Abstract
The first outbreak in Japan of GES-5 carbapenemase-producing Pseudomonas aeruginosa occurred in a long-term care facility in 2014. To assess the spread of GES-5 producing P. aeruginosa clinical isolates in medical settings in Japan, 1,476 carbapenem-resistant P. aeruginosa isolates obtained from 2012 to 2016 were characterized. Of these 1,476 isolates, 104 (7.0%) harbored blaGES-5. Southern blotting revealed that the blaGES-5 was located on the chromosome. The isolation rates of these GES-5 producers increased significantly every year, from 2.0% (6 of 295) in 2012 to 2.8% (8 of 283) in 2013 to 5.3% (16 of 303) in 2014 to 9.7% (29 of 300) in 2015 to 15.3% (45 of 295) in 2016. Of the 104 GES-5 producers, 102 belonged to clonal complex (CC) 235, including 99 belonging to ST235 and three belonging to ST2233). Whole genome sequence analysis revealed that CC235 P. aeruginosa harboring blaGES-5 spread in a clonal manner. These results indicate that these GES-5 producing CC235 P. aeruginosa clinical isolates have spread in medical settings throughout Japan.
Introduction
The dissemination of carbapenem-resistant Pseudomonas aeruginosa, known as P. aeruginosa high-risk clones, has become a serious problem in medical settings worldwide [1,2]. GES-type β-lactamases, belonging to class A extended-spectrum β-lactamases (ESBLs) [3], have been increasingly reported among Gram-negative pathogens, including P. aeruginosa, Enterobacter cloacae, Klebsiella pneumoniae, and Acinetobacter baumannii [4]. GES-1 was initially detected in a K. pneumoniae isolate in 1998 in France [5]. To date, 37 GES variants have been described (ftp://ftp.ncbi.nlm.nih.gov/pathogen/betalactamases/Allele.tab).
Some GES-type enzyme variants, including GESs-2, -4, -5, -6 and -14, have shown carbapenem-hydrolyzing activities [6–10]. GES-2 was the first GES-type enzyme with carbapenem-hydrolyzing activities to be identified, although its activities differ from GES-1 by a substitution of Gly170Asn, located inside the Ω-loop of the catalytic site [11]. GESs-4, -5, -6, and -14, with a substitution of Gly170Ser, exhibit higher carbapenem-hydrolyzing activities than GES-1, and GES-5 exhibit higher carbapenem-hydrolyzing activities than GES-5 [6–10].
ST235 multidrug-resistant P. aeruginosa co-producing IMP-type metallo-β-lactamases and AAC(6’)s was originally isolated in 2003 in Japan [12], and it was a cause of outbreaks in community hospitals throughout Japan [13]. The first outbreak of GES-5 producing P. aeruginosa in Japan occurred in 2014 [14]. The objective of this study was to characterize the spread of carbapenem-resistant GES-5 producing P. aeruginosa in Japan by analyzing 1476 clinical isolates obtained from 2012 to 2016.
Materials and methods
Bacterial strains
P. aeruginosa clinical isolates identified as carbapenem-resistant were isolated from 1,476 individual patients at hospitals located in all 47 prefectures throughout Japan between 2012 and 2016 (295 in 2012, 283 in 2013, 303 in 2014 300 in 2015 and 295 in 2016) in hospitals by BML Biomedical Laboratories R&D Center (Kawagoe, Saitama, Japan). Drug-susceptibility was tested using the microdilution method according to the criteria of the Clinical Laboratory Standards Institute (CLSI) criteria, with carbapenem-resistant P. aeruginosa defined as isolates resistant to imipenem or meropenem (MIC ≥ 8 μg/mL) [15].
Detection and sequencing of GES-type ESBLs
Carbapenem-resistant P. aeruginosa isolates were screened for blaGESs-type genes by PCR using the primers GES-F (5’-ATGCGCTTCATTCACGCAC-3’) and GES-R (5’-CTATTTGTCCGTGCTCAGG-3’), as described [16]. All PCR products were sequenced using a DNA sequencer (ABI PRISM 3130; Applied Biosystems, Foster City, CA).
Whole genome sequencing
Genomic DNAs of blaGES-5-positive isolates were extracted using DNeasy Blood and Tissue kits (Qiagen, Tokyo, Japan) and sequenced by a next generation sequencer (MiSeq; Illumina, San Diego, CA).
Phylogenetic analysis
To identify single nucleotide polymorphisms (SNPs) throughout the entire genomes of all 104 blaGES-5-positive isolates, all reads of each isolate were aligned against P. aeruginosa NCGM2.S1 (Gen Bank accession no. AP 012280) using CLC genomics workbench version 8.0.2 (CLC bio, Tokyo, Japan) [17]. SNP concatenated sequences were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/). Models and parameters used for the phylogenetic analyses were computed using j-Model Test-2.1.4. A maximum-likelihood phylogenetic tree was constructed from SNP alignment with PhyML 3.0 [18].
Drug resistance genes and MLST
Genes associated with resistance to β-lactams, aminoglycosides and quinolones were detected using ResFinder 3.0 (https://cge.cbs.dtu.dk/services/ResFinder/). Fluoroquinolone resistance has been associated with mutations in the quinolone resistance determining region, which includes the gyrA and parC genes that encode DNA gyrase and topoisomerase IV, respectively [19]. Multilocus sequence types (MLSTs) were deduced as described (http://pubmlst.org/paeruginosa/). Clonal complexes (CC) were determined by eBURST version 3 (http://eburst.mlst.net).
Pulsed-field gel electrophoresis and Southern blotting
DNA plugs of two blaGES-5 positive CC235 isolates, NCGM2108 (ST235) and NCGM2900 (ST2233), were digested with S1 nuclease, separated by pulsed-field gel electrophoresis (PFGE), and subjected to Southern blotting and hybridization using blaGES-5 probes.13
Statistical analysis
The yearly proportions of multidrug-resistant P. aeruginosa isolates positive for blaGES-5 were analyzed by the chi-square test.
Nucleotide sequence accession numbers
The whole genome sequences of all 104 blaGES-5-positive isolates have been deposited in GenBank as Accession Number DRA006450 (https://www.ncbi.nlm.nih.gov/sra/?term=DRA006450) (NCGM2012-2015) and DRA007009 (NCGM2016). The sequences of the genomic environment surrounding blaGES-5 in multidrug-resistant P. aeruginosa strains NCGM2100 (ST274), NCGM2900 (ST235) and NCGM3294 (ST1342) have been deposited in GenBank as Accession Numbers LC157846, LC155936 and LC360798, respectively.
Results
Identification and drug susceptibilities of blaGES-5-positive P. aeruginosa isolates
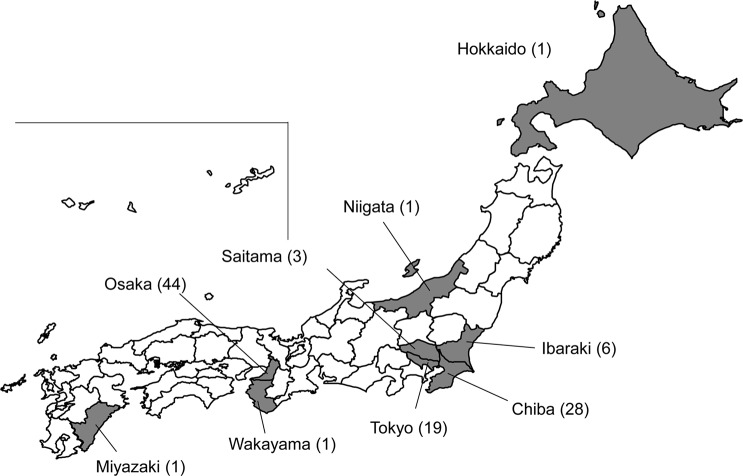
Of 1,476 carbapenem-resistant isolates positive P. aeruginosa isolates, 137 (9.3%) were positive for blaGESs. Sequence analysis revealed that, of these isolates, four (2.9%) were positive for blaGES-1, 104 (75.9%) for blaGES-5, two (1.5%) for blaGES-6, five (3.6%) for blaGES-15, one (0.7%) for blaGES-24, 15 (10.9%) for blaGES-26, one (0.7%) for blaGES-29, three (2.2%) for blaGES-30, and two (1.5%) for a novel variant of blaGES (GenBank accession no. LC385763). The proportion of blaGES-5-positive isolates increased significantly by year, being 2.0% (6/295) in 2012, 2.8% (8/283) in 2013, 5.3% (16/303) in 2014, 9.7% (29/300) in 2015 and 15.3% (45/295) in 2016 (p ≤0.01 for 2016 vs 2012). These 104 blaGES-5-positive isolates were obtained in nine regions of Japan, including 44 (42%) were in Osaka, 28 (27%) in Chiba, 19 (18%) in Tokyo, six (6%) in Ibaraki, three (3%) in Saitama, and one each (1.0%) in Hokkaido, Niigata, Wakayama, and Miyazaki (Fig 1). Of these isolates, 71 (68.3%) were from respiratory tracts, 22 (21.2%) from urinary tracts, seven (6.7%) from decubitus and four (3.8%) from pus.
Fig 1. Geographical distribution of the nine prefectures from which the blaGES-5-containing P. aeruginosa strains were isolated.
The numbers of the isolates in each prefecture were indicated in parentheses.
All 104 of these blaGES-5-positive isolates were resistant to imipenem and meropenem but sensitive to colistin (Table 1). Both MIC50 and MIC90 values were higher for meropenem than for imipenem. In addition, 50 isolates were resistant to amikacin, 28 to aztreonam, 19 to cefepime, 34 to ceftazidime and 103 to ciprofloxacin.
Table 1. Characterization of GES-5 producing P. aeruginosa isolates, including MIC ranges, MLST, drug-resistant factors and virulent factors.
| MLST | No. of isolatesa |
MIC Range against antibioticsb (mg/L) (% resistance)c |
Carbapenemase- and ESBL- encoding gene(s) |
Aminoglycoside resistance gene(s) |
Mutation in DNA gyrase |
Exotoxin gene(s) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | AZT | FEP | CAZ | CIP | CST | IMP | MEM | GyrA | ParC | |||||
| ST235 and ST2233 (CC235) |
102 | 4–64 (47%) |
2–64 (25%) |
4–128 (17%) |
4–512 (31%) |
2–64 (99%) |
0.063–0.5 (0%) |
16–64 (100%) |
32–512 (100%) |
blaGES-5 blaPAO blaOXA-488 |
aac(6')-Ib | T83I | S87L |
exoT exoU exoY |
| ST274 | 1 | 128 | 128 | 64 | 128 | 32 | 0.5 | 64 | 64 |
blaGES-5 blaPAO blaOXA-486 |
aac(6')-Ia | T83I | S87L |
exoS exoT exoY |
| ST1342 | 1 | 64 | 64 | 32 | 64 | 32 | 0.5 | 64 | 128 |
blaGES-5 blaPAO blaOXA-396 blaCMY-8 |
aac(6')-Ia | T83I | S87L |
exoS exoT exoY |
aOf 102 isolates belonging to CC235, 99 isolates belonged to ST235 and the remaining 3 did to ST2233.
bAMK: Amikacin, AZT: Aztreonam, FEP: Cefepime, CAZ: Ceftazidime, CIP: Ciprofloxacin, CST: Colistin, IMP: Imipenem, MEM: Meropenem.
cBreakpoints for resistance (mg/L): AMK; ≥32, AZT; ≥32, FEP; ≥32, CAZ; ≥32, CIP; ≥4, CST; ≥8, IMP; ≥8, and MEM; ≥8.
All 104 isolates harbored blaPAO. Of them, 102 (98.0%) harbored blaOXA-488, 1 had blaOXA-396 and 1 had blaOXA-486 (Table 1). Of the 104 blaGES-5-positive isolates, 102 (98.0%) harbored aac(6’)-Ib and the other two harbored aac(6’)-Ia (Table 1). The all 104 isolates were found to have point mutations in the quinolone-resistance-determining regions of gyrA and parC, consisting of the amino acid substitutions S83I in GyrA and S87L in ParC (Table 1). Of them, a quinolone-sensitive strain had the same mutations as the remaining quinolone-resistant strains, but it had amino acid substitutions in the efflux systems, including P363L in OprJ, R112Q in OprN, in addition to an OprD in-frame deletion. In-frame deletions of OprD were found in 54 of 104 isolates tested. The 99 isolates belonging to CC235 (ST235 and ST2233) harbored virulence genes, exoT, exoU and exoY, although the remaining two belonging ST274 and ST1342 did exoS, exoT and exoY (Table 1).
MLST and phylogenetic analysis
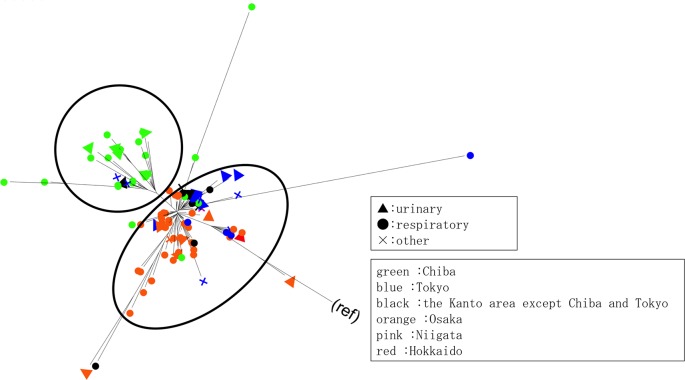
Of the 104 blaGES-5-positive isolates, 102 belonged to Clonal Complex (CC) 235, including 99 and 3 belonging to ST235 and ST2233, respectively. The individual isolates from Miyazaki and Wakayama prefectures belonged to ST274 (allelic profile: 23, 5, 11, 7, 1, 12, 7) and ST1342 (allelic profile: 1, 5, 26, 3, 1, 10, 3), respectively, and were unrelated to CC235. A maximum-likelihood phylogenetic tree constructed from the 102 CC235 isolates showed two major clades (Fig 2). Clade A mainly consisted of isolates from Hokkaido, Ibaraki, Niigata, Osaka, Saitama and Tokyo, whereas clade B mainly consisted of isolates from Chiba (Figs 1 and 2).
Fig 2. Molecular phylogeny of the 102 P. aeruginosa strains belonging to CC235.
A maximum-likelihood phylogenetic tree was constructed from these isolates. The GTR+I+G model was chosen as a nucleotide substitution model.
Genomic environments surrounding blaGES-5
In the 102 isolates belonging to CC235, blaGES-5 was present in a class 1 integron (accession no. LC155936) (Fig 3). The sequence of this integron from nt 1 to nt 6,647 was identical to the sequence from nt 10,786 to nt 17,432 of P. aeruginosa strain CH79 (C79) fosmid2CA-integron isolated in 2010 in Sydney, Australia (accession no. JF826499) [20]. The genomic environment surrounding blaGES-5 in the ST274 isolate from Miyazaki was aac(6’)-Ia-gcuG-blaGES-5- qacEΔ1-sulI-aac (accession no. LC157846) (Fig 3). The sequence of this genomic environment from nt 140 to nt 1,127, which contained aac(6’)-Ia-gcuG, was identical to the sequence from nt 980 to nt 1,967 of integron In831 in P. aeruginosa TUM4030 isolated in 2004 in Hokkaido, Japan (accession no. AB901039) [21], and the sequence from nt 1,297 to nt 4,135, containing the structure blaGES-5-qacEΔ1-sulI-aac, was more than 99% identical to the sequence from nt 18,969 to nt 21,807 of a plasmid pGES5 in Aeromonas hydrophilia WCHAH01 isolated in China (accession no. KR014105). The genomic environment surrounding blaGES-5 in the ST1342 isolate from Wakayama was tnpR-intI1-blaGES-5- aac(6’)-Ia-gcuG (accession no. LC360798) (Fig 3). The sequence of the genomic environment from nt 100 to nt 9,341, which contained tnpA-tnpR-intI1- blaGES-5, was identical to the sequence from nt 2,314 to nt 12,842 of P. aeruginosa CH79 fosmid2CA-integron (accession no. JF826499) [20], and the sequence from nt 9,499 to nt 10,486, which contained aac(6’)-Ia-gcuG, was identical to the sequence from nt 980 to nt 1,967 of integron In831 in P. aeruginosa TUM4030 (accession no. AB901039) [21].
Fig 3. Structure of the genomic environments surrounding blaGES-5.
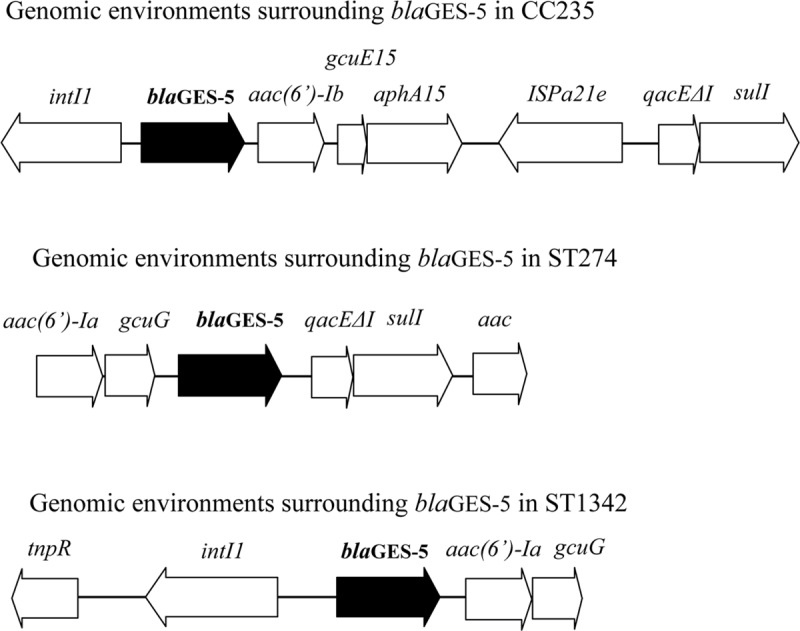
The 89,870 bp integrative conjugative element (ICE), including a class 1 integron with blaGES-5, was detected in CC235 GES-5 producing isolates by comparison with the sequence of PAO1 strain. The ICE was flanked by PA0747 (upstream region of the ICE) and PA2737 (downstream region of the ICE).
PFGE and Southern blotting showed that two blaGES-5-positive isolates belonging to CC235 (ST235 and ST2233) had no plasmid harboring blaGES-5 (data not shown).
Discussion
This study showed that carbapenem-resistant blaGES-5-positive P. aeruginosa isolates, first isolated in Japan in 2012, rapidly spread throughout Japan over the next several years. Outbreaks of blaGES-5-positive isolates occurred among hospitals in a region (Chiba prefecture), indicating that blaGES-5-positive isolates started to expand in a clonal manner within a specific region in Japan. Our previous study on whole genome analysis of 136 clinical isolates of ST235 multidrug-resistant P. aeruginosa producing IMP-type MBLs showed that these isolates fell within seven subclades, with each subclade having a characteristic genetic background confined to a geographic location. The previous study indicated that P. aeruginosa ST235 has become prevalent worldwide due to the antibiotics-selective pressures through mutations and acquisition of resistant-elements among local populations [2]. Our results suggest that clonal expansion is the driving force in generating the population structure of ST235 P. aeruginosa [22].
The primary reason for the emergence and spread of carbapenem-resistant blaGES-5-positive P. aeruginosa isolates in medical settings in Japan is the inability of microbiological laboratories in Japan, even those in tertiary hospitals, to detect blaGES-5-positive isolates. P. aeruginosa strains that produce Class B metallo-β-lactamases can be routinely detected in microbiological laboratories using several methods [23–25]. These methods, however, are unable to detect bacterial strains producing class A carbapenemases. The cross-transmission of blaGES-5 will be caused by the clonal nature of the outbreaks in medical settings in Japan. Microbiological laboratories therefore require methods to easily detect GES-5 producers, such as the Blue Carba test [26,27], the CIMTris [28] and a systematic blaGES PCR method.
Virulent factor ExoU will be associated with the antibiotic resistance and spreading of CC235 P. aeruginosa. Previous study reported that ExoU-positive P. aeruginosa strains had higher resistance to β-lactams and quinolones than ExoS-positive strains, because the ExoU-positive P. aeruginosa had more mutations in genes that were associated with β-lactams resistance and quinolone resistance [29]. The height of the mutation rate caused by ExoU may help to enhance the adaptability of CC235 P. aeruginosa to the environment.
GES-5 producing P. aeruginosa may spread in medical settings worldwide. A P. aeruginosa isolate producing GES-5 was first obtained in 2004 from the blood of a burn patient in China [16]. GES-5 producing P. aeruginosa strains have been isolated from patients in Brazil [30], Japan [14], Lithuania [31], South Africa [32], Spain [33] and Turkey [34]. Thus, it is important to monitor for GES-5 producing P. aeruginosa in medical settings worldwide.
Acknowledgments
This study was approved by the Biosafety Committee, Juntendo University (approval number: BSL2/29-1) and supported by grants of the Japan Society for the Promotion of Science (grant number 18K07120) and the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development (grant number 18fk0108061).
Data Availability
All relevant data, including GenBank accession numbers, are within the paper.
Funding Statement
The study was supported by grants from Research Program on Emerging and Re-emerging Infectious Diseases from AMED (18fk0108061) to TK (https://www.amed.go.jp/en/program/list/01/06/002.html) and JSPS KAKENHI Grant Number 18K07120 to TT. BML Inc. provided support in the form of salaries for author Masahiro Shimojima, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Oliver A, Mulet X, Lopez-Causape C, Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21–22: 41–59. 10.1016/j.drup.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 2.Treepong P, Kos VN, Guyeux C, Blanc DS, Bertrand X, Valot B, et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 2018;24: 258–266. 10.1016/j.cmi.2017.06.018 [DOI] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20: 440–58, table of contents. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naas T, Poirel L, Nordmann P. Minor extended-spectrum β-lactamases. Clin Microbiol Infect. 2008;14 Suppl 1: 42–52. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44: 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae IK, Lee YN, Jeong SH, Hong SG, Lee JH, Lee SH, et al. Genetic and biochemical characterization of GES-5, an extended-spectrum class A β-lactamase from Klebsiella pneumoniae. Diagn Microbiol Infect Dis. 2007;58: 465–468. 10.1016/j.diagmicrobio.2007.02.013 [DOI] [PubMed] [Google Scholar]
- 7.Bogaerts P, Naas T, El Garch F, Cuzon G, Deplano A, Delaire T, et al. GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob Agents Chemother. 2010;54: 4872–4878. 10.1128/AAC.00871-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnin RA, Nordmann P, Potron A, Lecuyer H, Zahar JR, Poirel L. Carbapenem-hydrolyzing GES-type extended-spectrum β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55: 349–354. 10.1128/AAC.00773-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vourli S, Giakkoupi P, Miriagou V, Tzelepi E, Vatopoulos AC, Tzouvelekis LS. Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol Lett. 2004;234: 209–213. 10.1016/j.femsle.2004.03.028 [DOI] [PubMed] [Google Scholar]
- 10.Wachino J, Doi Y, Yamane K, Shibata N, Yagi T, Kubota T, et al. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A β-lactamase, GES-4, possessing a single G170S substitution in the omega-loop. Antimicrob Agents Chemother. 2004;48: 2905–2910. 10.1128/AAC.48.8.2905-2910.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001;45: 2598–2603. 10.1128/AAC.45.9.2598-2603.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Fujino T, Kobayashi I, Morita K, et al. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6')-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob Agents Chemother. 2005;49: 3734–3742. 10.1128/AAC.49.9.3734-3742.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Kasai A, Mizuguchi Y, Araake M, et al. Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J Clin Microbiol. 2007;45: 979–989. 10.1128/JCM.01772-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanayama A, Kawahara R, Yamagishi T, Goto K, Kobaru Y, Takano M, et al. Successful control of an outbreak of GES-5 extended-spectrum β-lactamase-producing Pseudomonas aeruginosa in a long-term care facility in Japan. J Hosp Infect. 2016;93: 35–41. 10.1016/j.jhin.2015.12.017 [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 25th informational supplement CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne PA. 7th ed Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 16.Wang C, Cai P, Chang D, Mi Z. A Pseudomonas aeruginosa isolate producing the GES-5 extended-spectrum β-lactamase. J Antimicrob Chemother. 2006;57: 1261–1262. 10.1093/jac/dkl116 [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi-Akiyama T, Kuwahara T, Tada T, Kitao T, Kirikae T. Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J Bacteriol. 2011;193: 7010 10.1128/JB.06312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 19.Nakano M, Deguchi T, Kawamura T, Yasuda M, Kimura M, Okano Y, et al. Mutations in the gyrA and parC genes in fluoroquinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41: 2289–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez E, Marquez C, Ingold A, Merlino J, Djordjevic SP, Stokes HW, et al. Diverse mobilized class 1 integrons are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2012;56: 2169–2172. 10.1128/AAC.06048-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mano Y, Saga T, Ishii Y, Yoshizumi A, Bonomo RA, Yamaguchi K, et al. Molecular analysis of the integrons of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates collected by nationwide surveillance programs across Japan. BMC Microbiol. 2015;15: 41-015-0378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi-Akiyama T, Tada T, Ohmagari N, Viet Hung N, Tharavichitkul P, Pokhrel BM, et al. Emergence and spread of epidemic multidrug-resistant Pseudomonas aeruginosa. Genome Biol Evol. 2017;9: 3238–3245. 10.1093/gbe/evx243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simner PJ, Opene BNA, Chambers KK, Naumann ME, Carroll KC, Tamma PD. Carbapenemase detection among carbapenem-resistant glucose-nonfermenting Gram-Negative bacilli. J Clin Microbiol. 2017;55: 2858–2864. 10.1128/JCM.00775-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitao T, Miyoshi-Akiyama T, Tanaka M, Narahara K, Shimojima M, Kirikae T. Development of an immunochromatographic assay for diagnosing the production of IMP-type metallo-β-lactamases that mediate carbapenem resistance in Pseudomonas. J Microbiol Methods. 2011. [DOI] [PubMed] [Google Scholar]
- 25.Aktas E, Malkocoglu G, Otlu B, Copur Cicek A, Kulah C, Comert F, et al. Evaluation of the carbapenem inactivation method for detection of carbapenemase-producing Gram-negative bacteria in comparison with the RAPIDEC CARBA NP. Microb Drug Resist. 2017;23: 457–461. 10.1089/mdr.2016.0092 [DOI] [PubMed] [Google Scholar]
- 26.Pasteran F, Veliz O, Ceriana P, Lucero C, Rapoport M, Albornoz E, et al. Evaluation of the Blue-Carba test for rapid detection of carbapenemases in Gram-negative bacilli. J Clin Microbiol. 2015;53: 1996–1998. 10.1128/JCM.03026-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pires J, Novais A, Peixe L. Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2013;51: 4281–4283. 10.1128/JCM.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uechi K, Tada T, Shimada K, Kuwahara-Arai K, Arakaki M, Tome T, et al. A modified carbapenem inactivation method, CIMTris, for carbapenemase production in Acinetobacter and Pseudomonas species. J Clin Microbiol. 2017;55: 3405–3410. 10.1128/JCM.00893-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subedi D, Vijay AK, Kohli GS, Rice SA, Willcox M. Association between possession of ExoU and antibiotic resistance in Pseudomonas aeruginosa. PLoS One. 2018;13: e0204936 10.1371/journal.pone.0204936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picao RC, Poirel L, Gales AC, Nordmann P. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob Agents Chemother. 2009;53: 3908–3913. 10.1128/AAC.00453-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikucionyte G, Zamorano L, Vitkauskiene A, Lopez-Causape C, Juan C, Mulet X, et al. Nosocomial dissemination of VIM-2-producing ST235 Pseudomonas aeruginosa in Lithuania. Eur J Clin Microbiol Infect Dis. 2016;35: 195–200. 10.1007/s10096-015-2529-0 [DOI] [PubMed] [Google Scholar]
- 32.Labuschagne Cde J, Weldhagen GF, Ehlers MM, Dove MG. Emergence of class 1 integron-associated GES-5 and GES-5-like extended-spectrum β-lactamases in clinical isolates of Pseudomonas aeruginosa in South Africa. Int J Antimicrob Agents. 2008;31: 527–530. 10.1016/j.ijantimicag.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 33.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, et al. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother. 2009;53: 4930–4933. 10.1128/AAC.00900-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iraz M, Duzgun AO, Cicek AC, Bonnin RA, Ceylan A, Saral A, et al. Characterization of novel VIM carbapenemase, VIM-38, and first detection of GES-5 carbapenem-hydrolyzing β-lactamases in Pseudomonas aeruginosa in Turkey. Diagn Microbiol Infect Dis. 2014;78: 292–294. 10.1016/j.diagmicrobio.2013.12.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data, including GenBank accession numbers, are within the paper.