Abstract

Lipid coverage is crucial in comprehensive lipidomics studies challenged by high diversity in lipid structures and wide dynamic range in lipid levels. Current state-of-the-art lipidomics technologies are mostly based on mass spectrometry (MS), including direct-infusion MS, chromatography-MS, and matrix-assisted laser desorption ionization (MALDI) imaging MS, each with its pros and cons. Due to the need or favorability for measurement of isomers and isobars, chromatography-MS is preferable for lipid profiling. The ultra-high performance liquid chromatography-high resolution mass spectrometry (UHPLC-HRMS)-based nontargeted lipidomics approach and UHPLC-tandem MS (UHPLC-MS/MS)-based targeted approach are two representative methodological platforms for chromatography-MS. In the present study, we developed a high coverage pseudotargeted lipidomics method combining the advantages of nontargeted and targeted lipidomics approaches. The high coverage of lipids was achieved by integration of the detected lipids derived from nontargeted UHPLC-HRMS lipidomics analysis of multiple matrices (e.g., plasma, cell, and tissue) and the predicted lipids speculated on the basis of the structure and chromatographic retention behavior of the known lipids. A total of 3377 targeted lipid ion pairs with over 7000 lipid molecular structures were defined. The pseudotargeted lipidomics method was well validated with satisfactory analytical characteristics in terms of linearity, precision, reproducibility, and recovery for lipidomics profiling. Importantly, it showed better repeatability and higher coverage of lipids than the nontargeted lipidomics method. The applicability of the developed pseudotargeted lipidomics method was testified in defining differential lipids related to diabetes. We believe that comprehensive lipidomics studies will benefit from the developed high coverage pseudotargeted lipidomics approach.
Lipid plays a vital role in cellular barrier, signal conduction, substance transport, and energy storage.1 It has attracted increasing attention due to its unique biological significance. Lipidomics is a powerful analytical tool enabling one to provide detailed characterizations of lipid alterations in response to internal or external subtle perturbations in complex biological samples.2 It has been extensively used in biomarker discovery and mechanism studies related to cardiovascular disease,3 diabetes,4 cancer,5 and Alzheimer’s disease (AD).6
Lipids are not only highly complex in composition and structure but also widely dynamic in concentration. It was reported that cellular lipidome contains tens to hundreds of thousands of lipid molecular species, and their levels fluctuate from attomole to nanomole per mg of protein.7−9 Lipids are divided into classes and subclasses depending on the headgroup and the type of linkage between the headgroup and acyl chains.10 The acyl chains are varied with different lengths (i.e., different number of carbon atoms), various degrees of unsaturation, potential branches, etc. These varieties in headgroup and acyl chain of lipids contribute to the extremely huge number of individual lipid molecular species,11 making it impossible to fully characterize the whole lipidome by a single lipidomics approach. To this end, establishing a high coverage lipidomics method is a suitable compromise.
The mass spectrometry (MS)-based strategy is predominant for lipidomics due to its highly enhanced resolution and sensitivity.12 Current state-of-the-art lipidomics technologies mainly include direct-infusion MS, chromatography-MS, and matrix-assisted laser desorption ionization (MALDI) imaging MS, each with its pros and cons. Direct-infusion MS-based shotgun lipidomics has been a golden standard for high-throughput lipid quantitation widely used in many laboratories.13−15 MALDI imaging MS is powerful for studying the spatial distribution of lipids in tissues and enables one to provide molecular-histological maps.16 However, both approaches are not favorable for detection of isomers and isobars. In practice, MS coupled with ultra-high performance liquid chromatography (UHPLC) is frequently used for lipid profiling due to the need or favorability for measurement of lipid isomers and isobars, leading to realizable separation of isomers and isobars and reduced ion-suppression effects via a chromatographic separation prior to MS.17,18 The UHPLC-MS-based lipidomics strategies usually consist of nontargeted and targeted approaches, each having their own advantages and disadvantages.19
The nontargeted UHPLC-MS lipidomics approach enables one to cover many lipid classes as it is unbiased and able to detect “all” ion features in biological samples. To maximize detection efficiency, nontargeted lipidomics analyses are usually performed using high resolution MS (HRMS), such as time-of-flight (TOF), Orbitrap, or Fourier transform ion cyclotron resonance (FTICR) which can provide highly accurate masses. In these circumstances, especially for Orbitrap and FTICR, however, MS has to scan many ion features in a very short time. Limited by their scanning speed, these MS detectors are easily saturated and the scan time of every consecutive m/z is reduced, resulting in insufficient data-points for lipid peaks so that the linearity and repeatability are affected.20 Additionally, the data treatment in postacquisition of nontargeted lipidomics is also tedious. The complex peak assignment is easily affected by the parameter setting, bringing in bias or errors in data pretreatment.
Traditional UHPLC-tandem MS (MS/MS)-based targeted lipidomics analysis is often executed on triple quadrupole (QQQ) MS with selective MS scanning modes such as multiple reaction monitoring (MRM), product ion scanning (PIS), and/or neutral loss scanning (NLS). Such a targeted method greatly increases the detection sensitivity and is beneficial for quantification with the benefits of good linearity, good repeatability, and no complex peak assignment compared with the UHPLC-HRMS nontargeted lipidomics approach. However, its coverage is limited as it usually focuses on known lipids.
In this study, we established a high coverage pseudotargeted lipidomics method combining the advantages of nontargeted and targeted methods. First, UHPLC-HRMS nontargeted lipid analyses were performed in full scan and data dependent MS/MS (ddMS2) modes for multiple-matrix samples to generate as high coverage lipid profiling as possible. Second, lipids were assigned on the basis of MS/MS fragments, accurate masses, and retention time (tR). The unknown lipids without MS/MS fragments were speculated by comparing their accurate masses and tR with those of the known lipids and/or the online Lipid Maps (http://www.lipidmaps.org/). Third, the undetected but theoretically existing lipids, called extended lipids, were further studied. tR of the extended lipids were predicted according to the relationship between tR vs acyl chain carbon number (No.) or acyl chain double bond No. of the known lipids. Last but not least, lipid ion pairs were constructed on the basis of the characteristic fragmental ions and corresponding parent ions of both the detected and predicted lipids and were monitored in a scheduled MRM mode in a pseudotargeted lipidomics method. As a proof of applicability, the developed pseudotargeted lipidomics method was applied in serum differential lipid discovery related to diabetes, which is highly associated with lipid disturbance and has been a growing health burden worldwide.21
Experimental Section
Materials and Chemicals
HPLC-grade acetonitrile (ACN), methanol (MeOH), and isopropanol (IPA) were purchased from Merck (Darmstadt, Germany). HPLC-grade dichloromethane (CH2Cl2), tert-butyl methyl ether (MTBE), and ammonium acetate (AmAc) were purchased from Sigma–Aldrich (St. Louis, MO. USA). Ultrapure water was obtained by a Milli-Q system (Millipore, Billerica, MA). Lipid standards including phosphatidylcholine (PC) 19:0/19:0, lyso-phosphatidylcholine (LPC) 19:0, phosphatidylethanolamine (PE) 17:0/17:0, sphingomyelin (SM) d18:1/12:0, ceramide (Cer) d18:1/17:0, triacylglycerol (TG) 15:0/15:0/15:0, and fatty acid (FA) 16:0-d3 were purchased from Avanti Polar Lipids (Alabaster, AL).
Sample Collection
The bladder cancer cells were cultured in-house. Mouse brain and liver tissues were collected from Dalian Medical University in Dalian, China. Plasma samples from 20 healthy persons were collected from Dalian physical examination center in Dalian, China. These three sample matrices were specially used for pseudotargeted method development.
Serum samples from 30 patients with diabetes and 30 age-matched healthy controls were, respectively, collected from Suzhou University (Suzhou, China) and Dalian physical examination center (Dalian, China) and used for method application. Detailed information was provided in Table S1.
Sample Preparation
A total of ∼106 cells and ∼10 mg of fresh wet mouse brain and liver tissue as well as 40 μL of pooled plasma or serum were extracted by liquid–liquid extraction (LLE) with the MTBE/MeOH/H2O system, respectively. Notably, 30 μL of an internal standard (IS) mixture containing PC 19:0/19:0 at 6.7 μg/mL, LPC 19:0 at 3.3 μg/mL, PE 17:0/17:0 at 3.3 μg/mL, SM d18:1/12:0 at 1.7 μg/mL, Cer d18:1/17:0 at 1.7 μg/mL, TG 15:0/15:0/15:0 at 5.3 μg/mL, and FA 16:0-d3 at 6.7 μg/mL was added to each kind of sample matrix prior to lipid extraction except for the recovery experiment in method validation, in which the mixture of lipid ISs at three different levels was added to plasma either before or after lipid extraction.
Detailed descriptions on lipid extractions of cells, tissues, plasma, or serum were provided in the Supporting Information.
Nontargeted Lipidomics Analysis
An ACQUITY Ultra High Performance LC (UHPLC, Waters, Milford, MA, U.S.A.) system coupled via an electrospray ion source with a Q Exactive-HF MS system (Thermo Fisher Scientific, Rockford, IL, U.S.A.) was used for nontargeted lipid profiling in both full scan and ddMS2 modes to obtain lipid information including tR, accurate masses, and/or MS/MS fragments and so on.
LC conditions and MS parameters were described in detail in the Supporting Information.
Optimization of Pseudotargeted Lipidomics Analysis
An ACQUITY UHPLC system coupled with a hybrid QQQ-linear ion trap mass spectrometer, Q-Trap 5500 system, with a Turbo Ion Spray source (AB SCIEX/MDS-Sciex, Concord, ON, Canada) was used for UHPLC/QQQ MRM MS-based pseudotargeted lipidomics analysis. The LC conditions for pseudotargeted lipidomics analysis were the same as those in the nontargeted lipidomics analysis, and the MS parameters were provided in the Supporting Information.
The declustering potential (DP) and collision energy (CE) of lipid ion pairs were optimized to obtain the highest response for the mixture of lipid standards. The precursor ion (Q1), characteristic product ion (Q3), lipid name (ID), optimized DP, and CE were imported into the MS acquisition method.
Pseudotargeted Lipid Profiling of Serum Samples Related to Diabetes
A pooled quality control (QC) sample was prepared by mixing an equal aliquot of serum from all of the diabetic and control subjects enrolled in the method application. Lyophilized lipid extractions of QC samples were reconstituted in ACN/IPA/H2O (65:30:5, v/v/v/) containing 5 mM AmAc, and 5 μL was injected into the LC-MS system. Each was analyzed in triplicates by the UHPLC/QQQ MRM MS platform in two positive acquisition methods and a negative acquisition method. After QC sample measurement, lipid ion pair peaks were extracted from chromatograms to define which lipids existed in the QC samples. These lipid ion pairs detected in QC samples were used for 2 new UHPLC/QQQ MRM MS-based pseudotargeted lipidomics methods, respectively, in positive and negative modes for the subsequent measurement of serum samples from the diabetic and control subjects.
For the comparison of the coverage of nontargeted and pseudotargeted lipidomics methods, six aliquots of QC samples prepared in parallel were, respectively, analyzed by the established UHPLC/QQQ MRM MS-based pseudotargeted and UHPLC/LTQ-Orbitrap MS-based nontargeted lipidomics methods. LC conditions and MS parameters were provided in the Supporting Information.
The validation and application of the developed pseudotargeted lipidomics method were performed with UHPLC/QQQ MRM MS. A QC sample was regularly inserted into the analysis sequence after every eight samples when analyzing the real biological samples.
Data Processing and Statistical Analyses
The lipids detected by the UHPLC/LTQ-Orbitrap MS-based nontargeted lipidomics method were determined by a commercially available software LipidSearch (version 4.0) and extraction of ion chromatogram (EIC) of lipids based on an in-house lipid database. All of the detected lipids were quantified by Thermo TraceFinder EFS software (version 3.2).
The lipids monitored by the UHPLC/QQQ MRM MS-based pseudotargeted lipidomics method were processed with the Analyst software in Explore Mode and Quantitate Mode (version 1.6, AB SCIEX, Concord, ON, Canada). Normalization of the lipids by the appropriate lipid ISs for each sample was performed when necessary.
Statistical analyses in Wilcoxon, Mann–Whitney Test mode, and heatmap visualization were conducted by the open-source software MultiExperiment Viewer (MeV, version 4.9.0, Dana–Farber Cancer Institute, MA). The statistical significance was set at p < 0.05 and FDR < 0.01. Principal component analysis (PCA) was performed using SIMCA-p (Umetrics, Umeå, Sweden) software.
Results and Discussion
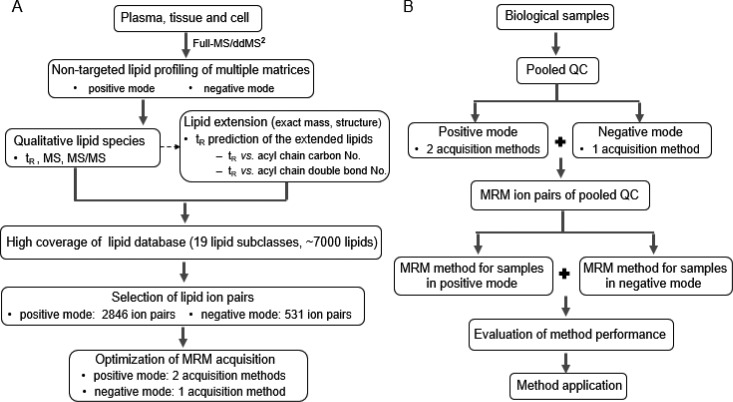
Comprehensive analysis of lipidome is of great importance for lipidomics studies. The present study aimed to establish a high coverage pseudotargeted lipidomics approach possessing the advantages of both targeted and nontargeted lipidomics approaches. The workflow of this study is shown in Figure 1.
Figure 1.
Workflow of pseudotargeted lipidomics method. (A) Scheme of acquiring high coverage of MRM lipid ion pairs. (B) Scheme of the real biological sample analysis by the pseudotargeted lipidomics method.
Detection of Lipids by Nontargeted Lipidomics Profiling
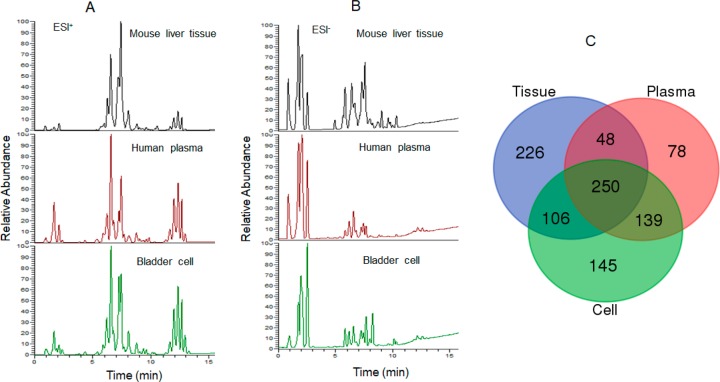
The Q Exactive-HF MS coupled with a UHPLC system was applied for nontargeted full scan and ddMS2 acquisition of lipids from multiple sample matrices to obtain as many lipids as possible. The representative chromatograms of multiple matrices are shown in Figure 2A (positive mode) and 2B (negative mode). Manual assignment was performed after lipid annotation derived from LipidSearch, and 515, 630, and 640 lipids were, respectively, identified in the pooled plasma, tissue, and cell samples (Figure 2C). Among them, a total of 955 unique lipids could be obtained with tR, accurate masses, and/or MS/MS fragments across 19 lipid subclasses.
Figure 2.
Representative chromatograms of multiple matrices by nontargeted lipidomics analyses in positive (A) and negative (B) modes. (C) Venn diagram of qualitative lipid species from nontargeted lipid profiling of multiple matrices by tR, MS, and MS/MS. These lipids were from 19 subclasses including 515 lipids in human plasma, 630 lipids in mouse liver tissue, and 640 lipids in cells.
It needs to be noted that such MS/MS fragment-based lipid identification would lose information on lipids which are unable to generate MS/MS fragments in ddMS2 due to low abundance, structure with a single acyl chain (i.e., fatty acids), or unsuitable collision energy. To compensate for such loss, EIC of lipids according to an in-house lipid database was performed. As expected, a large number of lipids could be extracted by this way. An extra 401, 492, and 438 lipid features were, respectively, found in the pooled plasma, tissue, and cell samples (Table S2, sheets 1 and 2). The identification of these extracted lipids was further confirmed by their predictively relative tR values compared to those of the detected lipids from the same lipid class according to the relationship between tR and acyl chain carbon No. or acyl chain double bond No. (Figure S1). Collectively, 508 unique lipids from three sample matrices were found. After combining the detected and extracted lipids, a total of 1463 lipids were defined with tR, accurate masses, and/or characteristic ions.
The 19 subclasses of the defined 1463 lipids could be summarized as fatty acids (FA), ceramides (Cer), hexaglycosylceramides (HexCer), dihexaglycosylceramides (Hex2Cer), sphingomyelins (SM), lyso-phosphatidylcholines (LPC), LPC with alkyl substituents (LPC-O), phosphatidylcholines (PC), alkyl and alkenyl substituent PCs (PC-O), phosphatidylethanolamines (LPE), LPE with alkyl substituents (LPE-O), phosphatidylethanolamines (PE), alkyl and alkenyl substituent PEs (PE-O), phosphatidylglycerols (PG), phosphatidylinositols (PI), phosphatidylserines (PS), diacylglycerols (DG), triacylglycerols (TG), and cholesterol esters (CE).
Prediction of Lipid Species
Although UHPLC-HRMS-based nontargeted lipidomics could provide rich lipid information, the frequently used HRMS detector (e.g., Orbitrap or FTICR) is easily saturated as it needs to scan too many ion features in a very short time. Thus, some lipid features, especially low abundant or the coeluted lipids, called the undetected but theoretically existing lipids, would be lost due to the limited MS scanning time per ion feature. It was essential to get the information on these lipids so that they could be covered in lipid detection by UHPLC/QQQ MRM MS with higher sensitivity and selectivity. With this consideration in mind, we extended the coverage of lipids via further prediction of lipid species based on their structure and retention relationship within a given lipid class. The prediction of the extended lipids was made on the basis of the following rules: for each kind of lipids, (1) the total No. of acyl chain carbon was defined considering the most frequently detected lipids in biological samples; (2) single lipid acyl chain carbon No. was considered to vary from 10 to 26, and acyl chain double bond No. was considered to vary from 0 to 6 as they are in mammals. It was summarized as (1) skeletons of sphingolipid: d16:0, d16:1, d18:0, d18:1, d18:2, d20:0, and d20:1; (2) ether-containing acyl chains: 16:0e, 16:0p, 18:0e, 18:0p, 18:1p, 18:2p, 20:0e, 20:0p, and 20:1p; (3) hydrophobic acyl chains: 10:0−26:0, 14:1-20:1, 22:1, 24:1, 26:1, 16:2, 17:2, 18:2, 20:2, 22:2, 18:3, 20:3, 22:3, 18:4, 20:4, 22:4, 20:5, 22:5, 22:6. Notably, the predicted lipids were presented with total acyl carbon No. and total acyl double bond No. When PC lipid species was taken as an example, the prediction was performed according to (1) the total No. of two acyl chain carbons varied from 20 to 48; (2) single acyl chain carbon No. varied from 10 to 26 and single acyl chain double bond No. varied from 0 to 6. For example, PC (20:X) would be PC (20:0) and PC (48:X) would include PC (48:0), PC (48:1), PC (48:2), PC (48:3), PC (48:4), PC (48:5), PC (48:6), PC (48:7), and PC (48:8). In this way, a total of 1914 lipids were predicted (Table S2, sheets 1 and 2).
Chromatographic tR is a very important parameter for lipid identification. It is well recognized that lipids from the same class/subclass have similar physicochemical properties. Therefore, the lipid structure and retention relationship could be well constructed within a specific class of lipids. When PC lipid class was taken as an example, Figure S1 displayed the structure and retention relationship constructed by tR versus acyl chain with different carbon No. or acyl chain with different double bond No. The tR of PCs with the same double bond No. was linearly increased with the increase of acyl chain carbon No. (Figure S1A). Furthermore, the tR of PCs with the same carbon No. was linearly decreased with the increase of acyl chain double bond No. (Figure S1C). These findings facilitated us to predict and identify more lipids. When PC (29:0) and PC (38:5) were taken as examples, the predicted tR of PC (29:0) was 5.75 min by the structure and retention relationship equation (black dots in black circle in Figure S1B). However, two peaks appeared at tR of 4.88 and 5.75 min in EIC of PC (29:0). The peak eluted at 5.75 min was regarded as PC (29:0). Similarly, tR of PC (38:5) was predicted to be 6.48 min (black dots in black circle in Figure S1D). In EIC of PC (38:5), there were three peaks eluted at tR of 6.48, 6.66, and 7.47 min, respectively. The peak at 7.47 min was excluded because of its larger difference as compared to predicted tR (6.48 min) by the structure and retention relationship equation. Peaks eluted at 6.48 and 6.66 min were regarded as two isomers of PC (38:5) and presented as PC (38:5)_1 and PC (38:5)_2. Lipid structure and retention relationships for all of the other classes of lipids were constructed in the same way (data not shown).
Construction of the Lipid Ion Pair Database
Lipid ion pairs were built up on the basis of the detected and predicted lipids for subsequent MRM. For each lipid class/subclass, lipid ion pairs were finalized by characteristic product ions with the highest intensity and the corresponding predominated precursor ions. Thus, the protonated ion ([M + H]+) was selected as precursor ions for PC, LPC, PC-O, LPC-O, PE, LPE, PE-O, SM, Cer, HexCer, and Hex2Cer; ammonium adducted ion ([M + NH4]+) for DG, TG, and CE; sodium adducted ion ([M + Na]+) for LPE-O; deprotonated ion ([M – H]−) for FA, PG, PS, and PI. For example, 184.1 was selected as a characteristic product ion for PC, LPC, PC-O, LPC-O, and SM; 241.0 for PI; 364.0, 392.0, 420.0, 390.0, 418.0, or 388.0, respectively, for the plasmalogen PE 16:0p_X, PE 18:0p_X, PE 20:0p_X, PE 18:1p_X, PE 20:1p_X, and PE 18:2p_X22 and so on (Table 1). Notably, isomers were not defined in most cases in the constructed lipid ion pair database. That means one given lipid ion pair might contain one or more lipid molecules due to many possible acyl chain combinations caused by different lengths of acyl chains or different acyl chain double bond No. (Table S3). A total of 3377 lipid ion pairs (Table S2 sheets 3 and 6, 2846 in positive and 531 in negative modes) with ∼7000 lipid molecular structures across 19 lipid subclasses were included in the lipid ion pair database.
Table 1. Summary of Detected Lipid Ion Pairs in Multiple Matrices from Plasma, Tissue, and Cell and Extended Lipid Ion Pairs.
| assignment |
||||||
|---|---|---|---|---|---|---|
| lipid classes | tR, MS, MS/MS | tR, MS | detected and extended ion pairs | ion adducts | fragmentation patterns | product ions |
| FA | 41 | 2 | 43 | [M – H]− | PIa | FA-H |
| LPC/PC | 118 | 76 | 260 | [M + H]+ | PI | 184.1 |
| LPC-O/PC-O | 64 | 54 | 160 | [M + H]+ | PI | 184.1 |
| LPE/PE | 74 | 49 | 239 | [M + H]+ | NLb | M + H+-141.0 |
| LPE-O | 4 | 3 | 9 | [M + Na]+ | NL | M + Na+-141.0 |
| PE-O | 61 | 50 | 305 | [M + H]+ | PI | 364.0/392.0/420.0/390.0/418.0/388.0 |
| PG | 24 | 37 | 156 | [M – H]− | PI | 153.0 |
| PI | 38 | 31 | 167 | [M – H]− | PI | 241.0 |
| PS | 35 | 42 | 165 | [M – H]− | NL | M – H–-87.0 |
| SM | 48 | 14 | 89 | [M + H]+ | PI | 184.1 |
| Cer/HexCer/Hex2Cer | 50 | 19 | 451 | [M + H]+ | PI | 238.3/236.2/266.3/264.3/262.2/312.3/310.3 |
| CE | 9 | 7 | 48 | [M + NH4]+ | PI | 369.4 |
| DG | 88 | 49 | 777 | [M + NH4]+ | NL | M + NH4+-17-FA |
| TG | 301 | 75 | 508 | [M + NH4]+ | NL | M + NH4+-17-FA |
| Sum | 955 | 508 | 3377 | |||
PI, product ion.
NL, neutral loss.
Construction of the Pseudotargeted Lipidomics Method
To efficiently monitor these constructed lipid ion pairs in MRM mode via the Q-Trap 5500 MS system, some important MS parameters (e.g., DP, CE, dwell time, etc.) were optimized. Optimal DP and CE values for 13 lipid standards in both positive and negative modes are shown in Table 2.
Table 2. Optimization of CE and DP Based on the Mixture of Lipid Standards at 1 μg/mL for Each Lipid.
| positive
mode |
negative
mode |
|||
|---|---|---|---|---|
| lipid classes | CE (eV) | DP (V) | CE (eV) | DP (V) |
| FA16:0(18:0)-d3 | –25 | –80 | ||
| LPC/PC | 40/40 | 100/110 | ||
| LPE/PE | 30/25 | 100/100 | ||
| PG | 25 | 110 | –45 | –80 |
| PS | 20 | 90 | –35 | –50 |
| PI | –60 | –70 | ||
| SM | 40 | 110 | ||
| Cer/HexCer/Hex2Cer | 40/45/45 | 100/100/100 | ||
| TG | 35 | 100 | ||
Other key factors for MRM acquisition performance are the appropriate dwell time and sufficient data-point. For our used Q-Trap 5500 MS system, the two parameters could be calculated by the equation of targeted time, which was equal to the No. of ion pairs in each detection window × (dwell time per ion pair + 5 ms). In this MS instrument, the minimum dwell time of one ion pair was 3 ms, that means ∼10 data-points would be acquired for a peak with 15 s of peak width when acquiring lipid ion pairs up to 200 in each detection window. Considering the peak width for most of the lipids was across the range of 0.2–0.3 min, ion pair No. of each detection window was suggested to be lower than 200. In the present developed method, the detection window was set as 60 s considering the existing tR shift during the chromatographic separation. Consequently, 2846 MRM transitions in positive mode were split into two MRM methods to get a combined demand of sensitivity and sufficient data acquisition rates (Table S2, sheets 4 and 5). To further ensure the rationality of this split, the input ion pair No. was investigated at a range from 25 to 400 during 7–8 min at a gradual increment of 50. It was found that the changes in the sensitivity and stability of MS could be neglected when the acquired lipid ion pair No. was lower than 200 in the 60 s detection window, which was in accord with the calculated result.
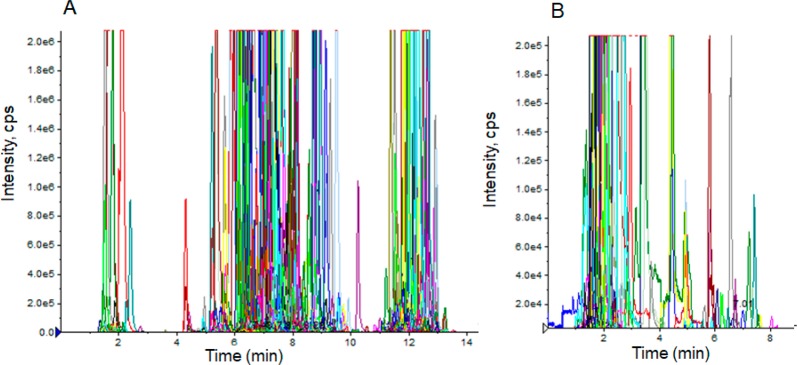
Furthermore, the constructed 3377 lipid ion pairs were monitored in 3 methods (Table S2, sheets 4, 5, and 7) for a QC serum sample from patients with diabetes and the healthy controls. The first and second acquisition methods contained 1423 ion pairs in positive mode, and the third acquisition method contained 531 ion pairs in negative mode. Typical chromatograms of the QC sample are shown in Figure S2. To reduce the random errors and the false positive results, each of the three acquisition methods was performed in triplicate. Additionally, the existing criterion of a lipid ion pair was set as the occurrence of a lipid ion pair being greater than 2/3 and the absolute value of time deviation being smaller than 0.5 min compared with the corrected tR achieved by the spiked lipid ISs.23 A total of 823 lipid ion pairs (758 for positive and 65 for negative modes, respectively) met the criteria and were used for the new UHPLC/QQQ MRM MS-based pseudotargeted lipidomics profiling to investigate the lipid disorder in diabetes. The typical chromatograms of the serum sample are shown in Figure 3.
Figure 3.
Representative MRM chromatograms of serum sample in the positive (A) and negative (B) modes based on ion pairs obtained from pooled QC of serum samples.
Additionally, for a reasonable comparison of the detection ability between the pseudotargeted and nontargeted lipidomics methods, the same QC sample was subjected to a UHPLC/LTQ-Orbitrap MS system for nontargeted lipid analysis. The result showed that 494 lipids were identified by UHPLC/LTQ-Orbitrap MS, 329 lipids less than the pseudotargeted method (Figure 4A). It could be attributed to (1) MRM analysis having higher selectivity and sensitivity in favor of detecting low abundant lipids. According to the LOD of 7 lipid ISs, pseudotargeted QQQ MS improved the absolute sensitivity of lipids up to 2–105 times compared to nontargeted HRMS except for LPC 19:0 (data not shown); (2) in the nontargeted method, the MS detector was easily saturated due to scanning too many ion features in a very short time, resulting in reduced data-points that would affect the detection of certain lipids; (3) the MS in MRM mode was able to separate some isomers (e.g., DGs, Cers, HexCers, and Hex2Cers in the present study) if different lipid ion pairs could be used. For example, two peaks were found for DG (34:2) (ion pair: 610.5/313.3, tR = 8.3; 610.5/311.3, tR = 8.3) and Cer (42:3) (ion pair: 646.6/264.3, tR = 9.2; 646.6/262.2, tR = 9.2), and three peaks were found for DG (38:6) (ion pair: 658.5/313.3, tR = 7.7; 658.5/337.3, tR = 7.6; 658.5/339.3, tR = 7.7) in pseudotargeted lipid profiling while only a single peak was found in nontargeted lipid profiling.
Figure 4.
(A) Venn diagram of detected lipids in a QC sample by pseudotargeted and nontargeted lipidomics methods. Reproducibility of common detected lipids in the positive (B) and negative (C) modes.
Validation of the Developed Pseudotargeted Lipidomics Method
Validation of the newly developed pseudotargeted lipidomics method was performed by taking plasma as an example. Linearity, precision, recovery, limit of detection (LOD), and repeatability were assessed.
As shown in Table 3, the linear regression coefficients (R2) were found to be better than 0.99 for all 7 lipid ISs (concentration range of 7 lipid ISs is provided in Table S4). The LODs were calculated at 3 times of signal-to-noise (S/N) ratio by the corresponding EICs. The LODs were satisfactory with values of 0.04–0.5 ng/mL in human plasma. Such low LODs were beneficial for the detection of the low abundant lipids with very important biological functions.
Table 3. Analytical Characteristics of the Pseudotargeted Lipidomics Method in Pooled Plasma.
| precision
(%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| linearity |
recovery
rate (%) |
low |
medium |
high |
|||||||||
| lipid IS | conc. range (μg/mL) | R2 | LOD (>3S/N) ng/mL | LOQ (>10S/N) ng/mL | low | medium | high | intra | inter | intra | inter | intra | inter |
| FA 16:0-d3 | 0.05–50 | 0.993 | 0.43 | 1.43 | 91 | 99 | 110 | 1.4 | 9.5 | 3.5 | 11.7 | 2.0 | 12.9 |
| LPC 19:0 | 0.02–100 | 0.999 | 0.34 | 1.14 | 84 | 85 | 107 | 2.8 | 4.8 | 3.1 | 6.6 | 1.4 | 6.1 |
| PC 19:0/19:0 | 0.05–125 | 0.994 | 0.5 | 1.67 | 99 | 102 | 92 | 2.9 | 13.6 | 2.7 | 12.9 | 0.8 | 4.6 |
| PE 17:0/17:0 | 0.02–100 | 0.991 | 0.18 | 0.59 | 84 | 93 | 102 | 3.2 | 10.8 | 4.3 | 10.9 | 1.5 | 5.5 |
| SM d18:1/12:0 | 0.01–50 | 0.999 | 0.04 | 0.13 | 98 | 85 | 115 | 1.7 | 11.1 | 3.3 | 9.4 | 1.1 | 2.4 |
| Cer d18:1/17:0 | 0.01–100 | 0.998 | 0.2 | 0.67 | 77 | 79 | 91 | 1.4 | 14.0 | 2.7 | 16.8 | 2.6 | 15.2 |
| TG 15:0/15:0/15:0 | 0.04–40 | 0.997 | 0.34 | 1.14 | 99 | 119 | 101 | 2.4 | 13.0 | 1.9 | 11.0 | 2.6 | 6.4 |
The intraday RSDs of 7 lipid ISs ranged from 0.8% to 4.3% with an average RSD of 2.3%; the interday RSDs of 7 lipid ISs ranged from 2.4% to 16.8% with an average RSD of 9.9%. The RSDs of 2.3% and 9.9% were acceptable for lipid profiling in complex biological samples, much lower than the maximum acceptable value of 15%. The low RSD for the interday precision indicated that the developed pseudotargeted method was very reproducible and could be applied for a large-scale sample analysis.
The recoveries of 7 lipid ISs were determined at the low, medium, and high concentration levels spiked before or after extraction (n = 3). Except for the Cer d18:1/17:0, the recoveries of other lipid ISs were in the range of 80–120% (Table 3), acceptable for complex biological matrices. The lower recoveries of Cer d18:1/17:0 at the low and medium concentration levels were caused by the limitation of the present extraction method. Our previous study proved that applying mild alkaline hydrolysis could improve the extraction of the sphingolipid.24
Repeatability is very important for lipidomics profiling. If a lipidomics method varied significantly, small but important changes between sample groups would be concealed.20 To further evaluate the adequacy of the present pseudotargeted method for lipidomics analysis, we investigated the repeatability of 494 lipids commonly detected in UHPLC/LTQ-Orbitrap and UHPLC/QQQ MS. RSDs of peak areas were calculated from 2 replicate injections of triplicate samples prepared in parallel, and the distribution of RSDs is shown in Figure 4. It can be seen from Figure 4B that 51% and 94% of lipids based on UHPLC/QQQ MRM detection showed RSDs less than 5% and 20%, whereas only 39% and 83% of lipids detected using UHPLC/LTQ-Orbitrap had RSDs less than 5% and 20%. In Figure 4C, 94% of lipids based on UHPLC/QQQ MRM showed RSDs less than 10%, whereas only 75% of lipids detected using UHPLC/LTQ-Orbitrap had RSDs less than 10%. Thus, 51% and 94% of the UHPLC/QQQ MRM MS-detected lipids had RSDs less than 5% and 20% in positive mode and 94% of the UHPLC/QQQ MRM MS-detected lipids had RSDs less than 10% in negative mode, indicating higher repeatability of this developed method. In addition, RSDs of the peak areas of the 823 lipids detected by the UHPLC/QQQ MRM MS method were calculated to evaluate the data quality. The result showed that 90% of the lipids had RSDs less than 20% in the positive mode and 98% of the lipids had RSDs less than 10% in the negative mode, indicating good repeatability of the pseudotargeted lipid profiling method (Figure S3).
Application of the Pseudotargeted Lipidomics Method
The applicability of the pseudotargeted lipidomics method was testified to discover serum lipid biomarker candidates for patients with diabetes. The serum lipid extracts of 30 patients with diabetes and 30 healthy controls were randomly analyzed by the UHPLC/QQQ MRM MS in the positive and negative modes. QC samples were regularly inserted into the batch analysis after running every 8 real samples for data quality control. After peak integration, 591 lipids with the RSD < 20% were selected for subsequent statistical analyses.
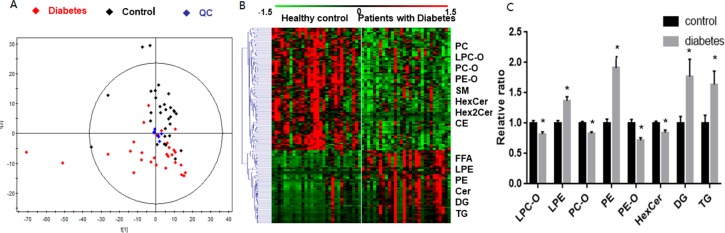
Unsupervised principal component analysis (PCA) showed that the QC samples were closely clustered to the center of the samples in the score plot, suggesting the good repeatability of the UHPLC/QQQ MRM MS method. Meanwhile, we noticed 2 outliers from the two groups, respectively, in PCA score plot. After checking clinical parameters and lipidome data, we found that 2 outliers in the control group had higher cholesterol than the mean value and 2 outliers in diabetic group displayed higher TG than the mean value. We have no scientific reason to exclude them. The patients with diabetes were distinctly separated from the healthy controls in the PCA score plot (Figure 5A), indicating that aberrant lipid metabolism occurred during the pathogenesis and development of diabetes.
Figure 5.
(A) PCA score plot of patients with diabetes, healthy controls, and QCs. (B) Heat map of differential lipid molecular species in diabetic patients as compared to the controls. (C) Relative value of each class of lipids in serum of patients with diabetes and healthy controls, analyzed by UHPLC/QQQ MRM MS (*p < 0.05; the data are expressed as group mean relative value ± SEM).
To find out the most important biomarker candidates distinguishing the patients with diabetes from the healthy controls, lipid metabolites with a p-value <0.05 and FDR < 0.01 were considered to be of interest. A total of 163 lipid metabolites across 15 lipid subclasses met the criteria (Table S5). The heat map of these significantly changed lipid metabolites is shown in Figure 5B.
To explore changes in lipid pool, the total contents of each lipid class/subclass were calculated for each of the samples. The student t test was performed for the patients diabetes versus the healthy controls, and a p-value <0.05 was considered to be of biological significance. The result is shown in Figure 5C. It can be observed that TGs, DGs, PEs, and LPEs were significantly increased, while HexCers, PE-Os, PC-Os, and LPC-Os were significantly decreased in patients with diabetes as compared to the healthy controls.
DGs are not only the functional components of membrane bilayers but also signaling molecules. It was reported that signaling of DG was mainly mediated by the recruitment and activation of the protein kinase C (PKC) isoforms to specific, membrane embedded DA species.25 PKC isoform nPKCθ and nPKCε could inhibit insulin signaling and ultimately glucose uptake.26−28 The increased DGs in patients with diabetes might indicate that PKC in diabetes was activated, which could influence the occurrence and development of diabetes.
Concerning the alteration of serum TGs, an interesting finding was that most of the polyunsaturated fatty acid-containing TGs were increased significantly in diabetes. Several prospective studies have identified dyslipidemia, particularly hypertriglyceridemia, as an independent predictor of incident type 2 diabetes mellitus.29,30 The increasing proportion of polyunsaturated fatty acid in TGs was attributed to insulin-mediated inhibition of hormone sensitive lipase: a decrease in saturated and monounsaturated fatty acid released from adipose tissue increased the relative amount of polyunsaturated fatty acids available to the liver for TGs assembly.31,32
Ether-containing glycerophospholipids, especially plasmalogens, have many roles in cellular function and are important components of the cell plasma membrane.33−35 It was reported that reduced plasmalogen content could be caused by peroxisomal dysfunction, oxidative stress, or phospholipase (particularly phospholipase A2) activation.36 In this study, PE-O lipids were significantly reduced while LPEs significantly increased in diabetes (Figure 5C). These findings might indicate that oxidative stress increased in diabetes, leading to the development of diabetes by damaging the islet β cells and reducing the sensitivity of peripheral tissues to insulin.37 This finding has been confirmed by Wang et al., who measured the indicator of oxidative states of 4-hydroxylalkenal.38 Wacker et al. have also verified the finding by measuring chlorinated lipids.39
Collectively, our results demonstrate that the UHPLC/QQQ MRM MS-based pseudotargeted lipidomics method possesses a high coverage and good characteristics of linearity, precision, recovery, and repeatability. It is very promising for discovering potential biomarkers of disease.
Conclusions
In the present study, a novel strategy for high coverage of lipid profiling was implemented, combining the advantages of both nontargeted and targeted lipidomics approaches. A total of 3377 ion pairs covering ∼7000 lipid molecules were derived from nontargeted UHPLC-full scan/ddMS2 lipid analysis of multiple biological matrices and lipid predictions according to the lipid structure and retention relationship within a given lipid class. To our knowledge, this is the first study using this strategy for high coverage of lipid profiling. It enabled the detection of 823 lipids from 40 μL of serum, displaying higher lipid coverage and better repeatability than the UHPLC/LTQ-Orbitrap MS-based nontargeted lipidomics method. Data processing was simple, and the efficiency was greatly improved. It showed high potential for defining the characteristics of lipids in complex biological matrices. Future studies will extend the method to biological matrices from different diseases. We firmly believe that the developed high coverage UHPLC/QQQ MRM MS-based pseudotargeted method will be beneficial for the lipidomics study.
Acknowledgments
This research was supported by the National Key Research and Development Program of China (2017YFC0906900), the foundations (21575140) and key foundation (21435006) from the National Natural Science Foundation of China, and the innovation program (DICP TMSR201601) of science and research from the DICP, CAS.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b01331.
Experimental details and additional information on the linear relationship between tR and number of acyl carbons or number of acyl double bonds; typical chromatograms of QC based on the pseudotargeted lipidomics method; reproducibility of the pseudotargeted lipidomics method; clinical parameters on 30 patients with diabetes and 30 healthy controls; one lipid ion pair corresponding to multiple molecular structures; concentration range of 7 lipid internal standards (PDF)
Information of the detected and predicted lipids (XLSX)
The most significantly changed 163 lipids contributing to the discrimination between patients with diabetes and the healthy controls (XLSX)
Author Contributions
# Q.X. and C.H. contributed equally to this manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Wenk M. R. Cell 2010, 143, 888–895. 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Brown H. A.; Murphy R. C. Nat. Chem. Biol. 2009, 5, 602–606. 10.1038/nchembio0909-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann C.; Pechlaner R.; Willeit P. J. Vasc Surg 2014, 60, 532. 10.1016/j.jvs.2014.06.095. [DOI] [Google Scholar]
- Rhee E. P.; Cheng S.; Larson M. G.; Walford G. A.; Lewis G. D.; McCabe E.; Yang E.; Farrell L.; Fox C. S.; O’Donnell C. J.; Carr S. A.; Vasan R. S.; Florez J. C.; Clish C. B.; Wang T. J.; Gerszten R. E. J. Clin. Invest. 2011, 121, 1402–1411. 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.; Wang Y. M.; Zhou D.; Li Z. L. Sci. Rep. 2015, 4, 5959. 10.1038/srep05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm M. O.; Michaelson D.; Hartmann T. J. Lipid Res. 2017, 58, 2083. 10.1194/jlr.R076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner L. B.; Chng C.; Guan X. L.; Lei Z.; Rozen S. G.; Wenk M. R. J. Lipid Res. 2014, 55, 1357. 10.1194/jlr.M049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M. S.; de Oliveira D. N.; de Oliveira R. N.; Allegretti S. M.; Catharino R. R. Anal. Chim. Acta 2014, 845, 62–69. 10.1016/j.aca.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Kishimoto K.; Urade R.; Ogawa T.; Moriyama T. Biochem. Biophys. Res. Commun. 2001, 281, 657–662. 10.1006/bbrc.2001.4404. [DOI] [PubMed] [Google Scholar]
- Qin Q.; Wang B. H.; Chang M. M.; Zhou Z. H.; Shi X. Z.; Xu G. W. J. Chromatogr A 2016, 1457, 125–133. 10.1016/j.chroma.2016.06.046. [DOI] [PubMed] [Google Scholar]
- Wang M.; Wang C.; Han R. H.; Han X. Prog. Lipid Res. 2016, 61, 83–108. 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gethings L. A.; Richardson K.; Wildgoose J.; Lennon S.; Jarvis S.; Bevan C. L.; Vissers J. P. C.; Langridge J. I. Rapid Commun. Mass Spectrom. 2017, 31, 1599–1606. 10.1002/rcm.7941. [DOI] [PubMed] [Google Scholar]
- May F. J.; Baer L. A.; Lehnig A. C.; So K.; Chen E. Y.; Gao F.; Narain N. R.; Gushchina L.; Rose A.; Doseff A. I.; Kiebish M. A.; Goodyear L. J.; Stanford K. I. Cell Rep. 2017, 18, 1558–1572. 10.1016/j.celrep.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. L.; Yang K.; Gross R. W. Mass Spectrom. Rev. 2012, 31, 134–178. 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. F.; Wang M.; Han X. L. Redox Biol. 2017, 12, 946–955. 10.1016/j.redox.2017.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildburger N. C.MALDI-Imaging Mass Spectrometry of Brain Lipids. In Lipidomics; Humana Press: New York, 2017. [Google Scholar]
- Sandra K.; Pereira A. S.; Vanhoenacker G.; David F.; Sandra P. J. Chromatogr A 2010, 1217, 4087. 10.1016/j.chroma.2010.02.039. [DOI] [PubMed] [Google Scholar]
- Cajka T.; Fiehn O. TrAC, Trends Anal. Chem. 2014, 61, 192–206. 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L. D.; Souza A. L.; Gerszten R. E.; Clish C. B.. Targeted Metabolomics. In Current Protocols in Molecular Biology, John Wiley & Sons, Inc.: 2012; Chapter 30, Unit 30.2, pp 30.2.1−30.2.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L.; Kong H. W.; Lu X.; Li Y. L.; Yin P. Y.; Zeng Z.; Xu G. W. Anal. Chem. 2013, 85, 8326–8333. 10.1021/ac4016787. [DOI] [PubMed] [Google Scholar]
- Zhou B.; Lu Y.; Hajifathalian K.; Bentham J.; Cesare M. D.; Danaei G.; Bixb H.; Cowan M. J.; Ali M. K.; Taddei C. Lancet 2016, 387, 1513. 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemski Berry K. A.; Murphy R. C. J. Am. Soc. Mass Spectrom. 2004, 15, 1499–1508. 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Fu Y. Q.; Zhou Z. H.; Kong H. W.; Lu X.; Zhao X. J.; Chen Y. H.; Chen J.; Wu Z. M.; Xu Z. L.; Zhao C. X.; Xu G. W. Anal. Chem. 2016, 88, 8870. 10.1021/acs.analchem.6b02482. [DOI] [PubMed] [Google Scholar]
- Li J.; Hu C. X.; Zhao X. J.; Dai W. D.; Chen S. L.; Lu X.; Xu G. W. J. Chromatogr A 2013, 1320, 103–110. 10.1016/j.chroma.2013.10.064. [DOI] [PubMed] [Google Scholar]
- Almena M.; Mérida I. Trends Biochem. Sci. 2011, 36, 593–603. 10.1016/j.tibs.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Li Y.; Soos T. J.; Li X. H.; Wu J.; Degennaro M.; Sun X. J.; Littman D. R.; Birnbaum M. J.; Polakiewicz R. D. J. Biol. Chem. 2004, 279, 45304. 10.1074/jbc.C400186200. [DOI] [PubMed] [Google Scholar]
- Erion D. M.; Shulman G. I. Nat. Med. 2010, 16, 400. 10.1038/nm0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. J.; Samuel V. T.; Petersen K. F.; Shulman G. I. Nature 2014, 510, 84–91. 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien K.; Cai T.; Hsu H.; Su T.; Chang W.; Chen M.; Lee Y.; Hu F. B. Diabetologia 2009, 52, 443–450. 10.1007/s00125-008-1232-4. [DOI] [PubMed] [Google Scholar]
- Kahn H. S.; Cheng Y. J.; Thompson T. J.; Imperatore G.; Gregg E. W. Ann. Intern. Med. 2009, 150, 741. 10.7326/0003-4819-150-11-200906020-00002. [DOI] [PubMed] [Google Scholar]
- Zhao X. J.; Peter A.; Fritsche J.; Elcnerova M.; Fritsche A.; Häring H. U.; Schleicher E. D.; Xu G. W.; Lehmann R. Am. J. Physiol-Endoc M 2009, 296, E384–393. 10.1152/ajpendo.90748.2008. [DOI] [PubMed] [Google Scholar]
- Parks E. J.; Hellerstein M. K. J. Lipid Res. 2006, 47, 1651–1660. 10.1194/jlr.R600018-JLR200. [DOI] [PubMed] [Google Scholar]
- Hu C. F.; Zhou J.; Yang S. S.; Li H. C.; Wang C. Y.; Fang X. L.; Fan Y. S.; Zhang J. D.; Han X. L.; Wen C. P. Free Radical Biol. Med. 2016, 101, 475–481. 10.1016/j.freeradbiomed.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Farooqui A. A.; Horrocks L. A. Neuroscientist 2001, 7, 232–245. 10.1177/107385840100700308. [DOI] [PubMed] [Google Scholar]
- Engelmann B. Biochem. Soc. Trans. 2004, 32, 147–150. 10.1042/bst0320147. [DOI] [PubMed] [Google Scholar]
- Nagan N.; Zoeller R. A. Prog. Lipid Res. 2001, 40, 199–229. 10.1016/S0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]
- Gross R. W. Biochemistry 1984, 23, 158–165. 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- Wang M.; Fang H. F.; Han X. L. Anal. Chem. 2012, 84, 4580–4586. 10.1021/ac300695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker B. K.; Albert C. J.; Ford B. A.; Ford D. A. Free Radical Biol. Med. 2013, 59, 92–99. 10.1016/j.freeradbiomed.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.