Abstract
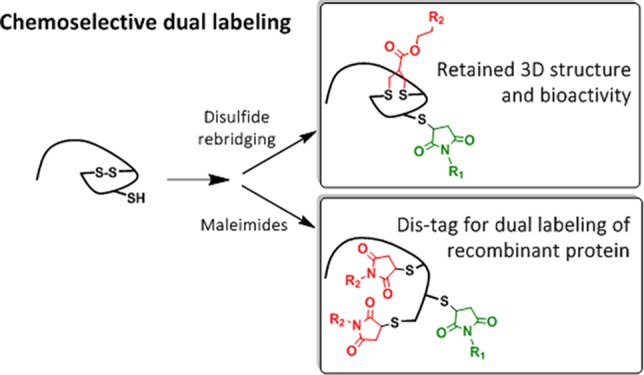
The attachment of two different functionalities in a site-selective fashion represents a great challenge in protein chemistry. We report site specific dual functionalizations of peptides and proteins capitalizing on reactivity differences of cysteines in their free (thiol) and protected, oxidized (disulfide) forms. The dual functionalization of interleukin 2 and EYFP proceeded with no loss of bioactivity in a stepwise fashion applying maleimide and disulfide rebridging allyl-sulfone groups. In order to ensure broader applicability of the functionalization strategy, a novel, short peptide sequence that introduces a disulfide bridge was designed and site-selective dual labeling in the presence of biogenic groups was successfully demonstrated.
Introduction
Site-selective modification of native proteins has grown as a vibrant field of research due to its diverse therapeutic and diagnostic applications.1,2 Desired functional properties can be introduced to the protein of interest through modifications by synthetic molecules.3 The field has since advanced beyond monofunctionalization, and the development of dual modification strategies for increasing the functionalities of proteins has received interest in recent years.4,5 In this context, Francis and Paavola et al. have reported pyridincarboxaldehyde as a tool for N-terminal protein modification and have demonstrated dual modification in combination with other bioorthogonal handles.6 Targeting cysteine (Cys) residues has also become a strategy of choice for introducing more than one functionality at distinct sites. Rathner et al. have explored dual functionalization of proteins based on the reactivity variations of different unpaired thiols.7 However, this approach requires identifying proteins bearing two unpaired Cys residues with sufficient nucleophilicity differences. Sonntag et al. have reported an orthogonal dual cysteine labeling strategy by the temporary protection of an N-terminal cysteine unit in the form of a thiazolidine ring.8 Likewise, Caddick et al. have demonstrated dual modification of green fluorescent protein (GFP) by the systematic development of dual Cys mutants to introduce two different thiols of desired reactivities.9 Despite the repertoire of reagents to functionalize proteins, site-selective addition of multiple functional groups onto a single protein still remains a significant challenge.10,11
Reagents addressing disulfides in proteins have been exploited for diverse site-specific protein functionalization strategies.12−16 It has been demonstrated previously that the proteins’ functions are retained after introduction of a disulfide rebridging reagent, e.g., for the site-selective incorporation of polyethylene glycol (PEG) polymers on native proteins.16 The abundant availability of surface accessible disulfide bonds in most proteins and minimum disturbance of the protein’s tertiary structure proved broad applicability of the disulfide rebridging technique.17,18 Recently, single step dual functionalization of disulfide in proteins has been achieved using aryloxymaleimide19 or the incorporation two orthogonal handles on the disulfide rebridging reagent.20,21
We have recently reported the synthesis of water-soluble allyl-sulfones that facilitate the site-selective functionalization at the disulfide of cyclic peptides or native proteins without the necessity to apply organic solvents.22 The allyl-sulfone has also been demonstrated to simultaneously introduce two different functionalities, e.g., a chromophore and an affinity tag at a single site by simple thiol-chemistry.22 However, the introduction of functionalities at two different sites is of particular interest for resolving the dynamics of peptides and proteins, e.g., by applying two chromophores that interact via Förster Resonance Energy Transfer (FRET). Relative positions of the chromophores can be detected at the single molecules level and allow elucidating protein folding or dynamic interactions that are associated with structural changes.
In this context, disulfide bonds could also be viewed as protected thiols that could be first reduced under mild conditions to “activate” their reactive thiols for functionalization, and such a pronounced difference in the reactivity of an unpaired cysteine and a “protected” disulfide can be capitalized upon for the dual functionalization of proteins at two distinct sites. Herein, we report the controlled dual modification of native and recombinant proteins using this approach. Stepwise functionalization is achieved first by modifying the unpaired Cys by maleimide conjugation, followed by disulfide reduction, which liberates two additional free thiols that can either react with a single allyl-sulfone rebridging reagent (Figure 1A) or with two maleimide groups (Figure 1B) to afford the corresponding dual labeled proteins. The functionalization strategy is demonstrated for interleukin-2 (IL-2) and EYFP possessing the respective functionalities and retained activity has been demonstrated for both the proteins. In case no accessible thiols or disulfides are present, a new, short peptide sequence of six amino acids (Dis-tag) has been designed that imparts an unpaired Cys or a disulfide group to the recombinant proteins.
Figure 1.
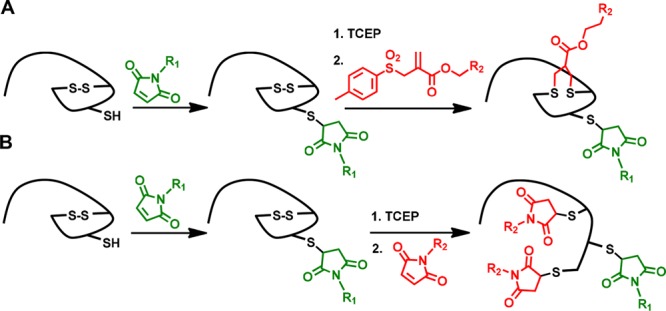
Dual labeling strategies based on protected (disulfide) and nonprotected thiols. (A) Sequential labelling of free thiol with maleimide followed by disulfide rebridging by allyl-sulfone reagent. (B) Maleimide-based sequential labeling introducing two new reagents at distinct sites.
Results and Discussion
IL-2 has been selected to demonstrate dual labeling of a native functional protein of high medical relevance. IL-2 is a 15.4 kDa cytokine with essential function for the immune system, consisting of 133 amino acids. Under steady state conditions, IL-2 is mainly secreted by CD4+ T cells yet secretion by CD4+ and CD8+ is highly induced upon activation of these cells.23 The IL-2 cytokine plays an essential role in the induction of T cell-mediated immune responses but is also indispensable for proper function of regulatory T cells that are important mediators of immune tolerance.24 Native IL-2 has three cysteines, a free thiol (Cys125) and a disulfide bond (Cys58-Cys105). Therefore, we propose applying the sequential conjugation of maleimide followed by disulfide rebridging strategy for the dual labeling of IL-2 (Figures 1A). First, the unpaired cysteine (Cys125) of IL-2 1 was reacted with 2 equiv of Cy3-MI (sulfo-cyanine-3-maleimide; Figure S1A) in 50 mM phosphate buffer (PB) at pH 7.4 and 15 °C overnight (Figure 2A). The conjugate IL2-Cy3 2 was purified by size-exclusion chromatography using Zeba filtration columns (MWCO 7000). The labeling efficiency was quantified (75%) by absorbance measurements in comparison to a Cy3-MI calibration curve and gel densitometry (Figures S3 and S4). The labeling efficiency is comparable to maleimide–thiol conjugation to other proteins such as BSA.25,26 The absorbance and emission spectra of the IL2-Cy3 2 reveal the characteristic Cy3 absorbance and emission bands at 550 and 565 nm, respectively (Figure 2B and C, red) indicating successful conjugation. For the second labeling step, the disulfide bond between Cys58-Cys105 in IL-2 was selected. To achieve this, the disulfide rebridging reagent allyl-sulfone-Cy5 was selected (Figures 1A and S1B; see also Scheme S1 in the Supporting Information for synthesis and characterization), which efficiently introduces the Cy5 chromophore site-specifically. In contrast to the attachment of maleimides, allyl-sulfone reagents preserve the tertiary structure of the protein by inserting a three carbon bridge (SCCCS) between the unpaired cysteines (Figure 2A). The disulfide bond was reduced with 2 equiv of TCEP for 30 min in 50 mM PB at pH 7.8 yielding the two deprotected thiol groups as reactive intermediates. Subsequently 2 equiv of the allyl-sulfone-Cy5 (Figure S1B) were added and the reaction mixture was stirred for 24 h at 15 °C in 50 mM PB at pH 7.8 yielding dual labeled IL2-Cy3-Cy5 3 (Figure 2B and C, orange).
Figure 2.
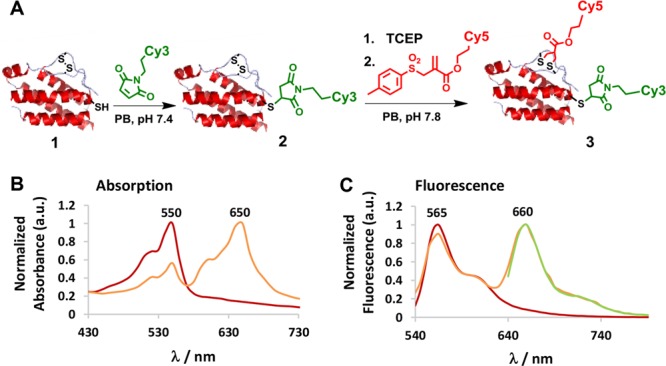
(A) Stepwise site-directed dual labeling of IL-2 yielding 2 in 75% and 3 in 55% yield. (B) Absorption spectra of IL2-Cy3 2 (red) and IL2-Cy3-Cy5 3 (orange). (C) Emission spectra of IL2-Cy3 2 (red) and IL2-Cy3-Cy5 3 (orange) excitation at 510 nm, and IL2-Cy3-Cy5 3 (green) excited at 610 nm.
The conjugate IL2-Cy3-Cy5 3 was purified by size exclusion chromatography using Zeba filtration column (MWCO 7000). A labeling efficiency of 55% was calculated for Cy5 from the absorption calibration curve and by gel densitometry (see Supporting Information, Figure S3 and S4). The dual functionalized product IL2-Cy3-Cy5 3 showed characteristic absorbance and emission bands of both chromophores Cy3 and Cy5 (Figure 2B and C, orange). When excited at 510 nm (Cy3 excitation maxima), IL2-Cy3-Cy5 3 revealed emission bands of both Cy3 (565 nm) and Cy5 (660 nm), displaying Förster resonance energy transfer (FRET) from Cy3 to Cy5 (Figure 2C, orange). Biological activity of the dual labeled IL-2 derivative was assessed on IL-2-dependent murine CTLL-2 cells (Figure 3A) according to a previously published procedure.27 IL-2-induced T cell proliferation was determined for IL2-Cy3 2 and IL2-Cy3-Cy5 3 in direct comparison with the unmodified IL-2 1 (Figure 3B). Similar T-cell (CTLL-2) proliferation was obtained for IL-2 1, IL2-Cy3 2, and IL2-Cy3-Cy5 3 (Figure 3B), indicating intact tertiary structure essential for IL-2 receptor binding and preserved bioactivity of the bioconjugates. It is noteworthy that Yamada et al. have demonstrated that by completely reducing the disulfide bond of human IL-2, there is a 90% loss in its bioactivity,28 while in our case, the bioactivity is preserved after rebridging. This approach is valuable when the disulfide bonds are essential to maintain the tertiary structures and bioactivities of proteins.29 As a control, we have demonstrated that the double addition of maleimide to the reduced disulfide in IL-2 did not lead to a significant change in activity compared to native IL-2 (for detailed experimental procedure, see Supporting Information, Figure S5).
Figure 3.
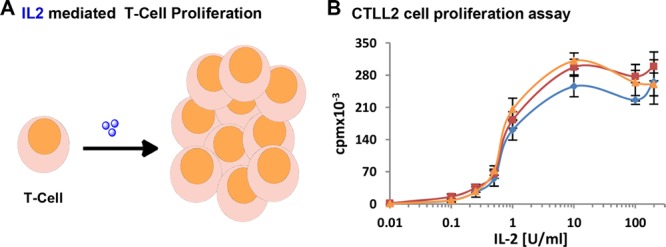
(A) Schematic diagram for IL-2-induced T-cell proliferation. IL-2 is depicted as blue spheres. (B) CTLL2 (T-cells) proliferation assay with titration of increasing concentrations of IL-2 1 (blue), IL2-Cy3 2 (red), and IL2-Cy3-Cy5 3 (orange).
In order to further broaden the applicability of the presented approach, a short peptide sequence has been developed containing the disulfide and unpaired Cys that could be inserted into the sequence of recombinant proteins for dual labeling purposes. Previously, systematic studies of the air oxidation of Cys-Xm-Cys peptides indicated more than 90% disulfide loop formation for m = 4–6.30 Therefore, a peptide sequence of a minimum of six amino acids should be sufficient to accommodate a single disulfide bond. The peptide was designed to accommodate the desired functionalities with minimum peptide length in order to avoid perturbation of protein activity.31 To demonstrate the potential of stepwise labeling, a model deca peptide CGGGCISTAC4 (Figure 4A, disulfide between Cys1 and Cys6 underlined) was designed. This model peptide contains the six amino acid sequence CISTAC, which forms the disulfide as well as a free cysteine separated from the disulfide tag by a short G spacer. The model peptide offers reaction optimization, purification, and characterization of each labeling step by MALDI-MS and purification by HPLC, which is more challenging for high molecular weight proteins. First, the free thiol of 4 was reacted with 1.5 equiv of 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarine (CPM; Figure S6) to afford the monolabeled product 5 in 61% yield after HPLC purification. In the next step, the disulfide between Cys1 and Cys6 was reduced by 2 equiv of TCEP and readily reacted with 4 equiv of CPM to generate the corresponding three coumarin-labeled peptide 6 in 72% yield. Both products 5 and 6 were purified via HPLC and characterized by MALDI-MS, indicating successful stepwise labeling of model peptide 4 (Figure 4B and C). Noteworthy, no dual conjugation product (m/z 1672.6022 [M+2CPM]) was observed in the MALDI-MS spectrum of monolabeled product 5 (Figure 4B) clearly indicating that the protecting disulfide was formed, remained stable during the first labeling step and could be reduced and labeled in the following reaction step.
Figure 4.
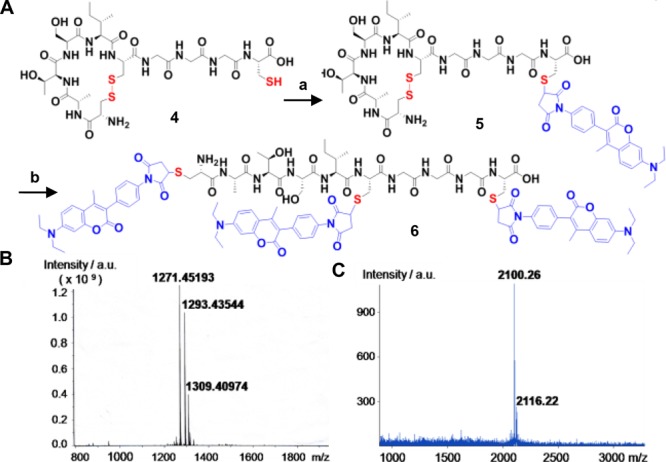
(A) Stepwise modification of model peptide 4. (a) CPM, 3 h RT. (b) TCEP, 30 min; CPM, 3 h at RT. Both reactions were accomplished in acetonitrile: phosphate buffer (1:1), pH 7.4 mixture. (B) MALDI-MS spectrum of 5 and (C) 6.
In order to further decrease the length of the peptide tag, the unpaired cysteine was integrated into the second position of the peptide, reducing the length of model peptide 4 (deca-peptide; CGGGCISTAC) (Figure 4A) to the hexapeptide “Dis-tag” (CISTCC) (Figure 5A). In the Dis-tag, Cys1, and Cys6 residues are separated by the required four intervening residues, which is essential for disulfide loop formation.30 In contrast, Cys2 with less than four residues between the neighboring cysteines (Cys1 and Cys6) does not participate in disulfide bond formation.
Figure 5.
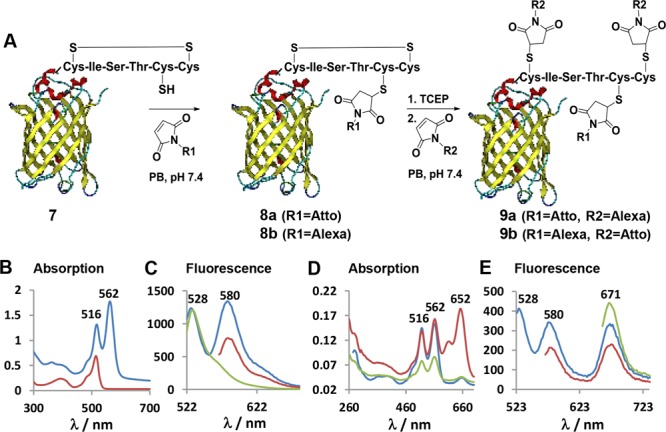
(A) Site-specific labeling of Dis-tag-EYFP 7 with two different fluorophores denoted as R1 and R2 in a generic fashion. (B) Absorption spectra of Dis-tag-EYFP 7 (red) and Atto550-EYFP 8a (blue) obtained after the first labeling step. (C) Emission spectra of the Atto550-EYFP 8a excited at 516 nm (blue), 562 nm (red), and Dis-tag-EYFP 7 excited at 514 nm (green). (D) Absorption spectra of the dual labeled Atto550-Alexa647-EYFP 9a (red) after the second labeling step and Atto550-EYFP 8a as control that has been incubated with Alexa647-maleimide without preincubation of TCEP (blue). Atto550-EYFP 8a (green) after first capping the additional free cysteine with N-(2-aminoethyl)maleimide and addition of Alexa647 without preincubation of TCEP (green). (E) Emission spectra of the Atto550-Alexa-EYFP 9a excited at 516 nm (blue), 562 nm (red), and 652 nm (green). Absorption and emission spectra of 8b and 9b are given in the Supporting Information Figure S11.
The Dis-tag was inserted into the N-terminus of the enhanced yellow fluorescent protein (EYFP) for dual modification on a reporter protein (see Supporting Information, Figure S7). EYFP offers characterization of retained structural integrity of each labeling step by simply recording the emission spectra after each chemical modification step. EYFP 7 containing the Dis-tag was sequentially labeled with two different dyes to yield a FRET pair (Figure 5A). Atto550-maleimide (Atto550-MI, 5 equiv) was first reacted to the Dis-tag-EYFP 7 at the unpaired cysteine via Michael addition for 3 h in PB at pH 7.4. The resulting monolabeled EYFP 8a was purified by size exclusion chromatography using Sephadex G-25 column. A labeling efficiency of 60% of the Atto550-EYFP 8a was obtained as determined from the absorbance calibration curve and gel densitometry (see Supporting Information, Figure S9 and S10A). The absorbance and emission spectra of Atto-EYFP 8a reveals the characteristic bands of EYFP (Ex/Em: 516/528) and Atto550 (Ex/Em: 562/580). The Atto550-EYFP conjugate 8a was excited at 516 nm and shows the two emission envelopes of EYFP and Atto550 (Figure 5C, blue) indicating energy transfer from EYFP to Atto550.
In the second labeling step, the disulfide bond in Atto550-EYFP 8a was reduced with 2 equiv of TCEP and reacted with 3 equiv Alexa647 maleimide (Alexa647-C2-MI; Figure S8) for 3 h in PB at pH 7.4 yielding dual labeled Atto550-Alexa647-EYFP 9a. The significant increase in absorbance at 652 nm reflects successful conjugation of the Alexa647 dye to the Atto550-EYFP 8a conjugate (Figure 5D, red). The final product 9a was purified by applying a Sephadex G-25 column and a labeling efficiency of 42% was calculated from the absorbance calibration curve and gel densitometry (see Supporting Information, Figure S9 and S10B). When the conjugate 9a was excited at 516 nm, all three characteristic emission peaks were observed at 528 nm (EYFP), 580 nm (Atto550), and 671 nm (Alexa647) demonstrating the energy transfer from EYFP to Atto550 and from Atto550 to Alexa647 (Figure 5E, blue). To rule out the possibility of unspecific disulfide cleavage during the first reaction step, we incubated Atto550-EYFP 8a with a large excess (10 equiv) of Alexa647 without reduction of the disulfide by TCEP. Only very weak absorbance at 652 nm (Figure 5D, blue) was visible, which was very minor and could also be due to unspecific adsorption of the Alex647 dye to EYFP due to the addition of a very large excess of this hydrophobic dye molecule (Figure 5D, blue).
As an additional control, the bioconjugate Alexa647-Atto550-EYFP 9b (Figure 5A) was synthesized, in which the respective positions of the chromophores was exchanged. Alexa647-MI (3 equiv) was first reacted with the unpaired Cys of Dis-tag-EYFP 7 for 3 h in PB at pH 7.4 (Figure 5A) and purified by Sephadex G-25 column to receive Alexa-EYFP 8b. A labeling efficiency of 47% was calculated from the absorbance (Supporting Information, Figure S11). Thereafter, the disulfide of Alexa-EYFP 8b was reduced applying an excess (10 equiv) of TCEP for 1 h, followed by reaction with 3 equiv of Atto550-MI in phosphate buffer at pH 7.4 for 3 h (Figure 5A). The resulting product 9b was purified by size exclusion chromatography using Sephadex G-25 and very high labeling efficiency of 94% of the Atto550 conjugation was calculated from the absorbance (Supporting Information, Figure S11). Thereafter, the energy transfer efficiency from the Atto550 energy donor chromophore to the Alexa647 acceptor dye was assessed in both protein conjugates 9a and 9b after excitation with 562 nm (e.g., Atto550 excitation) and the emission spectra were recorded (see Supporting Information Figure S12). Alexa647-Atto550-EYFP 9b revealed significantly enhanced (85%) emission compared to Atto550-Alexa647-EYFP 9a when excited under the same conditions (see Supporting Information Figure S12). The significantly increased emission of the Alexa647 acceptor chromophore in Alexa647-Atto550-EYFP 9b was attributed to the more effective energy transfer from the two Atto550 donor chromophores to the single Alexa647 acceptor dye in 9b (see Supporting Figure S12 A,B).
Both Atto550 and Alexa647 chromophores allow single molecule studies due to their overlapping absorption and emission spectra as well as their photostability. Single-molecule Förster Resonance Energy Transfer (smFRET) experiments have been accomplished to further support the site-specific attachment of the chromophores of Alexa647-Atto550-EYFP 9b and the formation of a single, distinct protein bioconjugates. SmFRET has grown as the most efficient tool to study complex FRET systems containing multiple fluorophores.32 To evaluate the energy transfer efficiency between the Atto (donor) and Alexa (acceptor chromophores) and EYFP, we performed the smFRET study of Alexa647-Atto550 -EYFP 9b following a procedure described previously.33 Briefly, the dual labeled bioconjugates 9b was loaded into a sample flow chamber and immobilized on the surface (Figure 6A). SmFRET experiments were performed using a frequency-doubled Nd:YAG laser (Newport Spectra-Physics) with an excitation wavelength of 532 nm. Emitted light of the Atto550 donor (550–610 nm) and the Alexa647 acceptor (660–760 nm) chromophores was separated using a dichroic mirror (Chroma) and detected using an electron multiplying charge-coupled device (EMCCD) camera. Fluorescence intensities of the donor and acceptor dyes of each EYFP protein were recorded with an integration time of 100 ms per frame for a total duration time of 15 s (Figures 6C and S13). A stable high smFRET signal between the two dyes was observed resulting in sharp single peaked histograms at a mean FRET efficiency of 97% (Figure 6C) clearly indicating that the labeling sites for the two organic dye molecules were in close vicinity (Figure 6B).
Figure 6.
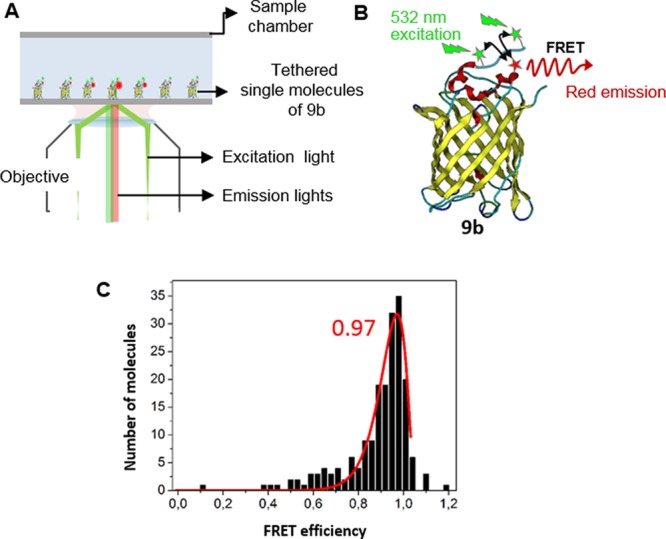
(A) Schematic for smFRET experiment. (B) Cartoon representation of a smFRET event on Alexa-Atto-EYFP 9b. Positions of Atto550 (green) and Alexa647 (red) are indicated by asterisks. (B) Histogram of smFRET measurements. Mean smFRET computed for 191 molecules with bin size 0.03. The mean smFRET efficiency of 97%.
Conclusion
In summary, we have presented the site-selective dual labeling of peptides and proteins utilizing protected and unprotected thiols and their corresponding reactivity differences. This methodology enables fast and efficient chemoselective stepwise labeling of native and recombinant proteins containing a free thiol and a disulfide bond. Two different functionalities were attached to the cytokine IL-2 of high clinical relevance. Bioactivity of the labeled IL-2 was preserved supporting the great value of this conjugation method to impart different functionalities of choice to a therapeutic protein.
In order to facilitate bioconjugation of proteins without cysteines, a novel, six amino acid peptide tag (“Dis-tag”) was designed and synthesized that introduces a new disulfide bridge and/or a free cysteine into the protein sequence applying molecular biology methods. Site-selective and stepwise functionalization in the presence of biogenic groups was successfully demonstrated on the model peptide tag. The recombinant protein EYFP was expressed with the Dis-tag and two distinct chromophores were introduced in a stepwise fashion yielding two new protein conjugates 9a,b. To support the applicability of our new approach for future applications, e.g., for single molecule studies of proteins, the labeled EYFP-Dis-Tag protein Alexa647-Atto550-EYFP 9b showed stable smFRET efficiency with a value corresponding to the short distance between the labeling sites of the tag thus proving that not more distant interactions or unspecifically adsorbed chromophore reagents were observed by smFRET. These results underline the great potential of our approach for studying the structure and dynamics of proteins and protein complexes at the single molecule level. Future applications could focus on site-specific protein modification with drug cargos and cell targeting groups for cell-type selective transport of biological and small molecules drug molecules.
Acknowledgments
The authors are grateful to the financial support of the German Research Foundation (DFG) CRC1066/A6/B6 and B11 (KS, SLK, TW), the ERC Synergy Grant 319130-BioQ, the Horizon2020 project “Hyperdiamond”, the Horizon2020 Marie Curie ITN ProteinConjugates and LGFG scholarship from Ulm University. MD was supported by a fellowship of the Max Planck Graduate Center (MPGC) Mainz. We thank L. Xu and M. Zegota for proofreading the manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.7b00675.
General experimental section; sequential modification of IL-2, model peptide 4, Dis-tag-EYFP; spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Krall N.; da Cruz F. P.; Boutureira O.; Bernardes G. J. (2015) Site-selective protein-modification chemistry for basic biology and drug development. Nat. Chem. 8, 103–113. 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- Kuan S. L.; Wang T.; Weil T. (2016) Site-Selective Disulfide Modification of Proteins: Expanding Diversity beyond the Proteome. Chem. - Eur. J. 22, 17112–17129. 10.1002/chem.201602298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozier J. K.; Distefano M. D. (2015) Site-Specific PEGylation of Therapeutic Proteins. Int. J. Mol. Sci. 16, 25831–25864. 10.3390/ijms161025831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruani A.; Richards D. A.; Chudasama V. (2016) Dual modification of biomolecules. Org. Biomol. Chem. 14, 6165–6178. 10.1039/C6OB01010E. [DOI] [PubMed] [Google Scholar]
- Rosen C. B.; Francis M. B. (2017) Targeting the N terminus for site-selective protein modification. Nat. Chem. Biol. 13, 697. 10.1038/nchembio.2416. [DOI] [PubMed] [Google Scholar]
- Crochet A. P.; Kabir M. M.; Francis M. B.; Paavola C. D. (2010) Site-selective dual modification of periplasmic binding proteins for sensing applications. Biosens. Bioelectron. 26, 55–61. 10.1016/j.bios.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Ratner V.; Kahana E.; Eichler M.; Haas E. (2002) A general strategy for site-specific double labeling of globular proteins for kinetic FRET studies. Bioconjugate Chem. 13, 1163–1170. 10.1021/bc025537b. [DOI] [PubMed] [Google Scholar]
- Sonntag M. H.; Ibach J.; Nieto L.; Verveer P. J.; Brunsveld L. (2014) Site-specific protection and dual labeling of human epidermal growth factor (hEGF) for targeting, imaging, and cargo delivery. Chem. - Eur. J. 20, 6019–6026. 10.1002/chem.201304090. [DOI] [PubMed] [Google Scholar]
- Nathani R. I.; Moody P.; Chudasama V.; Smith M. E. B.; Fitzmaurice R. J.; Caddick S. (2013) A novel approach to the site-selective dual labelling of a protein via chemoselective cysteine modification. Chem. Sci. 4, 3455–3458. 10.1039/c3sc51333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos N.; Francis M. B. (2011) Choosing an effective protein bioconjugation strategy. Nat. Chem. Biol. 7, 876. 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- Grunewald J.; Jones D. H.; Brock A.; Chiu H. P.; Bursulaya B.; Ng K.; Vo T.; Patterson P.; Uno T.; Hunt J.; Spraggon G.; Geierstanger B. H. (2014) Site-specific dual labeling of proteins by using small orthogonal tags at neutral pH. ChemBioChem 15, 1787–1791. 10.1002/cbic.201402204. [DOI] [PubMed] [Google Scholar]
- Hogg P. J. (2003) Disulfide bonds as switches for protein function. Trends Biochem. Sci. 28, 210–214. 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- Morgan R. E.; Chudasama V.; Moody P.; Smith M. E. B.; Caddick S. (2015) A novel synthetic chemistry approach to linkage-specific ubiquitin conjugation. Org. Biomol. Chem. 13, 4165–4168. 10.1039/C5OB00130G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudasama V.; Smith M. E. B.; Schumacher F. F.; Papaioannou D.; Waksman G.; Baker J. R.; Caddick S. (2011) Bromopyridazinedione-mediated protein and peptide bioconjugation. Chem. Commun. 47, 8781. 10.1039/c1cc12807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. E. B.; Schumacher F. F.; Ryan C. P.; Tedaldi L. M.; Papaioannou D.; Waksman G.; Caddick S.; Baker J. R. (2010) Protein Modification, Bioconjugation, and Disulfide Bridging Using Bromomaleimides. J. Am. Chem. Soc. 132, 1960–1965. 10.1021/ja908610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaunak S.; Godwin A.; Choi J. W.; Balan S.; Pedone E.; Vijayarangam D.; Heidelberger S.; Teo I.; Zloh M.; Brocchini S. (2006) Site-specific PEGylation of native disulfide bonds in therapeutic proteins. Nat. Chem. Biol. 2, 312–313. 10.1038/nchembio786. [DOI] [PubMed] [Google Scholar]
- Boutureira O.; Bernardes G. J. (2015) Advances in chemical protein modification. Chem. Rev. 115, 2174–2195. 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- Brocchini S.; Godwin A.; Balan S.; Choi J.; won J.-W.; Zloh M.; Shaunak S. (2008) Disulfide bridge based PEGylation of proteins. Adv. Drug Delivery Rev. 60, 3–12. 10.1016/j.addr.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Marculescu C.; Kossen H.; Morgan R. E.; Mayer P.; Fletcher S. A.; Tolner B.; Chester K. A.; Jones L. H.; Baker J. R. (2014) Aryloxymaleimides for cysteine modification, disulfide bridging and the dual functionalization of disulfide bonds. Chem. Commun. 50, 7139–7142. 10.1039/C4CC02107J. [DOI] [PubMed] [Google Scholar]
- Lee M. T. W.; Maruani A.; Baker J. R.; Caddick S.; Chudasama V. (2016) Next-generation disulfide stapling: reduction and functional re-bridging all in one. Chem. Sci. 7, 799–802. 10.1039/C5SC02666K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruani A.; Smith M. E. B.; Miranda E.; Chester K. A.; Chudasama V.; Caddick S. (2015) A plug-and-play approach to antibody-based therapeutics via a chemoselective dual click strategy. Nat. Commun. 6, 6645. 10.1038/ncomms7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Riegger A.; Lamla M.; Wiese S.; Oeckl P.; Otto M.; Wu Y. Z.; Fischer S.; Barth H.; Kuan S. L.; Weil T. (2016) Water-soluble allyl sulfones for dual site-specific labelling of proteins and cyclic peptides. Chem. Sci. 7, 3234–3239. 10.1039/C6SC00005C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek T. R. (2008) The Biology of Interleukin-2. Annu. Rev. Immunol. 26, 453–479. 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- Boyman O.; Sprent J. (2012) The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 12, 180–190. 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- Kostiainen M. A.; Szilvay G. R.; Lehtinen J.; Smith D. K.; Linder M. B.; Urtti A.; Ikkala O. (2007) Precisely defined protein-polymer conjugates: construction of synthetic DNA binding domains on proteins by using multivalent dendrons. ACS Nano 1, 103–113. 10.1021/nn700053y. [DOI] [PubMed] [Google Scholar]
- Janatova J.; Fuller J. K.; Hunter M. J. (1968) The Heterogeneity of Bovine Albumin with Respect to Sulfhydryl and Dimer Content. J. Biol. Chem. 243, 3612–3622. [PubMed] [Google Scholar]
- Frick S. U.; Domogalla M. P.; Baier G.; Wurm F. R.; Mailaender V.; Landfester K.; Steinbrink K. (2016) Interleukin-2 Functionalized Nanocapsules for T Cell-Based Immunotherapy. ACS Nano 10, 9216. 10.1021/acsnano.5b07973. [DOI] [PubMed] [Google Scholar]
- Yamada T.; Fujishima A.; Kawahara K.; Kato K.; Nishimura O. (1987) Importance of disulfide linkage for constructing the biologically active Human Interleukin-2. Arch. Biochem. Biophys. 257, 194–199. 10.1016/0003-9861(87)90558-3. [DOI] [PubMed] [Google Scholar]
- Brocchini S.; Godwin A.; Balan S.; Choi J. W.; Zloh M.; Shaunak S. (2008) Disulfide bridge based PEGylation of proteins. Adv. Drug Delivery Rev. 60, 3–12. 10.1016/j.addr.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Zhang R. M.; Snyder G. H. (1989) Dependence of formation of small disulfide loops in two-cysteine peptides on the number and types of intervening amino acids. J. Biol. Chem. 264, 18472–18479. [PubMed] [Google Scholar]
- Rashidian M.; Dozier J. K.; Distefano M. D. (2013) Enzymatic labeling of proteins: techniques and approaches. Bioconjugate Chem. 24, 1277–1294. 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R.; Hohng S.; Ha T. (2008) A practical guide to single-molecule FRET. Nat. Methods 5, 507–516. 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrecka J.; Lewis R.; Bruckner F.; Lehmann E.; Cramer P.; Michaelis J. (2008) Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 105, 135–140. 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


