC Company (Manassas, VA, USA).
The cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% inactivated fetal bovine serum. The medium was changed every 2 days up to 80% confluence. Next, the cells were washed twice with 1× phosphate-buffered saline and then detached from the flasks using 0.05% trypsin–0.1% EDTA. The cells were counted and re-suspended in fresh Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS, to a final concentration of 106 cells/mL, and used to perform all experiments.
RNA extraction, reverse transcription, and real-time PCR
As described previously,30–32 total RNA was extracted from colon cancer cells or from colon tissue using a DNA/RNA Mini kit from Qiagen NV (Venlo, The Netherlands) according to the manufacturer’s instructions. To quantify the RNA concentration and verify RNA purity and quality, we used the Agilent 2100 Bioanalyzer system and Agilent Small RNA analysis kit according to the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA) and a Nano-Drop8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthetized from 1 µg of each RNA sample with a high-capacity cDNA reverse transcription kit from Applied Biosystems (Thermo Fisher Scientific). The real-time PCR (qPCR) was performed as previously described30 using SYBR Green Master Mix from Bio-Rad Laboratories Inc. (Hercules, CA, USA) following the manufacturer’s instructions. The number of copies of the TLR2 gene was measured using the Applied Biosystems 7,500 Fast real-time PCR detection system. GAPDH served as the internal control. The primers used for real-time PCR are presented in Table S1. Each reaction was conducted in triplicate. The specificity of each primer pair was verified by the presence of a single melting temperature peak, and the results were analyzed using the 2−∆∆Ct (Livak) relative expression method.
Western blotting
Total proteins were extracted from different colon cancer cells (HCT-116, SW480, and Lovo) using lysis buffer (50 mM HEPES, pH 7.4; 1% (v/v) Triton X-100; 4 mM EDTA; 1 mM sodium fluoride; 0.1 mM sodium orthovanadate; 1 mM tetrasodium pyrophosphate; 2 mM phenylmethylsulfonyl fluoride; 10 µg/mL leupeptin; and 10 µg/mL aprotinin). Next, 20 µg of total protein was subjected to SDS polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% bovine serum albumin in Tween-20/Tris-buffered saline, and incubated overnight at 4°C with primary anti-TLR2 (1:250, Santa Cruz Biotechnology Inc., Dallas, TX, USA) and anti-β-actin (1:5,000) antibodies in blocking solution. After washing three times with Tween-20/Tris-buffered saline for 15 minutes, the membranes were incubated with anti-mousse secondary antibody (1:1,000 in Tween-20/Tris-buffered saline) for 1 hour at room temperature.
Protein detection was conducted by electrochemiluminescence (Amersham, GE Healthcare Biosciences, Little Chalfont, UK) solution and by means of a FujiFilm Image Reader LAS-1000 Pro (FujiFilm, Tokyo, Japan).
Immunohistochemistry
The paraffinized biopsied tissues from colon cancer and normal colon tissues were sectioned to a thickness of 1–2 µm. Hematoxylin and eosin staining was performed by routine methods as described by Semlali et al.32 Briefly, after deparaffinization and rehydration, the slides were incubated overnight at 37°C, followed by 30 minutes in a hot air oven at 60°C and processed in the automated immunohistochemistry slide staining system known as the Benchmark XT from Ventana Medical Systems, Inc. (Tucson, AZ, USA). Next, the slides were incubated with anti-human anti-TLR2 primary antibody from Santa Cruz Biotechnology (1:100, Santa Cruz) for 1 hour at 37°C and examined using the ultraview multimer detection system from Ventana Medical Systems Inc. The immunolocalized TLR2 proteins were visualized using a copper-enhanced DAB reaction from Ventana Medical Systems Inc. The slides were then stained with hematoxylin II from Ventana Medical Systems Inc. for 4 minutes and Bluing Reagent (Ventana Medical Systems Inc.) for 4 minutes, and coverslips were applied.
DNA extraction
For DNA extraction, 2–4 mL blood samples were collected in EDTA-containing tubes. Genomic DNA was isolated from all blood samples using the QIAmp DNA blood mini kit following the manufacturer’s instructions. The purity and concentration of each sample were quantified by a Nano Drop 8000 spectrophotometer (Thermo Fisher Scientific).
Genotyping of TLR2 polymorphisms
Three TLR2 SNPs (rs3804100, rs4696480, and rs3804099) were selected based on previous genetic association studies and their localization in regulatory areas of gene expression (Table S2) (promoter and exon). Briefly, 10–20 ng of DNA from each sample was analyzed using a Taq Man allelic discrimination assay (Applied Biosystems, Thermo Fisher Scientific) with allele-specific fluorogenic oligonucleotide probes as previously described.33 All genotypes were determined by endpoint reading on an ABI 7500 real-time PCR machine (Applied Biosystems, Thermo Fisher Scientific).
In silico analysis of the effect of nucleotide variation on splicing phenotype
Multiple in silico algorithms were used to investigate the silico analysis of the effect of nucleotide variation on splicing phenotype, a consequence of the nucleotide variation in SNP (rs3804100, rs4696480, and rs3804099) were determined using spliceman34,35 and MutationTaster (57), which predicts how likely mutations around annotated splice sites were to disrupt splicing. The Spliceman reports the L1 distance and percentile rank that correspond to the given mutation. This is the rank that the tool uses to predict how likely a mutation is to disrupt pre-mRNA splicing. The higher the percentile rank, the more likely the point mutation is to disrupt splicing. The MutationTaster analyzes and predicts the splicing effect using a neural network model. The “increased” or “gained” effect reported if the change in the confidence score is greater than 10%. While the “decreased” or “lost” effect reported defines in relative to intron/exon; the closer considered “real” splice change and sites distant from borders will be ignored.
Statistical analysis
For gene expression analysis, relative quantification changes as the relative fold-difference between the study groups were measured using the Student’s t-test for sample group comparisons. GAPDH served as an internal control (housekeeping gene). Each reaction was conducted in triplicate. The result was analyzed using the 2−∆∆Ct (Livak) relative expression method.
Genotypic and allelic frequencies were calculated and checked for deviation from Hardy–Weinberg equilibrium. The chi-squared test and odds ratios (ORs) with their 95% confidence intervals (CIs) were also calculated with Fisher’s exact test (two-tailed) and used to assess the association between TLR2 polymorphisms and the risk for colon cancer. Statistical analysis was performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). We considered P-values <0.05 as significant. Regional linkage disequilibrium (LD) plot was established using SNP Annotation and Proxy Search (http://www.broadinstitute.org/mpg/snap/ldplot.php).
Results
Clinical characteristics and investigation of the TLR2 differential gene expression in colon cancer tissues
A total of 115 patients with colon cancer and 102 controls were recruited in this study. The clinical characteristics of the patients, including age, nationality, family history, smoking habits, stage of colon cancer, medications, and presence of other diseases were collected and compared, as illustrated in Table 1. The study population age ranged from 45 to 88 years, and the mean age of the colon cancer patients was 57.04±14.37 years and that of the controls, 56.51±15.70 years. The mean ages did not differ considerably (Table 1). Of the patients with colon cancer, 57.4% were male, while 58.2% were controls, and 42.6% were female compared to 42.2% controls. The male to female ratio was not significantly different between cases and controls. All patients were classified as having cancer in the colon or the rectum regions. The rates of smokers and non-smokers in cancer patients were similar between the two study cohorts. However, the smokers formed 6.1% and nonsmokers formed 93.9% of the patients, whereas, in the controls, the smokers were 4.9% and nonsmokers were 95.1% of the total (Table 1).
Table 1.
Clinical characteristics of the subjects for genotyping
| Characteristic | Cancer (115) n (%) | Control (102) n (%) |
|---|---|---|
| Gender | ||
| Male | 66 (57.4%) | 60 (58.8%) |
| Female | 49 (42.6%) | 42 (41.2%) |
| Age | 56.04±14.37 | 52.84±15.88 |
| Localization | ||
| Colon | 76 (66.1%) | 0 (0%) |
| Rectum | 39 (33.9%) | 0 (0%) |
| Smoker/alcoholic | ||
| Smoker | ||
| Yes | 7 (6.1%) | 5 (4.9%) |
| No | 108 (93.9%) | 97 (95.1%) |
| Alcoholic | ||
| Yes | 2 (1.7%) | 0 (0%) |
| No | 113 (98.3%) | 102 (100%) |
| Therapy | ||
| Chemotherapy | ||
| Yes | 3 (2.6%) | 0 (0%) |
| No | 112 (97.4%) | 102 (100%) |
| Radiotherapy | ||
| Yes | 5 (4.3%) | 0 (0%) |
| No | 110 (95.7%) | 102 (100%) |
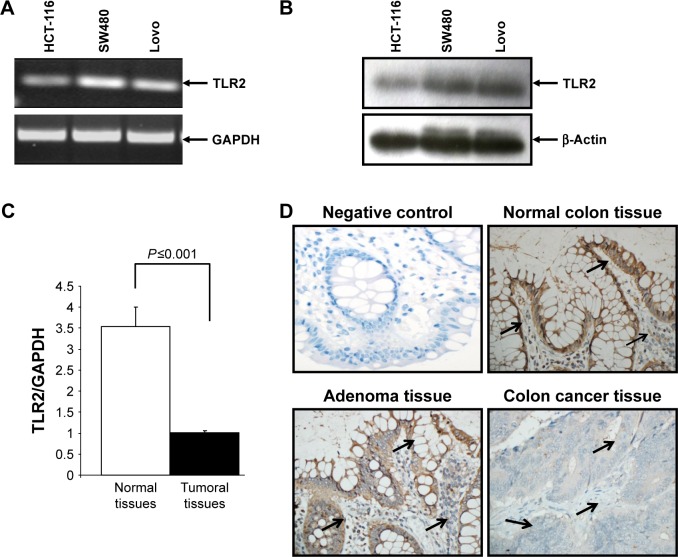
Since colon cancer predominantly affects colonic epithelial cells, we investigated TLR2 expression in various epithelial colon cancer cells. Our results show that TLR2 was expressed at high levels in HCT-116, Lovo, and SW480 cells, at both the mRNA and protein levels (Figure 1A and B).
Figure 1.
Toll-like-receptor 2 (TLR2) mRNA and protein expression in colon cancer cells and tissues. Expression of TLR2 in colon cancer cells at mRNA level (A). TLR2 expression in colon cancer cells at protein level (B). Total cellular RNA freshly extracted from matching normal and colon cancer tissues was reverse-transcribed into cDNA and then used to measure the TLR2 mRNA by real time PCR (C). Tissues were immunostained with specific TLR2 antibody in cancer and matching normal colon; 400×; arrows indicate expression level and localization of the protein (D). TLR2-positive cells in the tissues were estimated as follows: 0 points, no positive color; 1 point, <20% positive staining; 2 points, 21%–50% positive staining; 3 points, 51%–75% positive staining; and 4 points, >75% positive staining; 400×.
To compare the TLR2 differential expression in colon tissues, a quantitative real-time reverse transcription-PCR was performed using 40-colon cancer tissues and matched normal tissues. As shown in Figure 1C, the TLR2 mRNA decreases in the colon cancer tissues compared to the normal colon tissues. This result was confirmed at the protein level by immunohistochemistry (Figure 1D), In addition, TLR2 was prominent on the surface of the epithelium compared to the expression on most colon stromal cells and was generally highly observed in normal colon tissues when compared to tumor tissues. However, in adenoma tissues, the intensity of TLR2 expression was similar to that in normal tissues (Figure 1D).
TLR2 (rs3804100, rs4696480, and rs3804099) genotypic analysis: correlation between TLR2 polymorphism and risk of colon cancer in Saudi Arabian patients
To investigate the potential role of TLR2 polymorphisms on the susceptibility of the Saudi Arabian population to colon cancer, we selected three TLR2 SNPs and compared their allelic frequencies between 115 patients with colon cancer and 102 controls. The observed genotypic frequencies did not show any significant departure from Hardy–Weinberg expectations for all three polymorphic loci that were included in this study. The genotype distribution of the analyzed SNPs along with the corresponding odds ratios and significance are shown in Table 2. Only one of the three SNPs analyzed; TLR2 rs3804099, presents a statistically significant risk to develop colon cancer in this ethnic group. In addition, the genotype and allele frequencies between the patients and controls were exactly similar for SNPs rs3804100 and rs4696480 in the selected Saudi Arabian population. The frequencies of rs3804099 (Asn199Asn) genotypes in patients with colon cancer cases were 17% CC, 45% CT, and 38% TT, whereas in the controls, the frequencies were 26%, 47%, and 28%, respectively, for CC, CT, and TT. The homozygous “TT” genotype for the SNP rs3804099 showed more than a twofold higher risk for developing colon cancer compared to controls (OR, 2.13; CI, 1–4.54; P=0.04), and the T allele frequency was high in patients compared to controls (OR, 1.493; CI, 1.017–2.913; P=0.04) (Table 2).
Table 2.
Genotype and alleles frequencies of TLR2 gene polymorphism in colorectal cancer and controls
| SNP ID | Genotype | Colon | Controls | OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|
| rs3804099 | CC | 19 (0.17) | 27 (0.26) | Ref | |||
| CT | 50 (0.45) | 47 (0.47) | 1.51 | 0.74–3.07 | 1.31 | 0.25 | |
| TT | 42 (0.38) | 28 (0.27) | 2.13 | 1.00–4.54 | 3.89 | 0.04 | |
| CT + TT | 92 (0.83) | 75 (0.74) | 1.74 | 0.90–3.37 | 2.75 | 0.09 | |
| C | 88 (0.40) | 101 (0.49) | Ref | ||||
| T | 134 (0.60) | 103 (0.51) | 1.49 | 1.017–2.19 | 4.20 | 0.04 | |
| rs3804100 | CC | 2 (0.02) | 2 (0.02) | Ref | |||
| CT | 13 (0.11) | 19 (0.18) | 0.68 | 0.08–5.49 | 0.13 | 0.72 | |
| TT | 99 (0.87) | 82 (0.80) | 1.20 | 0.16–8.75 | 0.03 | 0.85 | |
| CT + TT | 112 (0.98) | 101 (0.98) | 1.10 | 0.15–8.01 | 0.01 | 0.92 | |
| C | 17 (0.08) | 23 (0.11) | Ref | ||||
| T | 211 (0.92) | 183 (0.89) | 1.56 | 0.80–3.01 | 1.78 | 0.18 | |
| rs4696480 | AA | 27 (0.26) | 25 (0.27) | Ref | |||
| AT | 49 (0.46) | 41 (0.45) | 1.10 | 0.55–2.19 | 0.08 | 0.77 | |
| TT | 30 (0.28) | 26 (0.28) | 1.06 | 0.50–2.27 | 0.03 | 0.86 | |
| AT + TT | 79 (0.74) | 67 (0.73) | 1.09 | 0.57–2.05 | 0.07 | 0.77 | |
| A | 103 (0.49) | 91 (0.49) | Ref | ||||
| T | 109 (0.51) | 93 (0.51) | 1.03 | 0.69–1.53 | 0.03 | 0.86 |
Note: Values in bold are significant.
Abbreviations: Ref, reference; SNP, single-nucleotide polymorphism.
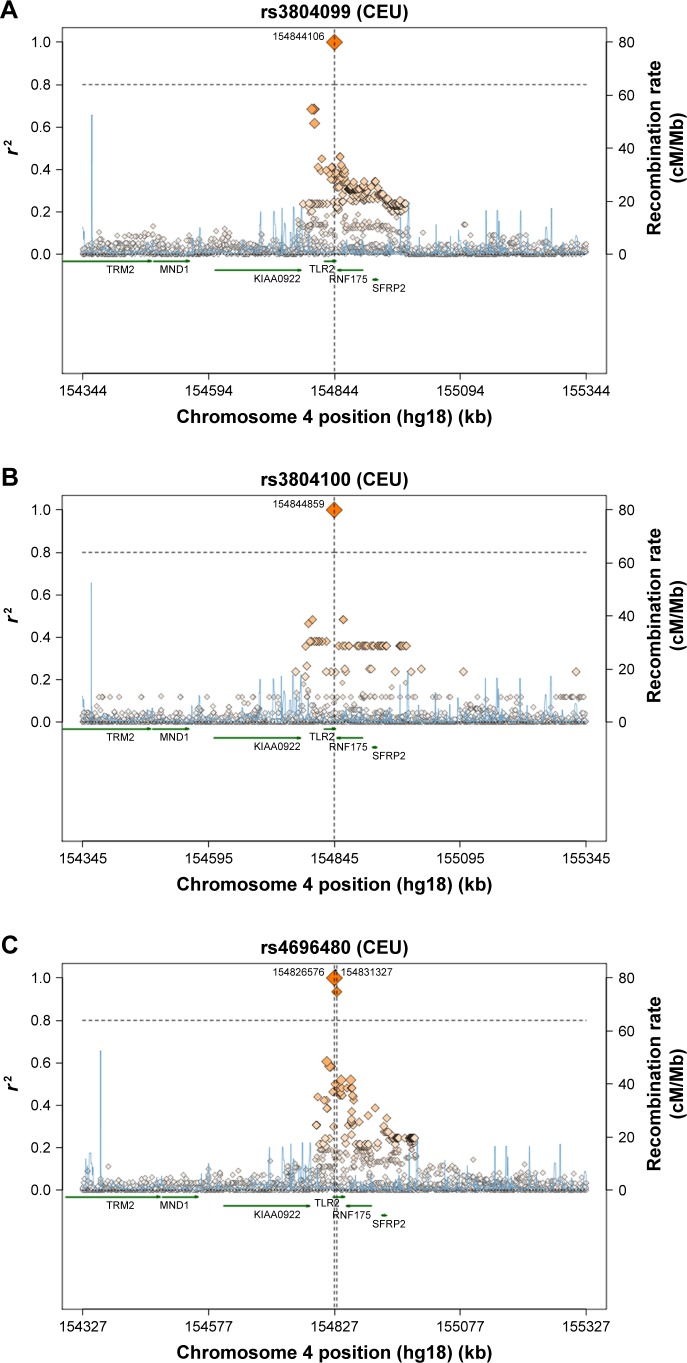
A comparison of the studied SNPs between the Saudi population and different populations was performed based on literature data, to determine the relationships that link these populations together (Table 3). In TLR2 rs3804100 SNP, Chinese (HCB) and Kenyan (MKK) populations were significantly different from our study population. However, only the Japanese (JPT) population showed different genotype distribution in terms of the TLR2 rs3804099 SNP. Surprisingly, TLR2 rs4696480 was different in our ethnic population compared to all other Hap-Map populations shown in Table 3. As shown in Figure 2, a regional linkage disequilibrium plot was established using SNP Annotation and Proxy Search (http://www.broadinstitute.org/mpg/snap/ldplot.php).
Table 3.
Allele and genotype distribution of TLR2 SNPs in the Riyadh region compared to other populations
| Population | Genotype frequency (N) | Allele frequency | χ2 | P-value | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| rs3804099 | TT | CT | CC | T | C | |||
| CRS (102) | 0.275 (28) | 0.461 (47) | 0.265 (27) | 0.505 | 0.495 | 0 | 0 | |
| CEU (224) | 0.268 (60) | 0.562 (126) | 0.170 (38) | 0.549 | 0.451 | 0.5479 | 0.4592 | |
| HCB (86) | 0.302 (26) | 0.651 (56) | 0.047 (4) | 0.628 | 0.372 | 2.8712 | 0.0902 | |
| JPT (172) | 0.535 (92) | 0.395 (68) | 0.070 (12) | 0.733 | 0.267 | 14.6067 | <0.005** | |
| YRI (226) | 0.159 (36) | 0.425 (96) | 0.416 (94) | 0.372 | 0.628 | 5.1193 | 0.0237* | |
| MEX (100) | 0.400 (40) | 0.500 (50) | 0.100 (10) | 0.65 | 0.35 | 4.3549 | 0.0369* | |
| MKK (286) | 0.112 (32) | 0.503 (144) | 0.385 (110) | 0.364 | 0.636 | 6.2142 | 0.0127* | |
| TSI (176) | 0.330 (58) | 0.477 (84) | 0.193 (34) | 0.568 | 0.432 | 1.0367 | 0.3086 | |
|
| ||||||||
| rs3804100 | TT | CT | CC | T | C | |||
| CRS (103) | 0.019 (2) | 0.184 (19) | 0.796 (82) | 0.888 | 0.112 | 0 | 0 | |
| CEU (226) | 0.850 (192) | 0.150 (34) | 0 | 0.925 | 0.075 | 1.2031 | 0.2727 | |
| HCB (86) | 0.372 (32) | 0.581 (50) | 0.047 (4) | 0.663 | 0.337 | 14.1407 | <0.005** | |
| JPT (166) | 0.578 (96) | 0.410 (68) | 0.012 (2) | 0.783 | 0.217 | 4.8500 | 0.0276* | |
| YRI (226) | 0.876 (198) | 0.124 (28) | 0 | 0.938 | 0.062 | 2.4384 | 0.1184 | |
| MEX (100) | 0.820 (82) | 0.180 (18) | 0 | 0.910 | 0.090 | 0.2620 | 0.6088 | |
| MKK (286) | 0.951 (272) | 0.049 (14) | 0 | 0.976 | 0.024 | 12.9344 | <0.005** | |
| TSI (176) | 0.875 (154) | 0.125 (22) | 0 | 0.938 | 0.062 | 2.1681 | 0.1409 | |
|
| ||||||||
| rs4696480 | AA | AT | TT | T | A | |||
| CRS (92) | 0.272 (25) | 0.446 (41) | 0.283 (26) | 0.505 | 0.495 | 0 | 0 | |
| CEU (110) | 0 | 0 | 1.000 (110) | 1.000 | 0 | 70.2188 | <0.005** | |
| HCB (88) | 0 | 0 | 1.000 (88) | 1.000 | 0 | 58.2447 | <0.005** | |
| JPT (82) | 0 | 0 | 1.000 (82) | 1.000 | 0 | 54.9141 | <0.005** | |
| YRI (120) | 0 | 0 | 1.000 (120) | 1.000 | 0 | 75.5660 | <0.005** | |
Notes: Data were obtained from The International HapMap Project39 (NCBI retired the HapMap resource on June 16, 2016).
P<0.05 and
P<0.005. Values in bold are significant.
Abbreviations: CEU, Utah residents with northern and western European ancestry from the CEPH collection; CEPH, Centre d’Etude du Polymorphisme Humain; CRS, central region population of Saudi Arabia; HCB, Han Chinese in Beijing, People’s Republic of China; JPT, Japanese in Tokyo, Japan; MEX, Mexican ancestry in Los Angeles, California; MKK, Maasai in Kinyawa, Kenya; TLR, Toll-like receptor; TSI, Toscani in Italia; YRI, Yoruba in Ibadan, Nigeria.
Figure 2.
Regional LD plots for (A) TLR2 rs3804099 SNP, (B) TLR2 rs3804100 SNP, and (C) TLR2 rs4696480 SNP.
Abbreviations: cM, centiMorgan; CEU, Caucasian; kb, kilobase; LD, linkage disequilibrium; Mb, megabase; SNP, single-nucleotide polymorphism; TLR, Toll-like receptor.
Prevalence of genotype and allele frequencies of TLR2 SNPs in colon cancer patients compared to that in normal control population according to clinical parameters
Further analysis of the TLR2 genotype distribution after correlation with the age, sex, and tumor location at colon cancer diagnosis revealed that the median age of onset of colon cancer was 57 years. To evaluate the association of the analyzed SNPs with age at colon cancer diagnosis, we classified the patients as ≤57 (n=56) or >57 (n=59) years of age. The genotype distributions for the individual SNP along with the statistical analysis are shown in Table 3. Interestingly, in the overall study, rs3804099 showed a significant association with the risk of colon cancer development among those diagnosed with this cancer at ≤57 years of age. The TT genotype frequency was 44% in patients and 21% in the controls. This genotype increases the risk of colon cancer development by more than fivefold in the Saudi Arabian population. The T allele frequency increased the risk of colon cancer by more than threefold in this population. Furthermore, rs3804100 also showed a significant risk in patients aged less than 57 years at age of diagnosis. However, the T allele was associated with a greater than 3.6-fold risk of colon cancer (Table 4). In addition, no association was observed in patients aged >57 years. All genotype and allele frequencies for the three SNPs of TLR2 were similar between normal and colon cancer patients (Table 4).
Table 4.
Genotype frequencies of TLR2 gene polymorphism in colorectal cancer cases
| SNP ID | Genotype | Colon, n (frequency) | Controls, n (frequency) | OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Below 57 years | |||||||
|
| |||||||
| rs3804099 | CC | 7 (0.12) | 19 (0.31) | Ref | |||
| CT | 24 (0.44) | 29 (0.48) | 2.25 | 0.81–6.24 | 2.47 | 0.12 | |
| TT | 24 (0.44) | 13 (0.21) | 5.01 | 1.67–15.03 | 8.80 | 0.003 | |
| CT + TT | 48 (0.88) | 42 (0.69) | 3.10 | 1.19–8.11 | 5.64 | 0.018 | |
| C | 38 (0.35) | 67 (0.55) | Ref | ||||
| T | 72 (0.65) | 55 (0.45) | 2.31 | 1.36–3.92 | 9.69 | 0.0018 | |
|
| |||||||
| rs3804100 | CC | 0 (0) | 2 (0.03) | Ref | |||
| CT | 5 (0.09) | 14 (0.23) | 1.90 | 0.08–46.11 | 0.69 | 0.40 | |
| TT | 51 (0.91) | 46 (0.74) | 5.54 | 0.26–118.36 | 2.17 | 0.14 | |
| CT + TT | 56 (1) | 60 (0.97) | 4.67 | 0.22–99.38 | 1.84 | 0.18 | |
| C | 5 (0.04) | 18 (0.15) | Ref | ||||
| T | 107 (0.96) | 106 (0.85) | 3.63 | 1.30–10.14 | 6.76 | 0.009 | |
|
| |||||||
| rs4696480 | AA | 10 (0.19) | 19 (0.35) | Ref | |||
| AT | 26 (0.49) | 19 (0.35) | 2.60 | 0.99–6.84 | 3.83 | 0.050 | |
| TT | 17 (0.32) | 16 (0.30) | 2.02 | 0.72–5.63 | 1.82 | 0.178 | |
| AT + TT | 43 (0.81) | 35 (0.65) | 2.33 | 0.96–5.66 | 3.60 | 0.058 | |
| A | 46 (0.43) | 57 (0.53) | Ref | ||||
| T | 60 (0.57) | 51 (0.47) | 1.46 | 0.85–2.50 | 1.89 | 0.17 | |
|
| |||||||
| Above 57 years | |||||||
|
| |||||||
| rs3804099 | CC | 12 (0.21) | 8 (0.20) | Ref | |||
| CT | 26 (0.47) | 18 (0.44) | 0.97 | 0.33–2.83 | 0.00 | 0.95 | |
| TT | 18 (0.32) | 15 (0.37) | 0.807 | 0.26–2.47 | 0.15 | 0.70 | |
| Supplementary 1: primers sequences used for real time PCR Supplementary 2 | |||||||
| CT + TT | 44 (0.79) | 33 (0.80) | 0.89 | 0.33–2.42 | 0.05 | 0.82 | |
| C | 50 (0.45) | 34 (0.41) | Ref | ||||
| 62 (0.55) | 48 (0.59) | 0.88 | 0.49–1.56 | 0.19 | 0.66 | ||
|
| |||||||
| rs3804100 | CC | 2 (0.03) | 0 (0) | Ref | |||
| CT | 8 (0.14) | 5 (0.12) | 0.31 | 0.01–7.74 | 1.15 | 0.28 | |
| TT | 48 (0.83) | 36 (0.88) | 0.27 | 0.01–5.705 | 1.47 | 0.22 | |
| CT + TT | 56 (0.97) | 41 (1) | 0.27 | 0.01–5.82 | 1.44 | 0.23 | |
| C | 12 (0.10) | 5 (0.06) | Ref | ||||
| T | 104 (0.90) | 77 (0.94) | 0.56 | 0.19–1.66 | 1.10 | 0.293 | |
|
| |||||||
| rs4696480 | AA | 17 (0.32) | 6 (0.16) | Ref | |||
| AT | 23 (0.43) | 22 (0.58) | 0.37 | 0.12–1.10 | 3.27 | 0.0703 | |
| TT | 13 (0.25) | 10 (0.26) | 0.46 | 0.13–1.59 | 1.53 | 0.22 | |
| AT + TT | 36 (0.68) | 32 (0.84) | 0.40 | 0.14–1.13 | 3.11 | 0.08 | |
| A | 57 (0.54) | 34 (0.45) | Ref | ||||
| T | 49 (0.46) | 42 (0.55) | 0.70 | 0.39–1.26 | 1.45 | 0.23 | |
Note: Bold values are significant.
Abbreviations: Ref, reference; SNP, single-nucleotide polymorphism.
We examined the association between colon cancer risk and individual SNPs based on the gender of the patient. The genotype distributions in male (n=65) and female (n=49) patients with colon cancer were compared with those of the respective male and female control subjects (Table 5). Only rs3804099 showed a significant association in female patients with colon cancer with the TT genotype (OR, 3.79; 95% CI, 1.17–12.30; P=0.02). In this group of females, the phenotype T was dominant in 62% of the patients as compared to 44% in the control (OR, 2.09; 95% CI, 1.15–3.79; P=0.01). The genotype and allele frequencies for rs4696480 and rs3804100 were similar between female patients and female controls (Table 5).
Table 5.
Genotype frequencies of TLR2 gene polymorphism in colorectal cancer male/female vs control
| SNP ID | Genotype | Colon, n (frequency) | Controls, n (frequency) | OR | 95% CI | χ2 value | P-value* |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Male vs control | |||||||
|
| |||||||
| rs3804099 | CC | 10 (0.16) | 13 (0.22) | Ref | |||
| CT | 31 (0.50) | 26 (0.45) | 1.55 | 0.58–4.11 | 0.78 | 0.38 | |
| TT | 21 (0.34) | 19 (0.33) | 1.43 | 0.51–4.03 | 0.48 | 0.49 | |
| CT + TT | 52 (0.84) | 45 (0.78) | 1.50 | 0.60–3.75 | 0.76 | 0.38 | |
| C | 51 (0.41) | 52 (0.45) | Ref | ||||
| T | 73 (0.59) | 64 (0.55) | 1.16 | 0.70–1.94 | 0.33 | 0.56 | |
|
| |||||||
| rs3804100 | CC | 1 (0.02) | 2 (0.04) | Ref | |||
| CT | 8 (0.12) | 9 (0.15) | 1.78 | 0.13–23.52 | 0.19 | 0.66 | |
| TT | 56 (0.86) | 48 (0.81) | 2.33 | 0.20–26.54 | 0.49 | 0.48 | |
| CT + TT | 64 (0.98) | 57 (0.86) | 2.25 | 0.20–25.43 | 0.45 | 0.50 | |
| C | 10 (0.08) | 13 (0.11) | Ref | ||||
| T | 120 (0.92) | 105 (0.89) | 1.49 | 0.63–3.53 | 0.81 | 0.37 | |
|
| |||||||
| rs4696480 | AA | 19 (0.32) | 14 (0.27) | Ref | |||
| AT | 25 (0.42) | 23 (0.44) | 0.80 | 0.33–1.96 | 0.24 | 0.63 | |
| TT | 15 (0.26) | 15 (0.29) | 0.74 | 0.27–1.99 | 0.36 | 0.55 | |
| AT + TT | 40 (0.68) | 38 (0.73) | 0.78 | 0.34–1.76 | 0.37 | 0.54 | |
| A | 63 (0.53) | 51 (0.49) | Ref | ||||
| T | 55 (0.47) | 53 (0.51) | 0.84 | 0.50–1.42 | 0.42 | 0.52 | |
|
| |||||||
| Female vs control | |||||||
|
| |||||||
| rs3804099 | CC | 9 (0.18) | 13 (0.31) | Ref | 0.46–3.748 | 0.25 | 0.62 |
| CT | 19 (0.39) | 21 (0.50) | 1.31 | 1.17–12.30 | 5.13 | 0.023* | |
| TT | 21 (0.43) | 8 (0.19) | 3.79 | 0.75–5.28 | 1.95 | 0.16 | |
| CT + TT | 40 (0.82) | 29 (0.69) | 1.99 | 1.163.79 | 6.03 | 0.014* | |
| C | 37 (0.38) | 47 (0.56) | Ref | ||||
| T | 61 (0.62) | 37 (0.44) | 2.09 | ||||
|
| |||||||
| rs3804100 | CC | 1 (0.02) | 0 (0) | Ref | 0.007–5.60 | 1.61 | 0.20 |
| CT | 5 (0.10) | 9 (0.21) | 0.19 | 0.02–10.97 | 0.76 | 0.38 | |
| TT | 43 (0.88) | 33 (0.79) | 0.43 | 0.02–9.59 | 0.87 | 0.35 | |
| CT + TT | 48 (0.98) | 42 (1) | 0.38 | 0.56–4.39 | 0.72 | 0.40 | |
| C | 7 (0.08) | 9 (0.11) | Ref | ||||
| T | 91 (0.92) | 75 (0.89) | 1.56 | ||||
|
| |||||||
| rs4696480 | AA | 8 (0.17) | 10 (0.26) | Ref | |||
| AT | 24 (0.51) | 18 (0.48) | 1.67 | 0.55–5.07 | 0.82 | 0.37 | |
| TT | 15 (0.32) | 10 (0.26) | 1.88 | 0.55–6.39 | 1.02 | 0.317 | |
| AT + TT | 39 (0.83) | 28 (0.74) | 1.74 | 0.61–4.97 | 1.09 | 0.30 | |
| A | 40 (0.43) | 38 (0.50) | Ref | ||||
| T | 54 (0.57) | 38 (0.50) | 1.35 | 0.74–2.48 | 0.94 | 0.33 | |
Note:
P<0.05.
Abbreviations: Ref, reference; SNP, single-nucleotide polymorphism.
We also investigated the possible association of colon cancer risk with individual SNPs based on tumor location. Our results revealed no significant association with tumor location status. Tumor location did not appear to affect the genotype frequencies in the three TLR2 gene polymorphisms (Table 6). Finally, we determined the prediction of the effects of mutations found on the SNPs on splicing phenotype and our results shows that using Spliceman the SNP rs3804099, rs3804100, and rs4696480, have 79%, 66%, and 56% probability respectively, to affect the TLR2 pre mRNA splicing, and using MutationTaster predictor it shows that, the SNP rs3804099 makes the existing splice site get stronger, while rs3804100 results show activation of an additional splice site with a score of 0.35 (Table 7).
Table 6.
Genotype frequencies of TLR2 gene polymorphism in colorectal cancer tumor located in
| SNP ID | Genotype | Colon, n (frequency) | Controls, n (frequency) | OR | 95% CI | χ2 value | P-value* |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Colon area vs control | |||||||
|
| |||||||
| rs3804099 | CC | 14 (0.21) | 27 (0.26) | Ref | |||
| CT | 27 (0.42) | 47 (0.47) | 1.11 | 0.50–2.47 | 0.06 | 0.80 | |
| TT | 24 (0.37) | 28 (0.27) | 1.65 | 0.71–3.85 | 1.37 | 0.24 | |
| CT + TT | 51 (0.79) (0.) | 75 (0.74) | 1.31 | 0.63–2.74 | 0.52 | 0.47 | |
| C | 55 (0.42) | 101 (0.49) | Ref | ||||
| T | 75 (0.58) | 103 (0.51) | 1.34 | 0.86–2.08 | 1.65 | 0.20 | |
|
| |||||||
| rs3804100 | CC | 2 (0.03) | 2 (0.02) | Ref | |||
| CT | 7 (0.10) | 19 (0.18) | 0.37 | 0.04–3.14 | 0.88 | 0.35 | |
| TT | 60 (0.87) | 82 (0.80) | 0.73 | 0.10–5.34 | 0.10 | 0.76 | |
| CT + TT | 67 (0.97) | 101 (0.98) | 0.66 | 0.09–4.83 | 0.17 | 0.68 | |
| C | 11 (0.08) | 23 (0.11) | Ref | ||||
| T | 127 (0.92) | 183 (0.89) | 1.45 | 0.68–3.08 | 0.95 | 0.33 | |
|
| |||||||
| rs4696480 | AA | 15 (0.25) | 25 (0.27) | Ref | |||
| AT | 25 (0.42) | 41 (0.45) | 1.02 | 0.45–2.27 | 0.00 | 0.97 | |
| TT | 20 (0.33) | 26 (0.28) | 1.28 | 0.54–3.05 | 0.32 | 0.57 | |
| AT + TT | 45 (0.75) | 67 (0.73) | 1.12 | 0.53–2.35 | 0.09 | 0.77 | |
| A | 55 (0.46) | 91 (0.49) | Ref | ||||
| T | 65 (0.54) | 93 (0.51) | 1.16 | 0.73–1.83 | 0.38 | 0.54 | |
|
| |||||||
| Rectum area vs control | |||||||
|
| |||||||
| rs3804099 | CC | 4 (0.12) | 27 (0.26) | Ref | |||
| CT | 18 (0.53) | 47 (0.47) | 2.59 | 0.79–8.43 | 2.60 | 0.11 | |
| TT | 12 (0.35) | 28 (0.27) | 2.89 | 0.83–10.09 | 2.92 | 0.09 | |
| CT + TT | 30 (0.82) | 75 (0.74) | 2.70 | 0.87–8.38 | 3.13 | 0.077 | |
| C | 26 (0.38) | 101 (0.49) | Ref | ||||
| T | 42 (0.62) | 103 (0.51) | 1.58 | 0.90–2.78 | 2.60 | 0.11 | |
|
| |||||||
| rs3804100 | CC | 0 (0.0) | 2 (0.02) | Ref | |||
| CT | 5 (0.15) | 19 (0.18) | 1.41 | 0.06–33.93 | 0.52 | 0.47 | |
| TT | 28 (0.85) | 82 (0.80) | 1.73 | 0.08–37.06 | 0.68 | 0.41 | |
| CT + TT | 33 (1) | 101 (0.98) | 1.65 | 0.08–35.24 | 0.65 | 0.42 | |
| C | 5 (0.08) | 23 (0.11) | Ref | ||||
| T | 61 (0.92) | 183 (0.89) | 1.53 | 0.56–4.20 | 0.70 | 0.40 | |
|
| |||||||
| rs4696480 | AA | 8 (0.23) | 25 (0.27) | Ref | |||
| AT | 19 (0.56) | 41 (0.45) | 1.45 | 0.55–3.80 | 0.57 | 0.45 | |
| TT | 7 (0.21) | 26 (0.28) | 0.84 | 0.27–2.67 | 0.09 | 0.77 | |
| AT + TT | 26 (0.77) | 67 (0.73) | 1.21 | 0.49–3.0 | 0.17 | 0.68 | |
| A | 35 (0.51) | 91 (0.49) | Ref | ||||
| T | 33 (0.49) | 93 (0.51) | 0.92 | 0.53–1.6 | 0.08 | 0.78 | |
Note:
P<0.05.
Abbreviations: Ref, reference; SNP, single-nucleotide polymorphism.
Table 7.
In silico prediction of TLR2 nucleotide variation on splicing phenotype
| TLR2 SNP | Point mutation | Sliceman | MutationTaster | |||||
|---|---|---|---|---|---|---|---|---|
| Wildtype (wt) | Mutation (mt) | L1 distance (AU) | Ranking (L1) (%) | Effect | Score | wt detection sequence | ||
| rs3804099 | cagaa(c/t)gtaag | aacgta | aatgta | 35,539 | 70 | Donor increased | wt: 0.26/mu: 0.41 | wt: CAGAATGTAAGTCAT mu: CAGAACGTAAGTCAT |
| rs3804100 | cacag(c/t)gtaac | cagcgt | cagtgt | 34,758 | 66 | Donor gained | 0.35 | mu: GAATACACAGCGTAA |
| rs4696480 | tctgg(a/t)gaggg | ctggag | ctggtg | 32,723 | 56 | VNA | VNA | VNA |
Note: L1: distance to its corresponding bin (ie, a comparison between two hexamers that resulted in low L1 distance would be assigned with a low percentile rank).
Abbreviations: VNA, applicable where the values were not obtained; SNP, single-nucleotide polymorphism.
Discussion
In previous studies,36 the association between cancer and inflammation was documented. Indeed, colon cancer is known to occur more frequently in people suffering from inflammatory bowel disease.36 In patients with ulcerative colitis, the risk of developing colon cancer is twofold higher than in normal patients.37 Cancer development is associated with a dysfunctional immune system, as patients with immunodeficiency present a twofold to threefold increased risk of cancer.38 Because TLR2 is known to be overexpressed in epithelial cells, which are part of the first line of defense against foreign antigens, it may be a major player in colon cancer development. In the present study, we confirm that TLR2 was prominent on the surface epithelium, but not in most colonic stromal cells. In addition, epithelial colon cancer cells showed higher expression of these receptors in their cytoplasmic membranes. Previous studies reported the expression of TLRs including TLR2 in the epithelium of various organs such as the cervix, uterus, and vagina under specific circumstances such as infection.39 However, our study demonstrated that TLR2 expression level is significantly decreased in colon tumor tissues compared to in matched normal colon tissues. This suggests that in cancerous tissues, epithelial cells have lost the defense and immune roles of TLR2, as its activation was found to protect against colitis-associated colon cancer as suggested by the inhibition of tumor development in mice.40 Several explanations have been suggested for the down-regulation of TLR2 expression in colon cancer tissues, such as epigenetic modifications, SNPs, or aberrant micro-RNA. Changes in TLRs that result from polymorphisms are often associated with susceptibility to various infectious diseases.41 In addition, TLR2 rs4696480 is located in the promoter region (Table S1) and therefore may be important in the transcriptional regulation of TLR2 expression. Although TLR2 rs3804099 and TLR2 rs3804100 are located in an exonic region, a mutation in this region may affect TLR2 expression. Polymorphisms in coding regions are currently thought to be relevant in disease mechanisms regulating gene expression. Based on our findings, further investigations should be conducted to determine the functional mechanism of these TLR2 SNPs during carcinogenesis. Recently, many studies have investigated the association between TLR polymorphisms and cancer risk,42 but the results are contradictory and inconclusive. On analyzing different studies on TLR activation, it appears that TLR activation can have two opposing effects, such as an anticancer effect; in a previous study, we demonstrated that activation of TLRs can inhibit carcinogenesis,43–46 and in contrast, TLR activation may lead to immunosuppression because of chronic inflammation, which can contribute to cancer predisposition and progression.47 Therefore, SNPs may affect TLR2 expression and TLR2 activity by altering transcription factor binding and mRNA stability of the sites.48 However, the mechanism for SNPs and TLR2 activity remains unclear.
In the current study, we found that TLR2 rs3804099 and TLR2 rs4696480 were closely associated with susceptibility to colon cancer, while TLR2 (rs3804100 C>T) showed no association in Saudi Arabian population. However, in our study sub-population as well for genotype and allele frequencies, the double mutant TT in TLR2 rs3804099 and TLR2 rs3804100 showed a significantly low frequency in normal control subjects compared to in colon cancer patients, indicating this genotype or allele is a risk factor in the Saudi population. Although, rs3804099 results in synonymous mutation (Asn199Asn), C>T nucleotide variation has a predicted 70% probability of affecting TLR2 mRNA splicing phenotype, and this results in changes in protein expression, conformation, and function. Genome wide association studies have shown that there is a substantial contribution of synonymous SNPs to human disease risk and other complex traits.49 Tchórzewski et al50 confirmed the expression of TLR2 and TLR4 receptor proteins on colon cancer cells using immunohistochemistry. They also reported that TLR4 might be responsible for tumor development under LPS stimulation.
Variant genotypes of TLR2 have been shown to be correlated with the susceptibility and protective action against inflammatory disease in various ethnic populations. TLR2 rs3804099 was reported to influence susceptibility to gastric cancer,51 hepatocellular carcinoma,41 papillary thyroid cancer, and clinicopathological features in a Korean population.26 To date, only one study has assessed the effect of TLR2 rs3804099 on the risk of colon cancer. Slattery et al15 reported that this SNP is involved in colon or rectal cancer in a population living in the Twin Cities metropolitan area. Slattery et al15 suggest that genetic variation in TLR2, TLR3, and TLR4 may influence the development of colon cancer as well as influence of survival after diagnosis with colon cancer. They reported that TLR2 rs7656411 and rs3804099 interact with NSAID use and cigarette smoking to enhance risk of colon cancer and few SNPs in TLR2 and TLR4 were significantly associated with colon cancer survival. The present study clearly showed that carriers of the CT or TT genotypes of this SNP had a 45% and 38% increased risk of developing colon cancer. Interestingly, TLR2 rs3804099 has been associated with the susceptibility to colon cancer in Saudi females aged less than 57 years. Additionally, this polymorphism was associated with pulmonary tuberculosis in female patients from South India.52 Why these genotypes contribute to the susceptibility to colon cancer in younger female patients remains unclear. We suggest that differences in the expression of sex hormones and genetic variability between males and females contribute to the differential incidence of colon cancer among these subjects. The TLR2 rs3804099 polymorphism is associated with colon cancer in women aged less than 50 years and is closely linked with increased levels of female sex hormones during the pre-menopausal period. In this regard, for the same SNP, previous studies have suggested that significant differences exist in tuberculosis incidence between males and females.53 The role of the estrogen and progesterone rates in protecting against colon cancer in women has been reported previously.54–57 In this context, polymorphisms should be investigated in pre- and post-menopausal women to clarify this phenomenon.
Interestingly, TLR2 rs3804100 was linked to early-onset colon cancer (before the age of 57 years) in this ethnic group. However, this study revealed no association between TLR2 rs4696480 and colon cancer development in Saudi Arabia. However, this SNP is involved in the development of other cancers.15,20,25,51 We concluded that TLR2 plays an active role in the innate immune system, and the presence of these genotypic alterations may lead to dysregulation of TLRs at the protein level. Such deregulation may contribute to decreasing epithelial cell innate immunity, leading to the initiation and development of cancer such as colon cancer.
Conclusion
Genetic variations in innate immunity genes may have a significant impact on cancer etiology. Thus, SNPs in TLR2 may promote colon cancer development as well as colon cancer tumor progression and metastasis. The new genetic markers identified in Toll-like receptors are potential therapeutic targets and may be useful indicators for both the prevention and therapy of colon cancer in the Saudi Arabian population. Further mechanistic studies on the role of SNPs and TLR activity are required. Furthermore, comparative studies of different populations may be helpful for determining whether TLR2 deregulation is specific to the Saudi Arabian population.
Supplementary materials
Table S1.
Primer sequences used for real time PCR
| Gene | Primer sequence (5′ to 3′) | Amp size (bp) | Tm (°C) |
|---|---|---|---|
| TLR2 | Fw: 5′-GCCTCTCCAAGGAAGAATCC-3′ Rv: 5′-TCCTGTTGTTGGACAGGTCA-3′ |
144 bp | 60 |
| GAPDH | Fw: 5′-GGTATCGTCGAAGGACTCATGAC-3′ Rv: 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ |
180 bp | 60 |
Abbreviations: Amp, amplicon; bp, base pair; FW, forward; Rv, reverse; Tm, temperature.
Table S2.
Characteristics of selected polymorphisms involved in the Toll-like receptor 2
| SNP ID | Chr/position | Nucleotide change | Region | Minor allele frequency (%) | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| rs3804099 Asn199Asn |
4/153703504 | c.597T>C | Exon | 60 | 55 |
| rs3804100 Ser450Ser |
4/153704257 | +1350T>C | Exon | 92 | 89 |
| rs4696480 | 4/153685974 | −16934T>A | Promoter | 51 | 51 |
Abbreviation: SNP, single nucleotide polymorphism.
Acknowledgments
This project was supported by funding from a research group program (RGP-VPP-260) from the Deanship of Scientific Research, King Saud University, Kingdom of Saudi Arabia.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2(8):675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124(4):823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Huang B, Zhao J, Unkeless JC, Feng ZH, Xiong H. TLR signaling by tumor and immune cells: a double-edged sword. Oncogene. 2008;27(2):218–224. doi: 10.1038/sj.onc.1210904. [DOI] [PubMed] [Google Scholar]
- 4.Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin Immunol. 2007;19(1):3–10. doi: 10.1016/j.smim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Huang B, Zhao J, Li H, et al. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005;65(12):5009–5014. doi: 10.1158/0008-5472.CAN-05-0784. [DOI] [PubMed] [Google Scholar]
- 6.Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111(7):927–930. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 7.Beutler B. Neo-ligands for innate immune receptors and the etiology of sterile inflammatory disease. Immunol Rev. 2007;220:113–128. doi: 10.1111/j.1600-065X.2007.00577.x. [DOI] [PubMed] [Google Scholar]
- 8.Harberts E, Gaspari AA. TLR signaling and DNA repair: are they associated? J Invest Dermatol. 2013;133(2):296–302. doi: 10.1038/jid.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald KA, Rowe DC, Barnes BJ, et al. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198(7):1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447(7147):972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 11.Bhide MR, Mucha R, Mikula I, et al. Novel mutations in TLR genes cause hyporesponsiveness to Mycobacterium avium subsp. paratuberculosis infection. BMC Genet. 2009;10(21):21. doi: 10.1186/1471-2156-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warner N, Núñez G. MyD88: a critical adaptor protein in innate immunity signal transduction. J Immunol. 2013;190(1):3–4. doi: 10.4049/jimmunol.1203103. [DOI] [PubMed] [Google Scholar]
- 13.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168(2):554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 14.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17(1):1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 15.Slattery ML, Herrick JS, Bondurant KL, Wolff RK. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer. 2012;130(12):2974–2980. doi: 10.1002/ijc.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semlali A, Reddy Parine N, Arafah M, et al. Expression and polymorphism of Toll-like receptor 4 and effect on NF-κB mediated inflammation in colon cancer patients. PLoS One. 2016;11(1):e0146333. doi: 10.1371/journal.pone.0146333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens VL, Hsing AW, Talbot JT, et al. Genetic variation in the toll-like receptor gene cluster (TLR10-TLR1-TLR6) and prostate cancer risk. Int J Cancer. 2008;123(11):2644–2650. doi: 10.1002/ijc.23826. [DOI] [PubMed] [Google Scholar]
- 18.de Oliveira JG, Silva AE. Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J Gastroenterol. 2012;18(11):1235–1242. doi: 10.3748/wjg.v18.i11.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne AN, Carneiro F, O’Morain C, Offerhaus GJ. Nature meets nurture: molecular genetics of gastric cancer. Hum Genet. 2009;126(5):615–628. doi: 10.1007/s00439-009-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tahara T, Arisawa T, Wang F, et al. Toll-like receptor 2 -196 to 174del polymorphism influences the susceptibility of Japanese people to gastric cancer. Cancer Sci. 2007;98(11):1790–1794. doi: 10.1111/j.1349-7006.2007.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priyadarshini A, Chakraborti A, Mandal AK, Singh SK. Asp299Gly and Thr399Ile polymorphism of TLR-4 gene in patients with prostate cancer from North India. Indian J Urol. 2013;29(1):37–41. doi: 10.4103/0970-1591.109982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing JJ, Li M, Yuan Y. Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms in cancer: a meta-analysis. Gene. 2012;499(2):237–242. doi: 10.1016/j.gene.2012.03.045. [DOI] [PubMed] [Google Scholar]
- 23.Georgel P, Macquin C, Bahram S. The heterogeneous allelic repertoire of human toll-like receptor (TLR) genes. PLoS One. 2009;4(11):e7803. doi: 10.1371/journal.pone.0007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011;2011:405310. doi: 10.1155/2011/405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal RK, George GP, Mittal RD. Association of Toll-like receptor (TLR) 2, 3 and 9 genes polymorphism with prostate cancer risk in North Indian population. Mol Biol Rep. 2012;39(7):7263–7269. doi: 10.1007/s11033-012-1556-5. [DOI] [PubMed] [Google Scholar]
- 26.Kim MK, Park SW, Kim SK, et al. Association of Toll-like receptor 2 polymorphisms with papillary thyroid cancer and clinicopathologic features in a Korean population. J Korean Med Sci. 2012;27(11):1333–1338. doi: 10.3346/jkms.2012.27.11.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caws M, Thwaites G, Dunstan S, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4(3):e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng WY, Yong Z, Yun Z, Hong H, Hai LL. Toll-like receptor 4 gene polymorphisms and susceptibility to colorectal cancer: a meta-analysis and review. Arch Med Sci. 2015;11(4):699–707. doi: 10.5114/aoms.2015.53288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Li J, Xie W, Zhang W, Chang Y. Toll-like receptor 2 gene polymorphisms and cancer susceptibility: a meta-analysis. Neoplasma. 2013;60(4):459–467. doi: 10.4149/neo_2013_060. [DOI] [PubMed] [Google Scholar]
- 30.Semlali A, Leung KP, Curt S, Rouabhia M. Antimicrobial decapeptide KSL-W attenuates Candida albicans virulence by modulating its effects on Toll-like receptor, human β-defensin, and cytokine expression by engineered human oral mucosa. Peptides. 2011;32(5):859–867. doi: 10.1016/j.peptides.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Semlali A, Jacques E, Plante S, et al. TGF-beta suppresses EGF-induced MAPK signaling and proliferation in asthmatic epithelial cells. Am J Respir Cell Mol Biol. 2008;38(2):202–208. doi: 10.1165/rcmb.2007-0031OC. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 33.Alanazi M, Pathan AA, Abduljaleel Z, et al. Association between PARP-1 V762A polymorphism and breast cancer susceptibility in Saudi population. PLoS One. 2013;8(12):e85541. doi: 10.1371/journal.pone.0085541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A. 2011;108(27):11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim KH, Fairbrother WG. Spliceman – a computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics. 2012;28(7):1031–1032. doi: 10.1093/bioinformatics/bts074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 37.Pohl C, Hombach A, Kruis W. Chronic inflammatory bowel disease and cancer. Hepatogastroenterology. 2000;47(31):57–70. [PubMed] [Google Scholar]
- 38.Lavolé A, Toper C, Belmont L, Ruppert AM, Wislez M, Cadranel J. Lung cancer and HIV infection. Rev Mal Respir. 2014;31(2):133–141. doi: 10.1016/j.rmr.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Chen C, Zibiao H, Ming Z, et al. Expression pattern of Toll-like receptors (TLRs) in different organs and effects of lipopolysaccharide on the expression of TLR 2 and 4 in reproductive organs of female rabbit. Dev Comp Immunol. 2014;46(2):341–348. doi: 10.1016/j.dci.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Lowe EL, Crother TR, Rabizadeh S, et al. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS One. 2010;5(9):e13027. doi: 10.1371/journal.pone.0013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junjie X, Songyao J, Minmin S, et al. The association between Toll-like receptor 2 single-nucleotide polymorphisms and hepatocellular carcinoma susceptibility. BMC Cancer. 2012;12:57. doi: 10.1186/1471-2407-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, Hickford JG, Fang Q, Lin YS. Allelic variation of the ovine Toll-like receptor 4 gene. Dev Comp Immunol. 2007;31(2):105–108. doi: 10.1016/j.dci.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Koido S, Homma S, Okamoto M, et al. Improved immunogenicity of fusions between ethanol-treated cancer cells and dendritic cells exposed to dual TLR stimulation. Oncoimmunology. 2013;2(8):e25375. doi: 10.4161/onci.25375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamoto M, Sato M. Toll-like receptor signaling in anti-cancer immunity. J Med Invest. 2003;50(1–2):9–24. [PubMed] [Google Scholar]
- 45.Sato S, Sugiyama M, Yamamoto M, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171(8):4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 46.Killeen SD, Wang JH, Andrews EJ, Redmond HP. Exploitation of the Toll-like receptor system in cancer: a doubled-edged sword? Br J Cancer. 2006;95(3):247–252. doi: 10.1038/sj.bjc.6603275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrade CF, Kaneda H, der S, et al. Toll-like receptor and cytokine gene expression in the early phase of human lung transplantation. J Heart Lung Transplant. 2006;25(11):1317–1323. doi: 10.1016/j.healun.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 48.Thomas KH, Meyn P, Suttorp N. Single nucleotide polymorphism in 5′-flanking region reduces transcription of surfactant protein B gene in H441 cells. Am J Physiol Lung Cell Mol Physiol. 2006;291(3):L386–L390. doi: 10.1152/ajplung.00193.2005. [DOI] [PubMed] [Google Scholar]
- 49.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 50.Tchórzewski M, Lewkowicz P, Dziki A, Tchórzewski H. Expression of toll-like receptors on human rectal adenocarcinoma cells. Arch Immunol Ther Exp. 2014;62(3):247–251. doi: 10.1007/s00005-013-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng HM, Pan KF, Zhang Y, et al. The correlation between polymorphisms of Toll-like receptor 2 and Toll-like receptor 9 and susceptibility to gastric cancer. Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45(7):588–592. [PubMed] [Google Scholar]
- 52.Selvaraj P, Harishankar M, Singh B, Jawahar MS, Banurekha VV. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis. 2010;90(5):306–310. doi: 10.1016/j.tube.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 53.Neyrolles O, Quintana-Murci L. Sexual inequality in tuberculosis. PLoS Med. 2009;6(12):e1000199. doi: 10.1371/journal.pmed.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marino M. Xenoestrogens challenge 17β-estradiol protective effects in colon cancer. World J Gastrointest Oncol. 2014;6(3):67–73. doi: 10.4251/wjgo.v6.i3.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barzi A, Lenz AM, Labonte MJ, Lenz HJ. Molecular pathways: estrogen pathway in colorectal cancer. Clin Cancer Res. 2013;19(21):5842–5848. doi: 10.1158/1078-0432.CCR-13-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Azzawi F, Wahab M. Estrogen and colon cancer: current issues. Climacteric. 2002;5(1):3–14. [PubMed] [Google Scholar]
- 57.Hendrickse CW, Jones CE, Donovan IA, Neoptolemos JP, Baker PR. Oestrogen and progesterone receptors in colorectal cancer and human colonic cancer cell lines. Br J Surg. 1993;80(5):636–640. doi: 10.1002/bjs.1800800531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primer sequences used for real time PCR
| Gene | Primer sequence (5′ to 3′) | Amp size (bp) | Tm (°C) |
|---|---|---|---|
| TLR2 | Fw: 5′-GCCTCTCCAAGGAAGAATCC-3′ Rv: 5′-TCCTGTTGTTGGACAGGTCA-3′ |
144 bp | 60 |
| GAPDH | Fw: 5′-GGTATCGTCGAAGGACTCATGAC-3′ Rv: 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ |
180 bp | 60 |
Abbreviations: Amp, amplicon; bp, base pair; FW, forward; Rv, reverse; Tm, temperature.
Table S2.
Characteristics of selected polymorphisms involved in the Toll-like receptor 2
| SNP ID | Chr/position | Nucleotide change | Region | Minor allele frequency (%) | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| rs3804099 Asn199Asn |
4/153703504 | c.597T>C | Exon | 60 | 55 |
| rs3804100 Ser450Ser |
4/153704257 | +1350T>C | Exon | 92 | 89 |
| rs4696480 | 4/153685974 | −16934T>A | Promoter | 51 | 51 |
Abbreviation: SNP, single nucleotide polymorphism.