Abstract
Stuttering is a communication disorder that affects approximately 1 % of the population. Although 5–8 % of preschool children begin to stutter, the majority will recover with or without intervention. There is a significant gap, however, in our understanding of why many children recover from stuttering while others persist and stutter throughout their lives. Detecting neurophysiological biomarkers of stuttering persistence is a critical objective of this study. In this study, we developed a novel supervised sparse feature learning approach to discover discriminative biomarkers from functional near infrared spectroscopy (fNIRS) brain imaging data recorded during a speech production experiment from 46 children in three groups: children who stutter (n = 16), children who do not stutter (n=16), and children who recovered from stuttering (n =14). We made an extensive feature analysis of the cerebral hemodynamics from fNIRS signals and selected a small number of important discriminative features using the proposed sparse feature learning framework. The selected features are capable of differentiating neural activation patterns between children who do and do not stutter with an accuracy of 87.5 % based on a five-fold cross-validation procedure. The discovered set cerebral hemodynamics features are presented as a set of promising biomarkers to elucidate the underlying neurophysiology in children who have recovered or persisted in stuttering and to facilitate future data-driven diagnostics in these children.
Keywords: stuttering, functional near-infrared spectroscopy (fNIRS), speech production, children, data mining, feature extraction and selection, biomarkers, mutual information, sparse modeling
I. INTRODUCTION
Stuttering is a communication disorder characterized by involuntary disruptions in the forward flow of speech. These disruptions, referred to as stuttering-like disfluencies, are recognized as repetitions of speech sounds or syllables, blocks where no sound or breath emerge, or prolongation of speech sounds. In recent years, there has been considerable progress toward understanding the origins of a historically enigmatic disorder. Past theories of stuttering attempted to isolate specific factors such as anxiety, linguistic planning deficiencies, or muscle hyperactivity as the root cause of stuttering (for review, see [1]). More recently, however, stuttering is hypothesized to be a multifactorial disorder. Atypical development of the neural circuitry underlying speech production may adversely impact the different cognitive, motor, linguistic, and emotional processes required for fluent speech production [2], [3].
The average age of stuttering onset is 33 months [4]. Although, 5–8 %, of preschool children begin to stutter, the majority (70–80 %) will recover with or without intervention [5], [4]. Given the high probability of recovery, parents often elect to postpone therapy to see if their child's stuttering resolves. However, delaying therapy in children at greater risk for persistence allows maladaptive neural motor networks to form that are challenging to treat in the future [6], [4]. The lifelong implications of stuttering are significant, impacting psychosocial development, education, and employment achievement [7], [8], [9], [10].
There is a significant gap in our understanding of why so many children recover while others persist in stuttering. Established behavioral risk factors for stuttering persistence include one or more of the following: positive family history, later age of onset (i.e. stuttering began after 36 months), time since onset, sex–boys are more likely to persist, and type and frequency of disfluencies [4]. Combining behavioral risk factors with objective, physiological biomarkers of stuttering may constitute a more powerful approach to help identify children at greater risk for chronic stuttering. Detecting such physiological biomarkers of stuttering persistence is a critical objective of our research [11], [12].
In our earlier study, Walsh et al. (2017) [13] recorded cortical activity during overt speech production from children who stutter and their fluent peers. During the experiment, the children completed a picture description task while we recorded hemodynamic responses over neural regions involved in speech production and implicated in the pathophysiology of stuttering including: inferior frontal gyrus (IFG), premotor cortex (PMC), and superior temporal gyrus (STG) with functional near-infrared spectroscopy (fNIRS), which is a safe, non-invasive optical neuroimaging technology that relies upon neurovascular coupling to indirectly measure brain activity. This is accomplished using near-infrared light to measure the relative changes in both oxygenated (Oxy-Hb) and deoxygenated hemoglobin (Deoxy-Hb), two absorbing chromophores in cerebral capillary blood [14]. fNIRS offers significant advantages including its relatively low cost and greater tolerance for movement, making it a more childfriendly neuroimaging approach. fNIRS has been used to assess the regional activation, timing, and lateralization of cortical activation for a diverse number of perceptual, language, motor, and cognitive investigations (for review, [15]).
Using fNIRS to assess cortical activation during overt speech production, we found markedly different speechevoked hemodynamic responses between the two groups of children during fluent speech production [13]. Whereas controls showed clear activation over left dorsal IFG and left PMC, characterized by increases in Oxy-Hb and decreases in Deoxy-Hb, the children who stutter demonstrated deactivation, or the reverse response over these left hemisphere regions. The distinctions in hemodynamic patterns between the groups may indicate dysfunctional organization of speech planning and production processes associated with stuttering and could represent potential biomarkers of stuttering.
Although different brain signal patterns can be observed for stuttering and control group in our previous studies, there is still a lack of reliable quantitative tools to evaluate stuttering treatment and recovery process based on brain activity patterns. In our previous studies, we have extensive research efforts on specialized machine learning (ML) and pattern recognition techniques for multivariate spatiotemporal brain activity pattern identification under different brain states [16], [17], [18], [19]. In this study, we aimed to detect neurophysiological biomarkers of stuttering using advanced ML techniques. In particular, we performed ML models for two experiments. In experiment (1), we made an extensive feature extraction from fNIRS brain imaging data of 16 children who stutter and 16 children in a control group collected in our previous study [13]. Next, we developed a novel supervised sparse feature learning approach to discover a set of discriminative biomarkers from a large set of fNIRS features, and construct a classification model to differentiate hemodynamic patterns from children who do and do not stutter. In experiment (2), we applied the constructed classification model on a novel test set of fNIRS data collected from a group of children who had recovered from stuttering and underwent the same picture description experiment. Using the novel test set with children's data that was not used to develop the initial algorithms allowed us to assess the model generalization with the discovered biomarkers from experiments (1) to (2). We elected to include children who had recovered from stuttering in the test group for theoretical and clinical bearings. Young children who begin to stutter are far more likely to recover than persist. It is important to assess the underlying neurophysiology of different stuttering phenotypes to learn, for example, whether recovered children's hemodynamic patterns would classify them with the group of controls or with the group of stuttering children. These proof-of-concept experiments represent a critical step toward identifying greater risk for persistence in younger children near the onset of stuttering.
The remainder of the paper is organized as follows: In Section 2, we present the methodology, including participant and data collection details, fNIRS data feature extraction and structured sparse feature selection models. In Section 3, we present the results of the pattern discovery of biomarkers as well as performance consistency on the novel test-set of data from recovered children. In section 4, we discuss the selected features and their interpretations in terms of brain regions of interest. Finally, we conclude the study in section 5.
II. METHOD
A. Participants, fNIRS Data Collection & Pre-processing
In experiment (1), fNIRS data from the 32 children who participated in the Walsh et al. (2017) study [13] was analyzed; 16 children who stutter (13 males) and 16 age- and socioeconomic status-matched controls (11 males). The participants were between the ages of 7–11 years (M = 9 years). Stuttering diagnosis and exclusionary criteria are provided in [13].
In experiment (2), a group of 14 children (10 males) between the ages of 8–16 years (M = 12 years) who recovered from stuttering was analyzed as an additional test group. All of the children completed a picture description experiment in which they described aloud different picture scenes (“talk” trials) that randomly alternated with “null” trials in which they watched a fixation point on the monitor. In order to compare hemodynamic responses among the groups of children, only fluent speech trials were considered in the analyses.
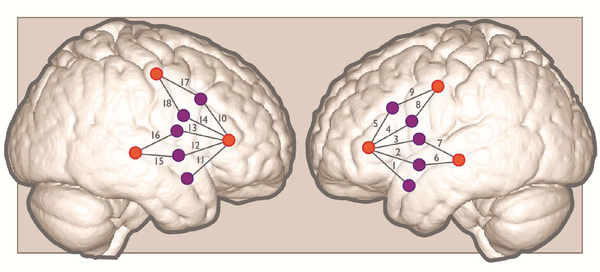
For each experiment, we recorded hemodynamic responses with a continuous wave system (CW6; TechEn, Inc.) that uses near-infrared lasers at 690 and 830 nm as light sources, and avalanche photodiodes (APDs) as detectors for measuring intensity changes in the diffused light at a 25-Hz sampling rate. Each source/detector pair is referred to as a channel. The fNIRS system acquired signals from 18 channels (9 over the left hemisphere and 9 over homologous right hemisphere regions) that were placed over ROIs relying on 10–20 system coordinates Figure (1).
Fig. 1:
Approximate positions of emitters (orange circles) and detectors (purple circles) are shown on a standard brain atlas (ICBM 152). The probes were placed symmetrically over the left and right hemisphere, with channels 1–5 spanning inferior frontal gyrus, channels 6–7 over superior temporal gyrus, and channels 8–9 over precentral gyrus/premotor cortex.
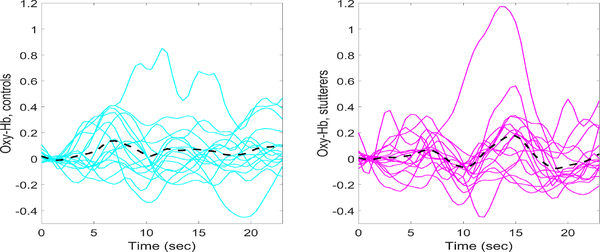
Data analysis is detailed in Walsh et al. [13]. Briefly, the fNIRS data was preprocessed using Homer2 software [20]. Usable channels of raw data were low-pass filtered at 0.5 Hz and high-pass filtered at 0.03 Hz. Concentration changes in Oxy-Hb and Deoxy-Hb were then calculated and a correlation-based signal improvement approach applied to the concentration data to reduce motion artifacts [21]. Finally, we derived each child's Oxy-Hb and Deoxy-Hb event-related hemodynamic responses from all channels from stimulus onset to the end of the trial. We then subtracted the average hemodynamic response associated with the null trials from the average hemodynamic response from the talk trials to derive a differential hemodynamic response for each channel [22]. The average Oxy-Hb and Deoxy-Hb hemodynamic response averaged over all 18 channels is plotted as a function of time for each child in Figure (2) and (3).
Fig. 2:
Oxy-Hb hemodynamic responses averaged over all 18 channels for each subject. Controls are plotted on the left (cyan curves) and stutterers on the right (magenta curves). The grand average hemodynamic response across all channels and subjects is represented by the black dashed curve.
Fig. 3:
Deoxy-Hb hemodynamic responses averaged over all 18 channels for each subject. Controls are plotted on the left (cyan curves) and children who stutter on the right (magenta curves). The grand average hemodynamic response across all channels and subjects is represented by the black dashed curve.
B. Feature Extraction
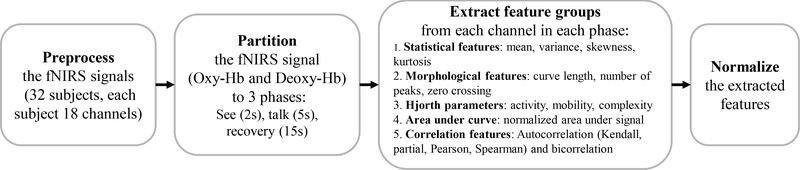
As shown in Figure (4), each experimental trial was partitioned into three phases: perception or the see-phase (0–2s, the children saw a picture on the monitor), the talk-phase (3–8s, the children described aloud the picture), and the recovery-phase (9–23s, the hemodynamic response returned to baseline) for measurements of Oxy-Hb and Deoxy-Hb. We extracted 21 features from each channel; 21 = 4 + 3 + 3 + 1 + (5 × 2(for 1 and 2 sec of delay)). These delays were implemented to account for correlation of the signal to its lagged values. The names of the feature group and subgroups are shown in Figure (4). Therefore, for each subject with 18 channels of fNIRS data, there were 378 extracted features from Oxy-Hb and Deoxy-hb measurements in each phase.
Fig. 4:
The process of feature engineering: pre-process input data, features extraction, post-process the features
The extracted groups of features are summarized in the following.
Statistical features capture descriptive information of the signals.
Morphological features comprised the number of peaks and zero crossings and measures of curve length.
Hjorth parameters capture signal variation over time expressed as activity, mobility, and complexity.. The three features are defined as: activity = Var(y(t)), ,
Normalized Area Under the Signal (NAUS) calculates the sum of values which have been subtracted from a defined baseline divided by the sum of the absolute values for the fNIRS signal.
Autocorrelation captured the linear relationship of the signal with its historical values considering 1 and 2 s delays Kendall, partial, Spearman and Pearson are four ways to compute autocorrelation.
Bicorrelation computes the bicorrelation on the time series Xv for given delays in τv. Bicorrelation is an extension of the autocorrelation to the third order moments, where the two delays are selected so that the second delay is twice the original , (i.e. x(t)x(t−τ)x(t−2τ)). Given a delay of τ and the standardized time series Xv with length n, denoted as Yv, the bicorr(τ) can be calculated as:
| (1) |
1) Personalized Feature Normalization: As illustrated in Figures (2) and (3) fNIRS signals vary dynamically across subjects, imposing a challenge to biomedical research. Because of inter-individual variability in signal features, it is difficult to build a robust diagnostic model to accurately discriminate between groups of participants. Outliers can further distort the trained model, thus impeding generalization. To tackle these issues, we applied a personalized feature normalization approach to standardize the extracted feature values of each subject onto the same scale to enhance feature interpretability across subjects.
To accomplish this, we calculated the upper and lower limits for each extracted feature using the formula Vl= max(minimum feature value, lower quartile + 1.5 × interquartile range) for the lower limit, and Vu= min(maximum feature value, upper quartile + 1.5 × interquartile range) for the upper limit. Feature values outside of this defined interval were considered to be outliers and mapped to 0 or 1. More details can be found in study [23]. Assuming the raw feature value was Fraw, the scaled feature value Fscaled was obtained by:
| (2) |
C. Integrated Structured Sparse Feature Selection using Mutual Information
Feature selection techniques are widely used to improve model performance and promote generalization in order to gain a deeper insight into the underlying processes or problem. This is accomplished by identifying the most important decision variables, while avoiding overfitting a model. Most feature selection techniques classify into three categories: embedded methods, wrapper methods, and filter methods [24]. Both embedded and wrapper methods seek to optimize the performance of a classifier or model. Thus, the feature selection performance is highly limited to the embedded classification models. Filter feature selection techniques assess the relevance of features by measuring their intrinsic properties. Widely used models include correlation-based feature selection [25], fast correlation-based feature selection [26], minimum redundancy features (biomarkers) maximum relevance (mRMR) [27] and information-theoreticbased feature selection methods [28].
Sparse modeling-based feature selection methods have gained attention owed to their well-grounded mathematical theories and optimization analysis. These feature selection algorithms employ sparsity-inducing regularization techniques, such as L1-norm constraint or sparse-inducing penalty terms, for variable selection. To construct more interpretable models, structured sparse modeling algorithms that consider feature structures have recently been proposed and show promising results in many practical applications including brain informatics, gene expression, medical imaging analysis, etc. [29], [30], [31], [32]. However, most of the current structured sparse modeling algorithms only consider linear relationships between response variables and predictor variables (features) in the analysis and may miss complex nonlinear relationships between features and response variables that may be present. On the other hand, although some filter or wrapper methods have the capability to capture nonlinear relationships between features and response variables, feature structures may not be optimally identified in the feature selection procedure. Constructing interpretable learning models with efficient feature selection remains an open and active research area in the machine learning community. Zhongxin et al. [33] proposed a feature selection algorithm based on mutual information (MI) and least absolute shrinkage and selection operator (LASSO) using L1 regularization with application to microarray data produced by gene expression. In our previous study, we also proposed a MI-based sparse feature selection model for EEG feature selection and applied it to epilepsy diagnosis [34]. However, feature structures were not considered during feature selection in both [33] and [34].
To consider both linear and nonlinear relationships between features and response variables, while acknowledging feature structures in feature selection, we propose a novel feature selection framework that integrates information theorybased feature filtering and structured sparse learning models to effectively capture feature dependencies and identify the most informative feature subset. There are two differences with respect to earlier studies [33] and [34]: (1) we did not use regularization techniques like LASSO as the second rank filtering; rather, we used sparse-inducing regularization to reveal the second-level feature-response relationships; (2) we applied structured feature learning by penalizing the feature groups. We implemented the proposed informationtheory-based structured sparse learning framework to identify the optimal feature subset as discriminant neurophysiological biomarkers of stuttering.
-
1)
Mutual Information for Feature Selection: MI is an index of mutual dependency between two random variables that quantifies the amount of information obtained about one random variable from the other random variable [35]. MI effectively captures nonlinear dependency among random variables and can be applied to rank features in feature selection problems [27]. The fundamental objective of MI-based filtering methods is to retain the most informative features (i.e., with higher MI) while removing the redundant or lessrelevant features (i.e., with low MI). The mutual information of two random variables X and Y, denoted by I(X,Y ), is determined by the probabilistic density functions p(x), p(y), and p(X,Y ):
| (3) |
-
1)Structured Sparse Feature Selection: A sparse model generates a sparse (or parsimonious) solution using the smallest number of variables with non-zero weights among all the variables in the model. One basic sparse model is LASSO regression, which employs L1 penalty-based regularization techniques for feature selection [36]. The LASSO objective function is formulated as follows:
where A is the feature matrix, Y is the response variable, λ1 is a regularization parameter and x is the weight vector to be estimated. The L1 regularization term produces sparse solutions such that only a few values in the vector x are nonzero. The corresponding variables with non-zero weights are the selected features to predict the response variable Y.(4)
Structured Features (Sparse Group LASSO (SGL)) The basic LASSO model, and many L1 regularized models, assume that features are independent and overlook the feature structures. However, in most practical applications, features contain intrinsic structural information, (e.g., disjoint groups, overlapping groups, tree-structured groups, and graph networks) [32]. The feature structures can be incorporated into models to help identify the most critical features and enhance model performance.
As outlined in section 2.2, the features we extracted from the raw fNIRS data are disparate; thus they can be categorized into disjoint groups. The sparse group LASSO regularization algorithm promotes sparsity at both the within- and betweengroup levels and is formulated as:
| (5) |
where the weight vector x is divided by g nonoverlapping groups: and is the weight i for group g. The parameter λ1 is the penalty for sparse feature selection, and the parameter λ2 is the penalty for sparse group selection (i.e. the weights of some feature groups will be all zeroes). In cases where feature groups overlap, the sparse overlapping group LASSO regularization can be used [37].
-
3)
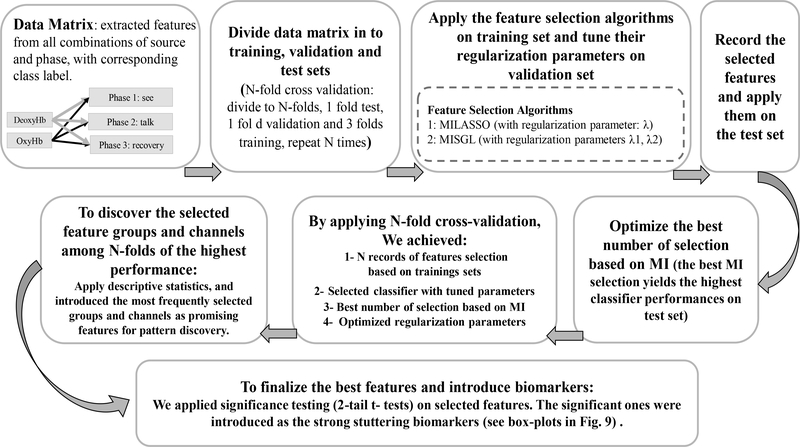
Integrated MI-Sparse Feature Selection Framework: The objective of our approach is to consider structured feature dependency while keeping the search process computationally efficient. To accomplish this, we employed the MI-guided feature selection framework outlined in Algorithm (1). Given a number of features k, the subset of top k features ranked by MI is denoted by S, and the subset of the remaining features is denoted by W. From S, the optimal feature subset is selected by exploring the k1 high-MI features which includes the iterative process of removal of highly-correlated features with 0.96 threshold. From W , the k2 sparse-model selected low-MI features. The final selected features subset is the set of (k1 + k2) features which are evaluated based on the crossvalidation classification performance. Enumeration of k1 starts from 1 and ascends until reaching the stopping criteria (i.e., when the cross-validation accuracy converges and cannot be further improved). MISS Algorithm (1) can be applied in two ways: (1) without group structure, which is a combination of mutual information and LASSO namely (MILASSO), (2) with group structure, which is a combination of mutual information and SGL namely (MISGL).
Algorithm 1.
Mutual Information Sparse Feature Selection (MISS)
| 1: | Rank all features based on mutual information |
| 2: | repeat |
| 3: | k1 = k1 + 1 |
| 4: | repeat |
| 5: | Divide sorted features to high-MI and low-MI |
| 6: | S ← high-MI |
| 7: | Remove redundant features from S |
| 8: | until k1 features remain after reduction |
| 9: | W ← low-MI |
| 10: | Apply sparsity learning to W |
| 11: | k2 ← number of selected features by SGL or LASSO |
| 12: | Build classifier model with k1 + k2 selected features |
| 13: | until classifier performance converges |
D. Machine Learning Algorithm Selection & Evaluation
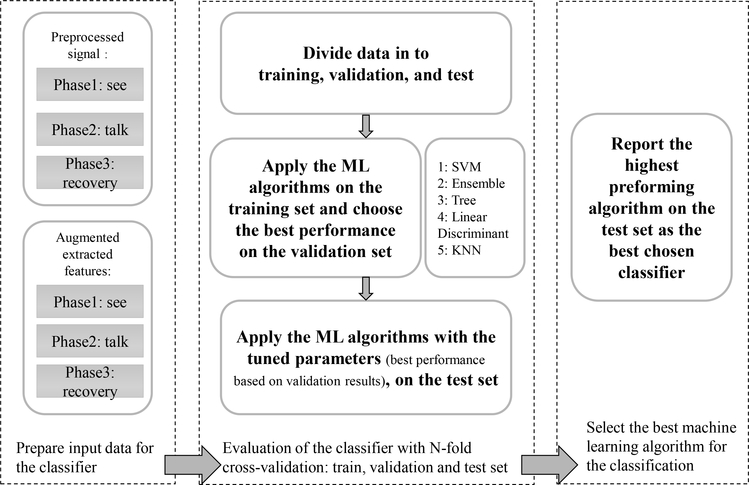
We applied established ML algorithms [38] (i.e., support vector machine (SVM), k-nearest neighbor (kNN), decision tree, ensemble, and linear discriminant) to assess whether cerebral hemodynamic features could accurately differentiate the group of children who stutter from controls. An overview of the steps involved in feature extraction and model evaluation is provided in Figure (6).
Fig. 6:
The process of choosing the most accurate ML classification algorithm with N-fold cross-validation and parameter tuning
-
1)
Support vector machines: SVM is considered to be a popular and promising approach among classification studies tuning [39]. It has been used in a variety of biomedical applications; for example, to detect patterns in gene sequences or to classify patients according to their genetic profiles, with EEG signals in brain-computer interface systems, and to discriminate hemodynamic responses during visuomotor tasks [40], [17], [41], [42]. In this study we applied Gaussian radial basis function (RBF) as the kernel which maps input data x to higher dimensional space.
-
2)
Bayesian Parameter Optimization: Parameters in each classifier significantly affect its performance. We applied Bayesian optimization, part of the Statistics and Machine Learning Toolbox in Matlab, to optimize hyper-parameters of classification algorithm [43]. By applying Bayesian optimization algorithm, we want to minimize a scalar objective function (f(x) = cross-validation classification loss) for the classifier parameters in a bounded domain.
-
3)
N-fold Cross-Validation: We applied N-fold cross-validation (N=5) for training and testing. First, we selected the features and optimized the parameters of the classification algorithm on the training set then applied the tuned model on the testing set, see Figure (6). Accuracy is defined as the ratio of correctly classified test subjects to the total number of subjects. Sensitivity is the ratio of children in the stuttering group correctly identified as stuttering to all of the children in the stuttering group. Specificity is the ratio of children correctly identified as controls to the total number of children in the control group. In this study, we used the average sensitivity and specificity values to measure binary classification accuracy for each ML model.
III. RESULTS
Classifier performance is reported for experiment (1) based on the outcome of the N-fold cross-validation procedure on the test-set, see Table (I). For experiment (2) classification performance was established on a novel test-set of 14 children who had recovered from stuttering, see Table (IV).
Table I:
Comparison among performance of various ML classifiers (before & after) feature extraction and application of feature selection
| Input data: fNIRS signal, from phase : Talk | ||||||
|---|---|---|---|---|---|---|
| Source: Oxy-Hb | Source: Deoxy-Hb | |||||
| Classifier | avg | sen | spe | avg | sen | spe |
| SVM | 0.75 | 0.75 | 0.75 | 0.725 | 0.6 | 0.85 |
| Ensemble | 0.7 | 0.65 | 0.75 | 0.65 | 0.7 | 0.6 |
| KNN | 0.7 | 0.7 | 0.7 | 0.675 | 0.5 | 0.85 |
| L Discr | 0.75 | 0.75 | 0.75 | 0.725 | 0.6 | 0.85 |
| Tree | 0.775 | 0.8 | 0.75 | 0.575 | 0.8 | 0.35 |
| Input data: extracted features from signal in phase (Talk) with application of MISS for feature selection | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source: Oxy-Hb | Source: Deoxy-Hb | |||||||||
| Classifier | MI num | Tot num | avg | sen | spe | MI num | Tot num | avg | sen | spe |
| SVM | 2 | 11 | 0.875 | 0.85 | 0.9 | 10 | 14 | 0.825 | 0.8 | 0.85 |
| Ensemble | 2 | 11 | 0.825 | 0.85 | 0.8 | 9 | 32 | 0.85 | 0.8 | 0.9 |
| KNN | 2 | 11 | 0.825 | 0.75 | 0.9 | 3 | 7 | 0.85 | 0.85 | 0.85 |
| L Discr | 1 | 10 | 0.825 | 0.8 | 0.85 | 10 | 33 | 0.775 | 0.7 | 0.85 |
| Tree | 3 | 13 | 0.675 | 0.8 | 0.55 | 6 | 29 | 0.75 | 0.8 | 0.7 |
sen: sensitivity, spe: specificity , avg: average of sen and spe, L Discr: linear discriminant
MI num: number of selected features based on MI
Tot num: total number of selection (based on MI and based on SGL or LASSO)
Table IV:
The best SVM performance on the additional testset (recovered samples)
| phase | source | Fsel M | Tot num | SRA |
|---|---|---|---|---|
| talk | Deoxy-Hb | MILASSO | 14 | 71.43 |
| talk | Oxy-Hb | MISGL | 11 | 71.43 |
Fsel M: feature selection method, SRA: stuttering recovery assessment Tot num: total number of selected features with MISS
A. Experiment (1): Choosing the best ML Algorithm
The most accurate ML algorithm on the raw fNIRS data was the tree classifier with 77.5 % accuracy. The highest accuracy obtained after feature extraction and application of feature selection (MILASSO) was SVM (with RBF kernel) that achieved 87.5 % accuracy, Table (I). The phase of the fNIRS trial that distinguished the groups of children was the talk interval and the source was Oxy-Hb. However in some cases performance using features derived from DeoxyHb measurements reached comparable accuracy as those from Oxy-Hb.
B. Experiment (1): Comparing Feature Selection Algorithms
In Table (II), we compared the performance of the proposed feature selection algorithm (MISS) with the popular existing MI-based method like mRMR and linear regularized methods like LASSO and SGL. MISS approach outperformed mRMR in feature selection by yielding higher SVM classification performance with the same number of selected features, (14 and 11 for measurement source of deoxy-Hb and oxy-Hb), approximately 7.5 and 27.5 % respectively. MISS approach outperformed LASSO and SGL in feature selection yielding higher classification accuracy approximately 2.5 to 12.5 %.
Table II:
Comparison among performance of various feature selection algorithms via SVM classification accuracy on the selected features with each approach
| Feature selection Method | Deoxy-Hb |
Oxy-Hb |
||
|---|---|---|---|---|
| Tot num feat | Avg(sen, spe) | Tot num feat | Avg(sen, spe) | |
| mRMR | 14 | 75 | 11 | 60 |
| LASSO | ~ 6.4 * | 80 | ~ 4.8 * | 75 |
| SGL | ~ 6.4 * | 77.5 | ~ 7 * | 78 |
| MISS(MILASSO, MISGL) | 14 | 82.5 | 11 | 87.5 |
Tot num feat: total number of selected features
indicates the average number of selected features among N-fold for LASSO and SGL methods Avg(sen, spe)= average of sensitivity and specificity (%)
C. Experiment (1): Selected Features
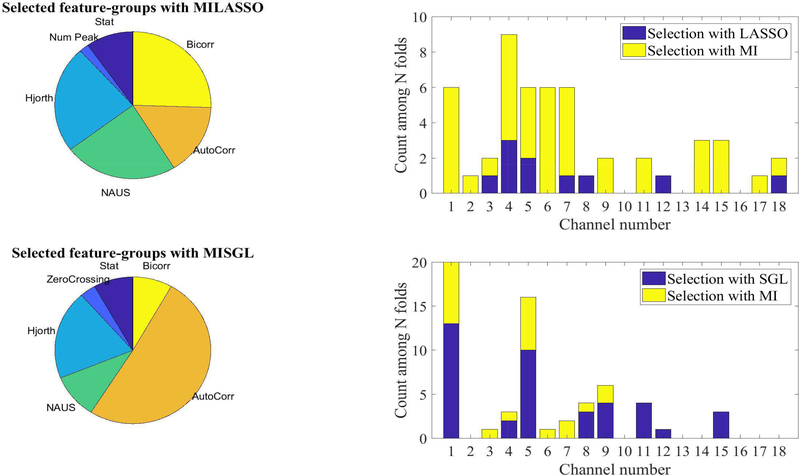
From an extended set of features, a subset that provided the highest classification accuracy was identified by MISGL and MILASSO in the SVM(RBF) model. This subset of features, shown in Figure (7), comprises statistical, NAUS, Hjorth parameters, autocorrelation and bicorrelation features. Channels that provided the highest discriminative power to differentiate between children who stutter and controls were localized to the left hemisphere; specifically, channels 1, 4, and 5 over left IFG.
Fig. 7:
Statistical summary of the selected feature groups and channels with MILASSO and MISGL in N-fold cross validation. In each fold, there was 11 to 14 selected features, from different channels and feature-groups. The pie charts illustrate the group that selected features most frequently came from. The histograms summarize the channel selection with MISGL and MILASSO. For example, from approximately 60 total features selected from 5 folds, 6 features were selected from channel 1, and 9 features from channel 4 (either based on MI ranking (yellow bar) or LASSO coefficients (blue bar) which are stacked for each channel).
The top 14 features from the entire feature set are listed inTable (III). These features, (2 based on MI and 12 based on LASSO), were extracted from the talk-phase with source OxyHb. We performed 2-tailed t-tests on these features. p-values ≤ 0.05, confirm a significant statistical difference between children who stutter and controls for a given feature.
Table III:
Top 14 features selected with MISS along with p-value (0.05 threshold for statistically significant t-test). With top 11 features, 87.5 % accuracy was achieved in N-fold cross-validation
| Feature rank | Feature name | p-value | Feature rank | Feature name | p-value |
|---|---|---|---|---|---|
| 1 | NAUS, ch 4 | 0.0001 | 8 | Hjorth mobility, ch 1 | 0.0014 |
| 2 | Hjorth mobility, ch 4 | 0.0022 | 9 | NAUS, ch 8 | 0.1095 |
| 3 | Hjorth activity, ch 1 | 0.2800 | 10 | AC partial 2s, ch 14 | 0.1745 |
| 4 | Bicorrelation 2s, ch 6 | 0.0225 | 11 | AC Spearman 1s, ch 6 | 0.9238 |
| 5 | NAUS, ch 5 | 0.0003 | 12 | Hjorth activity, ch 4 | 0.1792 |
| 6 | Variance, ch 9 | 0.5319 | 13 | Variance, ch 4 | 0.0605 |
| 7 | Bicorrelation 1s, ch 14 | 0.6252 | 14 | AC Spearman 1s, ch 7 | 0.6277 |
AC: autocorrelation, ZC: zero crossing, CL: curve length
NAUS: normalized area under signal, ch: channel 1 or 2s: 1 or 2 second of delay
-
1)
Feature Selection Optimization: The number of features selected by MILASSO or MISGL affects the performance of the classifier; a more sparse selection enhances model performance, promotes generalization, and facilitates the interpretation of results. During the enumeration process for MI selection, we learned that with less than 10 MI features (total features ≤ 15 − 22), the average classifier performance was approximately 80 %; with 15 to 30 MI features (25 − 35 ≤ total features ≤ 40) , performance was approximately 75 %; with more than 30 MI features, (total features ≥ 42 ), the accuracy decreased to 70 %. The highest accuracy with the least number of features came from 11 total features with the MILASSO approach, 2 MI and 9 LASSO and 12 total features with the MISGL approach, 8 MI and 4 SGL.
-
2)
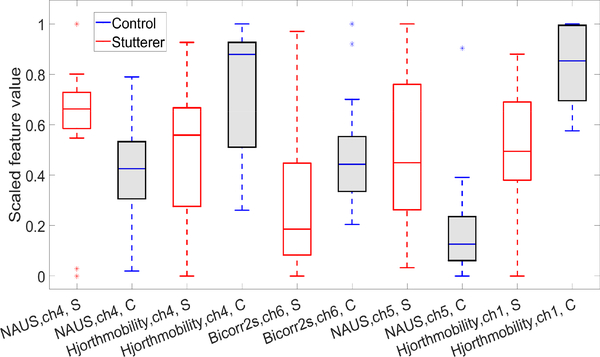
Biomarkers: The features in Table (III) that showed significant differences between children who stutter and controls are recognized as biomarkers. Box-plots of these features for the children who stutter and controls are plotted on a common scale in Figure (8). The discriminative features we detected in Figure (8) comprised significantly lower values of NAUS and slightly higher values of Hjorth mobility and bicorrelation with 2 sec of delay for children who stutter compared to controls.
Fig. 8:
Box-plot of top 5 significant features from talk-phase and source Oxy-Hb, ch: channel, (S: stutterer , C: control).
D. Experiment (2): Stuttering Recovery Assessment with Selected Features
In this section we report the performance of the classifier on the additional test-set (data from 14 children who recovered from stuttering), shown in Table (IV). We applied the best-performing algorithm based on the results from experiment (1): SVM with tuned parameters sigma = 1 and penalty = 0.001 on the entire dataset. We documented that 71.43 %, or approximately 10 out of 14 children who had recovered from stuttering, classified into the control group based on features derived from fNIRS signals derived from the talk-phase of the experiment. The same degree of stuttering recovery assessment (SRA) was achieved with both Oxy-Hb and Deoxy-Hb sources Table (IV).
IV. DISCUSSION
In experiment (1), we applied structured sparse feature learning models to previously collected speech-evoked fNIRS data from Walsh et al. [13] to explore whether neurophysiological biomarkers could accurately classify hemodynamic patterns from children who do and do not stutter. Following feature extraction and feature selection with MISS, the SVM achieved the highest classification accuracy of 87.5 %. With this model, classification performance was improved by 10 % using feature extraction and sparse MI-based features selection. This degree of accuracy was reached using features extracted during the talk interval of the trial from the source, Oxy-Hb (although features extracted from Deoxy-Hb reached comparable accuracy). A feature set comprising statistics, NAUS, Hjorth parameters, autocorrelation and bicorrelation features provided the highest discriminative power. Notably, nearly all of these features were extracted from channels localized to the left hemisphere (i.e. channels 1–9). The selected features may not be significant individually as shown in Table (III), thus they can be ignored or missed in basic statistical analyses used by many feature selection algorithms. The MISS approach is valuable to reveal clear discriminative patterns among features in a higher dimensional space, and to discover relevant multivariate biomarkers.
Features from channels 1, 4 and 5, which span left IFG, were identified as neurophysiological biomarkers that distinguished hemodynamic characteristics of children who stutter from controls. These included significantly reduced NAUS in left IFG channels 4 and 5 and increased Hjorth mobility parameters, denoting increased variability, in left IFG channels 1 and 4 in children who stutter.
In our earlier study [13], we found significantly reduced Oxy-Hb and increased Deoxy-Hb concentrations during the talk interval in channels over left IFG in the group of children who stutter. The left IFG comprising Broca's area is integral to speech production and may develop atypically in children who stutter. Neuroanatomical studies reveal aberrant developmental trajectories of white and gray matter of left IFG in children who stutter compared to controls [44], [45]. Moreover, there is evidence of reduced activation of IFG/Broca's area during speech production from fMRI studies with adults who stutter [46], [47]. In our earlier study [13], we hypothesized that this finding may represent a shift in blood flow to regions outside of our recording area to compensate for functional deficits in left IFG. An alternative possibility is a disruption in corticalsubcortical loops resulting in a net inhibition of this region. This is the first study to elucidate group-level differences by classifying individual children as either stuttering or not stuttering using features derived from their speech-evoked brain hemodynamics. Based on the sensitivity index from the final model, three children who stutter classified as controls (i.e., false negatives). Interestingly, two of these three children were considered to be mild stutterers when they participated and have since recovered from stuttering (determined via a followup visit or through parental report). It is tempting to speculate that the recovery process had already begun for these children when we recorded their hemodynamic responses during the initial study. However, longitudinal studies in younger children (i.e., near the onset of stuttering) are necessary to track the developmental trajectories of their hemodynamic responses as they either recover from or persist in stuttering to empirically assess this assumption.
Finally, we compared the consistency of the best-performing SVM classifier using N-fold cross-validation from experiment (1) with results achieved using the SVM classifier on a novel test-set of data from 14 children who had recovered from episodes of early childhood stuttering in experiment (2). We found that the majority of the recovered children, or 71.43 %, classified as controls, rather than children who stutter. This suggests that left-hemisphere stuttering biomarkers that dissociated stuttering children's speech-evoked hemodynamic patterns from controls, may indicate chronic stuttering, while recovery from stuttering in many of these children was associated with hemodynamic responses similar to those from children who never stuttered. Stuttering recovery may thus be supported, in part, by functional reorganization of regions such as left IFG that corrects anomalous brain activity patterns. Although this speculation warrants further study and replication, an fMRI study with adults who recovered from stuttering identified the left IFG as a pivotal region associated with optimal stuttering recovery [48].
A final point to consider is that although most of the recovered children had hemodynamic patterns similar to controls, four of these children classified into the stuttering group. Given that stuttering is highly heterogeneous, with multiple factors implicated in the onset and chronicity of the disorder [2], it is not surprising to find evidence suggesting that recovery processes may be different for some children. More research is clearly needed to substantiate the neural reorganization that accompanies both spontaneous and therapy-assisted recovery from stuttering.
V. CONCLUSION
In this final section, we present several suggestions regarding data preprocessing, feature selection and ML training and evaluation to guide future investigations in this line of research.
First, the personalized feature scaling approach facilitated the discovery of discriminative patterns by removing data outliers and reducing the variability in each feature. This was a critical step in our approach to address inherent interindividual differences in the physiological signals.
Second, the MISS approach yielded a final feature space that was both parsimonious and interpretable. In particular, MISGL, that considers feature group structures in sparse feature learning, and achieved the best classification performance with the least number of selected features. We compared our result from the MISS approach with commonly used feature selection techniques in Table (II), and the results proved that MISS outperformed the methods which solely applied either MI or regularized linear regression significantly. More importantly, MISS pinpointed specific left hemisphere channels that classified children as stuttering/nonstuttering with higher accuracy and corroborated findings from our earlier experiment [13].
In summary, the proposed MI-based structured sparse feature learning method demonstrates its effectiveness to discover the most discriminative features in a high dimensional feature space with a limited number of training samples, a common challenge for health care and medical data mining approaches. Compared to other methods, the proposed MISS approach offers a promising, interpretable solution to facilitate data-driven advances in clinical and experimental research applications.
Fig. 5:
Feature selection and tuning the regularization parameters via N-fold cross-validation in order to introduce the promising features (biomarkers).
ACKNOWLEDGMENT
Dr. Walsh was supported by NIH grant (NIH/NIDCD R03 DC013402) [13]
REFERENCES
- [1].Bloodstein O and Ratner NB A handbook on stuttering. Cengage Learning, Clifton Park, NY, 6 edition edition, October 2008. [Google Scholar]
- [2].Smith A and Weber C How Stuttering Develops: The Multifactorial Dynamic Pathways Theory. Journal of Speech, Language, and Hearing Research, pages 1–23, August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Smith A Stuttering: a unified approach to a multifactorial, dynamic disorder In Stuttering research and practice: bridging the gap. Psychology Press, 1999. [Google Scholar]
- [4].Yairi E and Ambrose NG Early childhood stuttering for clinicians by clinicians. Pro ed, Austin, Tex, 1 edition edition, November 2005.
- [5].Mnsson H Childhood stuttering. Journal of Fluency Disorders, 25(1):47–57, March 2000. [Google Scholar]
- [6].Guitar B Stuttering: An integrated approach to its nature and treatment. LWW, Baltimore, third edition edition, October 2006. [Google Scholar]
- [7].Blumgart E, Tran Y, and Craig A Social anxiety disorder in adults who stutter. Depression and Anxiety, 27(7):687–692, July 2010. [DOI] [PubMed] [Google Scholar]
- [8].Klein JF and Hood SB The impact of stuttering on employment opportunities and job performance. Journal of Fluency Disorders, 29(4):255–273, January 2004. [DOI] [PubMed] [Google Scholar]
- [9].OBrian S, Jones M, Packman A, Menzies R, and Onslow M Stuttering severity and educational attainment. Journal of Fluency Disorders, 36(2):86–92, June 2011. [DOI] [PubMed] [Google Scholar]
- [10].Iverach L and Rapee RM Social anxiety disorder and stuttering: Current status and future directions. Journal of Fluency Disorders, 40:69–82, 2014. Anxiety and stuttering. [DOI] [PubMed] [Google Scholar]
- [11].Usler E, Smith A, and Weber C A lag in speech motor coordination during sentence production is associated with stuttering persistence in young children. Journal of Speech, Language, and Hearing, 60(1):51–61, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mohan R and WeberFox C Neural systems mediating the processing of sound units of language distinguish recovery versus persistence in stuttering. Journal of Neurodevelopmental Disorders, 7(1), 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Walsh B, Tian F, Tourville JA, Ycel MA, Kuczek T, and Bostian AJ Hemodynamics of speech production: An fNIRS investigation of children who stutter. Scientific Reports, 7, June 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Villringer A and Chance B Non-invasive optical spectroscopy and imaging of human brain function. Trends in Neurosciences, 20(10):435–442, October 1997. [DOI] [PubMed] [Google Scholar]
- [15].Homae F A brain of two halves: insights into interhemispheric organization provided by near-infrared spectroscopy. NeuroImage, 85:354–362, January 2014. [DOI] [PubMed] [Google Scholar]
- [16].Chaovalitwongse WA, Pottenger RS, Wang S, Fan Y, and Iasemidis LD Pattern-and network-based classification techniques for multichannel medical data signals to improve brain diagnosis. IEEE Transactions on Systems, Man, and Cybernetics-Part A: Systems and Humans, 41(5):977–988, 2011. [Google Scholar]
- [17].Wang S, Zhang Y, Wu C, Darvas F, and Chaovalitwongse WA Online prediction of driver distraction based on brain activity patterns. IEEE Transactions on Intelligent Transportation Systems, 16(1):136–150, 2015. [Google Scholar]
- [18].Kam K, Schaeffer J, Wang S, and Park H A comprehensive feature and data mining study on musician memory processing using eeg signals In International Conference on Brain and Health Informatics, pages 138–148. Springer, 2016. [Google Scholar]
- [19].Puk K, Gandy KC, Wang S, and Park H Pattern classification and analysis of memory processing in depression using eeg signals In International Conference on Brain and Health Informatics, pages 124–137. Springer, 2016. [Google Scholar]
- [20].Huppert TJ, Hoge RD, Diamond SG, Franceschini MA, and Boas DA A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. NeuroImage, 29(2):368–382, January 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cui X, Bray S, and Reiss AL Functional near infrared spectroscopy (NIRS) signal improvement based on negative correlation between oxygenated and deoxygenated hemoglobin dynamics. NeuroImage, 49(4):3039–3046, February 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Plichta MM, Heinzel S, Ehlis A-C, Pauli P, and Fallgatter AJ Model-based analysis of rapid event-related functional near-infrared spectroscopy (NIRS) data: a parametric validation study. NeuroImage, 35(2):625–634, April 2007. [DOI] [PubMed] [Google Scholar]
- [23].Wang S, Gwizdka J, and Chovalitwongse WA Using wireless eeg signals to assess memory workload in the n-back task. IEEE Transactions on Human-Machine Systems, 46(3):424–435, June 2016. [Google Scholar]
- [24].Saeys Y, Inza I, and Larranaga P A review of feature selection techniques in bioinformatics. Bioinformatics, 23(19):2507–2517, 2007. [DOI] [PubMed] [Google Scholar]
- [25].Hall M Correlation-based feature selection for machine learning. PhD Thesis New Zealand: Department of Computer Science, Waikato University, 1999. [Google Scholar]
- [26].Yu L and Liu H Efficient feature selection via analysis of relevance and redundancy. Journal of Maching Learning Research, pages 1205–1224, 2004. [Google Scholar]
- [27].Peng H, Ding C, and Fulmi L Feature selection based on mutual information; criteria of max- dependency, max-relevance, and minredundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence, 27(8):1226–1238, 2005. [DOI] [PubMed] [Google Scholar]
- [28].Vergara JR and Estevez PA A review of feature selection methods´ based on mutual information. Neural Computing and Applications, 24(1):175–186, 2014. [Google Scholar]
- [29].Liu F, Wang S, Rosenberger J, Su J, and Liu H A sparse dictionary learning framework to discover discriminative source activations in eeg brain mapping. In AAAI, pages 1431–1437, 2017. [Google Scholar]
- [30].Xiao C, Wang S, Zheng L, Zhang X, and Chaovalitwongse WA A patient-specific model for predicting tibia soft tissue insertions from bony outlines using a spatial structure supervised learning framework. IEEE Transactions on Human-Machine Systems, 46(5):638–646, 2016. [Google Scholar]
- [31].Xiao C, Bledsoe J, Wang S, Chaovalitwongse WA, Mehta S, Semrud-Clikeman M, and Grabowski T An integrated feature ranking and selection framework for adhd characterization. Brain informatics, 3(3):145–155, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gui J, Sun Z, Ji S, Tao D, and Tan T Feature selection based on structured sparsity: A comprehensive study. IEEE transactions on neural networks and learning systems, 2017. [DOI] [PubMed] [Google Scholar]
- [33].Zhongxin W, Gang S, Jing Z, and Jia ZJ Feature selection algorithm based on mutual information and lasso for microarray data. volume 10, pages 278–286, 2016. [Google Scholar]
- [34].Wang S, Xiao C, Tsai J, Chaovalitwongse W, and Grabowski TJ A novel mutual-information-guided sparse feature selection approach for epilepsy diagnosis using interictal eeg signals In International Conference on Brain and Health Informatics, pages 274–284. Springer, 2016. [Google Scholar]
- [35].Cover TM and Thomas JA Elements of Information Theory 2nd Edition (Wiley Series in Telecommunications and Signal Processing). Wiley-Interscience, 2001. [Google Scholar]
- [36].Tibshirani R Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, Series B, 58:267–288, 1994. [Google Scholar]
- [37].Liu J and Ye J Moreau-yosida regularization for grouped tree structure learning. In Advances in Neural Information Processing Systems, pages 1459–1467, 2010. [Google Scholar]
- [38].Wu X, Kumar V, Ross Q J, Ghosh J, Yang Q, Motoda H, McLachlan GJ, Ng A, Liu B, Yu PS, Zhou Z, Steinbach M, Hand DJ, and Steinberg D Top 10 algorithms in data mining. Knowl. Inf. Syst, 14(1):1–37, December 2007. [Google Scholar]
- [39].Scholkopf B and Smola AJ¨ Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond (Adaptive Computation and Machine Learning). The MIT Press, December 2001. [Google Scholar]
- [40].Wang S, Lin CJ, Wu C, and Chaovalitwongse WA Early detection of numerical typing errors using data mining techniques. IEEE Transactions on Systems, Man, and Cybernetics-Part A: Systems and Humans, 41(6):1199–1212, 2011. [Google Scholar]
- [41].Brown M, Grundy W, Lin D, Cristianini N, Sugne C, Furey T, Ares M, , and D. Haussler. Knowledge-base analysis of microarray gene expressiondata by using support vector machines. PNAS, 97(1):262267, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Noble WS Support vector machine applications in computational biology, chapter 3 Computational molecular biology. MIT Press, 2004. [Google Scholar]
- [43].Gelbart MA, Snoek J, and Adams RP Bayesian Optimization with Unknown Constraints. ArXiv e-prints, March 2014.
- [44].Beal DS, Lerch JP, Cameron B, Henderson R, Gracco VL, and De Nil LF The trajectory of gray matter development in brocas area is abnormal in people who stutter. Frontiers in Human Neuroscience, 9:89, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chang SE, Zhu DC, Choo AL, and Angstadt M White matter neuroanatomical differences in young children who stutter. Brain, 138(3):694–711, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chang S, Kenney M, Loucks TMJ, and Ludlow CL Brain activation abnormalities during speech and non-speech in stuttering speakers. NeuroImage, 46(1):201–212, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Neef NE, Btfering C, Anwander A, Friederici AD, Paulus W, and Sommer M Left posterior-dorsal area 44 couples with parietal areas to promote speech fluency, while right area 44 activity promotes the stopping of motor responses. NeuroImage, 142(Supplement C):628–644, 2016. [DOI] [PubMed] [Google Scholar]
- [48].Kell CA, Neumann K, von Kriegstein K, Posenenske C, von Gudenberg AW, Euler H, and Giraud AL How the brain repairs stuttering. Brain, 132(10):2747–2760, 2009. [DOI] [PubMed] [Google Scholar]