Summary
Objective: To evaluate the feasibility of automated dose and adverse event information retrieval in supporting the identification of safety patterns.
Methods: We extracted all rabbit Anti-Thymocyte Globulin (rATG) reports submitted to the United States Food and Drug Administration Adverse Event Reporting System (FAERS) from the product’s initial licensure in April 16, 1984 through February 8, 2016. We processed the narratives using the Medication Extraction (MedEx) and the Event-based Text-mining of Health Electronic Records (ETHER) systems and retrieved the appropriate medication, clinical, and temporal information. When necessary, the extracted information was manually curated. This process resulted in a high quality dataset that was analyzed with the Pattern-based and Advanced Network Analyzer for Clinical Evaluation and Assessment (PANACEA) to explore the association of rATG dosing with post-transplant lymphoproliferative disorder (PTLD).
Results: Although manual curation was necessary to improve the data quality, MedEx and ETHER supported the extraction of the appropriate information. We created a final dataset of 1,380 cases with complete information for rATG dosing and date of administration. Analysis in PANACEA found that PTLD was associated with cumulative doses of rATG >8 mg/kg, even in periods where most of the submissions to FAERS reported low doses of rATG.
Conclusion: We demonstrated the feasibility of investigating a dose-related safety pattern for a particular product in FAERS using a set of automated tools.
Citation: Botsis T, Foster M, Arya N, Kreimeyer K, Pandey A, Arya D. Application of natural language processing and network analysis techniques to post-market reports for the evaluation of dose-related anti-thymocyte globulin safety patterns. Appl Clin Inform 2017; 8: 396–411 https://doi.org/10.4338/ACI-2016-10-RA-0169
Keywords: Natural language processing, network analysis, postmarketing product surveillance, information retrieval, data visualization
1. Introduction
Rabbit Anti-Thymocyte Globulin (or rATG) was first licensed for clinical use in April 16, 1984 (in Lyon, France) and plays an important role in the management of solid organ transplant patients and other therapeutic areas, such as the management of aplastic anemia [ 1 ]. Despite the benefits of rATG, concerns regarding the potential for infectious complications, malignancy, and post-transplant lymphoproliferative disorder (PTLD) have resulted in a shift toward lower rATG dosing to refine its risk-benefit balance [ 2 ]. PTLD has been reported in follow-up of patients who received rATG [ 3 ]. A number of studies have examined the empirical justification of the shift to lower rATG dosing. Marks et al. conducted a systematic review of papers published between 1999 and 2009 for trials of rATG in kidney and heart transplant recipients and concluded that higher cumulative doses showed no association with risk of PTLD [ 4 ]. A similar finding was reported after the analysis of a large-scale dataset from the TAILOR registry [ 5 ]. Mohty et al. performed an extensive review for rATG and stated that “the risk of PTLD and malignancy associated with modern rATG induction regimens may be less of a concern than during the high-dose era, but confirmatory data are required” [ 2 ]. Despite these findings, the theoretical risks associated with high dose rATG administration motivated us to explore its potential association with PTLD. We conducted this exploration using the post-market rATG reports from the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) for the lifecycle of this product and set of automated tools.
FAERS is a database which contains safety reports of adverse events (AEs) following exposure to drug and biologic products submitted to the FDA [ 6 ]. Each report contains structured fields that include patient demographics, dates, and other coded information, as well as free-text fields which further describe AEs. Important clinical information in the free-text field is coded using Preferred Terms (PTs) from the Medical Dictionary for Regulatory Activities (MedDRA) and leveraged by medical experts and epidemiologists for post-market safety surveillance.
FAERS provides minimal support for information retrieval by automated tools. Recovering detailed product information, such as dose, frequency, and duration of administration often requires considerable manual effort. If this information is missing, incorrectly coded, or represented by contradicting inputs in a structured field, the only way to retrieve the missing information is through manual review of the free-text narrative. While manual review is always an option, FAERS receives about 770,000 reports every year [ 7 ] and it is not feasible to use this method for processing large datasets. There is thus a clear need for automated strategies to assist medical reviewers and epidemiologists with detailed case review.
To address the need for automated review strategies, as well as to explore the potential association of rATG with PTLD, we analyzed all rATG reports submitted to FAERS using a set of automated tools. Specifically, we combined two natural language processing (NLP) tools to extract clinical, temporal, and medication information from the free-text narrative and analyzed this information using network analysis (NA) techniques.
2. Materials and Methods
We queried FAERS on February 8, 2016 for all rATG Individual Safety Reports (ISRs) from the product’s initial licensure to the date of the query. Three name variations for rATG were used in the query: “thymoglobuline”, “thymoglobulin”, and “anti-thymocyte globulin (rabbit) nos”. We processed these reports to retrieve the daily dose information from each ISR (2.1), determine the therapy start date (2.2), calculate the total dose per case (2.3), and evaluate the temporal patterns (2.4). This process is shown in ► Figure 1 .
Fig. 1.
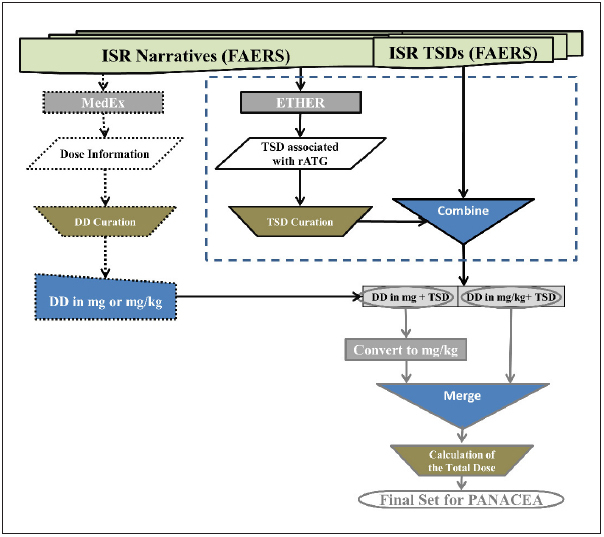
This flow chart shows the multiple steps followed for the automated processing of the Rabbit Anti-Thymocyte Globulin (rATG) reports submitted to the United States Food and Drug Administration Adverse Event Reporting System (FAERS). We initially processed the rATG reports with the Medication Extraction (MedEx) and the Event-based Text-mining of Health Electronic Records (ETHER) systems for the retrieval of the dose and the therapy start date (TSD) information, respectively. The dose information was curated and the daily doses (DD) in either mg or mg/kg were identified (trapezoid with the dotted outline). TSD was also retrieved for those ISRs by using either the values from the corresponding field in FAERS or the curated ETHER TSDs for the ISRs that did not have a TSD value in the corresponding FAERS field; this step of the analysis is depicted with the dotted rectangle and presented in detail in Figure 2. Daily dose information was coupled with the retrieved TSDs. We then converted all doses to mg/kg and merged the cases (only one ISR per case was used in and after this step). In the final step, we manually calculated the total doses and generated the final set that supported the subsequent network analysis in the Pattern-based and Advanced Network Analyzer for Clinical Evaluation and Assessment (PANACEA).
2.1 Daily Dose Retrieval
The first step in our study included the retrieval of the dose information from the rATG reports with a particular focus on the daily dose reported in each narrative (► Figure 1 ). We used the Medication Extraction (MedEx, version 1.3.3) NLP system to extract medication information from the ISR narratives and determine the daily dose for rATG in milligrams (mg) or milligram per kilogram body weight (mg/kg). We chose to use MedEx as it has been shown to perform well at extracting drug names and dose information from discharge summaries with F-measures ranging from 0.932 to 0.960 [ 8 ].
Before being processed by MedEx, each ISR narrative was split into sentences using the period as a separator, and each sentence was linked to the ISR Identification (ID) number. MedEx processed each sentence and generated one or more rows of output depending on the number of different drugs identified in the sentence. Each row in the output contained the following: sentence index, ISR ID and text, drug name, brand name, drug form, strength, dose amount, route, normalized frequency, duration, necessity, Unified Medical Language System (UMLS) Concept Unique Identifier (CUI), RxNorm RxCUI, RxNorm RxCUI for generic name, and generic name. The following is an example of an output row:
92039|12345678–2–20090118$From 21-Feb-2009 to 22-Jan-2009, patient received anti-thymocyte globulin (rabbit) at a dose of 75 mg per day|anti – thymocyte globulin ( rabbit )[73,105]|||75mg[119,124]|||per day[125,132]||||107044|107044|rabbit anti-human t-lymphocyte globulin
Fields are separated using the pipe (“|”) symbol and ISR ID is separated from the sentence text using the “$” symbol. The absence of any text between two pipes denotes that either no information existed (in the original sentence) or MedEx did not extract any value for the corresponding field, e.g. for route in the example above. The position of each string within the sentence is also shown in square brackets with the count starting from 0 (both characters and spaces are included). In the example above, position “0” is the first digit of the ISR ID as this is part of the long string that includes the actual sentence.
After processing in MedEx, we identified output rows that included dose information for rATG by searching the “drug name” and “drug name generic” fields for rATG generic and brand names. We retained all rows with at least one value for the strength, dose amount, frequency, or duration of administration fields.
Subsequently, two reviewers with backgrounds in pharmacology and two reviewers with backgrounds in medical informatics manually curated the dose information in each row by applying the following rules:
Only doses in mg or mg/kg were included; doses with miscellaneous units were excluded.
Only daily doses that were clearly reported or easily calculated were included.
When doses were reported in both mg/kg and mg in the text, all were retained.
When mg/kg was the dose unit, the corresponding value was considered as the daily dose.
The largest dose was used when ≥2 doses with the same unit appeared in the sentence.
The “at a dose” statement was considered equivalent to a daily dose.
The output of this step was a list of ISRs with a daily dose in mg, mg/kg, or both.
It should be noted that only individual sentences were reviewed in this phase, and the overall context of the ISR was not evaluated. Our primary goal in this phase was to automatically extract medication information (daily dose) and quickly refine it to support the next steps of the analysis.
2.2 Therapy Start Date Retrieval
In the second step of our study, we filled the missing Therapy Start Date (TSD) values for the ISRs with an identified daily dose that did not include this information in the corresponding FAERS field. To accomplish this, we processed the free-text narratives with the Event-based Text-mining of Health Electronic Records (ETHER) system and retrieved the rATG mentions with their corresponding exposure dates. ETHER is a rule-based NLP system that extracts clinical features and temporal associations from post-market safety surveillance reports as well as provides case summarization and information visualization [ 9 ]. ETHER’s ability to extract time information and identify temporal relationships in FAERS narratives was previously demonstrated by Wang et al. and is therefore an appropriate tool for generating missing TSD values [ 10 ].
In order to validate exposure dates derived by ETHER, two reviewers compared these dates with dates in the free-text narrative and corrected any discrepancies. After verification, the exposure dates were assigned as TSDs for each ISR. This process was only applied to ISRs with missing FAERS TSD values, and no comparison was made between ETHER derived exposure dates and existing FAERS TSDs. This phase resulted in ISRs containing either (i) a FAERS derived TSD, (ii) an ETHER derived and curated TSD, or (iii) no TSD, as a TSD could not be determined. Only ISRs with either a FAERS derived TSD or an ETHER derived TSD were used going forward.
2.3 Total Dose Calculation
In this step, we calculated the total rATG dose reported for the cases with an identified TSD. This process is complicated by the fact that multiple ISRs may exist for a single case, especially when there are updates following the initial submission. This is represented in FAERS by adding a numeric extension to the case number to specify subsequent submissions to the same case (e.g. CASE #-1, CASE#-2, etc.). The final submission (largest numeric extension) for a case is enriched with all submitted information and is considered as the “best representative” record for that case. To calculate the total dose we used the dose information only from the ISR with the largest extension.
For each of the “best representative” ISRs, we manually reviewed the free-text narrative to determine the total dose administered to a patient (in either mg or mg/kg). When applicable, the total dose was calculated based on the timeline described in the narrative. We then normalized the total doses in mg to mg/kg by dividing the total dose by the Body Weight structured field value in FAERS. The set of ISRs with total rATG dose in mg/kg (hereafter, final set) was then analyzed for temporal patterns.
2.4 Temporal Patterns
The total dose and TSDs in the final set were used to evaluate potential rATG dose-related PTLD patterns over time. The k-means clustering method was used to group TSD into distinct time periods. To select the number of clusters, the within-cluster sum of squares and between-cluster sum of squares were plotted and reviewed for k equals two to ten. k-means was applied using the Hartigan and Wong algorithm implemented in R with a choice of k=4 cluster centers and 100 random starts [ 11 , 12 ]. In the selected cluster assignment, a boundary between adjacent time periods was defined using the midpoint between the maximum observed TSD of the former cluster and the minimum observed TSD of the latter cluster. In the final time period, the date of the last observation was assigned as the ending date of the period.
We then evaluated the associations between the total rATG doses over time and the AEs using the FDA Pattern-based and Advanced Network Analyzer for Clinical Evaluation and Assessment (PANACEA) [ 9 ]. PANACEA is a NA tool that constructs graphs to represent the objects of interest and the relationships between those objects as nodes and edges connecting the nodes, respectively [ 9 ]. PANACEA supports the creation of “element networks” using drug (and other product) exposures and AEs as nodes. Nodes are connected with an edge when they co-appear in at least one report in a report set, and each edge is weighted based on the number of co-occurrences. PANACEA is equipped with multiple basic and advanced NA functionalities, such as the construction of various information visualizations and the evaluation of networks over time. These functionalities allow the processing of big datasets, the identification of safety patterns associated with the administration of a medical product, and the interactive presentation of results to the end users.
We created two groups of subnetworks in PANACEA using rATG doses and MedDRA PTs. The first group was built in a cumulative fashion with one subnetwork for each period containing all cases stamped with a TSD between licensure and the end of that period. The second group of subnetworks was completely disjointed in time and created in a non-cumulative fashion with each case appearing in only a single subnetwork depending on its TSD.
For all subnetworks, the curated total doses (in mg/kg) were split into four groups – [0, 4], (4, 8], (8, 12], and (12,∞) – represented by nodes named “a”, “b”, “c”, and “d”, respectively. The MedDRA PTs for the cases were also represented by nodes in the networks. A pair of nodes was connected when the corresponding elements (PTs and dose ranges) co-occurred in one or more cases. The weight of the connection, i.e. the “edge weight” was equal to the number of cases in which the elements co-occurred. To explore the AE patterns associated with different doses of rATG, we: (i) used two network layouts – island heights and principal components – that supported the visual evaluation of the AE patterns; and (ii) constructed plots with key network centrality metrics – weighted closeness versus weighted eigenvector centrality – that quantified the visual findings. For (ii), we imported the network edge lists into the igraph library, which supports the calculation of the weighted versions of closeness and eigenvector centrality [ 13 ].
The island layout is based on the idea of island heights for NA that has been described by the Pajek team [ 14 ]. PANACEA uses the heaviest edge weight of each node to assign an island height to that node, with the expectation that nodes with large edge weights will be more important within a network. In the visualization, the y-position of each node corresponds to its island height, while the x-position depends on the position of the other nodes it is connected to in the topology. The principal components layout attempts to place the nodes of the graph on meaningful horizontal and vertical axes. The horizontal axis differentiates nodes based on how central they are in the network. Very common and/or important nodes will appear to the left and rare, low importance nodes will appear to the right. The vertical axis differentiates nodes based on their various connections within the graph. The two most structurally different nodes will be placed at the top and bottom (although these two positions are interchangeable), and the remaining nodes are given a vertical height based on the end node to which they are most similar.
Closeness and eigenvector centrality are nodal metrics used to measure the importance of each node in a network. Closeness centrality is a measure of how easily the other nodes in a network can be reached from the selected node, while eigenvector centrality is a measure of a node’s connectedness to other highly connected nodes [ 15 ]. The representation of these nodal metrics in a plot quantified the visual observations and illustrated the most important nodes in each subnetwork.
3. Results
We retrieved 9,722 ISRs from FAERS, which included both initial submissions and follow-up information for 6,493 cases. There were a total of 312,285 (mean±SD: 32±29; range: 1–386) sentences, including some “sentences” with only a single digit, a period, or symbol depending on the way it was submitted to MedEx for processing. This processing retrieved medication information for 96,742 (mean±SD: 11±9; range: 1–132) sentences from 9,135 ISRs (for 5,958 cases). Since many sentences contained more than one drug, the total number of MedEx output rows was 198,863.
3.1 Daily Dose Retrieval
We searched the MedEx output for a list of rATG names provided by a medical expert in a 2-step process by applying the list to:
The “drug generic name” field in the MedEx output rows; 6,502 non-duplicate rows were retrieved from the MedEx output.
The “drug name” field in the remaining rows; 36,029 non-duplicate rows were retrieved from the MedEx output.
Of the 42,531 rows (21.4% of the total rows) containing rATG, 40,677 had at least one value in the dose-related parameters (strength, dose amount, frequency, and duration of administration). These rows included information from 8,651 ISRs that were submitted for 5,610 cases. The manual curation of the dose-related information resulted in 2,831 and 2,258 ISR narratives with daily dose information in mg and mg/kg, respectively.
3.2 Therapy Start Date Retrieval
FAERS included the TSD values for only 56% of the total rATG ISRs (5,457 out of the total 9,722 ISRs). We retrieved the exposure date of the rATG mentions in the narratives by processing each of the 9,722 ISRs with ETHER. We found 5,444 ISRs with at least one rATG stamped with an exposure date. Of these 5,444 ISRs, 4,296 had at least one FAERS TSD and 1,148 did not have any FAERS TSD (► Figure 2 ).
Fig. 2.
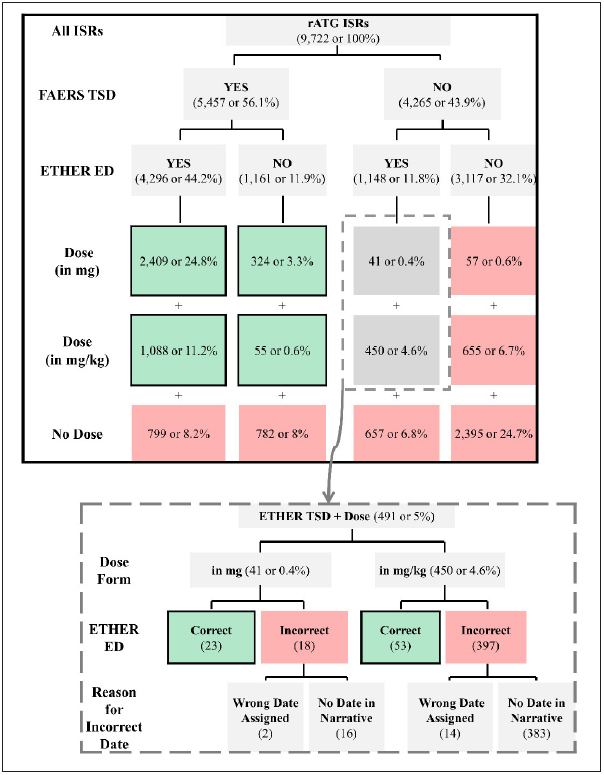
Comparison of the exposure dates (EDs) from the Event-based Text-mining of Health Electronic Records (ETHER) system with the therapy start dates (TSDs) from the United States Food and Drug Administration Adverse Event Reporting System (FAERS) for all individual safety reports (ISRs), and the ISRs with curated daily dose in mg or mg/kg. The ISRs without a FAERS TSD but an ETHER ED (N=491; 41 and 450 with dose information in mg and mg/kg, respectively) are marked with the dotted rectangle and are further analyzed in the embedded image at the bottom of the figure. Only the numbers of the ISRs in the green-shaded boxes were included in the analysis. All percentages have been calculated over the total number of ISRs (N=9,722).
The results for subsets of ISRs with daily dose information in mg and mg/kg are shown in Figure 2 . There were 491 ISRs (41 and 450 ISRs with daily dose information in mg and mg/kg, respectively) with at least one exposure date extracted by ETHER but no FAERS TSD. We reviewed the full narratives of the 491 ISRs to verify that the ETHER-based exposure dates could be substituted for the missing TSDs (► Figure 1 ). The curators found that the ETHER exposure dates were correct in 76 ISRs and incorrect in 415 ISRs. The “incorrect” class included ISRs with both incorrect and unclear statements on the rATG TSD. For the latter, the reviewers evaluated the overall context, decided that no TSD should be used, and marked the corresponding ETHER-based TSD as incorrect for 399 out of the 415 ISRs with daily dose information. The curation process resulted in the identification of 23 and 53 TSDs in addition to the 2,733 (2409+324; ► Figure 2 ) and 1,143 (1088+55; ► Figure 2 ) FAERS TSDs for the ISRs with daily dose information in mg and mg/kg, respectively.
3.3 Total Dose Calculation
In order to focus on specific cases (as opposed to ISRs, which may refer to the same case) we used the “best representative” ISR per case as described in section 2.3. Excluding cases with neither a FAERS TSD nor an ETHER derived TSD resulted in 1,562 and 646 cases with daily dose information in mg or mg/kg, respectively.
Of the 1,562 cases with dose information in mg, 1,154 cases included a body weight value in the corresponding FAERS field. We removed the cases with two different values or without a clear unit for body weight, and found 1,092 cases: 1,081 and 11 with body weight information in kilograms and pounds, respectively. We converted pounds to kilograms and calculated the daily dose in mg/kg for the 1,092 cases. After merging the two files with the MedEx derived and the calculated daily dose in mg/kg, we found 1,735 entries for 1,604 cases – 131 cases with two different entries in either the dose (N=126) or the TSD field (N=1) or both (N=4). The different entries per case were caused by the utilization of the full TSD and dose information contained in all the case ISRs. We selected the maximum dose and earliest exposure date for rATG in each case and created a list of 1,604 unique cases with a TSD and a daily dose in mg/kg.
Manual review of the 1,604 cases resulted in elimination of 188 cases in which it was not possible to determine the total dose due to: lack of duration data (N=59), incomplete dose information (N=73), multiple administrations of rATG over more than a year (N=3), and dose discontinuation without any information about the total dose received to that point (N=53).
In contrast, we included cases in which: rATG had been discontinued but contained information on total dose received to that point (N=35), rATG was administered on non-sequential days within a one month period (N=22), and multiple rATG administrations were described within a one year period (N=43).
Manual review resulted in 1,416 cases with total dose in either mg (N=938) or mg/kg (N=423), or both mg and mg/kg (N=55). For the cases with a total dose in both mg and mg/kg the latter was selected. For the cases with total dose in mg, we followed the same process as above and converted the dose to mg/kg if the corresponding weight values were found in FAERS. Missing weight values for 36 of these cases reduced the size of our final dataset to 1,380 cases containing both a specified rATG total dose in mg/kg and a TSD.
3.4 Temporal patterns
The yearly distribution of the rATG total doses in the final set is shown in ► Figure 3 . Some observations (N=17) were associated with the administration of cumulative doses of rATG ≥30 mg/kg (years 1997, 1999–2001, 2007, and 2009–2012), much higher than most other observations. We applied the k-means clustering method and plotted the within-cluster sum of squares and between-cluster sum of squares for k from two to ten (► Figure 4 ). The elbows at k=4 showed an improvement over k=3. Additionally, k=4 was small enough to facilitate subsequent NA and therefore we decided to use four clusters for our analysis. The time periods identified by the k-means algorithm were: (i) TSD on or before June 7, 2003 (N=71); (ii) TSD from June 8, 2003 to November 9, 2007 (N=201); (iii) TSD from November 10, 2007 to February 14, 2011 (N=636); and (iv) TSD from February 15, 2011 to December 31, 2015 (N=472). The highest rATG doses (“c” and “d” ranges) were mainly reported in the first and third periods (median doses were equal to 6.5 and 6 mg/kg, respectively). Lowest doses (“a” range) were reported in the second period (median dose=5.45 mg/kg). The left-skewed distribution in the most recent period (median dose=5 mg/kg) indicated a trend for the administration of rATG doses in the lower “a” and “b” ranges. We subsequently used PANACEA to analyze the temporal patterns in the four periods.
Fig. 3.
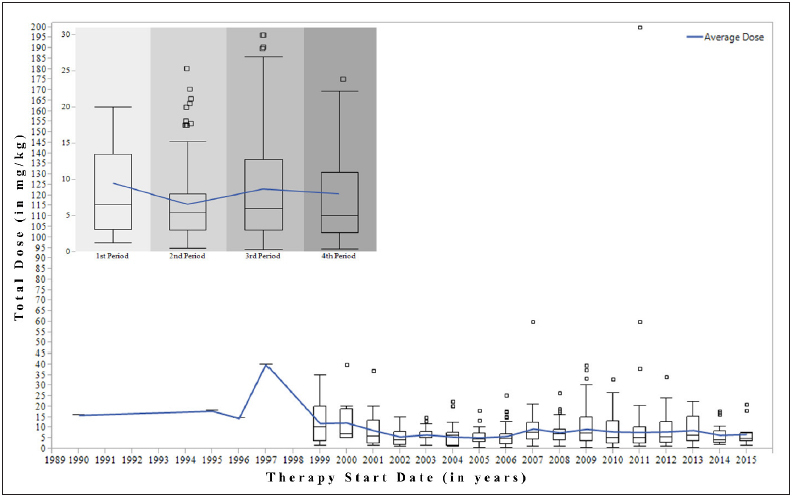
The box plots illustrate the distribution of the total doses in mg/kg since Rabbit Anti-Thymocyte Globulin (rATG) licensure. The therapy start date, either extracted by the Event-based Text-mining of Health Electronic Records (ETHER) system or retrieved from the United States Food and Drug Administration Adverse Event Reporting System (FAERS) has been used as the time parameter. The embedded plot (upper left corner) focuses on the dose distribution for the four periods. The line shows the rATG average dose fluctuations over the individual years and the four periods. 1st Period: from licensure until June 7, 2003; 2nd Period: from June 8, 2003 to November 9, 2007; 3rd Period: from November 10, 2007 to February 14, 2011; 4th Period: from February 15, 2011 to December 31, 2015.
Fig. 4.

The within-cluster sum of squares and between-cluster sum of squares for k from 2 to 10. Elbows at k=4 show an improvement over k=3.
We created the first group of subnetworks with nodes representing the MedDRA PTs and the four dose ranges. The subnetworks contained the cumulative sets of cases stamped with a TSD until the end of each period. As shown in ► Figure 5 and ► Figure 6 , the “lymphoproliferative disorder (ld)” and “post-transplant lymphoproliferative disorder (ptld)” nodes are among the top nodes in the first and third time periods which were associated with high rATG doses, in the “c” and “d” range.
Fig. 5.
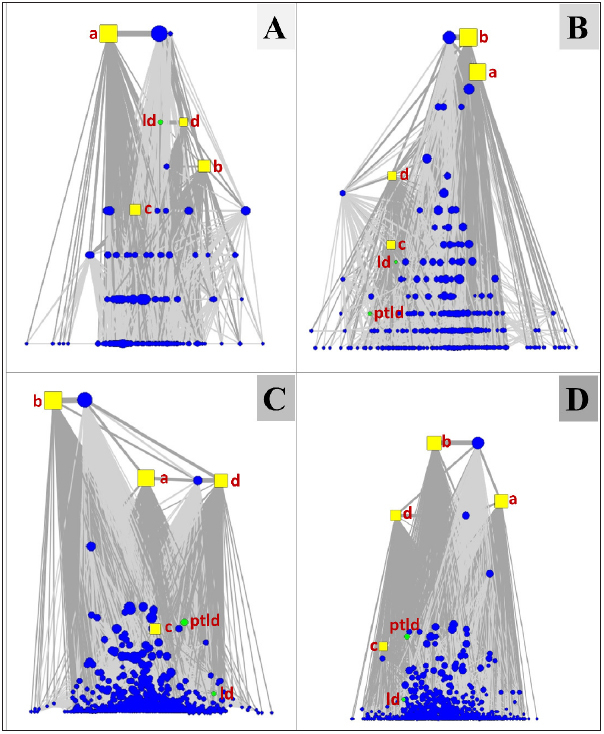
The subnetworks were constructed using the island layout for the four time periods in a cumulative fashion and illustrate the tight relationships between the high doses (“c” and “d” nodes) with “ptld” and “ld”. (A) the subnetwork for the first period with cases having a Therapy Start Date until June 7, 2003 (71 cases in total) – “ld” node is tightly connected to the “d” node and they both are among the top nodes verifying the tight association between “ld” and the highest rATG doses (>12mg/kg); (B) the subnetwork with the addition of more cases having a Therapy Start Date until November 9, 2007 (272 cases in total) – most of the cases were associated with doses ≤8mg/kg), which is indicated by the position of the “a” and “b” nodes in the topology, while “ld” is still associated with doses in the “c” range (>8mg/kg and ≤12mg/kg) and “ptld” does not play an important role in the topology; (C) the subnetwork based on all cases having a Therapy Start Date until February 14, 2011 (908 cases in total) – “ptld” is associated with the high dose “c” node and higher doses are reported, while the “ld” node is less important probably because the “ptld” term is more frequently used in recent years for coding purposes; (D) the full network with all rATG cases (N=1380) having complete dose information and known Therapy Start Date until the end of 2015 – small differences from the previous period are observed. ld: lymphoproliferative disorder; ptld: post-transplant Page 52 of 54 lympho-proliferative disorder; a: dose range 0–4.00 mg/kg; b: dose range 4.01–8.00 mg/kg; c: dose range 8.01–12.00 mg/kg; d: doses ≥12.01 mg/kg.
Fig. 6.

The quantification of the visual findings from the subnetworks shown in Figure 5. (A) the node metrics for the subnetwork for the first period with cases having a Therapy Start Date until June 7, 2003; (B) the node metrics for the subnetwork with the addition of more cases having a Therapy Start Date until November 9, 2007; (C) the node metrics for the subnetwork based on all cases having a Therapy Start Date until February 14, 2011; (D) the node metrics for the full network with all rATG cases until the end of 2015. “ld” and “ptld” were among the most central nodes in the subnetworks representing the first period and the most recent years (after 2007), respectively. They appeared to be less central in the second period when decreased rATG doses were administered. ld: lymphoproliferative disorder; ptld: post-transplant lymphoproliferative disorder; a: dose range 0–4.00 mg/kg; b: dose range 4.01–8.00 mg/kg; c: dose range 8.01–12.00 mg/kg; d: doses ≥12.01 mg/kg.
In the second group of subnetworks, representing the four time periods as disjoint networks, the tight connection of “ld” and “ptld” with the high dose nodes (“c” and “d”) is clearly shown in the principal component layouts shown in ► Figure 7 . Even in the second period where high doses were reported in fewer cases, both AE nodes were close to the corresponding nodes (panel B). In the third and fourth period, “ptld” is definitely more important than “ld” and still highly connected to the “c” node.
Fig. 7.
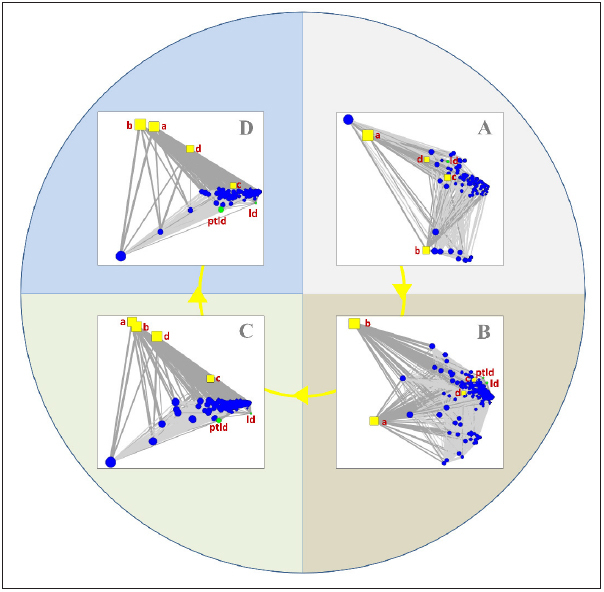
The second group of subnetworks was constructed using the principal component layout for the four time periods separately. The tight connections between the high doses (“c” and “d” nodes) with “pltd” and “ld” are clearer than in the previous, cumulative subnetworks. (A) the subnetwork for the first period with cases having a Therapy Start Date until June 7, 2003 – the “ld” node is tightly connected to the “d” node indicating a strong association between “ld” and high rATG doses in the first period; (B) the subnetwork for the second period with cases having a Therapy Start Date from June 8, 2003 to November 9, 2007 – the “ptld” node appears next to the “ld” node, and they are both tightly connected to the “c” and “d” nodes that represent the high rATG doses (>8mg/kg); (C) the subnetwork for the third period with cases having a Therapy Start Date from November 10, 2007 to February 14, 2011 – the “ptld” and “ld” nodes are mainly connected to the “c” node; (D) the subnetwork for the fourth period with cases having a Therapy Start Date from February 15, 2011 to December 31, 2015 – the “ptld” and “ld” nodes are connected to the “c” node as in the previous subnetwork. ld: lymphoproliferative disorder; ptld: post-transplant lymphoproliferative disorder; a: dose range 0–4.00 mg/kg; b: dose range 4.01–8.00 mg/kg; c: dose range 8.01–12.00 mg/kg; d: doses ≥12.01 mg/kg; rATG: rabbit Anti-Thymocyte Globulin.
4. Discussion
In this study, we explored whether the combined use of NLP and NA tools could support the evaluation of a dose-related AE pattern for rATG. For this purpose, we processed thousands of reports submitted to FAERS for rATG using a set of NLP and NA tools. Although we found that MedEx and ETHER supported the extraction of the appropriate information, manual curation was necessary to evaluate the context and improve the quality of the medication and time information. The subsequent analysis with PANACEA resulted in network representations of rATG and PTLD information over time and highlighted the tight association between PTLD and high rATG total doses (>8 mg/ kg). This finding was true even in periods when most of the submissions to FAERS reported low doses of rATG.
Our study has some limitations. First, the final set of cases (N=1,380) might not be considered representative of the original dataset (N=6,493), since it contained only 21% of the total cases. However, given the level of manual effort required to process the 1,380 cases, it was not possible to retrieve the complete dose and temporal information for the remaining 79%. Second, the manual curation of the MedEx and ETHER output was not in line with the development of a fully automated process. We felt though that the generation of an accurate dataset through a semi-automated process best supported the exploration of the AE pattern and its association with the rATG doses. This data-set could be potentially reused as the “gold standard” in future explorations. Third, we relied on the stored data and retrieved only the missing TSDs from the free-text narratives. We did not investigate data accuracy in the FAERS structured fields of interest with the exception of the weight information. For example, we did not perform a thorough evaluation of whether the exposure dates retrieved by ETHER were more or less accurate than the TSDs stored in the corresponding FAERS field, as this effort was outside the scope of this project.
We used two existing NLP platforms for the retrieval of medication and temporal information from the rATG reports. MedEx was previously found to perform well at identifying drug names and dose-related information, such as strength, route, and frequency [ 8 , 16 , 17 ]. ETHER’s text mining capabilities have been extensively evaluated as well with a demonstrated efficiency in the extraction of drug names and temporal information [ 10 , 18 ]. The NLP output was subsequently analyzed in PANACEA, a NA tool that supports the analysis of complex relationships in large datasets over time. PANACEA is equipped with certain capabilities for the analysis of post-market data and the recognition of safety patterns that were discussed in our previous work [ 19 , 20 , 21 ].
As highlighted above, earlier studies demonstrated the ability of the two NLP systems to efficiently retrieve certain named entities from clinical texts. Despite these findings, the manual curation performed in our work may raise some concerns about their applicability to more complex problems. For example, the reviewers concluded that no TSD should have been assigned by ETHER to many of the drug mentions. However, this should not characterize ETHER’s ability to support this task as it is related to its particular configuration: if a drug is not directly linked to an absolute time statement in the text, ETHER will (and has been set to) assign an exposure date according to either the submission date or other time information in the narrative [ 10 ]. ETHER’s configuration to avoid missing values can be easily changed to accommodate the requirements in other tasks.
Given the increasing volumes of data submitted to the FDA systems, automated processing and analysis of post-market safety surveillance data is a critical need [ 7 ]. Our work has demonstrated the feasibility of investigating a dose-related safety pattern for a particular product using a set of NLP and NA tools. It has illustrated areas for improvement as we continue to apply automated strategies for such tasks. In recent work, we described the tight integration of ETHER and PANACEA into a Decision Support Environment that has been built to contribute to the signal management process [ 9 ]. The Decision Support Environment is an ongoing project to assist the medical experts and epidemiologists at the FDA in performing post-market surveillance. As part of this effort, we are examining additional components or enhancements that can be implemented, such as the extraction of medication information. We have shown the combination of NLP and NA tools can support safety surveillance, and these methodologies may also be applicable to other types of clinical text to support similar activities outside FDA.
Multiple Choice Questions
What can be concluded if PTLD has a maximum edge weight N in an element network in PANACEA?
PTLD co-occurs with at least one other term in N reports.
PTLD occurs in at most N reports.
PTLD shares an edge with N other nodes in the element network.
PTLD has the heaviest edge in the network.
The correct answer is A. Edge weights are determined by co-occurrence of terms in reports. If PTLD has a maximum edge weight N, it co-occurs with at least one other term in N reports. B is incorrect because the total number of reports with PTLD can exceed N, the maximum number of reports with co-occurrence of PTLD and another term. C is incorrect because the number of nodes with which PTLD shares an edge is independent of the edge weight between PTLD and any given node (i.e. the number of unique terms co-occurring with PTLD cannot be determined by the number of reports with co-occurrence of a given term and PTLD). D is incorrect because the maximum edge weight of PTLD is not necessarily the maximum edge weight in the network.
Acknowledgements
This work was supported in part by the appointments of Nina Arya, Matthew Foster, Kory Kreimeyer, and Abhishek Pandey to the Research Participation Program administered by ORISE through an interagency agreement between the US Department of Energy and the US FDA.
Conflicts of Interest None declared.
Clinical Relevance Statement
• The automated retrieval of medication and other clinical information from the United States Food and Drug Administration Adverse Event Reporting System (FAERS) is critical for pharma-coepidemiological analysis.
• Natural language processing can be combined with other approaches, such as network analysis, to support the evaluation of safety patterns associated with medical product administration.
• The use of advanced techniques in the decision making process may assist medical experts and epidemiologists in performing their routine safety surveillance tasks.
Protection of Human and Animal Subjects
Human and/or animal subjects were not included in the project.
References
- 1.Gaber AO, Monaco AP, Russell JA, Lebranchu Y, Mohty M. Rabbit antithymocyte globulin (thymoglobulin): 25 years and new frontiers in solid organ transplantation and haematology. Drugs. 2010;70(06):691–732. doi: 10.2165/11315940-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Mohty M, Bacigalupo A, Saliba F, Zuckermann A, Morelon E, Lebranchu Y. New directions for rabbit anti-thymocyte globulin (Thymoglobulin®) in solid organ transplants, stem cell transplants and autoimmunity. Drugs. 2014;74(14):1605–1634. doi: 10.1007/s40265-014-0277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.3. THYMOGLOBULIN –Anti-thymocyte Globulin (rabbit) Injection, Powder, Lyophilized, for Solution. U.S. National Library of Medicine.National Institutes of Health 2016[cited December 12, 2016]. Available fromhttps://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=bbd8ab99–552e-4b81-aca4–6b0c7af8b9ae
- 4.Marks WH, Ilsley JN, Dharnidharka VR. Posttransplantation lymphoproliferative disorder in kidney and heart transplant recipients receiving thymoglobulin: a systematic review. Transplant Proc. 2011;43(05):1395–1404. doi: 10.1016/j.transproceed.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 5.Gaber AO, Matas AJ, Henry ML, Brennan DC, Stevens RB, Kapur S, Ilsley JN, Kistler KD, Cosimi AB. Thymoglobulin Antibody Immunosuppression in Living Donor Recipients I. Antithymocyte globulin induction in living donor renal transplant recipients: final report of the TAILOR registry. Transplantation. 2012;94(04):331–337. doi: 10.1097/TP.0b013e31825a7d1f. [DOI] [PubMed] [Google Scholar]
- 6.FDA.Questions and Answers on FDA‘s Adverse Event Reporting System (FAERS) 2016. Available from:http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/default.htm
- 7.Duggirala HJ. Use of data mining at the Food and Drug Administration. J Am Med Inform Assoc. 2016;23(02):428–434. doi: 10.1093/jamia/ocv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(01):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botsis T. Decision Support Environment for Medical Product Safety Surveillance. J Biomed Inform. 2016;64:354–362. doi: 10.1016/j.jbi.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Kreimeyer K, Woo EJ, Ball R, Foster M, Pandey A, Scott J, Botsis T. A new algorithmic approach for the extraction of temporal associations from clinical narratives with an application to medical product safety surveillance reports. J Biomed Inform. 2016;62:78–89. doi: 10.1016/j.jbi.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. Journal of the Royal Statistical Society Series C (Applied Statistics) 1979;28(01):100–108. [Google Scholar]
- 12.Team RC.R: A language and environment for statistical computing R Foundation for Statistical Computing; Vienna: Austria URLhttps://wwwR-projectorg/2013 [Google Scholar]
- 13.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006;1695(05):1–9. [Google Scholar]
- 14.de Nooy W, Mrvar A, Batagelj V, Granovetter M.Exploratory Social Network Analysis with PajekSecond ed.Cambridge University Press; 2011 [Google Scholar]
- 15.Newman M. Oxford university press; 2010. Networks: an introduction. [Google Scholar]
- 16.Doan S, Bastarache L, Klimkowski S, Denny JC, Xu H. Integrating existing natural language processing tools for medication extraction from discharge summaries. J Am Med Inform Assoc. 2010;17(05):528–531. doi: 10.1136/jamia.2010.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang M, Wu Y, Shah A, Priyanka P, Denny JC, Xu H.Extracting and standardizing medication information in clinical text –the MedEx-UIMA system AMIA Jt Summits Transl Sci Proc 2014. 2014:37–42. [PMC free article] [PubMed]
- 18.Botsis T, Buttolph T, Nguyen MD, Winiecki S, Woo EJ, Ball R. Vaccine adverse event text mining system for extracting features from vaccine safety reports. J Am Med Inform Assoc. 2012;19(06):1011–1018. doi: 10.1136/amiajnl-2012-000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball R, Botsis T. Can network analysis improve pattern recognition among adverse events following immunization reported to VAERS? Clin Pharmacol Ther. 2011;90(02):271–278. doi: 10.1038/clpt.2011.119. [DOI] [PubMed] [Google Scholar]
- 20.Botsis T, Ball R. Network analysis of possible anaphylaxis cases reported to the US vaccine adverse event reporting system after H1N1 influenza vaccine. Stud Health Technol Inform. 2011;169:564–568. [PubMed] [Google Scholar]
- 21.Botsis T, Scott J, Woo EJ, Ball R. Identifying Similar Cases in Document Networks Using Cross-Reference Structures. IEEE J Biomed Health Inform. 2015;19(06):1906–1917. doi: 10.1109/JBHI.2014.2345873. [DOI] [PubMed] [Google Scholar]


