Abstract
Mononuclear phagocytes (MP) consist of macrophages, dendritic cells (DCs), and monocytes. In all organs, including the lung, there are multiple subtypes within these categories. The existence of all these cell types suggest that there is a clear division of labor and delicate balance between the MPs under steady state and inflammatory conditions. Although great strides have been made to understand MPs in the mouse lung, and human blood, little is known about the MPs that exist in the human lung and lung- draining lymph nodes (LNs), and even less is known about their functional roles, studies of which will require a large number of sorted cells. We have comprehensively examined cell surface markers previously used in a variety of organs to identify human pulmonary MPs. In the lung, we consistently identify five extravascular pulmonary MPs and three LN MPs. These MPs were present in over 100 lungs regardless of age or gender. Notably, the human blood CD141+ DCs, as described in the literature, were not observed in non-diseased lungs or their draining LNs. In the lung and draining LNs, expression of CD141 was only observed on HLADR+ CD11c+ CD14+ extravascular monocytes (often confused in the LN as resident DCs based on the level of HLADR expression and mouse LN data). In the human lung and LNs there are at least two DC subtypes expressing HLADR, DEC205 and CD1c, along with circulating monocytes that behave as either antigen-presenting cells or macrophages. Furthermore, we demonstrate how to distinguish between alveolar macrophages and interstitial macrophage subtypes. It still remains unclear how the human pulmonary MPs identified here align with mouse MPs. Clearly, we are now past the stage of cell surface marker characterization, and future studies will need to move toward understanding what these cell types are and how they function. Our hope is that the strategy described here can help the pulmonary community take this next step.
Keywords: Human, Mononuclear phagocytes, Dendritic cells, Monocytes, Interstitial macrophages, Alveolar macrophages, Pulmonary, Lung
1. Introduction
In the lung, alveolar macrophages are the first line of defense against invading pathogens [1, 2]. If alveolar macrophages and incoming neutrophils are incapable of containing the invading pathogens, then an adaptive immune response is initiated to assist in its clearance [3, 4]. Underneath the epithelial cells are dendritic cells (DCs), interstitial macrophages (IMs), and monocytes, in addition to other leukocytes and non-hematopoietic cells [5, 6]. DCs link innate and adaptive immunity by acquiring, processing, and trafficking foreign and self-antigens to the draining lymph node (LN), where they present peptides on MHC molecules and activate cognate T cells [7–10]. In addition to macrophages and DCs, monocytes can also contribute to the clearance of microbes by acquiring a macrophage-like phenotype and induce adaptive immunity by acquiring a DC-like phenotype. Lastly, we were unable to identify CD303+ or CD123+ cells in the lung or draining LNs; hence, why plasmacytoid DCs are not discussed here.
For decades monocytes have been viewed as precursors to macrophages. However, we now know that monocytes continuously traffic through nonlymphoid and lymphoid tissue without becoming a bona fide macrophage or dendritic cell. In mice and humans, extravascular lung and LN monocytes are as abundant as DCs in the steady state and are even more abundant during inflammation [11, 12]. The role of monocytes in adaptive immunity is underappreciated, and although it has been shown that they can induce T cell proliferation, how they selectively activate lymphocytes is unclear [12]. In addition to monocytes, DCs, and tissue- specific alveolar macrophages, there are interstitial macrophages (IM). In mice we have identified three unique IM subtypes in the lung and other organs, albeit what they functionally do is still unclear [13]. In this chapter, we outline how to identify and isolate human pulmonary MPs from whole lungs en bloc with the hopes that in the near future we can functionally align these cell types with the well-characterized murine MPs.
All in all, we observe consistency and reproducibility when we strictly adhere to the six cautionary steps (see Notes 1–6) for the isolation and use of human pulmonary MPs.
2. Materials
2.1. Human Lung and Lymph Nodes
Non-diseased human lungs and lung-draining LNs were acquired from three sources as stated in the acknowledgments (also see ref. [14]).
2.2. Bronchoalveolar Lavage (BAL)and Media
PBS: 1× Phosphate-buffered saline (PBS) without calcium or magnesium for perfusion and lavage. Make 1 L per lobe.
PBS/ETDA buffer: 1× PBS and 3 mM ethylenediaminetetraacetic acid (EDTA, from 0.5 M stock solution pH 8.0). Make 500 mL per lobe.
BSS-B buffer: 132 mM NaCL, 5 mM KCl, 0.5 mM NaH2PO4, 2 mM Na2HPO4, 10 mM HEPES, 1 g/L Dextrose, 1.9 mM CaCl, 1.3 mM MgSO4, pH 7.4. Make 500 mL per lobe.
Scissors.
Large serrated forceps.
2–3 pairs of hemostats.
60 mL syringe.
200 mL plastic beakers.
500 mL plastic beakers.
100 μm filter membrane.
1/3 and 1 cm diameter PVC tubing: for perfusion in the pulmonary veins and inflation into the bronchus.
Various sized pipet tips.
FACS buffer: 1× PBS with 1 mM EDTA, 0.15% bovine serum albumin (BSA); keep at 4 °C.
2.3. Tissue Digestion for Lung MPs
Elastase buffer: 4.2 U elastase/mL in BSS-B. Approximately 150–250 mL is required for one lobe from an average adult (see Note 7).
1 gallon plastic bags: strong and durable (e.g., small biohazard bag).
String (to tie off plastic bag).
Water bath at 37 °C.
Rolls or sheets of nylon filter membrane: 350 and 100 μm.
Cheesecloth.
Vegetable strainer.
Two 1 L beakers.
Heat inactivated fetal calf serum (FCS).
Large tissue culture dishes to mince tissue and 1 L plastic beakers.
Blender (Ninja Professional Blender).
Chilled centrifuge for 250 mL centrifuge tubes.
50 mL conical tubes.
Kreb/HEPES buffer: 0.9% NaCL, PO4, KCl, HEPES.
Optiprep reagent density 1: 1.080 g/mL (heavy) by dilution of Optiprep reagent in Kreb/HEPES buffer in 50 mL conicals.
Optiprep reagent density 2: 1.040 g/mL (light) by dilution of Optiprep reagent in Kreb/HEPES buffer in 50 mL conicals.
Scissors.
Large serrated forceps.
2–3 pairs of hemostats.
FACS buffer: 1× PBS with 1 mM EDTA, 0.15% bovine serum albumin (BSA); keep at 4 °C.
2.4. Nonenzymatic Cell Crawl-Out Method for Lymph Node MPs
Scissors.
Forceps.
Culture media: RPMI 5% FCS, penicillin, streptomycin, fungi-zone, L-glutamine.
150 × 25 mm tissue culture plate.
Tissue culture incubator, 37 °C, 5% CO2.
100 μm filter.
50 mL conical tubes and centrifuge.
FACS buffer: 1× PBS with 1 mM EDTA, 0.15% bovine serum albumin (BSA); keep at 4 °C.
Cell scraper.
2.5. Enrichment for Myeloid Cells
FACS buffer.
Enrichment using anti-CD11c-biotin (alternatively use CD64- biotin for macrophages) and anti-CD1c PE conjugated (see Table 1 for antibodies) for positive selection, either STEMCELL or Miltenyi kits can be used.
Table 1.
Antibody clones used for pulmonary MP identification and isolation
| Antigen | Clone | Conjugate |
|---|---|---|
| CD1a | Ancell | FITC |
| CD1c | REA694 | PE |
| CD3 | UCHT1 | PB |
| CD11b | ICRF44 | PerCP |
| CD11c | 3.9 | PE-Cy7, biotin |
| CD14 | MΦP9 | V500 |
| CD15 | W6D3 | PB |
| CD20 | 2H7 | PB |
| CD26 | BA5b | PE, PE-Cy7 |
| CD36 | AC106 | APC |
| CD43 | eBio84–3C1 | FITC |
| CD45 | H130 | BUV395 |
| CD56 | TULY56 | PB |
| CD64 | 10.1 | PE |
| CD206 | 15–2 | PerCP |
| DEC205 | HD30 | APC |
| HLA-DR | L243 | APC-Cy7 |
2.6. Staining for FACS Analysis and MP Identification
FACS buffer.
Pooled human serum.
FACS buffer with human serum: 1 mM EDTA, 0.15% bovine serum albumin (BSA), 20% pooled human serum; keep at 4 °C.
Fluorochrome conjugated antibodies (see Table 1).
Antibody cocktail: 3–10 μL of fluorochrome conjugated antibodies per 100 μL of FACS buffer with human serum.
4’,6-Diamidine-2’-phenylindole dihydrochloride (DAPI) working solution: 30 μg/mL DAPI in PBS.
Flow cytometers: analyzers (BD LSR II and Fortessa); sorter (BD ARIA fusion).
FlowJo software: for flow cytometric analyses.
3. Methods
3.1. Preparation of Lung Tissue for Alveolar Macrophages: Tissue Dissection, Perfusion, and Bronchoalveolar Lavage (BAL)
Starting with the trachea. Identify individual lobes and corresponding large bronchus. Select one lobe for BAL and tissue dissection.
Dissect paratracheal, subcarinal, and carinal LNs. LNs are darker, more dense tissue nodules immediately surrounding the trachea and early bronchial branches (see Note 8).
Identify pulmonary arteries supplying the lobe of interest. Use a 60 mL syringe attached to 1 cm PVC tubing with appropriately sized pipet tip to follow arterial branches. Fill 60 mL syringe with PBS, and insert pipet tip into the large and small pulmonary arteries. Perfuse the lobe, and repeat several times with fresh PBS. Continue to perfuse until no more blood drains out of the pulmonary veins. The tissue should be visibly whiter.
Cannulate the large bronchus with 1 cm PVC tube. Secure the tubing in place with a piece of string.
- To lavage for alveolar macrophages, fully inflate the lobe with PBS/EDTA buffer via the cannulated bronchus. Upend the tissue to drain the lavage fluid into a beaker, and gently massage the fluid out of the lobe. Drain as much fluid as possible before inflating the lung again. Lavage six times using this sequence of buffers (see Note 9):
- Twice with PBS/ETDA buffer
- Twice with 1× PBS
- Twice with BSS-B buffer
Pass all lavage fluid through 100 μm filters to remove mucus and centrifuge at 250 × g-force for 10 min. Resuspend pellet in FACS buffer.
3.2. Preparation of Lung Single-Cell Suspension by Elastase Digestion
Cut excess tissue surrounding the lobe. Make sure not to pass the fissures between lobes to avoid leakage out of the lobe of interest. Transfer the lavaged lobe into a clean 1 gallon plastic bag for digestion.
Fully inflate the lobe with prepared elastase buffer (see Note 7). After inflation with elastase buffer, use hemostat to cross clamp the bronchus to avoid leakage of buffer.
Incubate in a water bath at 37 °C for 40 min.
Remove from water bath and transfer to a large dish. Cut away un-inflated tissue and poorly perfused patches. Remove the cannula and surrounding upper airway tissue. Cut the remaining lung tissue into large chunks (~1 × 1 × 1 inches).
Transfer lung pieces and fluid into a blender containing 75 mL of heat inactivated FCS and 150 mL of BSS-B buffer. Blend for two short (5 s) pulses (see Note 10). Large undigested pieces of cartilage will remain after pulse blending. These large pieces will be poured onto the cheesecloth in the following step 6.
The pulse-blended lobe is passed through a series of filters to remove undigested pieces and create a single-cell suspension. First, pour lung homogenate through a cheesecloth-lined vegetable strainer into a 1 L beaker. This will catch a large amount of undigested matter. Increase cell yield by washing out the blender with BSS-B buffer and pouring this over the cheesecloth. Squeeze the cheesecloth gently to drain buffer and cells.
Then pass the homogenate through a 350 μm filter into a second 1 L beaker and lastly through 100 μm filter to remove smaller cell clumps. Cells are passed through another 100 μm filter before centrifugation.
Centrifuge filtered lung cells at 250 × g-force for 10 min.
Resuspend the single-cell suspension in 40 mL of FACS buffer. Overlay 10 mL onto each of four prepared Optiprep density separation tubes.
Centrifuge at 500 × g-force for 20 min, make sure centrifuge brake is turned off, or placed on the lowest setting.
Collect cells within the light/heavy interface, avoiding lighter dead cells and denser red blood cells.
Wash once with a large volume of FACS buffer to dilute out Optiprep and then resuspend in FACS buffer for further enrichment.
3.3. Preparation of LN Single-Cell Suspension by Overnight Crawl Out
Collect LNs and carefully remove and discard surrounding perinodal fat tissue.
In 1.5 mL eppendorf tube, use scissors and forceps to finely mince LNs in tissue culture media.
Transfer minced LNs into a (220 × 25 mm) TC dish and add 50 mL of fresh culture media. Only place ~2–3 LNs per dish to avoid cell overcrowding.
Incubate overnight (16–20 h) at 37 °C with 5% CO2.
The following morning, collect all single cells from the plate by pipetting off and saving all the culture media. Next gently wash adherent cells from the dish using FACS buffer and a cell scraper. Filter-collected sample through a 100 μm filter into a 50 mL conical before centrifugation.
Centrifuge at 250 × g-force for 5 min. Resuspend in FACS buffer.
Alternatively (instead of steps 1–6), LNs can be finely chopped, digested in collagenase D for 30 min at 37 °C, filtered through a 100 μm filter, collected, and resuspended in FACS buffer [7–10].
3.4. Enrichment of Myeloid Cells by CD11c+ Selection for Lung MPs and LN MPs
To improve MP purity, first block non-specific antibody binding by preincubating lung or LN cell suspensions with FACS buffer with human serum for at least 10 min.
Incubate with anti-CD11c-biotin for 15 min on ice.
Incubate with biotin selection cocktail for 15 min on ice.
Incubate with magnetic nanoparticles/beads for 15 min on ice.
Wash once with FACS buffer.
Resuspend cell pellet in 5 mL of FACS buffer and place tube into the EasySep magnet. Allow cells to bind magnet for 5 min, and then decant unbound cells, as described by STEMCELL. For Miltenyi isolation use LS columns and Miltenyi microbeads. Both STEMCELL or Miltenyi enrichments work in this protocol (see Notes 11 and 12).
Repeat step 6 to increase purity for STEMCELL isolation.
3.5. Staining for FACS Analysis and MP Identification
Resuspend 2–5 × 106 cells per sample from BAL, enriched lung, or LN in 100 μL of FACS buffer with human serum.
Add 100 μL of antibody cocktail, vortex, and incubate for 45 min on ice.
Wash once in FACS buffer and resuspend in 250 μL of FACS buffer. Place cells on ice for flow sorting or analysis.
Immediately prior to acquiring samples on the flow cytometer or sorter, add 50 μL DAPI working solution to 250 μL of cells. Dead cell exclusion is essential for further analysis of lung and LN MP populations (see Note 13). Use control samples to optimize FACS parameters.
3.6. Gating and Analysis of Lung and LN MPs by Flow Cytometry
3.6.1. Lung MPs (Figs. 1 and 2)
Fig. 1.
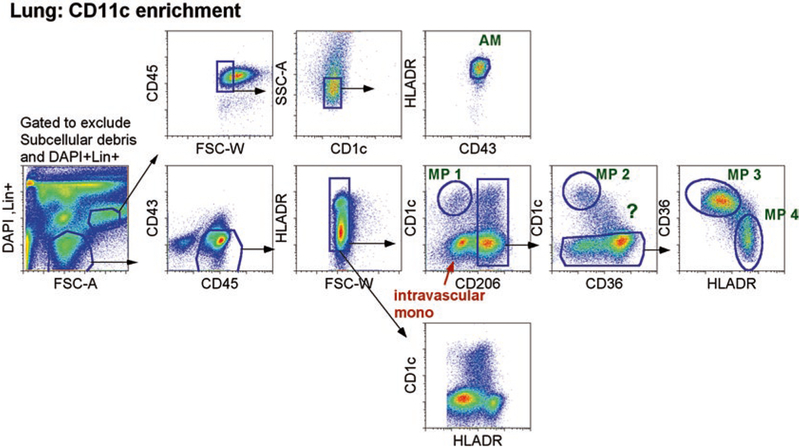
CD11c enrichment for pulmonary MP identification. Left figure depicts enriched CD11c+ cells from single- cell suspension of an entire digested lobe. Cells were first gated on live cells, which excluded dead cells (DAPI+), lineage+ cells, and subcellular debris. Then, single, CD45+ cells were gated. Top row, SSChiHLADRhiCD43+ cells were AMs (that were also CD14− not depicted here, in ref [14]). Bottom row, gates on SSCintCD45+CD43− cells, which were plotted as CD1c versus CD206 to identify extravascular cells: CD1c+CD206− and CD206+ cells. One extravascular CD1c+ MPs was identified: CD206−CD1a+ MP (designated as lung MP1). Three other extravascular MPs were CD206+ and distinguished by CD1c, CD36, and HLADR. One was CD1c+CD1aint MP (designated as lung MP2), and the other two were CD1c−CD36+HLA-DR+ and CD1c−CD36lo/-HLA-DR++ (designated as lung MP3 and MP4). Previously, we demonstrated in healthy individuals that intravascular circulating monocytes were CD206−, whereas extravascular monocytes/MPs were CD206+ [14]. Therefore, CD206 can distinguish between intra- and extravascular MPs
Fig. 2.
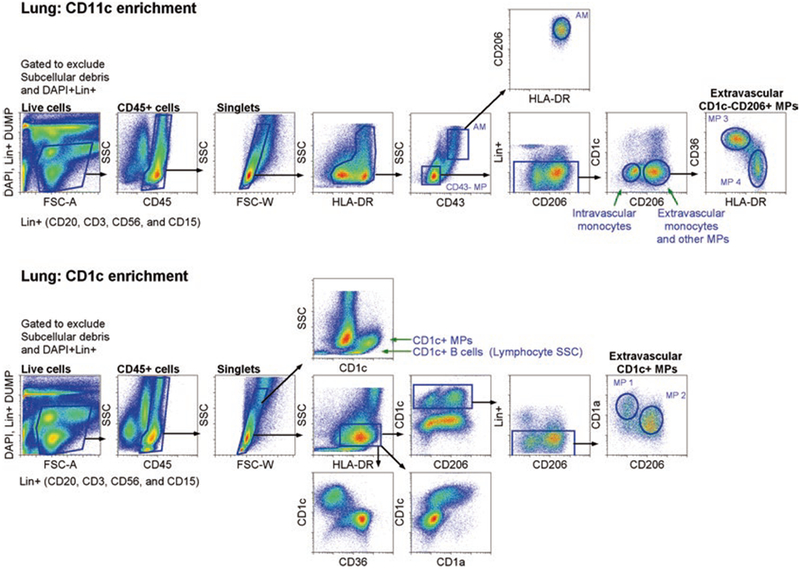
CD11c and CD1c enrichment for pulmonary MP sorting: five extravascular MPs in the lung designated AM and MP 1–4. Top figure depicts enriched CD11c+ cells, and bottom figure depicts enriched CD1c+ cells from single-cell suspension of an entire digested lobe. For both enrichments, cells were first gated on live cells, which excluded dead cells (DAPI+), lineage+ cells, and subcellular debris. Then, single, CD45+ cells were gated. For CD11c+ cell enrichment, SSChi/intHLADRhi cells were gated to identify and sort CD43+SSChi AMs (that were also CD14− not depicted here, in ref [14]) and two other CD1c−CD206+ pulmonary MPs. CD1c−CD206+ MPs were distinguished by CD36 and HLA-DR (designated as lung MP3 and MP4). For CD1c+ cell enrichment, SSCintHLADR+ cells were gated to exclude CD1c+ B lymphocytes. To further exclude Lin+ contaminating cells (including B cells), which were not as bright as DAPI, another gate was used to exclude any additional Lin+ cells that were not excluded in the first live cell gate. Two CD1c+ MPs were identified: CD206−CD1a+ MP (designated as lung MP1) and CD206+ CD1aint (designated as lung MP2). Previously, we demonstrated in healthy individuals that intravascular circulating monocytes were CD206−, whereas extravascular monocytes/MPs were CD206+ [14]. Therefore, CD206 can distinguish between intra- and extravascular MPs
Begin with the exclusion of dead cells and small debris using DAPI and forward scatter area (FSC-A) (see Figs. 1 and 2).
Use CD45, side scatter area (SSC-A), and FSC-width (W) to gate on single, hematopoietic cells. CD11c could also be used here to identify all myeloid cells.
Alveolar macrophages are SSChi, CD14−CD43+ cells that are uniformly CD206+ and HLA-DR+ (see ref. [14]). These tend to represent the majority of the SSChi population in the lung. SSChi CD43−HLADR− cells are mostly neutrophils (not shown), which can be confirmed by staining with CD15 or CD16.
- From the SSCintCD43− gate, we plot CD206 vs. CD1c to separate out four different populations of lung MPs as well as intravascular blood monocytes (Fig. 1).
- MP 1: CD1c+CD206− cells represent an HLA-DR+, CD1a+ dendritic cells.
- MP 2: CD1c+CD206+ (transcriptome data suggest this MP is a DC).
- MP 3: CD1c−CD206+CD36+ (transcriptome data suggest this is an interstitial macrophage, see Note 15).
- MP 4: CD1c−CD206+CD36−HLADR+ (transcriptome data suggest this is an interstitial macrophage, see Note 15).
- MP ?: CD1cintCD206+CD36+ (sorting, not shown- it is unclear whether this MP is more DC-like or Macrophage-like).
3.6.2. Lymph Node MPs
Begin by excluding dead cells and small debris, then doublets using a combination of DAPI and FSC-A, then FSC-W and SSC-A.
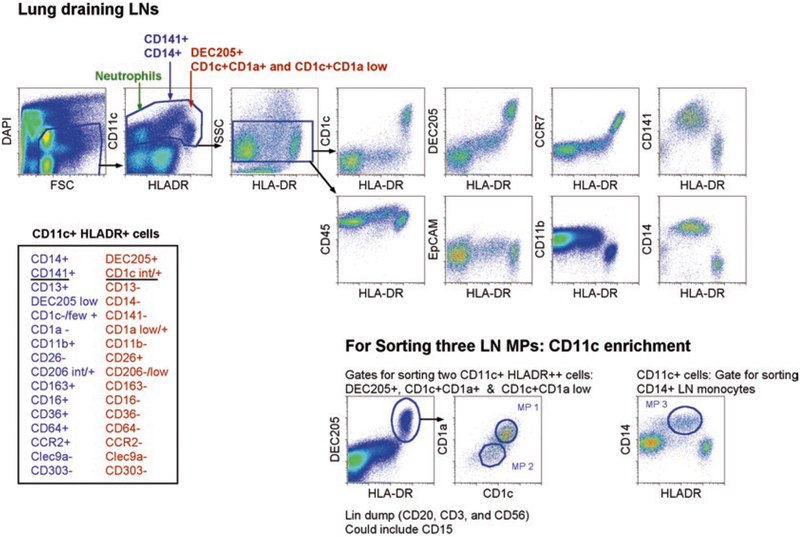
- CD11c, SSC, and HLA-DR can be used to identify three distinct populations (Fig. 3).
- CD11c+HLA-DR+/++ cells: monocytes (HLADR+) and DCs (HLADR++)
- CD11c+ HLA-DR− SSChi cells: Neutrophils
- CD11c−SSClo cells: Lymphocytes
- Taking the CD11c+HLA-DR+/++ gate, one can repeatedly observe three MPs (Fig. 3):
- MP1: HLADR++DEC205+CD1c+CD1a+
- MP2: HLADR++DEC205+CD1c+CD1alow
- MP3: HLADR+CD141+CD14+ monocytes (which also express CCR2, CD36, CD206, CD11b, CD64, and CD163)
Fig. 3.
CD11c enrichment for lung-draining lymph node (LN) MP identification and sorting: three extravascular MPs in the lung-draining LN designated MP 1–3. Top figure depicts non-enriched cells, and bottom figure depicts enriched CD11c+ cells from the single-cell suspension of lung-draining LNs. First cells were gated on live cells, which excluded dead cells (DAPI+), lineage+ cells, and subcellular debris. Then, single, CD45+ cells were gated on. For non-enriched cells, SSCintCD11c+, HLADR−/+ cells were gated. CD11c+HLADR− cells were neutrophils (green arrow). CD11c+HLADRint were CD14+CD141+ monocytes (blue arrow), and CD11c+HLADR++ cells (red arrow) contained two DEC205+CD1c+ LN MPs. The right, top rows illustrate various stains for CD11c+ cells on the y-axis (CD1c, DEC205, CCR7, CD141, CD45, EpCAM, CD14, and CD11b) with the x-axis constant for the expression of HLA-DR [14]. For sorting, CD11c expressing cells were enriched, and three MPs were identified and sorted. Two were CD11c+HLADR++, DEC205+CD1c+ MPs that were either CD1a+CD1c+ or CD1alowCD1c+/int (designated as lymph node MP1 and MP2). The third LN MP (designated as LN MP3) was CD11c+HLADR+, CD14+CD141+ monocytes. Inserted table on the bottom left summarized all stains used to analyze the two overarching LN MPs: CD14+CD141+ and DEC205+CD1c+/int MPs [14]
Acknowledgments
The authors would like to thank Drs. William Janssen and Robert Manson for collaborating and assisting in the acquisition of non- diseased human lungs from any of the following three sources: National Disease Research Interchange (Philadelphia, PA), the International Institute for the Advancement of Medicine (Edison, NJ), and University of Colorado Donor Alliance. This human subject research falls under federal exemption # 4. Grant support: C.V.J. NIH R01-HL115334 and R01-HL135001.
4 Notes
First major cautionary step during the isolation procedure of pulmonary MPs: The time to acquire, process, and isolate pulmonary MPs, followed by sorting for experiments or analyses, is very long. For this reason, many investigators may choose to save the MPs for later use by either fixing or freeze-thawing the cells. In our opinion, it is extremely important to analyze or examine the function of MPs right after isolation. From our experience, if MPs are fixed either with 1% PFA or formalin and analyzed the following day, the data quality is significantly less. This is most likely due to the following three reasons: first, fixed cells shrink thus altering their forward/side scatter (FSC/SSC) properties; second, autofluorescence due to fixation leads to cells shifting in fluorescent channels making antibody stains less distinguishable; and lastly, fixed cells eliminate the ability to exclude dead cells, which is vital when studying human MPs.
As for freeze-thaw, unlike alveolar macrophages or self- renewing non-hematopoietic structural cells, IMs, monocytes, and DCs do not remain viable after freezing and thawing. Therefore, we do not recommend freezing and thawing MPs. Perhaps the lack of viability is because DCs and monocytes are relatively short lived and do not self-renew or clonally expand like lymphocytes. In addition, IMs, extravascular monocytes, and DCs live within a complex extracellular matrix that cannot be replicated in vitro, and therefore, survival signals and cross talk with other cells are lacking, which most likely results in the initiation of their death when extracted from this environment.
Second major cautionary step during the isolation procedure of pulmonary MPs: Do not use logarithmic FSC/SSC and instead take advantage of linear FSC/SSC parameters to clearly exclude subcellular debris that can be DAPI negative and known to bind non-specifically to antibodies. For instance, in the figures illustrated here, CD1c+ MPs are bigger in SSC than CD1c+ B cells, thus allowing for the exclusion of not only sub-cellular debris but also lymphocytes. Of course it is important to distinguish live cells not only based on FSC/SSC but also with DAPI (dead cell exclusion dye) and lymphocyte and granulocyte stains for lineage dump. In our case, we exclude both dead cells and lineage cells in the same channel. Therefore, it is important to note that DAPI exclusion is higher on the log scale than lineage + (Lin+) antibodies used to exclude lymphocytes and granulocytes (see Figs. 1 and 2), and thus a second gate to exclude lineage cells should be used later in the sorting strategy. Lastly, autofluorescence, particularly for alveolar macrophages, will always be there, so try not to exclude these cells if desired for sorting or analyzing; even if they overlap with Lin+ cells (see Figs. 1 and 2), there are other ways to exclude contaminating cells from this population.
Third major cautionary step during the isolation procedure of pulmonary MPs relates to digestion and filtering. Liberase TM, collagenase D, and elastase all cleave away cell surface molecules used to identify pulmonary MPs. AMs are lavaged, which can then be directly sorted. Tissue MPs are digested through the alveolar epithelium to preserve the cell surface molecules on the MPs since the digestive enzymes are working directly on the epithelium rather than directly on interstitial cells. Although, we do not outline how to digest a small piece of lung tissue, small pieces of tissue can be finely minced and digested in low concentration of collagenase D [14], in a gentle shaker at 37 °C for 25 min. For LNs, due to the rapid cleaving of cell surface molecules by direct digestion, we allow LN MPs to crawl out overnight.
Fourth major cautionary step during the isolation procedure of pulmonary MPs: Overall, in our experience, there are some cell surface markers that are not readily cleaved by digestive enzymes, which include CD14, CD11b, HLADR, and CD11c. However, markers such as CD141, CD303, Clec9a, CD1c, and CD1a are highly susceptible to digestive cleavage (unpublished data).
Fifth major cautionary step during the isolation procedure of pulmonary MPs: Frequent filtering is vital to acquire and maintain cell yield and viability. The presence of fat cells in a single-cell suspension promotes clumping that also traps live cells. Particularly after centrifugation, it is impossible to disassociate live cells from clumped fat/dead cells. So to avoid clumping, or a major loss of cells, always filter before centrifugation, even after staining cells for flow and right before running samples on the cytometer. Lastly, make sure to use 100 μm filters at first, as 70- and 40-micron filters are too small to readily allow large MPs to pass through.
Studying LN MPs requires the use of relatively clean, healthy LNs. Thus, LNs not contaminated with excessive smoking or pollution particulates are preferred. To date, we are unable to recover any MPs from black LNs.
Sixth major cautionary step during the isolation procedure of pulmonary MPs is to be aware that intravascular and extravascular MPs exist within a piece of tissue even after extensive perfusion [13]. Therefore identifying intra- and extravascular cells in the human lung is important, and how to do this was illustrated in our previous publication [14].
The volume required for digestion is best determined during the lavage by measuring the volume required to fully inflate the lobe. 4.2 U elastase/mL BSS-B buffer x volume of lobe. On average a right middle lobe from a middle-aged person requires 150–250 mL of buffer.
Make sure not to cut too deep into tissue, as the lobe will be inflated with lavage and enzymatic fluid for pulmonary MP isolation.
EDTA is included only in the first two lavages to help detach adherent macrophages from the airways epithelia. However, efficient removal of EDTA is essential to avoid subsequent inhibition of enzymatic digestion.
Avoid overprocessing the lung; the shear force of the blender damages cells and will result in greatly increased cell death.
Enrichment protocols provided by the manufacturer serve as a good starting point for titrating cell/antibody/bead ratios for optimizing enrichment efficiency based on downstream requirements.
Enriching LN cells using CD11c for sorting is required if optimal RNA seq analyses and functional assays are being performed. If cells are not enriched, sorting will take too long, and the viability of the LN MPs will diminish.
For microRNA or messenger RNA extraction from MPs, add lymphocyte and granulocyte lineage stains in addition to DAPI to insure proper exclusion during cell sorting.
CD206 is expressed on monocyte-derived cells and other MPs upon extravasation into tissue. CD1c+CD206− cells represent HLA-DR+, CD1a+ dendritic cells, as this was clearly shown using time-lapse video (see ref. [14]). Although we hypothesize that there are two DCs, we have yet to clearly identify the second DC in the lung, which may be the CD1c+CD206+ MP, as LN CD11c+HLADR++CD20− cells all express DEC205 and CD1c, which are divided by CD1a+ and CD1alow (see Fig. 3).
The CD1c−CD206+ population further splits into two populations. One population that expresses high level of CD36 and second population that expresses lower CD36 and more HLA-DR.
References
- 1.Janssen WJ, Bratton DL, Jakubzick CV, Henson PM (2016) Myeloid cell turnover and clearance. Microbiol Spectr 4(6). https://doi.org/10.1128/microbiolspec.MCHD-0005-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bharat A, Bhorade SM, Morales-Nebreda L, Mc Quattie-Pimentel AC, Soberanes S, Ridge K, DeCamp MM, Mestan KK, Perlman H, Budinger GR, Misharin AV (2015) Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol 54:147–149. https://doi.org/10.1165/rcmb.2015-0147LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLean JA, Xia W, Pinto CE, Zhao L, Liu HW, Kradin RL (1996) Sequestration of inhaled particulate antigens by lung phagocytes. A mechanism for the effective inhibition of pulmonary cell-mediated immunity. Am J Pathol 148(2):657–666 [PMC free article] [PubMed] [Google Scholar]
- 4.Yu YA, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, Kraft M, Hollingsworth JW, Gunn MD, Tighe RM (2015) Flow cyto-metric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol 54:13–24. https://doi.org/10.1165/rcmb.2015-0146OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt PG (2005) Pulmonary dendritic cells in local immunity to inert and pathogenic antigens in the respiratory tract. Proc Am Thorac Soc 2(2):116–120. https://doi.org/10.1513/pats.200502-017AW [DOI] [PubMed] [Google Scholar]
- 6.Sung SS, Fu SM, Rose CE Jr, Gaskin F, Ju ST, Beaty SR (2006) A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol 176(4):2161–2172. doi:176/4/2161 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Vermaelen KY, Carro-Muino I, Lambrecht BN, Pauwels RA (2001) Specific migratory dendritic cells rapidly transport antigen from the airways to the thoracic lymph nodes. J Exp Med 193(1):51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakubzick C, Tacke F, Llodra J, van Rooijen N, Randolph GJ (2006) Modulation of dendritic cell trafficking to and from the airways. J Immunol 176(6):3578–3584. doi:176/6/3578 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Jakubzick C, Helft J, Kaplan TJ, Randolph GJ (2008) Optimization of methods to study pulmonary dendritic cell migration reveals distinct capacities of DC subsets to acquire soluble versus particulate antigen. J Immunol Methods 337(2):121–131. https://doi.org/10.1016/j.jim.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desch AN, Randolph GJ, Murphy K, Gautier EL, Kedl RM, Lahoud MH, Caminschi I, Shortman K, Henson PM, Jakubzick CV (2011) CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell- associated antigen. J Exp Med 208(9):1789–1797. https://doi.org/10.1084/jem.20110538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakubzick C, Bogunovic M, Bonito AJ, Kuan EL, Merad M, Randolph GJ (2008) Lymph- migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J Exp Med 205(12):2839–2850. https://doi.org/10.1084/jem.20081430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakubzick CV, Randolph GJ, Henson PM (2017) Monocyte differentiation and antigen- presenting functions. Nat Rev Immunol 17:349–362. https://doi.org/10.1038/nri.2017.28 [DOI] [PubMed] [Google Scholar]
- 13.Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, Leach SM, Bratton DL, Henson PM, Janssen WJ, Jakubzick CV (2017) Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol 57:66–76. https://doi.org/10.1165/rcmb.2016-0361OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, Slansky JE, Jacobelli J, Mason R, Ito Y, Messier E, Randolph GJ, Prabagar M, Atif SM, Segura E, Xavier RJ, Bratton DL, Janssen WJ, Henson PM, Jakubzick CV (2015) Flow cytometric analysis of mononuclear phagocytes in non- diseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med 193:614–626. https://doi.org/10.1164/rccm.201507-1376OC [DOI] [PMC free article] [PubMed] [Google Scholar]