Abstract
The endosomal sorting complexes required for transport (ESCRT) pathway mediates cellular membrane remodeling and fission reactions. The pathway comprises five core complexes: ALIX, ESCRT-I, ESCRT-II, ESCRT-III, and Vps4. These soluble complexes are typically recruited to target membranes by site-specific adaptors that bind one or both of the early-acting ESCRT factors: ALIX and ESCRT-I/ESCRT-II. These factors, in turn, nucleate assembly of ESCRT-III subunits into membrane-bound filaments that recruit the AAA ATPase Vps4. Together, ESCRT-III filaments and Vps4 remodel and sever membranes. Here, we review recent advances in our understanding of the structures, activities, and mechanisms of the ESCRT-III and Vps4 machinery, including the first high-resolution structures of ESCRT-III filaments, the assembled Vps4 enzyme in complex with an ESCRT-III substrate, the discovery that ESCRT-III/Vps4 complexes can promote both inside-out and outside-in membrane fission reactions, and emerging mechanistic models for ESCRT-mediated membrane fission.
Keywords: ESCRT pathway, membrane remodeling, membrane fission, ESCRT-III, Vps4, AAA ATPase
1. THE ESCRT PATHWAY
1.1. ESCRT-Dependent Membrane Fission Reactions
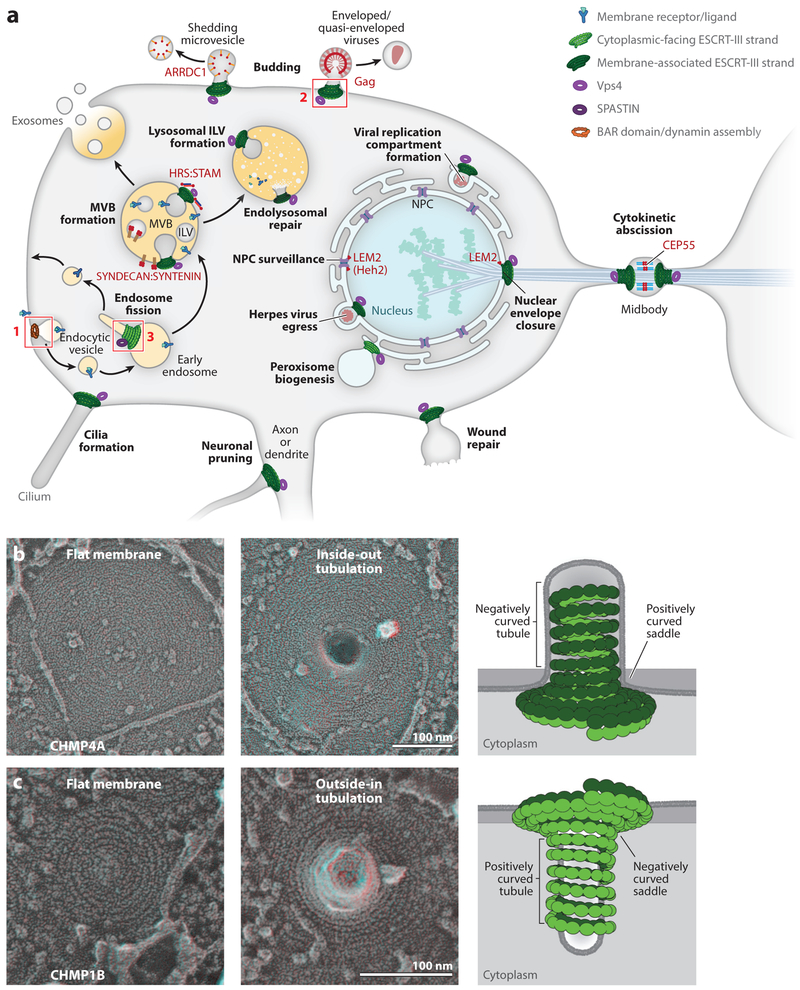
The ESCRT pathway mediates membrane fission reactions throughout the cell (Christ et al. 2017, Frankel & Audhya 2018, Lippincott-Schwartz et al. 2017, Schöneberg et al. 2017, Scourfield & Martin-Serrano 2017, Stoten & Carlton 2018) (Figure 1a). Cytoplasmic protein complexes like the ESCRT machinery can separate a single continuous lipid bilayer into two discontinuous bilayers via one of two reciprocal orientations: one in which the opposing membranes are drawn together to create a membrane neck that encircles cytoplasm (here termed inside-out membrane fission) and another in which the membrane neck is surrounded by cytoplasm (outside-in membrane fission). During outside-in fission, the cytoplasmic machinery binds and stabilizes positively curved membrane tubules and negatively curved saddles, whereas in inside-out fission, the cytoplasmic machinery binds negatively curved tubules and positively curved saddles (McMahon & Boucrot 2015) (Figure 1b,c). Endocytic vesicle formation exemplifies an outside-in fission reaction (Figure 1a, box 1), in which the bud neck is surrounded by cytoplasm and the well-characterized BAR/dynamin family fission machineries bind outside and constrict the positively curved membrane necks to form budded vesicles. By contrast, ESCRT-III/Vps4 complexes are the only well-characterized cytoplasmic assemblies that can mediate inside-out membrane fission. Enveloped virus budding exemplifies this process (Figure 1a, box 2), in which the bud neck encircles cytoplasm and ESCRT-III filaments bind within the membrane neck and draw the opposing membranes toward the fission point. This unusual but essential activity explains why the ESCRT machinery has evolved to participate in so many different reactions of this orientation throughout the cell (Figure 1a).
Figure 1. ESCRT-dependent membrane fission reactions.
(a) Cellular processes proposed to be ESCRT dependent are labeled in bold, cellular structures are labeled in normal font, membrane-specific adaptor proteins that recruit early-acting ESCRT factors are labeled in red (with human protein names in all caps and yeast protein names, where provided, in parentheses), and a stylized membrane receptor and its ligand are shown in blue/turquoise. Sites of ESCRT membrane remodeling are denoted by stylized double-stranded ESCRT-III filaments (green, with the membrane-associated strand in dark green) and rings of Vps4 (violet) or the related meiotic clade AAA ATPase, SPASTIN (dark purple). Box 1 shows the site of endocytic vesicle formation, with stylized BAR domain/dynamin assemblies (orange) formed about the neck of an endocytic vesicle. Box 2 shows the site of enveloped virus budding, with double-stranded ESCRT-III filaments stabilizing a negatively curved membrane tubule (see panel b). Box 3 shows a site of endosomal vesicle formation, with double-stranded ESCRT-III filaments stabilizing a positively curved membrane tubule (see panel c). Abbreviations: ILV, intralumenal vesicle; MVB, multivesicular body; NPC, nuclear pore complex. (b) Filaments containing the ESCRT-III protein CHMP4A can stabilize flat membranes and negatively curved plasma membrane tubules and thereby promote inside-out membrane fission reactions. The left and middle panels show electron micrographs of human cells overexpressing CHMP4A under conditions of reduced VPS4 ATPase activity (Hanson et al. 2008, McCullough et al. 2015). These electron micrographs show that cytoplasmic filaments containing CHMP4A can assemble on flat membranes (left panel) and can promote membrane tubule extrusion and coat the interior of the negatively curved tubules, consistent with a role in inside-out fission reactions (middle panel). The right panel shows a schematic depiction of the middle panel (side view), (c) Filaments containing the ESCRT-III protein CHMP1B can stabilize flat membranes and positively curved plasma membrane tubules and thereby promote outside-in membrane fission reactions (Cashikar et al. 2014, Hanson et al. 2008, McCullough et al. 2015). The three subpanels in panel c are analogous to those in panel b, except that CHMP1B promotes membrane tubule invagination and coats the exterior of the positively curved tubules. Panels b and c are adapted from McCullough et al. (2015) and reprinted with permission from AAAS.
1.2. Inside-Out ESCRT-Dependent Membrane Fission Reactions
The ever-growing list of cellular membrane remodeling reactions performed by the ESCRT pathway has been reviewed extensively (Christ et al. 2017, Lippincott-Schwartz et al. 2017, Schôneberg et al. 2017, Scourfield & Martin-Serrano 2017, Stoten & Carlton 2018). Here, we briefly summarize cellular ESCRT functions, highlighting how the machinery functions across a remarkable range of temporal and spatial dimensions and can mediate membrane fission in both possible orientations.
The ESCRT machinery was first characterized in yeast, owing to its essential role in the formation of intralumenal vesicles (ILVs) (Bryant & Stevens 1998, Henne et al. 2013). ILVs carry cargo into the lumen of late endosomes, where they can either be degraded if the late endosome fuses with the vacuole/lysosome (Frankel & Audhya 2018) or be released from the cell as exosomes if the endosome fuses with the plasma membrane (Jackson et al. 2017, Juan & Fürthauer 2018) (Figure 1a). ILVs are small (~25–30 nm in diameter), and their bud necks are narrow (<10 nm). A recent study has defined the global parameters and coordinated assembly of ESCRT-III/Vps4 activity during yeast ILV formation by showing that budding sites act within a short time window (3—45 s) and comprise several hundred copies of different ESCRT-III subunits [e.g., 75–200 copies of Snf7 (CHMP4) and 15–50 copies of Vps24 (CHMP3)] and two or more Vps4 hexamers (Adell et al. 2017). The budding of enveloped viruses from the plasma membrane appears to be an analogous vesiculation process, as the bud neck is again narrow and the ESCRT-III/Vps4 machinery accumulates and is released rapidly (Baumgartel et al. 2011, Bleck et al. 2014, Feng et al. 2013, Jouvenet et al. 2011). Other analogous ESCRT-dependent vesiculation processes include ILV budding directly into the vacuole (the yeast equivalent of the lysosome) (Zhu et al. 2017, Caspi & Dekker 2018), shedding microvesicle release from the plasma membrane (Choudhuri et al. 2014, Matusek et al. 2014, Nabhan et al. 2012), and possibly also nuclear egress of the Herpes viral core particle (Lee et al. 2012) and vesicle secretion at the ciliary transition zone (Diener et al. 2015, Wood et al. 2013).
The primordial function of the ESCRT pathway appears to have been in cytokinetic abscission, and the pathway still performs this function in animals, in some archaea, and possibly also in plants (Carlton & Martin-Serrano 2007, Ettema & Bernander 2009, Gao et al. 2017, Lindas et al. 2008, Morita et al. 2007, Samson et al. 2008). During animal cell cytokinesis, the cleavage furrow ingresses to create the midbody, a thin intercellular membrane bridge that connects the two daughter cells. The ESCRT machinery is recruited to the midbody by the CEP55 adaptor, which binds both ESCRT-I and ALIX (Carlton & Martin-Serrano 2007, Lee et al. 2008, Morita et al. 2007). ESCRT-III filaments form on either side of the central Flemming body and constrict the midbody to points of fission located 0.7–1 μm away (Elia et al. 2011, Guizetti et al. 2011). The cytokinetic abscission reaction differs considerably from ILV formation in both scale and timing because the ESCRT machinery accumulates and acts over a much longer time window (~80 min) and the ESCRT-III-lined bud neck within the midbody is nearly 1 μm in length and 1.25 μm in diameter (Elia et al. 2011, 2012). Moreover, the ESCRT-III filaments within the midbody appear to be much wider (~17 nm) than those formed during vesiculation reactions (~5 nm) and may therefore comprise bundles of ESCRT-III filaments and potentially other proteins (Guizetti et al. 2011, Mierzwa et al. 2017).
The ESCRT machinery can also seal membrane fenestrations at several sites, including the nuclear envelope (Denais et al. 2016, Olmos et al. 2015, Raab et al. 2016, Vietri et al. 2015), the plasma membrane (Jimenez et al. 2014, Scheffer et al. 2014), and endolysosomes (Skowyra et al. 2018). The nuclear membrane has two opposed bilayers, and holes that cross both membranes are therefore filled with cytoplasm and are topologically equivalent to ILV bud necks. Specialized ESCRT-III/Vps4 complexes can seal openings in the nuclear envelope that are produced by a series of processes, including reformation of the postmitotic nucleus (Gu et al. 2017; Olmos et al. 2015, 2016; Vietri et al. 2015), damage due to shear stress (Denais et al. 2016, Raab et al. 2016), and removal of embedded complexes like the nuclear pore complex (Webster et al. 2014, 2016) and the spindle pole body (Gu et al. 2017). ESCRT-dependent wound repair of plasma membranes and endolysosomal membranes differs from nuclear membrane sealing because these membranes are single bilayers. Here, the ESCRT machinery is presumed to act by drawing the membranes together to extrude and release the damaged region, much like the vesicle release reactions. Analogous ESCRT-dependent reactions may also be used to prune neuronal axons and dendrites (Loncle et al. 2015).
Finally, there appear to be several cases in which ESCRT-III membrane remodeling activities occur without a subsequent fission step. These include the formation of CUPs (compartments for unconventional protein secretion), which are cup-shaped endoplasmic reticulum (ER)-derived vesicles involved in noncanonical protein secretion (Curwin et al. 2016), and the invagination of ER membranes to form the replication compartments of some positive-stranded RNA viruses (Barajas et al. 2009, Diaz et al. 2015).
1.3. Outside-In ESCRT-Dependent Membrane Fission
A surprising recent development is the discovery that some ESCRT-III filaments can stabilize tubules of positive membrane curvature, apparently en route to outside-in membrane fission reactions that release vesicles from the surface of the endosome and possibly also the ER (Allison et al. 2013, Mast et al. 2018, McCullough et al. 2015) (Figure 1). Despite the important caveat that these systems are not yet as well characterized as the inside-out reactions, there is increasing evidence that two ESCRT-III proteins, IST1 (CHMP8) and CHMP1B, can act together with the Vps4-related AAA ATPase SPASTIN to facilitate vesicle budding from tubular endosomes (Allison et al. 2013, McCullough et al. 2015). Other components, including microtubules and SNX4, have also been implicated in this process, which apparently carries membrane proteins like the transferrin receptor from endosomes back to the plasma membrane (Allison et al. 2013, Traer et al. 2007). As shown in Figure 1b, overexpression of the canonical ESCRT-III subunit, CHMP4A, in a human cell lacking VPS4 activity induces the extrusion of negatively curved membrane tubules from the plasma membrane that are filled by thin (~5-nm), spiraling ESCRT-III filaments. By contrast, overexpression of CHMP1B alone in a cell lacking VPS4 activity induces the invagination of positively curved membrane tubules from the plasma membrane (Figure 1c). In the latter case, the thin ESCRT-III filaments wrap around the tubule exterior. In both cases, the filaments also assemble on flat patches of the membrane that surround the tubule. This experiment implies that different ESCRT-III filaments can stabilize membrane tubules that curve in opposite directions (as well as flat membrane patches). In the following sections, we review what is known about the structures and activities of ESCRT-III subunits and filaments and of the Vps4 ATPase and discuss how they might work together to remodel membranes and promote membrane fission.
2. ESCRT-III
Assembly of the ESCRT machinery initiates when site-specific adaptors bind one (or more) of the early-acting ESCRT factors: ALIX, ESCRT-I/ESCRT-II, or the ESCRT-II/ESCRT-III hybrid protein CHMP7. As reviewed elsewhere (Christ et al. 2017, Schöneberg et al. 2017), these early-acting factors can bind adaptors, ubiquitinated cargoes, and membranes, and their critical functions include concentrating vesicular cargoes, initiating membrane bending, and nucleating the assembly of ESCRT-III subunits into membrane-bound filaments. Humans express 12 related ESCRT-III proteins, which can be grouped into eight different families, termed CHMP1–7 and IST1 (CHMP8) (Christ et al. 2017, Schöneberg et al. 2017). Several of the human protein families contain two or three homologous gene products (e.g., CHMP4A–C), whereas Saccharomyces cerevisiae encodes just a single gene for each family. At present, we have only a partial understanding of the biological rationale for the existence of so many different ESCRT-III proteins. In yeast, Snf7 (CHMP4), Vps24 (CHMP3), and Vps2 (CHMP2) appear to form the essential filaments, whereas the other subunits have been assigned more specialized roles, including filament initiation [Vps20 (CHMP6)], regulation [Did2 (CHMP1), Vps60 (CHMP5), and Ist1 (IST1/CHMP8)], and site-specific roles like nuclear membrane remodeling [Chm7 (CHMP7)]. Inside-out ESCRT-dependent reactions in human cells also appear to require at least one CHMP4 and CHMP2 family member, although an absolute requirement for CHMP3 in vivo is less clear. Other subunits again appear to play accessory roles or to function in more specialized roles such as nuclear envelope closure [CHMP7 (Vietri et al. 2015)] and maintenance of the abscission checkpoint [CHMP4C (Carlton et al. 2012)]. As discussed below, it is also possible that different ESCRT-III subunits favor different degrees of filament curvature, with an extreme example being specialized roles for CHMP1B and IST1 in stabilizing positively curved membrane tubules.
2.1. Subunit Structures
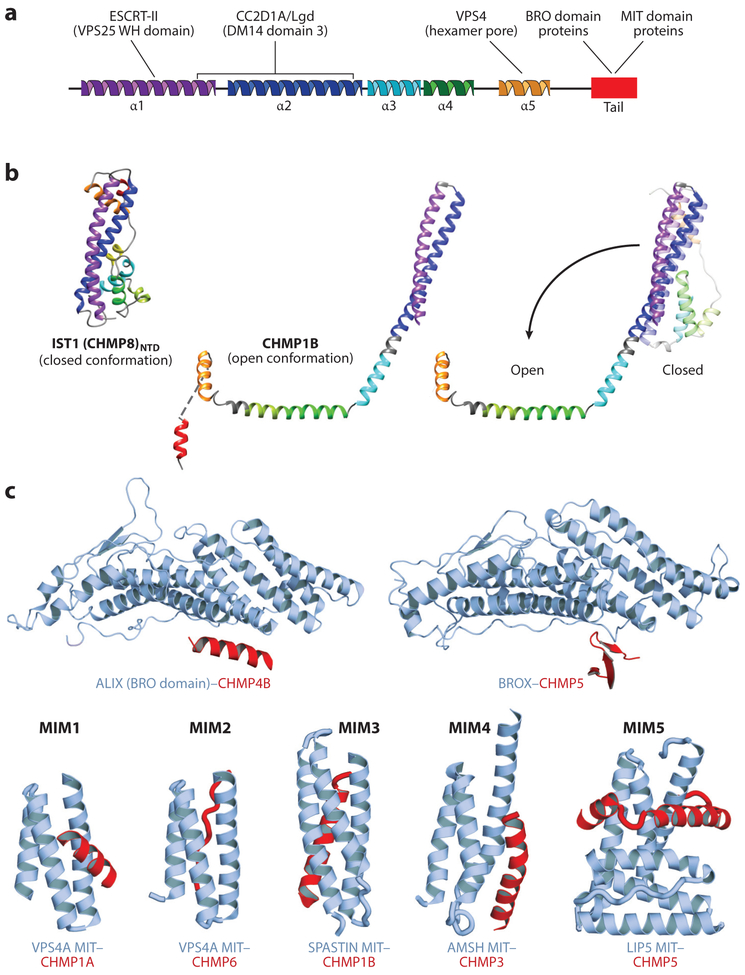
Many ESCRT-III subunits can interconvert between two conformations: a closed, monomeric soluble state and an open, polymeric, membrane-bound state (Lata et al. 2008a, Lin et al. 2005, McCullough et al. 2015, McMillan et al. 2016, Tang et al. 2015) (Figure 2a,b). Crystal structures of CHMP3 and IST1 demonstrate that the closed state is organized by a long N-terminal helical hairpin that spans the entire domain (Bajorek et al. 2009, Muziol et al. 2006, Xiao et al. 2009). Helices 3 and 4 pack against the open end of the hairpin and are connected via a linker of variable length to helix 5, which packs against the closed end of the helical hairpin. In IST1, the variable linker contains short helices (named A, B to maintain a common numbering scheme for the conserved core helices). Sequences beyond helix 5 typically lack persistent structure but contain a series of different ligand binding sites, including elements for binding MIT domain proteins (most ESCRT-III subunits) and BRO domain proteins (CHMP5 and CHMP4) (Figure 2c).
Figure 2. Structures and binding partners of the ESCRT-III proteins.
(a) Secondary structure showing the five conserved helices that organize ESCRT-III proteins in the closed conformation. The terminal ligand-binding tail (red) is helical in most ESCRT-III complexes but can alternatively adopt a β-strand secondary structure in some cases (see panel c). ESCRT-III ligands and their approximate binding sites are shown above the secondary structure, (b) Structures of ESCRT-III subunits in their open and closed configurations. (Left to right) Crystal structure of the ESCRT-III subunit IST1NTD in the closed conformation (from Bajorek et al. 2009), cryo-EM structure of CEIMP1B in the open conformation (from McCullough et al. 2015), and superposition of the closed (lighter shades, modeled) and open (darker shades) conformations of CHMP1B (from McCullough et al. 2015, Talledge et al. 2018). The N-terminal helical hairpin remains intact (and is extended upon opening), while the remaining helices either pack against the hairpin (closed conformation) or open to pack against other subunits in the CHMP1B filament (open conformation; see Figure 3b). (c) Structures of the C-terminal tails of ESCRT-III proteins (red) in complex with their two major classes of binding partners (blue-gray): BRO domain proteins such as ALIX and BROX (above) and MIT domain proteins such as VPS4, SPASTIN, AMSH, and LIP5 (Vta1) (below). Note the variety of distinct ways in which different ESCRT-III tails can bind BRO and MIT domains [denoted MIT-interacting motifs (MIMs) 1–5 in the MIT case]. Structures above are from McCullough et al. (2008) (left) and Mu et al. (2012) (right). Structures below (from left to right) are from Stuehell-Brereton et al. (2007), Kieffer et al. (2008), Yang et al. (2008), Solomons et al. (2011), and Skalieky et al. (2012).
ESCRT-III subunits open dramatically to create a polymerization-competent, membrane-binding conformation (Figure 2b). Open conformations have been visualized in structures of human CHMP1B (McCullough et al. 2015, Talledge et al. 2018) and truncated CHMP4 homologs from S. cerevisiae (Snf7) and Drosophila (Shrub) (McMillan et al. 2016, Tang et al. 2015). The open conformations resemble an arm, with closed helices 1–3 rearranging to form an extended helical hairpin that defines the upper arm; helix 4 and helix A combining to form the helical forearm; and helix 5 (missing in the two CHMP4 structures) forming the hand. The joints between the final three helices correspond to the elbow and the wrist. Although the structure of a single ESCRT-III subunit is not yet available in both the open and closed states, we have generated a structure-based homology model for the CHMP1B closed state and have used it to visualize the conformational changes that accompany subunit opening and polymerization (McCullough et al. 2015). During this transformation, the N-terminal helical hairpin remains intact (but extends), whereas the helix 5 “hand” is displaced by ~ 100 Å. Remarkably, this dramatic movement requires only three local rearrangements: an opening of the elbow angle and conversion of the loops that connect helix 2/3 and helix 4/A into helical elements that create longer, continuous helices that extend the upper arm to the elbow and create the forearm (Figure 2b). The open conformation lacks an extensive hydrophobic core but is stabilized by intersubunit interactions within polymeric ESCRT-III filaments (described below).
2.2. ESCRT-III Filament Nucleation
ESCRT-III filament formation can be promoted by membrane curvature (Lee et al. 2015); by binding partners; and by modifications that favor the open state [or removal of modifications that restrict assembly (Crespo-Yàñez et al. 2018)], concentrate the subunits, and/or properly orient subunits on a target membrane. The two best characterized nucleators of ESCRT-III polymerization, ALIX and ESCRT-II, appear to employ these principles, although their detailed mechanisms are not yet fully understood. Both ALIX and ESCRT-II ultimately recruit CHMP4, which participates in all known inside-out ESCRT-dependent membrane fission reactions. The Y-shaped ESCRT-II complex displays two EAP20 (Vps25) subunits at the ends of two arms (Hierro et al. 2004, Teo et al. 2004). These EAP20 subunits bind CHMP6 (Vps20), which in turn binds CHMP4 (Snf7) (Im et al. 2009). The myristoylated CHMP6 (Vps20) subunit preferentially adopts an open conformation, consistent with its role in filament nucleation (Fyfe et al. 2011). In the ALIX case, the N-terminal BRO domain binds directly to a C-terminal CHMP4 helix located immediately downstream of the ordered ESCRT-III core domain (Figure 2). ALIX (Bro1) binding may thereby weaken autoinhibitory interactions, consistent with the observation that CHMP4 autoinhibition can be relieved by removing the terminal helix (Lata et al. 2008b, 2009; Tang et al. 2015, 2016). ALIX can dimerize (Pires et al. 2009), and both ESCRT-II and ALIX may, therefore, help activate ESCRT-III subunits to form two filaments or even double-stranded filaments (Henne et al. 2012, McCullough et al. 2015, Mierzwa et al. 2017, Teis et al. 2010). This activity is reminiscent of the activities of actin and microtubule nucleators, which bind and orient two actin monomers (Rottner et al. 2017) or heterodimeric tubulin subunits (Kollman et al. 2011).
2.3. ESCRT-III Filament Structures
Structures are available for model ESCRT-III filaments visualized in two different contexts: (a) a high-resolution cryo-EM reconstruction of the double-stranded heteropolymeric filaments formed by CHMP1B and the ordered N-terminal ESCRT-III domain of IST1 (IST1NTD) (both free and in complex with nucleic acids) (McCullough et al. 2015, Talledge et al. 2018), and (b) linear filaments of truncated homologs of CHMP4 from S. cerevisiae (Snf7) and Drosophila (Shrub) that crystallized with similar lattice packing interactions, both of which recapitulate interactions seen in the CHMP1B strand of the IST1NTD/CHMP1B filament (McMillan et al. 2016, Tang et al. 2015). Both sets of structures are informative but also suffer from some limitations. The double-stranded IST1NTD/CHMP1B filament structure is more complete than the crystalline filaments, but this filament stabilizes positive membrane curvature and therefore presents a challenge for generalizing to conventional ESCRT-III filaments. The CHMP4 filaments, by contrast, are composed of a subunit that is universally involved in stabilizing negative membrane curvature, but the truncated constructs that were crystallized lack the C-terminal forearm, hand, and downstream Bro1 (ALIX) and MIT-interacting motifs (MIMs), and their filaments are necessarily homopolymeric and linear in the crystal lattice. Fortunately, the two different model structures share important similarities, reinforcing the idea that they provide insights into the conserved organizational principles of ESCRT-III filaments (Figure 3).
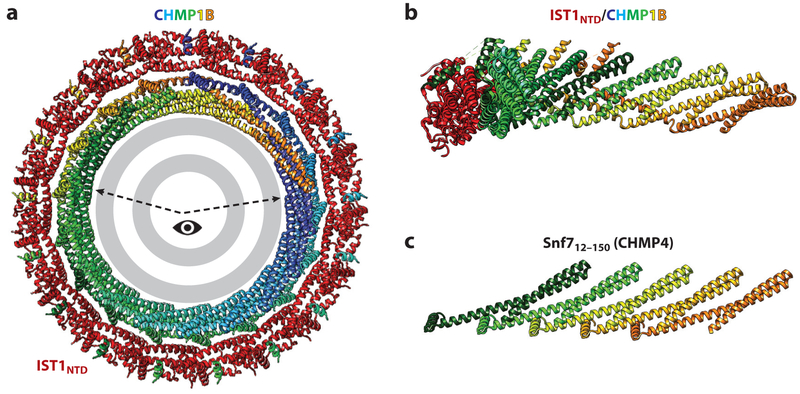
Figure 3. ESCRT-III filament structures.
(a) End-on view of a turn of the N-terminal ESCRT-III domain of an IST1NTD/CHMP1B filament surrounding a stylized lipid bilayer. IST1NTD subunits are shown in red, CHMP1B subunits are shown in rainbow colors, and the lipid bilayer is shown in gray. The structure is from McCullough et al. (2015). (b) Side view of a segment of the IST1NTD/CHMP1B filament (viewed from the membrane and corresponding to the wedge highlighted in panel a). Seven interacting CHMP1B subunits are shown, with just a single associated ISTNTD subunit shown for clarity. (c) Equivalent view showing a linear strand of Snf712–150 and emphasizing the equivalent packing of N-terminal helical hairpins in the two ESCRT-III strands. The structure is from Tang et al. (2015).
In the double-stranded IST1NTD/CHMP1B filament, closed IST1NTD subunits form the outer strand, and open CHMP1B subunits form the inner strand (McCullough et al. 2015). The double-stranded filament is ~7 nm thick, and it wraps up into a regular helix with an external diameter of 24 nm and 17 subunits per turn. Adjacent turns of the helix stack primarily through ionic interactions, with a highly basic side of the filament contacting an acidic side. The double-stranded nature of the filament was unexpected, as was the observation that IST1NTD can polymerize in its closed conformation. It remains to be determined whether all biologically relevant ESCRT-III filaments exhibit these properties. The closed IST1NTD subunits exhibit limited homomeric contacts, interacting end to end through contacts between the N-terminal end of helix 2 and the C-terminal end of an adjacent helix 2. By contrast, subunits along the open-state CHMP1B strand interact extensively. Each CHMP1B molecule interacts with eight other highly intercalated CHMP1B subunits. Hairpins from each subunit pack side by side at an angle of ~30° relative to the filament axis, and they cross the forearm of the first subunit at four different sites (Figure 3a,b). In addition, the helix 5 hand contacts the shoulder of the hairpin four subunits away, making a domain-swapped contact that is analogous to the intrasubunit hand-shoulder interaction observed in the closed ESCRT-III conformation (Figure 2b). The two different strands interact along the extended helix 4 of CHMP1B, with every copy of CHMP1B helix 4 binding three different segments of IST1NTD helix 1. An emerging cryo-EM analysis of the IST1NTD/CHMP1B assembly at higher resolution has further revealed that the CHMP1B tail “snorkels” from the inner strand to the outer strand, where the short MIM helix packs antiparallel against helix 5 on the surface of the closed IST1 subunit (Talledge et al. 2018) (Figure 3). This interaction further stabilizes the IST1NTD/CHMP1B filament and matches an interaction seen previously in the crystal structure of yeast Ist1 bound to the homologous Did2 (CHMP1) MIM peptide (Xiao et al. 2009). Thus, the inner and outer strands of IST1 and CHMP1B are woven together, and the MIM-containing tails of both subunits are arrayed around the tube exterior, where they are available to bind MIT domains.
As illustrated in Figure 3b,c, truncated Snf7 and Shrub (CHMP4) constructs form a very similar open N-terminal hairpin in which the residues that form the second and third helices and intervening loop in the closed conformation associate into a single extended second helix of the N-terminal helical hairpin (corresponding to the CHMP1B forearm). These helical hairpins again pack side to side, recapitulating the intermolecular hairpin stacking interactions seen in the open CHMP1B strand. Importantly, CHMP4 filament formation can be blocked by Lgd (CC2D1A/B), a negative regulator of the multivesicular body (MVB) pathway (Drusenheimer et al. 2015, Martinelli et al. 2012, Troost et al. 2012, Usami et al. 2012), and a recent crystal structure has shown how the third DM14 domain of Lgd binds the isolated Shrub hairpin to occlude polymerization sterically (McMillan et al. 2017). Thus, the similarity of open subunit structures and interactions seen in the different ESCRT-III filaments, as well as the conserved mechanisms of the Lgd (CCD1) inhibitors (Martinelli et al. 2012, McMillan et al. 2017), indicate broad evolutionary conservation of the opening, polymerization, and regulatory mechanisms (McCullough et al. 2015, McMillan et al. 2016, Tang et al. 2015).
2.4. Filament Flexibility, Curvature, and Membrane Binding
Flexibility appears to be an intrinsic property of ESCRT-III filaments. The highly interlocked and domain-swapped nature of the CHMP1B strand of the IST1NTD/CHMP1B filament—in which each monomer interacts with eight neighbors by virtue of a CHMP1Bi to CHMP1Bi+4 domain swap—suggests that this strand will be unusually robust to bending (Chiaruttini & Roux 2017, McCullough et al. 2015, Mierzwa et al. 2017). By contrast, actin also polymerizes through noncovalent interactions, but the F-actin filament contains interactions between globular nearest neighbors only. Bending forces that disrupt these nearest-neighbor interactions therefore sever the filaments, whereas the longer-range domain-swapped interactions should make the CHMP1B strand more tolerant to bending through a range of curvatures (McCullough et al. 2015). Moreover, the elbow and wrist joints within the CHMP1B strand provide several potential sites of flexibility, which may also be accommodated by limited homomeric IST1NTD interactions in the outer strand. Consistent with these arguments, CHMP4 filaments exhibit unusually high flexibility and much shorter persistence lengths than does actin (300–800 nm versus ~ 17 μm) (Chiaruttini & Roux 2017, Chiaruttini et al. 2015, Shen et al. 2014).
Filaments with a preferred membrane-binding surface can induce membranes to follow their trajectories. As for all polymeric filaments, the angle between nearest-neighbor CHMP1B subunits determines the trajectory and magnitude of filament curvature. For helical IST1NTD/CHMP1B filaments with ~17 subunits per turn, the average intersubunit angle is ~21°. These helical tubes bind very highly curved membranes within their lumens (McCullough et al. 2015), which is consistent with their postulated role in binding positively curved endosomal tubules and facilitating outside-in vesicle formation. In this case, membrane binding is mediated by CHMP1B helix 1, whose highly basic face lines the tube interior. The membrane-binding site on CHMP4 (Snf7/Shrub) filaments is currently less certain than the membrane-binding site on the CHMP1B surface. Mutational analyses point to a role for helix 2 in Snf7 membrane binding (Buchkovich et al. 2013, Tang et al. 2015). However, the basic character of helix 1 is conserved across different ESCRT-III subunits, and this helix has been postulated to form a common membrane-binding surface for other ESCRT-III subunits, as observed for CHMP1B (Buchkovich et al. 2013, McCullough et al. 2015, Muziol et al. 2006).
2.5. Reconciling Roles for ESCRT-III Filaments in Stabilizing Positive and Negative Membrane Curvature
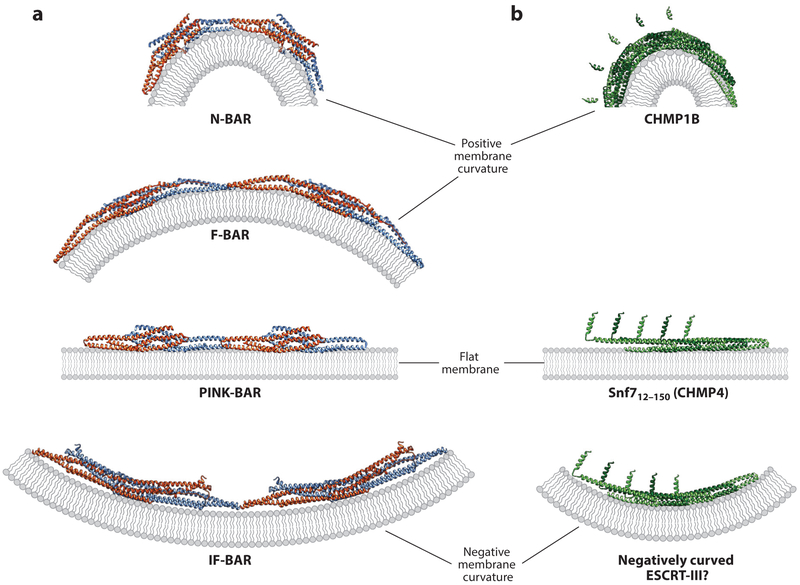
The discovery that different ESCRT-III filaments can stabilize positive, negative, and zero membrane curvature was unexpected, but not without precedent. Different members of the BAR domain family of proteins can also stabilize membranes that curve in both directions (Cannon et al. 2017, McMahon & Boucrot 2015, Mim & Unger 2012, Simunovic et al. 2015) (Figure 4a). In all cases, similar BAR domain dimers associate end to end to form analogous helical filaments, which bind membranes through the same basic surface. BAR domain–driven membrane bending is partly a function of the intrinsic shape of the dimer, and this property is amplified by polymerization. Membrane curvature is dictated by the trajectory of the BAR domain polymers, which in turn is determined by the angle between adjacent dimeric subunits. As illustrated in Figure 4b, we envision that a similar situation could explain the properties of different ESCRT-III filaments, where differences in the preferred trajectory of the filament could alter the direction and magnitude of membrane curvature. Alternatively, ESCRT-III filaments that stabilize positive versus negative curvature could bind membranes by using opposite surfaces of the filament (e.g., helix 1 in CHMP1B versus helix 2 in CHMP4). That is not what happens in the BAR domain case, however, and we find it less plausible in the case of ESCRT-III polymers.
Figure 4. Models for stabilization of curved and flat membranes by BAR domain and ESCRT-III proteins.
(a) Illustrations showing how changing the angle between the end-associated BAR domain dimers (blue and orange subunits) can stabilize a continuum of differentially curved membranes (gray). Pairs of dimers from continuous BAR domain assemblies (end-on views) are shown in each case. Structural models are based on Mim & Unger (2012) and Mim et al. (2012) for the N-BAR case, Shimada et al. (2007) and Frost et al. (2008) for the F-BAR case, Guerrier et al. (2009) and Sporny et al. (2017) for the IF-BAR case, and Pykalainen et al. (2011) for the PINK-BAR case, (b) Illustrations showing how changes in intrinsic filament curvature could similarly allow ESCRT-III filaments (green) to stabilize a continuum of differentially curved membranes. (Top to bottom) A CHMP1B strand from the IST1NTD/CHMP1B filament bound to a stylized membrane (from McCullough et al. 2015), structure of the linear strand of Snf712–150 (CHMP4) from a crystal lattice (from Tang et al. 2015) bound to a stylized membrane, and a hypothetical strand of Snf712–150 (CHMP4) subunits bound to a negatively curved membrane. The Snf712—150 (CHMP4) strand in the bottom panel was modeled by altering the angle between each successive subunit in the strand shown in the middle panel.
2.6. Outstanding Issues
As discussed above, the IST1NTD/CHMP1B structure appears to explain how that particular ESCRT-III filament can stabilize positively curved membrane tubules, but we lack a similar understanding of how CHMP4-containing ESCRT-III filaments stabilize negatively curved tubules. Moreover, the complexity of the different subunit interactions seen in IST1NTD/CHMP1B filaments begs additional questions. First, are all ESCRT-III filaments double stranded and composed of a mixture of open and closed subunits? Answering this question will likely require a high-resolution structure of a curved filament containing CHMP4 subunits. Second, can all ESCRT-III subunits change conformation, or do some play more specialized roles that require them to be constitutively open or closed? There is considerable biochemical evidence that many ESCRT-III subunits can open reversibly (Bajorek et al. 2009, Lata et al. 2008a, Lin et al. 2005, Zamborlini et al. 2006), and one small-angle scattering study demonstrated that CHMP3 can change conformations with changing solution conditions (Lata et al. 2008a), but there are also indications that some ESCRT-III subunits, such as CHMP6, may prefer just one conformation (Schuh et al. 2015). Third, a related question is whether there are other preferred pairs of ESCRT-III subunits, in addition to the IST1/CHMP1B pair. Additional examples exist (Effantin et al. 2013, Lata et al. 2008b), but the redundancy of the pairing code and precisely which ESCRT-III subunits work together for different tasks are not yet known.
3. Vps4 AND RELATED AAA ATPASES
3.1. Overview
Vps4 enzymes drive the dynamic exchange of subunits into and out of ESCRT-III filaments and ultimately recycle the subunits back into the cytoplasm, thereby harnessing the energy of ATP hydrolysis to power ESCRT-dependent membrane fission reactions (Monroe & Hill 2016). Vps4 enzymes contain three different structural elements: (a) an N-terminal MIT domain that binds the tails of ESCRT-III proteins (Figures 2c and 5a); (b) a central ATPase cassette comprising large and small domains that mediate hexamerization and ATP hydrolysis; and (c) a β-domain insert within the small ATPase domain that binds LIP5 (Vta1), an ATPase activator and ESCRT-III-binding protein. Our understanding of the enzyme’s structure and molecular mechanism has increased dramatically with recent high-resolution cryo-EM structures of Vps4 proteins in their active, hexameric ring conformations (Han et al. 2017, Monroe et al. 2017, Su et al. 2017, Sun et al. 2017), particularly structures of the yeast Vps4 enzyme in complex with an ESCRT-III substrate (Han et al. 2017, Monroe et al. 2017). These structures revealed that Vps4 forms an asymmetric ring hexamer that binds the exposed tails of ESCRT-III subunits and translocates the subunits through the constricted central pore of the hexamer. Importantly, this mechanism may be conserved across AAA ATPases of the meiotic clade, including SPASTIN, KATANIN, and FIDGETIN, which are best characterized as microtubule-severing enzymes that bind the tails of tubulin subunits and extract them from the polymer (Monroe & Hill 2016).
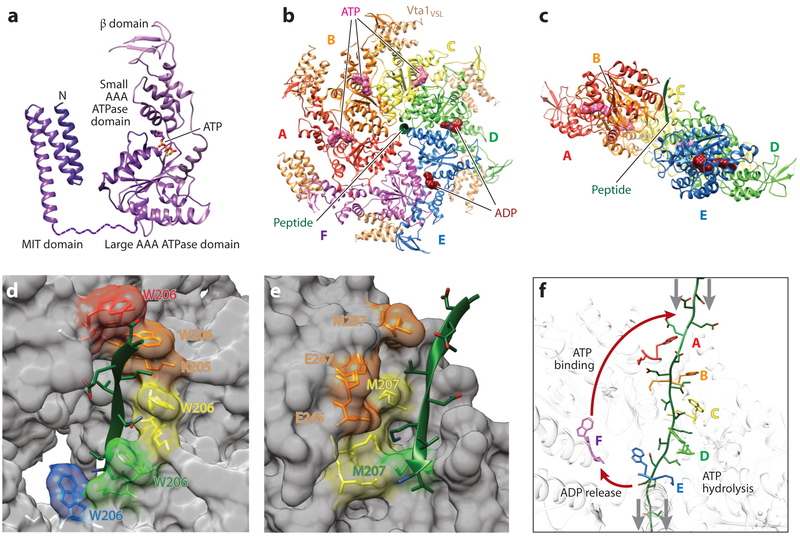
Figure 5. Structure, assembly, and mechanism of the Vps4 ATPase.
(a) Domain structure of a Vps4 monomer in complex with ATP. Structures are reproduced from Han et al. (2017) (ATPase cassette) and Obita et al. (2007) (MIT domain), (b) Top view of the asymmetric yeast Vps4 ring hexamer (subunits A–F) with associated dimeric VSL domains from Vta1 (beige), ESCRT-III peptide (green), and nucleotides (ATP, pink; ADP, dark red), (c) Same as panel b, viewed from the lower side and with subunit F and the VSL domains removed for clarity. (d) Type 1 binding pockets for alternating odd-numbered substrate amino acids, formed along the central channel of the Vps4 hexamer. Bound substrate amino acid side chains are shown explicitly, as are the Vps4 residues that compose the pocket (fully labeled in the second pocket). Note that the four intact amino acid binding sites form a helix around the bound ESCRT-III substrate in the central channel. Vps4 structure and color coding are the same as in panels b and c. (e) Type 2 binding pockets for alternating even-numbered substrate amino acids, formed along the central channel of the Vps4 hexamer. Bound substrate amino acid side chains are shown explicitly, as are the Vps4 residues that compose these pockets. Note that these binding sites form a second helix around the bound ESCRT-III substrate in the central channel. Vps4 structure and color coding are the same as in panels b and c. (f) Proposed mechanism of ESCRT-III translocation by Vps4. This panel shows the central translocation pore with representative Vps4 residues from the type 1 pocket (W206) and the type 2 pocket (M207) and with the ESCRT-III substrate (green) passing through the pore. Substrate translocation is proposed to occur as Vps4 subunits progress through states A to E while maintaining contacts with their respective substrate dipeptides. ATP hydrolysis at subunit D destabilizes the D/E interface and promotes displacement of subunit E toward the transitioning subunit F configuration, which allows for displacement of ADP and full release of the F subunit from the substrate (lower red arrow). Subsequent ATP binding allows subunit F to rejoin to the top of the helix (upper red arrow), where it packs against subunit A, binds the next substrate dipeptide, and assumes the subunit A configuration. Gray arrows show the relative direction of substrate peptide translocation. Panels b–f adapted from Han et al. (2017).
3.2. Vps4 Assembly and Nucleotide Binding
Vps4 hexamerizes only weakly, and the cytoplasmic protein is monomeric or dimeric until locally concentrated by binding an ESCRT-III filament, at which point Vps4 presumably hexamerizes. Vps4 hexamerization is also promoted by the binding of six dimers of VSL domains from the LIP5/Vta1 activator, which link the β and small ATPase domains of adjacent Vps4 subunits around the ring exterior (Monroe et al. 2017, Su et al. 2017) (Figure 5b). The Vps4 ring is an asymmetric hexamer, with five of the six subunits (A–E) forming a continuous helix that surrounds the central ESCRT-III polypeptide substrate. The E subunit is displaced slightly from its idealized helix position, and the sixth (F) Vps4 subunit sits outside the helix and is fully disengaged from the substrate (Figure 5). The nucleotide state of each subunit in the active hexamer has not been determined with absolute certainty, but it appears that subunits A–C bind ADP•BeFx in an ATP-like configuration and D and E bind ADP, while the F subunit appears to lack a nucleotide (Han et al. 2017). Nucleotides bind at subunit interfaces and are canonically coordinated by (a) a Walker A motif, which contacts the adenosine base, sugar, and first two phosphates; (b) a Walker B motif, which chelates a magnesium ion and catalytic water; and (c) two arginine finger residues from an adjacent subunit, which coordinate the gamma phosphate and help promote ATP hydrolysis (Wendler et al. 2012). Intersubunit interfaces are formed primarily by large domain–to–large domain and large domain–to–small domain contacts, except in the transitioning F subunit, where the large domain–to–large domain contacts are broken on both sides.
3.3. Substrate Binding
Filament binding is initially mediated by the Vps4 MIT domains, which bind the exposed C-terminal MIM tails of ESCRT-III subunits (Hurley & Yang 2008). MIT domains are three-helix bundles that bind different ESCRT-III tails in a remarkable variety of ways (Figure 2c). ESCRT-III substrate engagement activates the enzyme by relieving autoinhibitory MIT domain interactions (Babst et al. 2011, Han et al. 2015, Merrill & Hanson 2010). The Vps4 hexamer can then assemble about ESCRT-III elements located upstream of the terminal MIT binding site (Han et al. 2015, 2017; Monroe et al. 2017; Shim et al. 2008), yielding an active holoenzyme.
Figure 5 shows the structure of yeast Vps4 bound to an eight-amino-acid segment located upstream of the terminal MIM element in the ESCRT-III protein Vps2. The ESCRT-III substrate binds in an extended (β-strand) conformation (Han et al. 2017). Alternating side chains project from opposite sides of the strand, creating a helical array of repeating dipeptide units. Vps4 accommodates the two amino acids of each dipeptide in two distinct types of binding pockets (type 1 and 2 pockets) formed by residues from the two conserved, central Vps4 pore loops (pore loops 1 and 2). The four intact type 1 amino acid binding pockets surround one side of the helical substrate, binding alternating odd-numbered side chains (Figure 5d). These pockets are flanked by two pore loop 1 Trp residues from adjacent Vps4 subunits and a pore loop 1 Lys side chain from the first Vps4 subunit. The pocket is otherwise open to solvent, allowing the translocation channel to accommodate different side chains. Four type 2 Vps4 binding pockets similarly engage the remaining alternating even-numbered amino acid substrate side chains. These pockets are created by Vps4 residues from both pore loops 1 and 2 and are again highly solvated (Figure 5e). Thus, two distinct types of substrate binding pockets form a double helix within the central pore of the Vps4 hexamer that binds four substrate dipeptides. Remarkably, the substrate polypeptide is uniquely oriented in the translocation channel (from N to C from top to bottom in Figure 5c) by a repeating pattern of main-chain hydrogen bonds between Vps4 pore loop 1 and the substrate.
3.4. Mechanism of ESCRT-III Substrate Translocation
The structure of the substrate-bound Vps4 complex suggests a conveyor belt mechanism in which each subunit sequentially transitions through the five different positions of the Vps4 helix (positions A–E in diagrams in Figure 5) before translocating (position F) from the bottom of the helix and subsequently rejoining the top of the helix (position A). As each Vps4 subunit passes through these transitions, it carries a substrate dipeptide down the central pore, releases the dipeptide at the bottom, and then reengages a new dipeptide at the top. The linked Vps4 subunits can, therefore, be thought of as walking along the polypeptide substrate in 12 amino acid steps (as viewed from the perspective of the polypeptide), which is equivalent to translocating the substrate through the central pore of the ring (when viewed from the perspective of the ring). Vps4 subunits hydrolyze ATP as they move through the C/D transition, release ESCRT-III substrates as they pass through the E/F transition, exchange ADP for ATP along the E/F transition, and subsequently rebind the substrate through the E/F/A transitions. ATP hydrolysis weakens the intersubunit interfaces in the Vps4 hexamer, promoting the disengagement of subunits E and F. This cycle of ATP binding, hydrolysis, and release thereby provides the energy required to unfold and extract ESCRT-III subunits from filaments.
A similar mechanism appears to be employed by other single- and double-ring AAA+ ATPases that act on polypeptide substrates. This generalized model is supported by conservation of pore loop residues and other key structural elements (Monroe & Hill 2016, Sauer & Baker 2011) and by a series of recent structures of other AAA+ ATPases in complex with mixed polypeptide substrates (Deville et al. 2017, Gates et al. 2017, Puchades et al. 2017, Ripstein et al. 2017). These structures collectively represent a long-awaited breakthrough in our understanding of this large and important class of cellular machines (Harrison 2004), whose well-known representatives include essential activities like the 19S proteasome, NSF, and p97/Cdc48.
3.5. Outstanding Issues
Despite considerable recent progress, our understanding of Vps4 mechanism and function remains incomplete. At a detailed level, we do not yet fully understand the allosteric coupling of nucleotide binding/hydrolysis/release, substrate binding/release, and intersubunit interface interactions. For example, the existing Vps4 structures do not obviously explain why ATP is hydrolyzed as subunits pass through the C/D transition even though the A–D subunits adopt similar conformations. A detailed mechanistic model for allosteric coupling was recently proposed on the basis of the structure of a nonhydrolyzing mutant of the AAA+ protease YME1 bound to ATP/ADP and a polypeptide substrate (Puchades et al. 2017). This model is similar to that described above for Vps4 but differs in the proposed site of ATP hydrolysis (which is uncertain owing to use of an inactive enzyme for YME1 and a nonhydrolyzable nucleotide for Vps4).
At a more macroscopic level, it is not yet clear whether Vps4 can translocate peptides bidirectionally or can accommodate peptide loops, as has been proposed for other AAA ATPases that act on polypeptide substrates (Sauer & Baker 2011). The activities of Vps4 on ESCRT-III subunits in native filaments also remain to be investigated in mechanistic detail, although the enzyme has been shown to act processively and to unfold ESCRT-III subunits entirely in some contexts (Yang et al. 2015). Even more fundamentally, there is now good evidence that Vps4 activity is required for membrane fission (Baumgartel et al. 2011, Jouvenet et al. 2011), not just for ESCRT-III subunit recycling, but as discussed below, it is not yet clear how the Vps4 unfolding activity is used to promote membrane fission.
4. MEMBRANE REMODELING, CONSTRICTION, AND FISSION
A series of models have been proposed to explain how ESCRT-III filaments and Vps4 together catalyze membrane remodeling, constriction, and fission (Adell et al. 2017, Chiaruttini & Roux 2017, Fabrikant et al. 2009, Hanson et al. 2008, Henne et al. 2013, Johnson et al. 2018, Lenz et al. 2009, Peel et al. 2011, Saksena et al. 2009, Schoeneberg et al. 2018, Schöneberg et al. 2017) (Figure 6). A consensus model has not emerged, however, and different aspects of the divergent models may need to be combined to explain how the ESCRT machinery can remodel membranes across such a variety of spatial scales and membrane geometries.
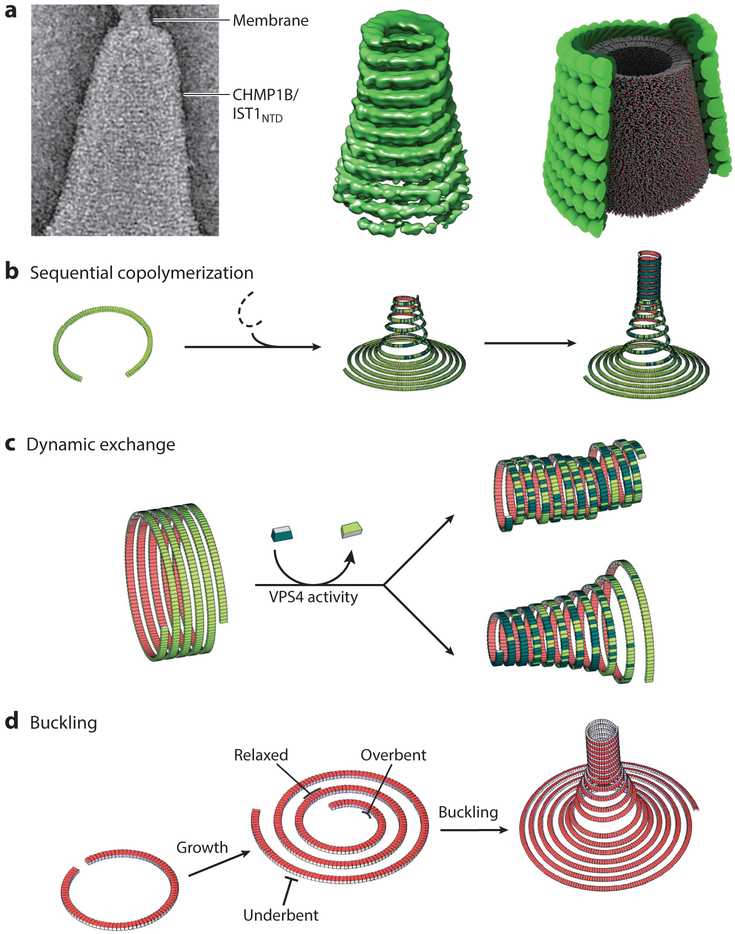
Figure 6. ESCRT-III filament topologies and membrane remodeling.
(a) Cone formation by double-stranded filaments formed by the N-terminal ESCRT-III domain of IST1 (IST1NTD) and CHMP1B. (Left to right) Double-stranded IST1NTD/CHMP1B filaments wrapping about a conical membrane; reconstructed cone comprising double-stranded IST1NTD/CHMP1B filaments; schematic depiction of the structures shown in the left panel, with the IST1NTD strand shown in light green, the CELMP1B strand shown in dark green, and the internal lipid bilayer shown in gray, (b–d) Illustrations of how membrane deformation, constriction, and fission could be driven by heteromeric and dynamic ESCRT-III filaments that adopt different topologies in response to a variety of conditions, including (b) sequential copolymerization of different ESCRT-III subunits with distinct intrinsic curvatures, (c) dynamic exchange of subunits with high intrinsic curvature into tubes of less curved filaments, and (d) out-of-plane buckling induced by the accumulation of elastic stress arising from growth of a spiraling filament beyond its preferred radius of curvature. For simplicity, panels b–d show single long filaments, but analogous principles could also apply to arrays of shorter, close-packed filaments. Illustrations in panel a were adapted from McCullough et al. (2015) and reproduced with permission from AAAS. Illustrations in panels b–d were adapted from Chiaruttini & Roux (2017).
4.1. Filament Dynamics
Recent studies have demonstrated that ESCRT-III subunits exchange rapidly at sites of filament action, although the role of Vps4 in mediating this exchange is debated (Adell et al. 2017, Mierzwa et al. 2017). Vps4 activity is required for virus budding (Baumgartel et al. 2011), abscission (Mierzwa et al. 2017), and MVB vesicle formation (Adell et al. 2017), and the enzyme appears to perform an essential role in promoting the fission reaction (rather than simply recycling ESCRT-III subunits after fission). In vitro, Vps4-mediated subunit exchange facilitates subunit exchange into highly dynamic arrays of growing, shrinking, and spiraling ESCRT-III filaments (Mierzwa et al. 2017). However, ESCRT-III subunit exchange can continue at sites of yeast MVB formation even in the absence of an active Vps4 enzyme (Adell et al. 2017). Regardless, filament remodeling and dynamics also likely play important roles in ESCRT-mediated membrane fission reactions.
4.2. Models for Membrane Constriction and Fission
A central question is how ESCRT-III filaments draw membranes together toward the point of fission. One idea is that membrane-associated ESCRT-III filaments could initially form membrane-associated helical tubules that have a greater radius of curvature than is preferred energetically. An extreme example of this is during cytokinesis, where the preformed midbody is initially greater than 1 μm in diameter. In this situation, filament underbending could provide a driving force toward forming constricting spirals rather than regular helices, and such spiraling filaments could constrict the associated membranes. Pure recombinant IST1/CHMP1B filaments can indeed form conical spirals, both in solution and around positively curved membranes (McCullough et al. 2015) (Figure 6a). This general theme has been further embellished by a series of different proposals. First, Vps4 could sever and release such underbent spiraling filaments from their anchoring adaptors, allowing them to translocate along the midbody and to constrict the membrane as they convert into tighter spirals (Elia et al. 2011, 2012; Goliand et al. 2017). Second, underbent filaments could constrict as they polymerized by preferentially incorporating other types of ESCRT-III subunits with greater intrinsic degrees of curvature (Figure 6b). Third, in the extreme, a regular helix comprising one type of ESCRT-III subunit (e.g., CHMP4) could be capped by a more tightly spiraling filament composed of other subunits (e.g., CHMP2/CHMP3), and the resulting dome could draw the membranes together to a fission point at the apex (Fabrikant et al. 2009). Fourth, Vps4 could exchange ESCRT-III subunits that favor higher curvature into a preformed, wider base. This activity would either constrict the helix symmetrically or create a cone, depending upon whether the tighter ESCRT-III subunits were added evenly along the tube or preferentially at one end (Figure 6c). Finally, analogous constriction mechanisms could also operate in double-stranded filaments if, for example, the second strand had a greater intrinsic curvature and therefore tightened the first. Any of these mechanisms could, in principle, constrict membranes to the point where they were closely apposed in a high-energy configuration. Fission could then occur spontaneously if Vps4 depolymerized the filamentous scaffold, either locally or globally. Support for the latter idea has recently come from the observation that ESCRT-III and Vps4 proteins are released from sites of virus budding sites ~20 s prior to membrane scission (Johnson et al. 2018).
In an alternative class of models, membrane extrusion and fission are driven by filament buckling and unbuckling (Chiaruttini & Roux 2017, Chiaruttini et al. 2015, Lenz et al. 2009, Schoeneberg et al. 2018). As illustrated in Figure 6d, this elegant model envisions that ESCRT-III filaments initially spiral on a flat planar membrane, as is seen in the EM images in Figure 1b,c. The filaments will have a preferred radius of curvature and will therefore store energy and exert an axial force as they polymerize at greater radii where they are underbent. Similarly, filaments will store overbending energy as they polymerize toward smaller circumferences (and also push against concentrated cargoes). These straining forces could be relieved by the out-of-plane buckling of filaments at the center of the array. This buckling transition would define the direction of vesiculation by creating membrane tubules that extrude either inside out (if the filaments buckle toward the membrane) or outside in (if the filaments buckle away from the membrane).
Importantly, the energy required for the subsequent membrane fission reaction could be provided through an ATP-dependent reversal of the transition from tubular back to planar filaments (Carlson et al. 2015). In this model, extruded membranes would be drawn together and a cargo-filled vesicle released from the end of the tubule when the ESCRT-III filaments pulled on the tubule base by retracting back out of the tubule to readopt a planar configuration. This idea is supported by the recent demonstration that ESCRT-III/Vps4 complexes can assemble within the necks of synthetic membrane tubules and convert the energy of ATP hydrolysis into axial pulling forces that can sever the tubules (Schoeneberg et al. 2018). As in coning models, changes in the magnitude and direction of filament tension could be dictated by altering subunit compositions during polymerization or by Vps4-dependent changes such as filament bundling, subunit exchange, depolymerization, or severing.
4.3. Outstanding Issues
Distinguishing and refining models for ESCRT-dependent membrane remodeling and fission will require characterizing cellular systems with ever greater temporal and spatial resolution, as well as in reconstituted reactions in purified systems, in which high-resolution imaging and force measurements can be used to discern the components, structures, timing, and physical properties of each step. Once the fundamental reaction mechanism is understood, ever-more-precise measurements and simulations should shed light on important details such as the roles of different lipids, the structures and energetics of protein-lipid interfaces, and the relevance and contributions of different possible reaction lipid-mixing intermediates. These studies should thereby provide a mechanistic understanding of one of the most fascinating, important, and evolutionarily ancient membrane remodeling systems.
ACKNOWLEDGMENTS
We thank the following colleagues for critical conversations and generous help with figures: Janet Iwasa (Figures 1, and 4), Han Han and Chris Hill (Figure 5), and Nicolas Chiaruttini and Aurélien Roux (Figure 6). We also thank Chris Hill and members of our laboratories, especially Nathaniel Talledge and Henry Nguyen, for helpful discussions and for sharing their unpublished observations. This work was supported by NIH grants P50 082545 (to A.F.), R01 GM112080 and R37 AI51174 (to W.I.S.), and 1DP2GM110772-01 (to A.F.). A.F. is further supported by a Faculty Scholar grant from the HHMI and is a Chan Zuckerberg Biohub investigator.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Adell MAY, Migliano SM, Upadhyayula S, Bykov YS, Sprenger S, et al. 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 6:e31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R, Lumb JH, Fassier C, Connell JW, Ten Martin D, et al. 2013. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol 202:527–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Davies BA, Katzmann DJ. 2011. Regulation of Vps4 during MVB sorting and cytokinesis. Traffic 12:1298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, et al. 2009. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol 16:754–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLOS Pathog 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, et al. 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol 13:469–74 [DOI] [PubMed] [Google Scholar]
- Bleck M, Itano MS, Johnson DS, Thomas VK, North AJ, et al. 2014. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. PNAS 111:12211–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. 1998. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev 62:230–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich NJ, Henne WM, Tang S, Emr SD. 2013. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell 27:201–14 [DOI] [PubMed] [Google Scholar]
- Cannon KS, Woods BL, Gladfelter AS. 2017. The unsolved problem of how cells sense micron-scale curvature. Trends Biochem. Sci 42:961–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Shen QT, Pavlin MR, Hurley JH. 2015. ESCRT filaments as spiral springs. Dev. Cell 35:397–98 [DOI] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. 2012. ESCRT-III governs the Aurora B–mediated abscission checkpoint through CHMP4C. Science 336:220–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908–12 [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Shim S, Roth R, Maldazys MR, Heuser JE, Hanson PI. 2014. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife 3:e02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi Y, Dekker C. 2018. Dividing the archaeal way: the ancient Cdv cell-division machinery. Front. Microbiol 9:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, et al. 2015. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163:866–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, Roux A. 2017. Dynamic and elastic shape transitions in curved ESCRT-III filaments. Curr. Opin. Cell Biol 47:126–35 [DOI] [PubMed] [Google Scholar]
- Choudhuri K, Llodra J, Roth EW, Tsai J, Gordo S, et al. 2014. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature 507:118–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci 42:42–56 [DOI] [PubMed] [Google Scholar]
- Crespo-Yàñez X, Aguilar-Gurrieri C, Jacomin AC, Journet A, Mortier M, et al. 2018. CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLOS Genet 14(6):e1007456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwin AJ, Brouwers N, Alonso YAM, Teis D, Turacchio G, et al. 2016. ESCRT-III drives the final stages of CUPS maturation for unconventional protein secretion. eLife 5:e16299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, et al. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science 352:353–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville C, Carroni M, Franke KB, Topf M, Bukau B, et al. 2017. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv 3:e1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Zhang J, Ollwerther A, Wang X, Ahlquist P. 2015. Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLOS Pathog 11:e1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Lupetti P, Rosenbaum JL. 2015. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol 25:379–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusenheimer N, Migdal B, Jackel S, Tveriakhina L, Scheider K, et al. 2015. The mammalian orthologs of Drosophila Lgd, CC2D1A and CC2D1B, function in the endocytic pathway, but their individual loss of function does not affect Notch signalling. PLOS Genet 11:e1005749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G, Dordor A, Sandrin V, Martinelli N, Sundquist WI, et al. 2013. ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell. Microbiol 15:213–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J 2012. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys. J 102:2309–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. PNAS 108:4846–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJ, Bernander R. 2009. Cell division and the ESCRT complex: a surprise from the archaea. Commun. Integr. Biol 2:86–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. 2009. Computational model of membrane fission catalyzed by ESCRT-III. PLOS Comput. Biol 5:e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, et al. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel EB, Audhya A. 2018. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol 74:4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, et al. 2008. Structural basis of membrane invagination by F-BAR domains. Cell 132:807–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe I, Schuh AL, Edwardson JM, Audhya A. 2011. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J. Biol. Chem 286:34262–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Shen J, Jiang L. 2017. Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci 22:986–98 [DOI] [PubMed] [Google Scholar]
- Gates SN, Yokom AL, Lin J, Jackrel ME, Rizo AN, et al. 2017. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357:273–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliand I, Dadosh T, Elia N. 2017. Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM imaging. bioRxiv 194613 https://doi.org/10.1101/194613 [DOI] [PubMed] [Google Scholar]
- Gu M, LaJoie D, Chen OS, von Appen A, Ladinsky MS, et al. 2017. LEM2 recruits CHMP7 for ESCRTmediated nuclear envelope closure in fission yeast and human cells. PNAS 114:E2166–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, et al. 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138:990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, et al. 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331:1616–20 [DOI] [PubMed] [Google Scholar]
- Han H, Monroe N, Sundquist WI, Shen PS, Hill CP. 2017. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. eLife 6:e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Monroe N, Votteler J, Shakya B, Sundquist WI, Hill CP. 2015. Binding of substrates to the central pore of the Vps4 ATPase is autoinhibited by the microtubule interacting and trafficking (MIT) domain and activated by MIT interacting motifs (MIMs). J. Biol. Chem 290:13490–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol 180:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. 2004. Whither structural biology? Nat. Struct. Mol. Biol 11:12–15 [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD. 2012. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151:356–71 [DOI] [PubMed] [Google Scholar]
- Henne WM, Stenmark H, Emr SD. 2013. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol 5:a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Sun J, Rusnak AS, Kim J, Prag G, et al. 2004. Structure of the ESCRT-II endosomal trafficking complex. Nature 431:221–25 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Yang D. 2008. MIT domainia. Dev. Cell 14:6–8 [DOI] [PubMed] [Google Scholar]
- Im YJ, Wollert T, Boura E, Hurley JH. 2009. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell 17:234–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CE, Scruggs BS, Schaffer JE, Hanson PI. 2017. Effects of inhibiting VPS4 support: a general role for ESCRTs in extracellular vesicle biogenesis. Biophys. J 113:1342–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. 2014. ESCRT machinery is required for plasma membrane repair. Science 343:1247136. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Bleck M, Simon SM. 2018. Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. eLife; 7:e36221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol 13:394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan T Fürthauer M. 2018. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol 74:66–77 [DOI] [PubMed] [Google Scholar]
- Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, et al. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15:62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kollman JM, Merdes A, Mourey L, Agard DA. 2011. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol 12:709–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, et al. 2008a. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol 378:818–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, et al. 2008b. Helical structures of ESCRT-III are disassembled by VPS4. Science 321:1354–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Solomons J, Pires R, Gottlinger HG, Weissenhorn W. 2009. Structure and function of ESCRT-III. Biochem. Soc. Trans 37:156–60 [DOI] [PubMed] [Google Scholar]
- Lee CP, Liu PT, Kung HN, Su MT, Chua HH, et al. 2012. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLOS Pathog 8:e1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. 2008. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 322:576–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Kai H, Carlson LA, Groves JT, Hurley JH. 2015. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. PNAS 112:15892–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz M, Crow DJ, Joanny JF. 2009. Membrane buckling induced by curved filaments. Phys. Rev. Lett 103:038101. [DOI] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. 2005. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem 280:12799–809 [DOI] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. 2008. A unique cell division machinery in the Archaea. PNAS 105:18942–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Freed EO, van Engelenburg SB. 2017. A consensus view of ESCRT-mediated human immunodeficiency virus type 1 abscission. Annu. Rev. Virol 4:309–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Agromayor M, Martin-Serrano J, Williams DW. 2015. An ESCRT module is required for neuron pruning. Sci. Rep 5:8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, et al. 2012. CC2D1A is a regulator of ESCRT-III CHMP4B. J. Mol. Biol 419:75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast FD, Herricks T, Strehler KM, Miller LM, Saleem RA, et al. 2018. ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol https://doi.org/10.1083/jcb.201706044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusek T, Wendler F, Poles S, Pizette S, D’Angelo G, et al. 2014. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature 516:99–103 [DOI] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, et al. 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350:1548–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. PNAS 105:7687–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. 2015. Membrane curvature at a glance. J. Cell Sci 128:1065–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan BJ, Tibbe C, Drabek AA, Seegar TCM, Blacklow SC, Klein T. 2017. Structural basis for regulation of ESCRT-III complexes by Lgd. Cell Rep 19:1750–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan BJ, Tibbe C, Jeon H, Drabek AA, Klein T, Blacklow SC. 2016. Electrostatic interactions between elongated monomers drive filamentation of Drosophila Shrub, a metazoan ESCRT-III protein. Cell Rep 16:1211–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill SA, Hanson PI. 2010. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem 285:35428–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, Konig J, et al. 2017. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol 19:787–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, et al. 2012. Structural basis of membrane bending by the N-BAR protein endophilin. Cell 149:137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM. 2012. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci 37:526–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe N, Han H, Shen PS, Sundquist WI, Hill CP. 2017. Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. eLife 6:e024487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe N, Hill CP. 2016. Meiotic clade AAA ATPases: protein polymer disassembly machines. J. Mol. Biol 428:1897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, et al. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26:4215–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu R, Dussupt V, Jiang J, Sette P, Rudd V, et al. 2012. Two distinct binding modes define the interaction of Brox with the C-terminal tails of CHMP5 and CHMP4B. Structure 20:887–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, et al. 2006. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10:821–30 [DOI] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. 2012. Formation and release of arrestin domain-containing protein 1–mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. PNAS 109:4146–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, et al. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449:735–39 [DOI] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. 2015. ESCRT-III controls nuclear envelope reformation. Nature 522:236–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Perdrix-Rosell A, Carlton JG. 2016. Membrane binding by CHMP7 coordinates ESCRT-IIIdependent nuclear envelope reformation. Curr. Biol 26:2635–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. 2011. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem. Sci 36:199–210 [DOI] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, et al. 2009. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17:843–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchades C, Rampello AJ, Shin M, Giuliano CJ, Wiseman RL, et al. 2017. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358:eaao0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pykalainen A, Boczkowska M, Zhao HX, Saarikangas J, Rebowski G, et al. 2011. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat. Struct. Mol. Biol 18:902–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, et al. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352:359–62 [DOI] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. 2017. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife 6:e25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E. 2017. Actin assembly mechanisms at a glance. J. Cell Sci 130:3427–35 [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. 2008. A role for the ESCRT system in cell division in archaea. Science 322:1710–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SauerRT Baker TA. 2011. AAA+proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem 80:587–612 [DOI] [PubMed] [Google Scholar]
- Scheffer LL, Sreetama SC, Sharma N, Medikayala S, Brown KJ, et al. 2014. Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun 5:5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeneberg J, Yan S, Righini M, Pavlin MR, Lee I-H, et al. 2018. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. bioRxiv; https://doi.org/10.1101/262170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J, Lee IH, Iwasa JH, Hurley JH. 2017. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol 18:5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AL, Hanna M, Quinney K, Wang L, Sarkeshik A, et al. 2015. The VPS-20 subunit of the endosomal sorting complex ESCRT-III exhibits an open conformation in the absence of upstream activation. Biochem. J 466:625–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scourfield EJ, Martin-Serrano J. 2017. Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem. Soc. Trans 45:613–34 [DOI] [PubMed] [Google Scholar]
- Shen QT, Schuh AL, Zheng Y, Quinney K, Wang L, et al. 2014. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol 206:763–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Merrill SA, Hanson PI. 2008. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19:2661–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, et al. 2007. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129:761–72 [DOI] [PubMed] [Google Scholar]
- Simunovic M, Voth GA, Callan-Jones A, Bassereau P. 2015. When physics takes over: BAR proteins and membrane curvature. Trends Cell Biol 25:780–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, et al. 2012. Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport. J. Biol. Chem 287:43910–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra ML, Schlesinger PH, Naismith TV, Hanson PI. 2018. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 359:aar5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomons J, Sabin C, Poudevigne E, Usami Y, Hulsik DL, et al. 2011. Structural basis for ESCRT-IIICHMP3 recruitment of AMSH. Structure 19:1149–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporny M, Guez-Haddad J, Kreusch A, Shakartzi S, Neznansky A, et al. 2017. Structural history of human SRGAP2 proteins. Mol. Biol. Evol 34:1463–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoten CL, Carlton JG. 2018. ESCRT-dependent control of membrane remodelling during cell division. Semin. Cell Dev. Biol 74:50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–44 [DOI] [PubMed] [Google Scholar]
- Su M, Guo EZ, Ding X, Li Y, Tarrasch JT, et al. 2017. Mechanism of Vps4 hexamer function revealed by cryo-EM. Sci. Adv 3:e1700325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Li L, Yang F, Wang X, Fan F, et al. 2017. Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution. Nat. Commun 8:16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talledge N, McCullough J, Wenzel D, Nguyen HC, Lalonde JM, et al. 2018. The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids. bioRxiv 386532. https://doi.org/10.1101/386532 [Google Scholar]
- Tang S, Buchkovich NJ, Henne WM, Banjade S, Kim YJ, Emr SD. 2016. ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis. eLife 5:e15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, et al. 2015. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife 4:e12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Judson BL, Emr SD. 2010. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J 29:871–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell 7:559–69 [DOI] [PubMed] [Google Scholar]
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, et al. 2007. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol 9:1370–80 [DOI] [PubMed] [Google Scholar]
- Troost T, Jaeckel S, Ohlenhard N, Klein T. 2012. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J. Cell Sci 125:763–76 [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Weiss ER, Vriesema-Magnuson C, Calistri A, Gottlinger HG. 2012. Regulation of CHMP4/ESCRT-III function in human immunodeficiency virus type 1 budding by CC2D1A. J. Virol 86:3746–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, et al. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522:231–35 [DOI] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jäger J, Lusk CP. 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159:388–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Thaller DJ, Jäger J, Ochmann SES, Borah S, Lusk CP. 2016. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J 35:2447–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Ciniawsky S, Kock M, Kube S. 2012. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 1823:2–14 [DOI] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, Rosenbaum JL. 2013. The cilium secretes bioactive ectosomes. Curr. Biol 23:906–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. 2009. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 20:3514–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. 2015. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol 22:492–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoise B, Lippincott-Schwartz J, Blackstone C, Hurley JH. 2008. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol 15:1278–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. PNAS 103:19140–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Jorgensen JR, Li M, Chuang YS, Emr SD. 2017. ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. eLife 6:e026403. [DOI] [PMC free article] [PubMed] [Google Scholar]