Abstract
Purpose
The purpose of this study was to directly compare mutation profiles in multiple single CTCs and cfDNA isolated from the same blood samples taken from patients with metastaic breast cancer (MBC). We aimed to determine whether cell-free DNA would reflect the heterogeneity observed in 40 single CTCs.
Experimental design
CTCs were enumerated by Cellsearch. CTC count was compared with the quantity of matched cfDNA and serum CA15-3 and alkaline phosphatase (ALP) in 112 patients with metastatic breast cancer. In 5 patients with ≥100 CTCs, multiple individual EpCAM-positive CTCs were isolated by DEPArray and compared with matched cfDNA and primary tumour tissue by targeted next generation sequencing (NGS) of ~2200 mutations in 50 cancer genes.
Results
In the whole cohort, total cfDNA levels and cell counts (≥5 CTCs) were both significantly associated with overall survival, unlike CA15-3 and ALP. NGS analysis of 40 individual EpCAM-positive CTCs from 5 patients with MBC revealed mutational heterogeneity in PIK3CA, TP53, ESR1 and KRAS genes between individual CTCs. In all 5 patients cfDNA profiles provided an accurate reflection of mutations seen in individual CTCs. ESR1 and KRAS gene mutations were absent from primary tumour tissue and therefore likely reflect either a minor sub-clonal mutation or were acquired with disease progression.
Conclusion
Our results demonstrate that cfDNA reflects persisting EpCAM-positive CTCs in patients with high CTC counts and therefore may enable monitoring of the metastatic burden for clinical decision-making.
Keywords: Liquid biopsy, circulating free DNA, circulating tumor cells, targeted next-generation sequencing
Introduction
Breast cancer is the most common cancer in women and ranks as second in the world in terms of cancer deaths. Recent studies using next generation sequencing (NGS) have shown that cancer evolves in the patient (1–3), and therapies can induce the evolution of clones of cells that are refractory to the treatment (4, 5). Much research has been devoted to developing bioassays that can help in the selection of therapies as breast cancer evolves, including characterisation of circulating tumor cells (CTCs) and the tumour derived fraction of circulating cell-free DNA (cfDNA) termed circulating tumour DNA.
Although a number of commercial platforms are under investigation for isolation and characterisation of CTCs without the use of a surface marker, CellSearch (Janssen Diagnostics), which enumerates CTCs of epithelial origin (CD45-, EpCAM+, and cytokeratins (CK) 8, 18+, and/or 19+ positive cells), is currently the only FDA approved platform in clinical use. EpCAM-positive CTCs as measured by CellSearch are rarely detected in patients with primary breast cancer and many patients with metastatic disease also have few EpCAM-positive CTCs (0-5 per 7.5ml blood). When detected, the number of EpCAM-positive CTCs reflects both the effects of systemic therapy (6–8) and prognosis (9). In contrast, circulating tumour DNA is detected in early and late stages in a high proportion of cases (10–12) and can persist for many years after supposedly successful therapy (13) despite no evidence of overt distant metastases. In metastatic breast cancer, the dynamics of circulating tumour DNA compare favourably to serum CA15-3 and CTC counts determined by CellSearch in reflecting changes in tumor burden (14).
Metastases are heterogeneous, both between metastatic sites and also between the cells that compose each metastasis. Early metastases are comprised of a greater proportion of cells with EMT and ‘stem-like’ characteristics; whereas larger metastases are dominated by more differentiated, heterogeneous patterns of cells, more closely reflecting the heterogeneity seen in primary tumors (15). Taking all these considerations into account, together with the well-established phenomenon of clonal evolution (16, 17), it is evident that a single conventional tissue biopsy will not necessarily provide accurate information regarding appropriate therapy. An additional layer of complexity is suggested by the role of the immune system in patients with cancer, since immune destruction of persisting micrometastases could mean that the circulating tumour DNA is predominantly a reflection of destroyed cancer cells; whereas persisting CTCs could be envisaged as a more accurate guide to appropriate therapy.
Previous studies have compared mutations in cfDNA to either biopsies of metastases or CTCs (2, 18). We reported emergence of ESR1 mutations in cfDNA and in a single patient with a very high CTC count (>3000 EpCAM-positive CTCs in 7.5ml blood) the ESR1 p.D538G mutation was detected in cfDNA and matched CTCs (2). Since cfDNA is in principal easier to obtain than CTCs for molecular analysis, it is essential to determine whether mutations in cfDNA are the same as those seen in individual CTCs, since heterogeneity is frequently observed in metastatic biopsies (19, 20). Here, we compared total cfDNA levels and CellSearch CTC counts with serum CA15-3 and alkaline phosphatase (ALP) levels in patients with metastatic breast cancer. We performed deep amplicon sequencing of 50 genes and compared mutation profiles in cfDNA and individual EpCAM-positive CTCs isolated from the same blood sample in 5 patients with high CTC counts (≥100 CTCs in 7.5ml blood). Results show that cfDNA mutation profiles reflect mutational heterogeneity seen across individual CTCs; and that total cfDNA levels and ≥5 CTCs are significantly associated with overall survival. Circulating tumour DNA is a useful substitute for metastasis biopsy (18). These data suggest that circulating tumour DNA reflects persisting EpCAM- positive CTCs in patients with high CTC counts and could enable monitoring of the metastatic burden, but requires confirmation in larger groups of patients.
Material and Methods
Patients and samples
The study protocol was approved by the Riverside Research Ethics Committee (Imperial College Healthcare NHS Trust; REC reference number: 07/Q0401/20) and conducted in accordance with Good Clinical Practice Guidelines and the Declaration of Helsinki. All patients gave written informed consent prior to participation. We recruited 112 patients with radiologically-confirmed metastatic breast cancer (Table 1). 20 ml of blood was taken into EDTA- tubes (BD Biosciences) and processed to plasma for cfDNA (21) and 7.5ml blood was taken into CellSave preservative tubes for CTCs capture and enumeration with the CELLSEARCH® CTC Test (Janssen Diagnostics) as described previously (22). CA15-3 and ALP results were obtained from patient notes.
Table 1. Clinicopathological features of 112 patients with metastatic breast cancer.
*Record of PR testing not available. **Date of diagnosis predates the introduction of HER2 testing in the UK.
| Clinicopathologic feature | Detail | N |
|---|---|---|
| Age | ≤ 40 | 9 (8%) |
| > 40 | 103 (92%) | |
| Median | 59 | |
| Range | 25 - 88 | |
| ER | Positive | 96 (86%) |
| Negative | 14 (12%) | |
| Unknown | 2 (2%) | |
| PR | Positive | 65 (58%) |
| Negative | 27 (24%) | |
| Unknown* | 20 (18%) | |
| HER2 | Positive | 21 (19%) |
| Negative | 73 (65%) | |
| Not tested** | 18 (16%) | |
| CA15-3 | ≤ 30U/ml | 40 (36%) |
| > 30U/ml | 72 (64%) | |
| Median | 51 | |
| Range | 1 - 3613 | |
| ALP | ≤ 100 IU/L | 72 (64%) |
| > 100 IU/L | 40 (36%) | |
| Median | 88 | |
| Range | 30 - 555 | |
| CTCs/7.5 ml blood | 0 | 61 (54%) |
| 1-4 | 26 (23%) | |
| ≥5 | 25 (23%) | |
| Range | 0 - 701 |
Extraction and quantitation of DNA
CfDNA was isolated from 3 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen) according to manufacturer’s instructions. Isolation of DNA from lymphocytes and quantitation of total cfDNA was as described previously (21). cfDNA levels (ng/ml) were converted to copies/ml plasma assuming 3.3 pg DNA per haploid genome. FFPE tissue blocks with matching H&E were retrieved from the histopathology archive and reviewed by a consultant histopathologist. Two separate regions of tumour were then cored from the block using a 1 mm tissue microarray needle. DNA was extracted from the FFPE tissue core using the Qiagen GeneRead kit according to manufacturer’s instructions. CTC isolation by DEPArray, QC to identify those samples with a genomic integrity index (GII) of 3 or 4 suitable for sequencing (20) and Ampli1™ whole genome amplification were carried out as a service by Silicon Biosystems. Details of the exact numbers of single cells recovered and analysed for each patient are given in Supplementary Table 1.
Targeted next generation sequencing
Ampliseq reactions were set up using 10 ng WGA DNA, an average of 8 ng cfDNA and 10 ng FFPE tumour tissue DNA. All samples with sufficient DNA (including all individual cell samples) were sequenced using 2 AmpliSeq panels: an in-house 30 amplicon panel, designed using Ion AmpliSeq designer software (https://www.ampliseq.com), covering mutation hotspots in ESR1, PIK3CA, TP53 and ERBB2 (Supplementary Table 2) and the Custom Cancer Hotspot Panel v2 for Ampli1™ WGA DNA (49 genes, over 2200 COSMIC mutations) (Silicon Biosystems, see http://www.siliconbiosystems.com/fee-for-service and Supplementary Table 3). Three genes (PIK3CA, TP53 and ERBB2) were on both panels making a total of 50 genes surveyed for hotspot mutations. Overlapping mutations were reviewed in IGV if they were only detected on one panel. In total the two panels surveyed ~2200 COSMIC ID mutations.
Sequencing on a 316 chip using the Ion PGM and data analysis was as described previously (2). In brief, sequencing data was accessed through the Torrent Suite v.4.2.0. Reads were aligned against the human genome (hg19) using Alignment v4.0-r77189 and variants called using the coverageAnalysis v4.0-r77897 and variantCaller v4.0-r76860, respectively. Due to the sensitivity of the in house panel, for cfDNA and FFPE tumour tissue variantCaller was configured to call on high stringency, somatic variants with down sampling set to 6000 and gen_min_coverage set to 6. COSMIC IDs were obtained using COSMIC v71 (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/ - last accessed 31st February 2016). Variants in individual CTCs and WBCs were analysed using a pre-set high stringency germline threshold, as recommended by the manufacturer, which is optimised for high-frequency variants and minimal false positive calls. The allelic dropout rate (ADR) and false positive rate (FDR) was calculated as described by Leung et al. (23). All mutations with a quality score below 25 were omitted and all variants detected in the first or last 10 bases of an amplicon were omitted as likely mispriming events (2, 24). ANNOVAR (25) was used to annotate all private mutations with refGene ID, functional consequence (e.g., non-synonymous), and functional predictions (using SIFT (26), Polyphen-2 (27), and MutationTaster (28)). All variants detected were also manually confirmed across all samples using the Integrated Genomics Viewer 2.3 (29, 30). The mutant allele frequency (termed AF) was calculated as the proportion of total reads at a site, which contained the variant allele (e.g. if you have 200 reads in total and 8 of them have the variant, then the AF is (8/200) x100 = 4%).
Digital droplet PCR (ddPCR)
Droplet digital PCR was used to validate selected mutations in ESR1 p.Y537C/N/S, p.D538G; PIK3CA p.E545K and H1047R, and KRAS p.G12D. Each assay was performed in duplicate using a Bio-Rad QX200 digital droplet PCR system as described previously (31). Inventoried assays were used for detection of PIK3CA p.E545K (assay numbers dHsaCP2000075 and dHsa2000076) and p.H1047R (assay numbers dHsaCP2000077 and dHsaCP2000078) and KRAS p.G12D (assay numbers dHsaCP2000001 and dHsaCP2000002) (Bio-Rad Laboratories) according to manufacturers’ instructions. For the ESR1 mutations, a dual labelled LNA probe strategy was used (see Supplementary Table 4 for sequences). Thermal cycling conditions for the ESR1 assays were: 10 min hold at 95 °C, 40 cycles of 95 °C for 15 s then 60 °C for 60 s. Raw fluorescence amplitude was analysed using the Quantasoft version 1.6.6.0320 software and used to obtain the fractional abundance for a given mutation. This was reported as the allele frequency (AF) to be consistent with NGS data. Calculation of the allele fraction (AF) was performed as described previously (32, 33). The total number of droplets (with and without DNA) was used to calculate DNA copies/μl, then dividing the number of mutant copies by the number of total DNA copies (mutant plus wild-type) and multiplying by 100 to give the percentage (allele fraction) of mutant DNA copies based on Poisson distribution of positive to empty droplets (33).
Statistical analysis
Correlation analysis of total cfDNA levels, CA15-3, ALP and CTC counts was performed using a non-parametric Spearman’s correlation. P values were two-tailed and considered significant if P < 0.05. Survival analysis was performed using multiple Cox-regression as described previously (14) with each biomarker as a continuous time-dependent variable (Supplementary Table 5). Briefly, for each variable a model was constructed using a counting process notation (i.e. start, end, event). In our study, start was taken as the date of the baseline blood sample, and the end was taken as an arbitrary cut-off of 01/12/2015.
Results
This was a prospective study set up to identify patients with high CTC counts (≥100 CTCs per 7.5 ml blood) suitable for isolation of individual CTCs by DEPArray. The threshold of ≥100 CTCs per 7.5 ml blood was based on a previous study by Polzer et al. (20), that analysed individual CTCs from patients with similar high CTC counts but did not compare individual CTCs with matched cfDNA. We recruited 112 patients with metastatic breast cancer to the study (Table 1). Each had undergone multiple lines of treatment. Sixty-one patients were CTC negative and 51 patients had ≥1 CTCs measured by CellSearch (median, 5; range 1 – 701). Serum CA15-3 levels were elevated (>30 U/ml (34)) in 72 patients (64%), ALP was elevated (>100 IU/L) in 40 patients (36%) and both were elevated in 31 (28%). Total cfDNA was obtained for all samples (median, 2757 copies/ml of plasma; range 30 copies – 115724 copies/ml of plasma). The markers were significantly correlated (Spearman’s correlation analysis; Supplementary Table 6), except cfDNA and CA15-3 (P = 0.058). High CTC count (≥5 CTCs) (HR, 2.8, 95% CI, 1.4 to 5.7; P = 0.005) and total cfDNA level (hazard ratio (HR), 2.2; 95% CI, 1.3 to 3.6; P = 0.03) were significantly associated with poorer overall survival (Supplementary Table 6).
cfDNA and individual CTCs have overlapping mutation profiles
At the time of the study we had established that adequate cells could be obtained from the DEPArray system for patients with high CTC counts (≥100 CTCs per 7.5ml blood) based on a previous study (20). We therefore chose five patients with high EpCAM-positive CTCs for single CTC isolation by DEPArray: 4 had ERα positive metastatic disease and one was triple negative. Clinical details for the 5 patients are given in Supplementary Data 1 and treatment timelines are in Supplementary Figure 1. Formalin fixed paraffin embedded (FFPE) primary tumour tissue was available for 4 patients. Four had serial cfDNA and 1 had serial CellSearch CTC samples (Figure 1). Individual CTCs were recovered successfully for each patient. In total 42 individual EpCAM-positive CTCs, 16 individual white blood cells (WBCs) as germ line and QC controls, 5 CTC pools and 6 WBC pools were isolated by DEPArray. Of these 40 individual CTCs, 15 individual WBCs and all cell pools passed QC checks (genomic integrity index of ≥ 3 – Supplementary Table 1) for sequencing (20).
Fig. 1. Study workflow.
112 patients with metastatic breast cancer were recruited to the study. Blood was collected and analysed for CTC count and total cfDNA levels in all 112 patients. In 5 patients with high CTC count individual CTCS were isolated by DEPArray and subjected to targeted NGS in comparison with matched cfDNA and primary tumour tissue where available.
All CTC and WBC samples, matching cfDNA and FFPE tumour tissue DNA were analysed by deep amplicon sequencing (Figure 1) using two panels covering ~2200 COSMIC mutations in 50 cancer genes (for details of the custom 30 amplicon panel see Supplementary Table 2, for full details of the Ampli1 custom cancer hotspot panel, see http://www.siliconbiosystems.com/fee-for-service). A high average read depth was obtained with both panels: 929x for the Custom Cancer Hotspot Panel v2 for Ampli1™ WGA DNA (Silicon Biosystems (20)) and 4537x for the 30 amplicon custom panel (Supplementary Table 7). The allelic dropout rate (ADR) across individual CTCs and WBCs, and the false positive rate (FPR) for SNVs were 25.01%; SEM 2.68%, and 0.6%; SEM 0.32% respectively (Supplementary Table 8), within the range reported previously (23). Mutations were only called in individual CTCs and WBCs if these were present in 2 or more single cells (23). Overall only 3 individual CTCs had no coverage at a specific mutated base (Table 2). In each case, the mutation was called in other CTCs and the matched plasma cfDNA.
Table 2. Mutations identified in matched FFPE, cfDNA and individual CTCs.
AF; Allele frequency; -----; no mutation detected; NC, no coverage at this amplicon. ------/NC; PIK3CA mutation not detected/No coverage at ESR1 mutation site. Shown are COSMIC mutations detected in individual CTCs (C1, 2), CTC pools (Cpool) matched plasma cfDNA and two regions of primary tumor. Patient CTCM167 had two serial samples for both CTCs and plasma cfDNA. All data are summarized in Supplementary Tables 10-13.
| Sample ID | Receptor status | CTC | cfDNA (AF) | Primary tumor region 1 (AF) | Primary tumor region 2 (AF) | |
|---|---|---|---|---|---|---|
| CTCM155 | ER+/PR+/HER2- | C1 | PIK3CA p.E545K; ESR1 p.E380Q | PIK3CA p.E545K (23.7%); ESR1 p.E380Q (3.9%) | PIK3CA p.E545K (67.8%) | PIK3CA p.E545K (72.4%) |
| C2 | PIK3CA p.E545K; ESR1 p.E380Q | |||||
| C3 | PIK3CA p.E545K; ESR1 p.E380Q | |||||
| C4 | PIK3CA p.E545K; ESR1 p.E380Q | |||||
| C5 | PIK3CA p.E545K | |||||
| Cpool | PIK3CA p.E545K; ESR1 p.E380Q | |||||
| CTCM138 | ER+/PR+/HER2- | C1 | -----/NC | PIK3CA p.H1047R (31%); ESR1 p.E380Q (23.8%); TP53 p.R175H (0.3%); ESR1 Y537C (0.43%) | PIK3CA p.H1047R (14.2%); TP53 p.R175H (1.9%) | PIK3CA p.H1047R (36%); TP53 p.R175H (0.99%) |
| C2 | PIK3CA p.H1047R; ESR1 p.E380Q | |||||
| C3 | PIK3CA p.H1047R; ESR1 p.E380Q | |||||
| C4 | ----- | |||||
| C5 | ----- | |||||
| Cpool | PIK3CA p.H1047R; ESR1 p.E380Q | |||||
| CTCM105 | ER-/PR-/HER2- | C1 | KRAS p.G12D | KRAS p.G12D (3.8%); TP53 p.P278R (2.3%) | ----- | ----- |
| C2 | KRAS p.G12D | |||||
| C3 | KRAS p.G12D | |||||
| C4 | ----- | |||||
| C5 | KRAS p.G12D | |||||
| C6 | KRAS p.G12D | |||||
| C7 | NC | |||||
| C8 | KRAS p.G12D | |||||
| C9 | ----- | |||||
| C10 | KRAS p.G12D | |||||
| C11 | NC | |||||
| Cpool | KRAS p.G12D | |||||
| CTCM292 | ER+/PR+/HER2- | C1 | PIK3CA p.E545K; ESR1 p.D538G | PIK3CA p.E545K (0.17%); ESR1 p.D538G (0.27%) | None available | None available |
| C2 | PIK3CA p.E545K; ESR1 p.D538G | |||||
| CTCM167.1 | ER+/PR+/HER2- | C1 | ----- | ----- | ----- | ----- |
| C2 | ----- | |||||
| C3 | ----- | |||||
| C4 | ----- | |||||
| C5 | ----- | |||||
| Cpool | ----- | |||||
| CTCM167.2 | C1 | ----- | ----- | ----- | ----- | |
| C2 | ----- | |||||
| C3 | ----- | |||||
| C4 | ----- | |||||
| C5 | ----- | |||||
| C6 | ----- | |||||
| C7 | ----- | |||||
| C8 | ----- | |||||
| C9 | ----- | |||||
| C10 | ----- | |||||
| C11 | ----- | |||||
| C12 | ----- | |||||
| Cpool | ----- | |||||
Although we examined mutation hotspots in 50 genes across 2 amplicon panels by NGS, mutations were only detected by NGS in 4 genes in ESR1, PIK3CA, TP53 and KRAS. Two of these PIK3CA, TP53 were common to both panels. Droplet digital PCR was used as validation for ESR1 p.Y537C/N/S, p.D538G; PIK3CA p.E545K and H1047R, and KRAS p.G12D when sufficient remaining DNA was available.
Overall, 4 patients had mutations detected in PIK3CA, TP53, ESR1 and KRAS genes in cfDNA and individual EpCAM-positive CTCs (Fig. 2; Table 2). Levels of circulating tumour DNA varied. The mean plasma mutant allele frequency (AF) ranged from 0.3% to 32% and a rise in levels of circulating tumour DNA generally accompanied a rise in total cfDNA (Fig. 2). Mutational heterogeneity was seen between individual EpCAM-positive CTCs isolated from the same blood sample with the majority of CTCs having 1, 2 or no mutations detected. All mutations detected in individual EpCAM-positive CTCs were detected in the matched cfDNA (Table 2). PIK3CA and TP53 gene mutations were also present in the primary tumour DNA; whereas ESR1 and KRAS mutations were unique to cfDNA and EpCAM-positive individual CTCs (Table 2) suggesting potential clonal evolution. Private mutations of unknown significance were also detected in individual CTCs and WBCs (Supplementary Table 9); however these were excluded as likely amplification artefacts as they were not detected in the matched CTC pool, WBC pool or primary tumour (Supplementary Tables 10-13).
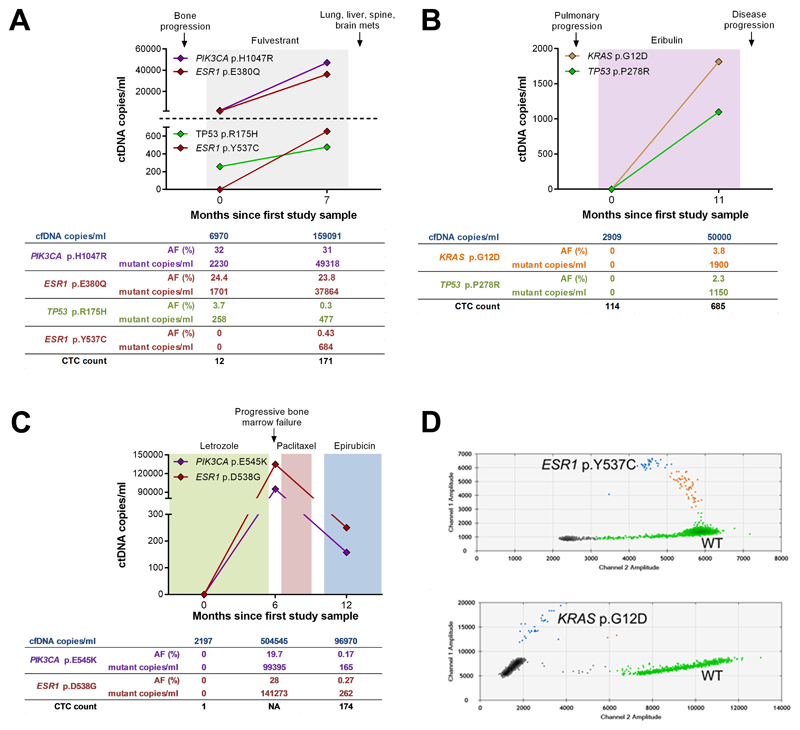
Fig. 2. Longitudinal follow up of circulating tumor DNA in 3 patients with high CTC counts.
(A - C) Total cfDNA and circulating tumor DNA levels. (A) patient CTCM138; (B) patient CTCM105; (C) patient CTCM292. Endocrine or cytotoxic therapies are indicated by colored shading. The number of cfDNA copies obtained from the blood sample, mutant allele frequency (AF), number of mutant copies and CTC count is given below each graph. NA = Not available or CellSearch failed. (D) Digital PCR detection of low frequency mutations in circulating tumour DNA. ESR1 p.Y537C mutation, patient CTCM138 (top) and KRAS p.G12D mutation, patient CTCM105 (bottom). Green dots represent HEX-labelled wild-type (WT), blue dots represent FAM-labelled mutant DNA and brown dots represent double positive droplets containing WT and mutant DNA. Grey dots represent empty droplets.
Patient CTCM155 (ER positive, HER2 negative) had two blood samples taken 11 months apart (Supplementary Fig. 1). Plasma cfDNA was only available for the first sample, as insufficent blood was obtained at the second time point for both cfDNA and CTCs. EpCAM-positive CTCs were detected in the second sample after progressing on exemestane and everolimus combination therapy. The primary tumour and baseline cfDNA had a PIK3CA mutation (p.H1047R, AF 70% and 23%, respectively). A second lower frequency ESR1 mutation (p.E380Q, AF 3.9%) was also detected in cfDNA suggesting either a subclonal origin or that this mutation was acquired on treatment. All 5 individual CTCs and the CTC pool had both mutations (Table 2, Supplementary Table 10).
Patients with rising circulating tumour DNA
Patient CTCM138 (ER positive, HER2 negative) had a PIK3CA (p.H1047R) mutation (AF 25%) and low frequency sub-clonal TP53 (p.R175H) mutation (AF 1.5%) in primary tumour DNA. She was recruited to the study in 2014 some 10 years after diagnosis of breast cancer and having received multiple lines of endocrine therapy. Both mutations seen in the primary tumour were detected in baseline cfDNA (PIK3CA AF 32%, TP53 AF 3.7%) as well as an acquired ESR1 (p.E380Q) mutation (AF 24.2%). The second blood sample was taken 7 months later just prior to clinical disease progression. Levels of circulating tumour DNA increased from baseline, the 3 mutations persisted and a second ESR1 mutation (p.Y537C) was acquired at a low level (AF 0.43%) (Fig. 2a and d; Supplementary Table 11). Five individual CTCs were isolated from the second blood sample, 2 of which had the PIK3CA (p.H1047R) and ESR1 (p.E380Q) mutations, but not the lower frequency ESR1 p.Y537C or TP53 p.R175H mutations (Table 2), although these could have been missed by sampling 5 individual CTCs.
Patient CTCM105 (Triple negative) was recruited to the study in March 2014 with progressive disease. No mutations were detected across the 50 genes in either the primary tumour or baseline cfDNA sample. A second blood sample was taken 11 months later. Six of 9 individual CTCs isolated from this sample had a KRAS (p.G12D) mutation that was also detected in the CTC pool (Fig. 2b, d; Table. 2; Supplementary Table 12). Matched cfDNA had the KRAS p.G12D mutation (AF 3.8%) (Fig. 2d) and a low-frequency mutation in TP53 (p.P278R) (AF 2.3%). The low frequency TP53 mutation might be missed by sampling 9 CTCs; however, it is also possible that 2 metastatic clones were shedding DNA in to plasma, one reflected in CTCs, one not. As neither mutation was detected in the FFPE primary tumour tissue these may either have been acquired on treatment or have arisen from a minor subclone missed by tumour sampling.
Patient CTCM292 (ER positive, HER2 negative) presented with bone metastases in 2012 and was treated with tamoxifen. She subsequently relapsed and commenced letrozole, which was then switched to chemotherapy with paclitaxel, then epirubicin. Three serial blood samples were taken over a 12 month period commencing upon letrozole. No mutations were detected in the baseline cfDNA sample, but 2 mutations were detected 6 months later, while she was on letrozole (ESR1 p.D538G AF 28% and PIK3CA p.E545K 19.7%) and then reduced at 12 months on paclitaxel (AF 0.27% and 0.17%, respectively) (Fig. 2c). Both mutations were also present in individual EpCAM-positive CTCs isolated from the third blood sample (Fig. 2c, Table 2; Supplementary Table 13).
Lastly, the fifth patient (CTCM167: ER positive, HER2 negative) was monitored over an 18 month period and had a marked rise in total cfDNA levels and CTC counts at disease progression (Supplementary Fig. 2). No mutations were identified in the 50 genes analysed in FFPE primary tumour DNA. Moreover, there was no evidence for acquisition of mutations across the 50 genes in either cfDNA or individual CTCs from two serial blood samples (Table 2).
Discussion
CTCs are an established prognostic indicator in metastatic breast cancer (7, 9, 35). A recent study has shown that it is possible to interrogate both cfDNA and pools of mutation-bearing CTCs from the blood of patients with metastatic breast cancer (36). Here, we present results of mutations in multiple individual CTCs and matched cfDNA in patients with metastatic breast cancer and high CTC counts. First, in 4 patients we observed heterogeneity in mutations between individual EpCAM-positive CTCs. In all 4 patients cfDNA profiles provided an accurate reflection of mutations see in individual CTCs, and in 2 patients cfDNA had more mutations than those found in CTCs. ESR1 and KRAS gene mutations were present in individual CTCs and cfDNA but were absent from primary tumour tissue. Therefore, these likely reflect either a minor sub-clonal mutation or were acquired with disease progression. Of note, the primary tumour, serial cfDNA samples and multiple individual CTCs from 1 of the 5 patients (CTCM167) had no hotspot mutations detected across the 50 genes. Further analysis, such as whole exome sequencing or array CGH is needed to identify mutations and copy number variation in these samples.
Second, in 2 patients, mutations were detected in cfDNA (in TP53 (p.P278R) and in TP53 (p.R175H) and ESR1 (p.Y537C)) that were not seen in individual CTCs or the CTC pool isolated from the same blood sample. As these were low frequency (and likely sub-clonal) mutations these may have been missed by sampling small numbers of individual CTCs. It is also possible that these were missed due to allelic distortion/dropout, which had an average frequency of 25.46% across the samples analysed, similar to a previous single cell genomics study (23). Of note, in a previous study (2), we also identified an ESR1 gene mutation unique to cfDNA that was not seen in a matched pool of >3000 EpCAM-positive CTCs suggesting that 2 or more metastatic clones were shedding DNA in to plasma, one reflected in CTCs, one not.
Third, although we detected private mutations in single CTCs and single WBCs (mutations observed exclusively in single cells (37)) these were excluded as likely polymerase error during WGA (38, 39) or sequencing error as all were absent from matched FFPE tissue, cfDNA and CTC/WBC pools. Previous studies have suggested “census-based” sequencing of more than one CTC to distinguish private mutations from polymerase errors (38, 39); whereas other studies have compared results with ultra-deep sequencing of the primary and/or metastatic tumor (40) both approaches were performed here.
Although we sequenced ~2200 COSMIC mutations across 50 genes in 5 patients, mutations were only detected in 4 genes (PIK3CA, ESR1, TP53, and KRAS) and only 1 or 2 mutations were detected in the majority of samples. This is consistent with a recent study, which used the same platform and a 50 gene panel (41). TP53 and PIK3CA are the most frequently mutated genes in breast cancer; however, a large number of other genes are less commonly mutated (42). Whole exome or whole genome analysis of the primary tumour/metastatic tissue would be required to determine additional mutations not covered by the targeted sequencing approach. However, metastatic tissue is frequently impossible to obtain in this group of patients.
ESR1 mutations have been linked to increased invasive and metastatic behaviour (43). The ESR1 p.D538G mutation leads to ligand-independent activation of ERα, and is acquired in patients who have received aromatase inhibitors. The p.E380Q mutation has also been reported; but according to Li et al. (44), remains sensitive to anti-oestrogens. In one patient we saw evidence of mutational heterogeneity in ESR1, with p.E380Q at a higher level than p.Y537C in cfDNA, suggesting heterogeneous clonal responses to treatment, as reported in a recent study (45). The PIK3CA p.E545K mutation affects the helical region of the P110alpha catalytic domain (46); whereas the p.H1047R mutation affects the kinase domain, and induces a multi-potent genetic programme in normally lineage-restricted populations at an early stage of tumor development leading to intra-tumoral heterogeneity (47).
Some studies suggest that ERα-positive/PIK3CA mutant tumors or ERα-positive cancers with aberrant PI3K signalling are less responsive to anti-oestrogens than wild-type tumors (48, 49), but other studies have linked these mutations to improved outcome (50, 51), regardless of the type of endocrine therapy used (52). However, PIK3CA mutations have been significantly associated with lower rates of pathogenic response to anti-HER2 therapies (53). Of note the patient who had triple negative breast cancer had different mutations, in TP53 and KRAS in cfDNA and KRAS only in individual CTCs, both of which have been implicated in resistance to cytotoxic chemotherapy.
It has been suggested that targeted sequencing of cfDNA could be used for monitoring of cancer patients during treatment or while in remission, and that single CTC analysis may then be needed to guide more appropriate therapy upon relapse (54). Here, we describe the relationship between mutations in multiple single EpCAM-positive CTCs and cfDNA isolated from the same blood sample in patients with high CTC counts (≥100 CTCs per 7.5ml blood). Results from 5 patients with high CTC counts show that cfDNA sequencing finds more mutations than CTCs as well as mutations not seen in the primary tumour. Our CTC data highlights the mutational heterogeneity of single EpCAM-positive CTCs, supporting other recent studies (19, 20). In 2 out of the 4 patients with mutations detected in CTCs mutations were only detected in about half of the CTCs sequenced (Table. 2). Together these results suggest that if EpCAM-positive CTCs were to be used, concurrent analysis of multiple CTCs may be necessary for clinical decision-making, particularly when metastatic biopsy is not possible. In aggregate our results suggest that cfDNA reflects persisting EpCAM-positive CTCs in patients with high CTC counts and could enable monitoring of the metastatic burden for clinical decision-making. Lastly, our results also suggest that total cfDNA levels are also an independent indicator of overall survival (P = 0.03), but a confirmatory study is needed in a larger series of patients. Further studies are ongoing to examine the relationship between CTCs and cfDNA using Parsortix filtration (55), an alternative marker independent approach for CTC isolation.
Supplementary Material
Translational relevance.
Next generation sequencing is a key approach for monitoring tumour genomic alterations in circulating cell free DNA (cfDNA). High circulating tumor cells (CTCs) count (≥5 EpCAM-positive CTCs per 7.5ml blood as measured by Cellsearch) is a poor prognostic indicator, but thus far individual CTCs have not been directly compared with matched cfDNA by mutation profiling. However, concurrent analysis of individual CTCs and matched cfDNA has the potential for comprehensive characterisation of tumour derived genetic alterations in blood by“liquid biopsy”. In this article we show that individual CTCs have heterogeneous mutations, and that cfDNA isolated from the same blood sample provides an accurate reflection of mutations seen in individual CTCs. We also show that the total cfDNA level, like CTC counts, is an independent prognostic marker in patients with metastatic breast cancer. Overall, our results suggest that cfDNA reflects persisting EpCAM-positive CTCs in patients with high CTC counts and could potentially enable monitoring of the metastatic burden for clinical decision-making when CTCs cannot be obtained.
Acknowledgments
Single cell isolation was performed by Silicon Biosystems as a per fee service. We thank Dr David Moore for reviewing tissue sections, the Imperial and Leicester ECMCs, the Imperial BRC and the Imperial College Tissue Bank for supporting patient recruitment.
Funding: Supported by a Cancer Research UK Clinical and Translational Research Committee programme award (C14315/A13462X).
Footnotes
Author contributions: J.A.S., S.A., J.S. and R.C.C. designed the study. A.H. perfomed CellSearch analyses and coordinated DEPArray single cell sorting. B.R., K.S.G., O.O. and L.W. enrolled the patients. D.S.G. and K.P. performed next generation sequencing and analyzed the data. R.K.H. and J.L. supported bioinformatics and D.F.G. performed all statistical analyses. J.A.S., R.C.C. and D.S.G. wrote the manuscript with assistance and final approval from all authors.
Competing interests: All authors declare that they have no competing interests.
References
- 1.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science translational medicine. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 2.Guttery DS, Page K, Hills A, Woodley L, Marchese SD, Rghebi B, et al. Noninvasive Detection of Activating Estrogen Receptor 1 (ESR1) Mutations in Estrogen Receptor-Positive Metastatic Breast Cancer. Clinical chemistry. 2015;61:974–82. doi: 10.1373/clinchem.2015.238717. [DOI] [PubMed] [Google Scholar]
- 3.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 4.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature genetics. 2013;45:1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nature genetics. 2013;45:1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rack B, Schindlbeck C, Juckstock J, Andergassen U, Hepp P, Zwingers T, et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. Journal of the National Cancer Institute. 2014;106 doi: 10.1093/jnci/dju066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 8.Smith BM, Slade MJ, English J, Graham H, Luchtenborg M, Sinnett HD, et al. Response of circulating tumor cells to systemic therapy in patients with metastatic breast cancer: comparison of quantitative polymerase chain reaction and immunocytochemical techniques. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2000;18:1432–9. doi: 10.1200/JCO.2000.18.7.1432. [DOI] [PubMed] [Google Scholar]
- 9.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. The New England journal of medicine. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Bonnefoi H, Diebold-Berger S, Lyautey J, Lederrey C, Faltin-Traub E, et al. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 1999;5:2297–303. [PubMed] [Google Scholar]
- 11.Shaw JA, Smith BM, Walsh T, Johnson S, Primrose L, Slade MJ, et al. Microsatellite alterations plasma DNA of primary breast cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6:1119–24. [PubMed] [Google Scholar]
- 12.Silva JM, Dominguez G, Garcia JM, Gonzalez R, Villanueva MJ, Navarro F, et al. Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer research. 1999;59:3251–6. [PubMed] [Google Scholar]
- 13.Shaw JA, Page K, Blighe K, Hava N, Guttery D, Ward B, et al. Genomic analysis of circulating cell-free DNA infers breast cancer dormancy. Genome Res. 2012;22:220–31. doi: 10.1101/gr.123497.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. The New England journal of medicine. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 15.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–5. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Molecular oncology. 2015;9:783–90. doi: 10.1016/j.molonc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nature medicine. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 20.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO molecular medicine. 2014;6:1371–86. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PloS one. 2013;8:e77963. doi: 10.1371/journal.pone.0077963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. Journal of oncology. 2010;2010 doi: 10.1155/2010/617421. 617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung ML, Wang Y, Waters J, Navin NE. SNES: single nucleus exome sequencing. Genome biology. 2015;16:55. doi: 10.1186/s13059-015-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCall CM, Mosier S, Thiess M, Debeljak M, Pallavajjala A, Beierl K, et al. False positives in multiplex PCR-based next-generation sequencing have unique signatures. The Journal of molecular diagnostics : JMD. 2014;16:541–9. doi: 10.1016/j.jmoldx.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 29.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical chemistry. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaver JA, Jelovac D, Balukrishna S, Cochran RL, Croessmann S, Zabransky DJ, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:2643–50. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nature methods. 2013;10:1003–5. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keshaviah A, Dellapasqua S, Rotmensz N, Lindtner J, Crivellari D, Collins J, et al. CA15-3 and alkaline phosphatase as predictors for breast cancer recurrence: a combined analysis of seven International Breast Cancer Study Group trials. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2007;18:701–8. doi: 10.1093/annonc/mdl492. [DOI] [PubMed] [Google Scholar]
- 35.Bidard FC, Peeters DJ, Fehm T, Nole F, Gisbert-Criado R, Mavroudis D, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. The Lancet Oncology. 2014;15:406–14. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 36.Strauss WM, Carter C, Simmons J, Klem E, Goodman N, Vahidi B, et al. Analysis of tumor template from multiple compartments in a blood sample provides complementary access to peripheral tumor biomarkers. Oncotarget. 2016 doi: 10.18632/oncotarget.8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auer M, Heitzer E, Ulz P, Geigl JB, Speicher MR. Single circulating tumor cell sequencing for monitoring. Oncotarget. 2013;4:812–3. doi: 10.18632/oncotarget.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–85. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Wang S, Hou C, Xing Y, Cao J, Wu K, et al. Genome-wide detection of copy number variations among diverse horse breeds by array CGH. PloS one. 2014;9:e86860. doi: 10.1371/journal.pone.0086860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer research. 2013;73:2965–75. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 41.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, et al. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4586–96. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, et al. D538G mutation in estrogen receptor-alpha: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer research. 2013;73:6856–64. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 44.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports. 2013;4:1116–30. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Science translational medicine. 2015;7:313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. The Biochemical journal. 2008;409:519–24. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 47.Van Keymeulen A, Lee MY, Ousset M, Brohee S, Rorive S, Giraddi RR, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–23. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 48.Ellis MJ, Lin L, Crowder R, Tao Y, Hoog J, Snider J, et al. Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast cancer research and treatment. 2010;119:379–90. doi: 10.1007/s10549-009-0575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller TW, Hennessy BT, Gonzalez-Angulo AM, Fox EM, Mills GB, Chen H, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. The Journal of clinical investigation. 2010;120:2406–13. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 51.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10208–13. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabine VS, Crozier C, Brookes CL, Drake C, Piper T, van de Velde CJ, et al. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:2951–8. doi: 10.1200/JCO.2013.53.8272. [DOI] [PubMed] [Google Scholar]
- 53.Loibl S, von Minckwitz G, Schneeweiss A, Paepke S, Lehmann A, Rezai M, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:3212–20. doi: 10.1200/JCO.2014.55.7876. [DOI] [PubMed] [Google Scholar]
- 54.Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome medicine. 2013;5:70. doi: 10.1186/gm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hvichia GE, Parveen Z, Wagner C, Janning M, Quidde J, Stein A, et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. International journal of cancer Journal international du cancer. 2016 doi: 10.1002/ijc.30007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.