Abstract
In Escherichia coli thymidylate synthase (EcTS), rate-determining hydride transfer from the cofactor, 5,10-methylene-5,6,7,8-tetrahydrofolate, to the intermediate 5-methylene-2’-deoxyuridine 5’-monophosphate occurs by hydrogen tunneling, requiring precise alignment of reactants and a closed binding cavity, sealed by the C-terminal carboxyl group. Mutations that destabilize the closed conformation of the binding cavity allow small molecules such as β-mercaptoethanol (β-ME) to enter the active site and compete with hydride for addition to the 5-methylene group of the intermediate. The C-terminal deletion mutant of EcTS produced the β-ME adduct in proportions that varied dramatically with cofactor concentration, from 50% at low cofactor concentrations to 0% at saturating cofactor conditions, suggesting communication between active sites. We report the 2.4Å X-ray structure of the C-terminal deletion mutant of E. coli TS in complex with substrate and a cofactor analog, CB3717. The structure is asymmetric, with reactants aligned in a manner consistent with hydride transfer in only one active site. In the second site, the CB3717 has shifted to a site where the normal cofactor would be unlikely to form 5-methylene-2’-deoxyuridine 5’-monophosphate, consistent with no formation of β-ME adduct. The structure shows how the binding of cofactor at one site triggers hydride transfer and borrows needed stabilization from substrate binding at the second site. It indicates pathways through the dimer interface that contribute to allostery relevant to half-sites reactivity.
Thymidylate synthase (TS) (EC 2.1.1.45), is a critical enzyme in the sole de novo biosynthetic pathway of thymidine, a nucleic acid essential for DNA synthesis, which converts 2’-deoxyuridine 5’-monophosphate (dUMP) and 5,10-methylene-5,6,7,8-tetrahydrofolate (mTHF) to thymidine 5’-monophosphate (dTMP) and dihydrofolate (DHF). TS is also one of the most conserved enzymes, a testament to its essential function in biology, with a conserved mechanism that uses proton tunneling to transfer hydrogen in the rate-determining step1. It is an obligate two-fold symmetric dimer in which residues from both protomers contribute to each substrate-binding site2. E. coli TS is a half-the-sites reactive enzyme3, 4, yet shows only minor cooperativity in binding substrate and cofactor5, 6. This suggests energy is transferred between subunits to favor the ligand alignment and dynamics required for the critical hydride transfer step at one of the active sites over the second. Understanding the nature of this communication between protomers requires a crystal structure of an asymmetric state with one site primed. However since EcTS binds ligands to its two active sites with similar affinity most crystal structures represent two-fold symmetric, stable intermediates along the reaction path7.
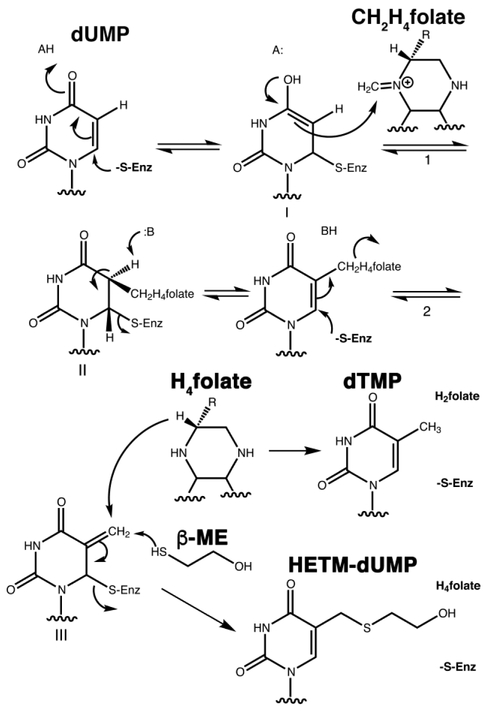
Scheme 1 shows the currently accepted mechanism8. The cofactor, mTHF, donates a methyl group to C5 of dUMP to produce covalent intermediate II (Scheme 1, step 1). An analog of this intermediate, TS•5-fluoro-dUMP•CH2H4folate, is stable and its two-fold symmetric structure has been determined by X-ray crystallography9, 10. This structure and structures of other analogs of II show that extensive conformational changes accompany formation of ternary complexes7. These serve to close down the active site, and sequester the reactants from solvent.
Scheme 1.
Thymidylate synthase mechanism
Elimination of H4folate from II gives exocyclic methylene intermediate III and THF (Scheme 1, step 2). The rate-determining step in the thymidylate synthase reaction is hydride transfer of H6 from the cofactor to intermediate III11. Hydride transfer occurs by tunneling, and prior to hydride transfer, protein conformational changes are required to preorganize the active site into a reactive complex that can access the tunnel-ready state through nanosecond to picosecond fluctuations of the protein12. Mutations that alter the structure or stability of this reactive complex impair hydride transfer. A destabilized intermediate III can admit small thiols to the active site, which compete with hydride for addition to the exocyclic methylene intermediate, producing aberrant products such as HETM-dUMP (Scheme 1)12–14. Thus small thiols have been used as probes of the stability of intermediate III in E. coli and L. casei thymidylate synthase mutants12, 15. Formation of thiol adducts by mutant TSs indicates the mutants are competent to undergo the initial steps in the reaction up to formation of intermediate III, but cannot efficiently undergo hydride transfer because the reactive conformation of III is destabilized.
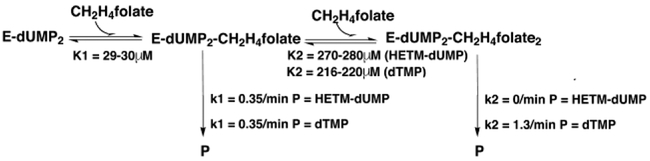
One of the more dramatic conformational changes during TS ternary complex formation is the shift of the C-terminus into the active site where it forms hydrogen bonds with residues on the opposite wall of the active site cavity, thereby ‘sealing’ the cavity shut. Variath et al.15 assessed the importance of the C-terminus to the integrity of the active site by measuring the partitioning of intermediate III between dTMP and HETM-dUMP under saturating dUMP conditions for three L. casei TS C-terminal mutants and the E. coli TS mutant I264Am, which has one-amino acid at the C-terminal deleted. Surprisingly, the partition coefficient varied as a function of mTHF concentration, even though the two products emanated from the same ternary complex intermediate, III.
This result could be explained by cooperativity between the active sites. Briefly, binding of a single molecule of mTHF to the dUMP-saturated enzyme leads to a higher KmmTHF for the other site. Under increasing mTHF concentrations, a second molecule of mTHF binds and changes the relative rates of product and HETM-dUMP production in one or both active sites.15 (Scheme 2). In the case of all four mutants tested, as cofactor concentration increased, proportionately more dTMP was produced. This implies that binding a second molecule of cofactor to a complex that already has one molecule of cofactor bound stabilizes the reactive conformation of the enzyme at one or both active sites of the dimer.
Scheme 2.
Kinetics for E. coli TS I264Am from Variath et al.15
Changes in partitioning between dTMP and HETM-dUMP after a second molecule of cofactor bound to the enzyme were most pronounced for E. coli TS I264Am, the mutant in which the C-terminal carboxyl and the residue that presents it for closing the active site, were deleted. With two molecules of dUMP and one molecule of cofactor bound, I264Am produced approximately equal amounts of dTMP and HETM-dUMP, indicating reduced stability of the reactive complex. Binding a second molecule of cofactor increased the rate of dTMP production 4-fold to ~.25% of wild type kcat12, and completely eliminated production of HETM-dUMP. To understand the structural basis for this behavior we determined the structure of I264Am with dUMP and an mTHF analog, CB3717, bound at both active sites.
One possible explanation consistent with the kinetics is that binding the second molecule of mTHF stabilizes intermediate III at both active sites, preventing HETM-dUMP from forming. However our structure suggests a different model. The crystal structure reveals an asymmetric dimer in which one protomer does not attain a conformation required for formation of intermediate III, thus during the reaction neither product nor HETM-dUMP would be produced at this site. The other site is well ordered and resembles wild type EcTS ternary complexes; during the reaction it would be expected to produce only the product, dTMP. The structure is consistent with the kinetic results and contributes to an understanding of how cofactor binding to one active site can better align ligands in the other for proton tunneling between cofactor and substrate methylene group.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification and Crystallization.
E. coli TS I264Am was prepared, expressed and purified as described previously15. The protein was dialyzed against 20 mM potassium phosphate (pH = 7.5) containing 1 mM EDTA and 2 mM DTT before use. The protein-ligand solution was prepared by mixing the I264Am (6.5 mg/mL) with 2 mM dUMP, 2 mM CB3717 and 10 mM DTT. Crystals were grown using the hanging-drop method combined with micro seeding in 4-μL hanging-drops with a 1:1 ratio of protein solution and the crystallization solution. The crystallization solution contains 100 mM HEPES (pH 8.0 – 8.3), 200 mM NH4Ac, 5 mM DTT, and 32% PEG 4000 (w/v). In order to prepare the crystal seeds, 5 mM MgCl2 was added to the protein-ligand solution. The day-old seeds were transferred to new drops (prepared in absence of MgCl2) and grew another day to reach an optimal shape.
X-Ray Diffraction Data Collection.
Data were collected at beamline 9–1 at the Stanford Synchrotron Radiation Laboratory under cryo conditions. The crystal was transferred from the drop directly to the liquid nitrogen steam without treating with additional cryoprotectant. Data to 2.4Å resolution were collected on a MAR detector with 2° oscillation scans per frame. Data indexing, integration, scaling, and merging were performed using the programs DENZO and SCALEPACK 16.
Structure Solution and Refinement.
The structure was determined by molecular replacement using the program AMoRe 17 using the dimer of the wild type E. coli thymidylate synthase in complex with dUMP and CB3717 18 with ligands and C-terminal residues omitted, as the search model. dUMP and CB3717 were modeled into the structure using Quanta (Accelrys Inc., San Diego, CA). Repeated cycles of conjugate gradient minimization in CNS 19 and manual rebuilding using Quanta were performed followed by automatic water-picking and individual B-factor refinement using CNS. Hydrogen atoms were added to the structure in riding positions before final rounds of refinement against a maximum likelihood target using phenix.refine 20. The final model from the 2.4Å data has a crystallographic R factor of 18% and a free R factor of 23%. The crystallographic and refinement data are shown in Table 1. The coordinates and structure factors have been deposited in the Protein Data Bank with accession code 6CDZ.
Table 1.
Data collection and refinement statistics.
| E. coli TS I264Am, PDB 6CDZ | |
|---|---|
| Wavelength (Å) | 0.9700 |
| Resolution range (Å) | 30.0 - 2.4 (2.44 - 2.4) |
| Space group | C 1 2 1 |
| Unit cell (Å,°) | 67.7 80.4 94.1 90 103.5 90 |
| Total reflections | 55027 |
| Data Cutoff (σ) | −3.0 |
| Unique reflections | 16256 (720)1 |
| Multiplicity | 3.4 (3.3) |
| Completeness (%) | 84.3 (76.4) |
| Mean I/sigma(I) | 10.5 (2.2) |
| Wilson B-factor (Å2) | 28.40 |
| R-merge (%) | 11.1 (56.3) |
| Reflections used in refinement | 16237 |
| Reflections used for R-free | 853 |
| R-work | 0.18 (0.26) |
| R-free | 0.23 (0.29) |
| Number of non-hydrogen atoms | 4595 |
| macromolecules | 4340 |
| ligands | 90 |
| Protein residues | 526 |
| RMS(bonds) (Å) | 0.003 |
| RMS(angles) (°) | 0.56 |
| Ramachandran favored (%) | 97 |
| Ramachandran allowed (%) | 3 |
| Ramachandran outliers (%) | 0.38 |
| Rotamer outliers (%) | 1.1 |
| Clash score | 2.43 |
| Average B-factor (Å2) | 34.2 |
| Macromolecules (protomers A,B) | 32.0, 36.3 |
| Ligands (protomers A,B) | 36.9, 38.6 |
| solvent | 32.87 |
Statistics for the highest-resolution shell are shown in parentheses.
RESULTS and DISCUSSION
Deletion of the C-terminal COO- breaks the symmetry of the obligate dimer.
The I264Am-dUMP-CB3717 complex crystallized in space group C2 with a dimer in the asymmetric unit. The two protomers (chains A and B) have similar overall conformations. Active sites in both protomers contain one molecule of substrate dUMP and one molecule of cofactor analog CB3717. However, the active site of one of the protomers (chain A) is more open than that of the other (chain B) and the CB3717 and substrate dUMP in the open active site A, adapting to the shifted positions of the ligand binding residues, are shifted away from the catalytic cysteine, Cys146. Crystallization of this asymmetric structure indicates that in solution the asymmetry is homogeneous.
The asymmetry produces a state in which only one active site adopts an arrangement compatible with catalysis.
In order to understand the conformational changes relating the two active sites we identified structural domains conserved between the two protomers using difference distance matrices, as implemented in the program Rapido 21, 22. There is a structural core of ~125 contiguous residues whose inter-Cα distances are the same +/− 0.5Å in the two protomers and a smaller conserved domain of 68 residues. The relative positions of these two domains in the two protomers are related by a rigid body motion, which opens and closes the active site cavity. Loops bordering the active site cavity are not part of conserved domains and their conformations are significantly different between the two protomers. The conformational changes relating protomer A to active-state protomer B resemble the conformational changes wild type E. coli TS undergoes on going from the open, binary dUMP complex to the closed ternary complex with substrate and cofactor 23.
The large conserved core of the I264Am protomers, which comprises most of the central β-sheet and helices that pack against the sheet, is also conserved in wild type TS binary and ternary complexes, allowing us to compare the conformations of the I264Am protomers to the conformations of these wild type EcTS complexes. After alignment of the conserved cores of each I264Am protomer with their wild type counterparts, the rmsds for Cαs were calculated and are reported in Table 2. From these values, it is clear that the structure of I264Am chain A most closely resembles the structure of the wild type E. coli TS-dUMP binary complex, which has an apo enzyme-like conformation, while chain B is more similar to the subsequently formed active-like state seen in wild type E. coli TS ternary complexes. In summary, even though I264Am lacks the C-terminal residue, which is thought to be critical for closing the active site in the ligand-bound enzyme, it is able to attain the same closed, conformation seen in wild type ternary complexes, but in only protomer B. Thus by slightly destabilizing the energetics, deletion of the C-terminal carboxyl allowed us to capture an asymmetric state of EcTS in which only one protomer appears to be competent for catalysis.
Table 2.
RMSDs (Å) for Protomers of I264Am and Wild type Complexes.
| Complex, chain | WT-dUMP- CB3717, A |
WT-dUMP- CB3717, B |
WT-dUMP (A=B) |
I264Am-dUMP- CB3717, B |
|---|---|---|---|---|
| I264Am-dUMP-CB3717, A | 0.77 | 0.79 | 0.47 | 0.63 |
| I264Am-dUMP-CB3717, B | 0.39 | 0.43 | 0.68 | – |
Active Site B Resembles the Wild Type Ternary Complex.
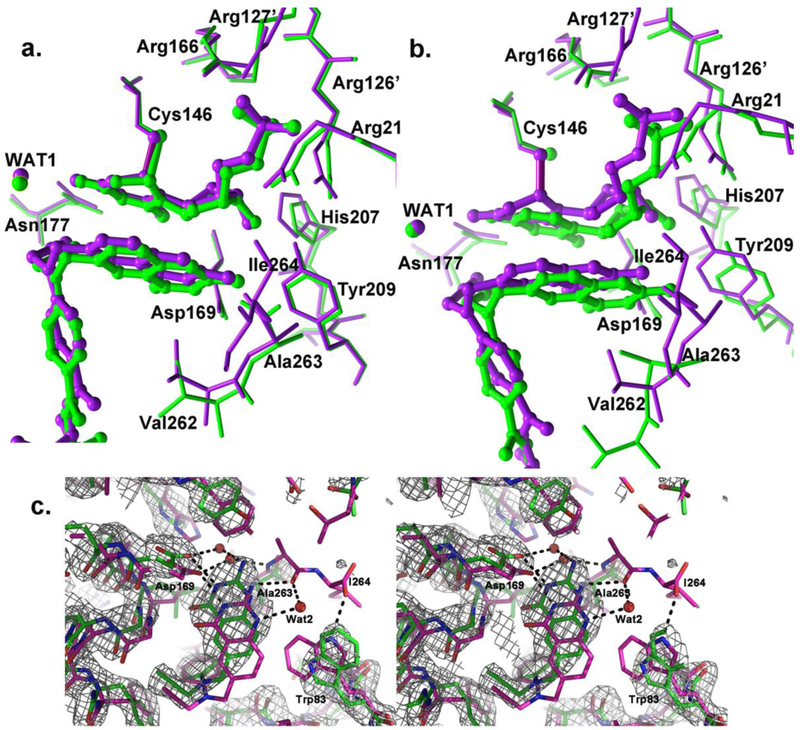
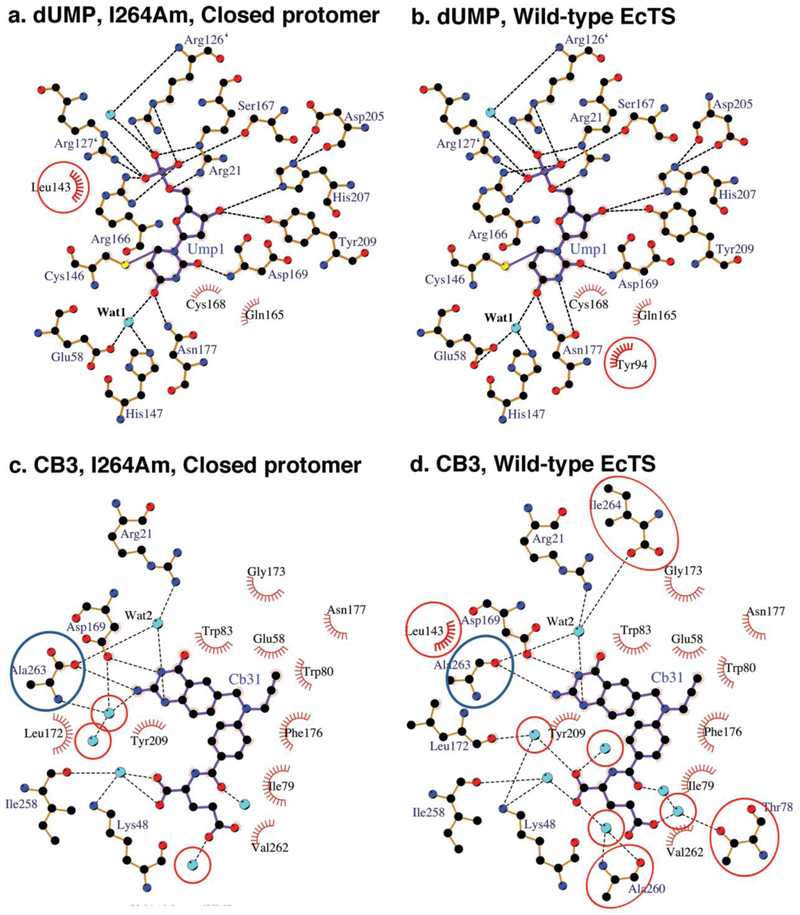
Ligand-binding modes in protomer B are very similar to those in fully loaded substrate- and cofactor-analog-bound, wild type ecTS-dUMP-CB3717 (Figures 1a, 2a, 3). The substrate dUMP is covalently bound to the catalytic Cys146, as in the reaction intermediates and as seen in the wild type complex, and hydrogen bond interactions between dUMP and protein residues are also conserved (Figure 1a, 3a, Table S1). After alignment of the I264Am protomer and wild type protomer structures, the rmsd for dUMP is only 0.42 Å, which is less than twice the 0.28Å coordinate error estimated in PHENIX by maximum likelihood methods24 (Figure 1a).
Fig. 1.
Comparison of ligand binding in the two protomers of wild type E. coli TS-dUMP-CB3717 (purple) and I264Am-dUMP-CB3717 (green) highlighting the shift of CB3717 in the open protomer of I264Am. a.) Comparison to closed protomer (chain B). Ligands dUMP and CB3717 (shown in ball-and-stick rendition) and residues that make hydrogen bonds to these ligands (shown as sticks, and labeled) in wild type EcTS are overlaid with those from protomer B of I264Am. b.) Same type of plot as in (a.) comparing active sites of wild type EcTS and the open protomer (chain A). c.) Overlay of CB3717 and interacting residues in wild type EcTS-dUMP-CB3717 with those of the open protomer of the I264Am structure, viewed looking into the face of the quinazoline ring and shown in divergent-eyes stereo. Dashed lines depict hydrogen bonds. Atoms are color-coded: carbon, green for I264Am, purple for wild type; oxygen, red; nitrogen, blue. Water molecules are shown as red spheres. Likelihood-weighted 2Fo-Fc density for the I264Am complex contoured at 1.2 is shown
Fig. 2.
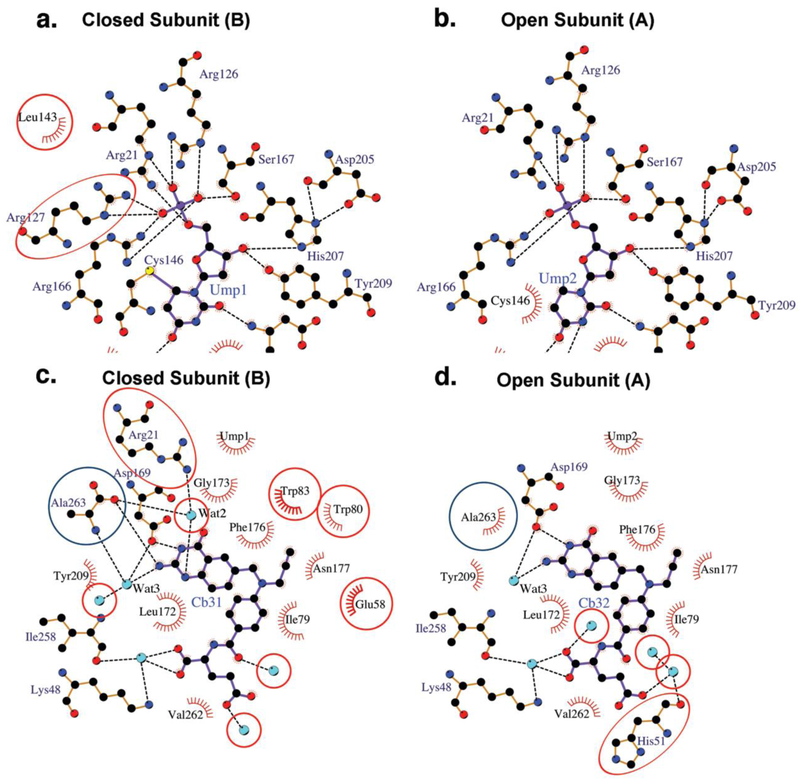
Schematic drawings showing hydrogen bond and hydrophobic interactions to dUMP and CB3717 in the two active sites of the I264Am ternary complex. Dashed lines indicate hydrogen bonds and the eyelash motifs indicate hydrophobic interactions. Cyan spheres are water molecules. Atoms are colored by atom type: carbon, black; oxygen, red; nitrogen, blue. a. and b.) dUMP in the closed protomer and open protomer, respectively. Interactions not conserved between the two protomers are circled. c. and d.) CB3717 in the closed protomer and open protomer, respectively. Interactions not conserved between the two protomers are circled. Ala263, the C-terminal residue of I264Am, is circled in blue.
Fig. 3.
Schematic drawings showing hydrogen bond and hydrophobic interactions to dUMP and CB3717 in the active sites of the I264Am and wild type ternary complexes. Dashed lines indicate hydrogen bonds and the eyelash motifs indicate hydrophobic interactions. Cyan spheres are water molecules. Atoms are colored by atom type: carbon, black; oxygen, red; nitrogen, blue. a. and b.) dUMP in the closed protomer of I264Am and the wild type EcTS ternary complex, respectively. Interactions not conserved between the two protomers are circled. c. and d.) CB3717 in the closed protomer of I264Am and wild type EcTS ternary complex, respectively. Interactions not conserved between the two protomers are circled. Ala263, the C-terminal residue in I264Am, and the penultimate residue in wild type thymidylate synthase, is circled in blue.
CB3717, an inhibitor designed to mimic the cofactor but to trap the enzyme before the chemical steps of catalysis, is mostly unchanged from its position in the wild type complex, with a slight shift away from dUMP (0.3 Å for the quinazoline ring and 0.7 Å for the PABA-Glu moiety) (Figure 1a). After alignment of the mutant and wild type structures, the overall rmsd for CB3717 is 0.63Å, and the rmsd for the quinazoline ring alone is 0.3 Å, approximately the same as the estimated coordinate error. Interactions between CB3717 and the protein are largely the same in site B as in wild type ecTS-dUMP-CB3717 (Figure 3, Table S2). However, the lack of the carboxy terminal residue in the I264Am mutant abolishes several hydrogen bonds that stabilize the closed complex. In the wild type complex, the terminal carboxylate of Ile264 forms a water-mediated hydrogen bond to N1 of CB3717 via conserved water Wat2. It also accepts hydrogen bonds from two key ligand-binding residues on either side of the active site cavity: Arg21, which interacts with the phosphate group of dUMP, and Trp83, which packs against the quinazoline ring of CB3717. The network of hydrogen bonds to the C-terminus helps to seal closed the active site cavity in the wild type ternary complex. The lack of these interactions presumably destabilizes the complex just enough to allow determination of a substrate-induced conformation showing the enzyme en route between singly-bound state and the fully-bound symmetric state.
Ligands in Active Site A presage the conformation aligned for catalysis.
The structure of active site A mimics the first intermediate in the multistep reaction, the dUMP-bound enzyme. It deviates significantly from the wild type EcTS-dUMP-CB3717 active site, which closely indicates the ‘closed’ state around substrate and cofactor poised for catalysis (Figure 1b,c). The rmsds of the ligands between the wild type and I164Am site A complexes are 0.8 Å for dUMP and 1.4 Å for CB3717, which are 3 and 5 times, respectively, the estimated coordinate error of the structure (Figure 1b,c). The active site residues are shifted by an average of 1.0 Å (main chain) from their positions in wild type EcTS-dUMP-CB3717, underlining a dramatic difference in state, which is mirrored in the half-sites reactive kinetics.
The pyrimidine ring of dUMP is planar and there is no covalent bond between Cys146-Sγ and dUMP as there would be after the cofactor-binding step of the normal reaction. Cys146 adopts a different rotamer conformation that places Cys146-Sγ 3.5 Å from dUMP-C6. Although dUMP is shifted from a productive binding mode, it engages in almost all of the protein-dUMP hydrogen bond interactions seen in the wild type complex (Figure 2b, Table S1). Only the hydrogen bonds between Arg127’ (a “ ‘ “ following a residue indicates the residue is in the opposite protomer) and the dUMP phosphate moiety are absent, because the Arg127’ guanidinium group has rotated out of the phosphate-binding site. Importantly, a water molecule that links catalytic acid Glu58 to dUMP O4 in the wild type complex is absent. We propose this water has a catalytic role of proton transfer and is present in both the wild type ternary complex active sites and in site B of the I264Am complex25.
CB3717 in site A exhibits the largest conformational change among the ligands in both active sites. The quinazoline ring shifts by 1.2 Å towards the space that is occupied by the C-terminal residues in the wild type active site (Figures 1b,c). One of the segments that shifts to close the active site in wild type EcTS ternary complexes contains Asp169, which is the only residue whose side chain directly hydrogen bonds to the cofactor. In protomer A, Asp169 Oδ2 accepts a hydrogen bond from N1 of the quinazoline ring of CB3717, as in wild type EcTS-dUMP-CB3717, but since Asp169 has not shifted towards the active site, CB3717 is displaced from its binding site, and out of alignment with dUMP (Figure 1b,c). The dihedral angle between the quinazoline and PABA rings is changed from 61° in the wild type active site and 62° in site B to 70° in I264Am site A.
As for the protein, the most prominent difference between site A and the wild type site (and site B) is the conformation of the C-terminus. Besides lacking Ile264, the C-terminal residues move away from the active site and Ala263 does not make direct or water-mediated hydrogen bonds to the quinazoline ring of the cofactor analog. Wat2, which coordinates a hydrogen bond network that stabilizes the closed conformation of the wild type ternary complex, is absent in site A. The absence of Ile264 and Wat2 and the movement of Ala263 in site A abolish five hydrogen bonds that were present in the wild type complex (Figure 2d, Table S2).
The displacement of CB3717 in active site A also changes the pattern of van der Waals interactions with protein residues, particularly for the quinazoline moiety (Table S3). The quinazoline ring and two tryptophans in the cofactor-binding site, Trp80 and Trp83, move away from each other and Trp83 adopts a different rotamer than in the wild type ternary complex (Figure 1c). These changes abolish the hydrophobic interactions between CB3717 and these two residues. In contrast, the movement of the quinazoline ring towards the C-terminus in site A allows new contacts with Ala263 and Asp169. Phe176 also adopts a rotamer conformation in site A that is different from that in site B and in the wild type ternary complex active site. As a result, instead of interacting with the PABA ring, Phe176 packs against the propargyl moiety in site A.
The average overall B-factor for protomer A is lower than for protomer B (Table 1, Figure S1). However, the two protein segments that show the most dramatic movement upon formation of EcTS ternary complexes with dUMP and cofactor analogs, namely the phosphate-binding loop and C-terminus, have approximately 20% higher B-factors (~60Å3) than the same segments in protomer B (Figure S1). The high thermal motion of these segments reflects the fact that they are not fully integrated into the hydrogen bond network that normally stabilizes ternary complexes of EcTS.
In summary, in our asymmetric structure one active site closely resembles the wild type catalytically productive complex, except for absence of the C-terminus, consistent with it producing dTMP exclusively in the face of competition by a probe (β-ME) of its instability. The second active site is not in a closed conformation, as would be required for activity, and the cofactor analog CB3717 is shifted away from the active site. This explains its lack of reactivity with the stability probe. A cofactor molecule in the same position would likely not proceed through the steps required to make the exocyclic methylene intermediate III, thus no HETM-dUMP or dTMP would be produced at this site. The structure is in line with current thinking about the TS structural mechanism: the closed conformation of the active site is required for proper alignment of ligands, and precise alignment of ligands is in turn critical for the hydride transfer7, 12, Of note is that even the slight 0.3-Å shift of CB3717 in protomer B is associated with a kcat that is 400-fold less than the wild type EcTS kcat determined by steady-state kinetics. The structural results are different than originally predicted by Variath et al., since it was proposed that both HETM-dUMP and dTMP would be produced at both active sites, albeit at different rates15. Based on the structure, we suggest the rates at one of the active sites are essentially zero, which is still consistent with the kinetic results.
The asymmetric dimer provides clues to the communication pathways between active sites in wild type TS.
The dissociation constant, K1, for binding mTHF to the I264Am-dUMP2 binary complex is 30 μM while there is an almost 10-fold higher dissociation constant for binding a second molecule of mTHF to the complex (K2= 220mM) (Scheme 2). We sought to understand how energy of binding one mTHF molecule transferred across the dimer interface to affect binding in the opposite protomer.
There is evidence from NMR spectroscopy that structural changes resulting from substrate binding propagate to and across the dimer interface in wild type EcTS as well 5, 26. During titration of EcTS with the mechanism-based di-ligand inhibitor 5-fluoro-dUMP•mTHF, resonances not present in symmetric apo or doubly bound EcTS for several residues at or near the dimer interface could be assigned to a singly bound state5. Further evidence is that enthalpies of cofactor analog binding to the C146S mutant of EcTS, as measured by isothermal titration calorimetry, were different between apo and singly bound enzyme6. However, the cofactor-induced changes to the dimer interface had only a small effect on binding affinity for the second molecule of cofactor analog, explaining why crystal structures of wild type EcTS ternary complexes are two-fold symmetric6.
We analyzed the dimer interface of I264Am to identify regions of asymmetry that might explain the asymmetric kinetic constants. The conformational changes that close the active site of EcTS upon ternary complex formation do not propagate to a significant extent to the central β-sheet that comprises the main part of the dimer interface. Indeed, most of the central β-sheet comprises the structural core conserved in EcTS complexes. Thus, the dimer interface of the I264Am ternary complex is approximately two-fold symmetric even though the dimer as a whole is asymmetric. However there are two regions in the interface where symmetry breaks down and which may bear on the large difference between K1 and K2 (Scheme 2)15.
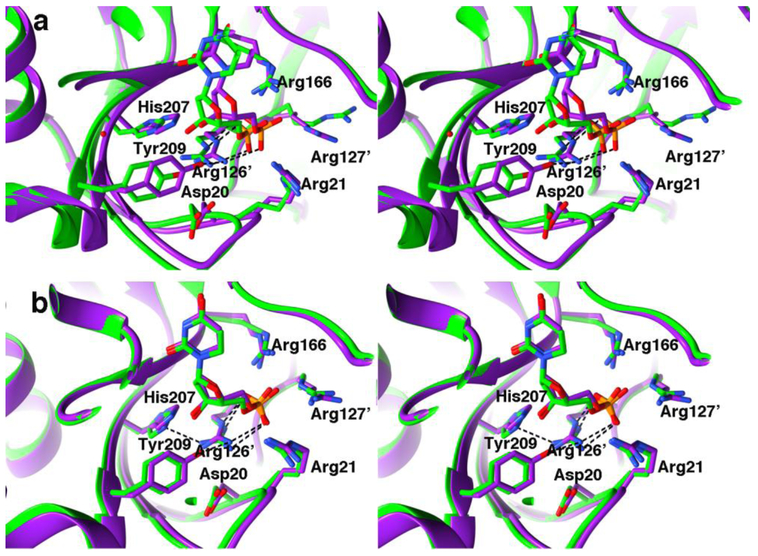
First, the phosphate-moiety of dUMP binds at an edge of the dimer interface where arginine side chains from each protomer contribute to phosphate binding. The dimer interface here is made up of the conserved, phosphate-binding, arginine-containing loops that are connected to the β-strands of the central β-sheet together with β-sheet residues His207 and Tyr209, which make hydrogen bonds to the dUMP ribose hydroxyl group. The dimer interface at the phosphate-binding sites of the I264Am complex is asymmetric, reflecting the different binding modes of dUMP in sites A and B (Figure 4).
Fig. 4.
Divergent-eyes stereo cartoon rendition of the dimer interface at the two phosphate-binding sites in the asymmetric I264Am-dUMP-CB3717 dimer. The phosphate-binding site is comprised of four arginines, two from each protomer, whose side chains are plotted along with the other major interface residues at the phosphate site. The two arginines contributed from the opposite protomer to that whose active site is shown have primes following their residue number (Arg126’, and Arg127’). I264Am is shown with green carbons. For comparison, the symmetric wild type ternary complex has been superimposed and its two protomers shown in purple cartoon rendition, with same side chains as were plotted for I264Am also shown. a) Protomer A (open protomer) b) Protomer B (closed protomer)
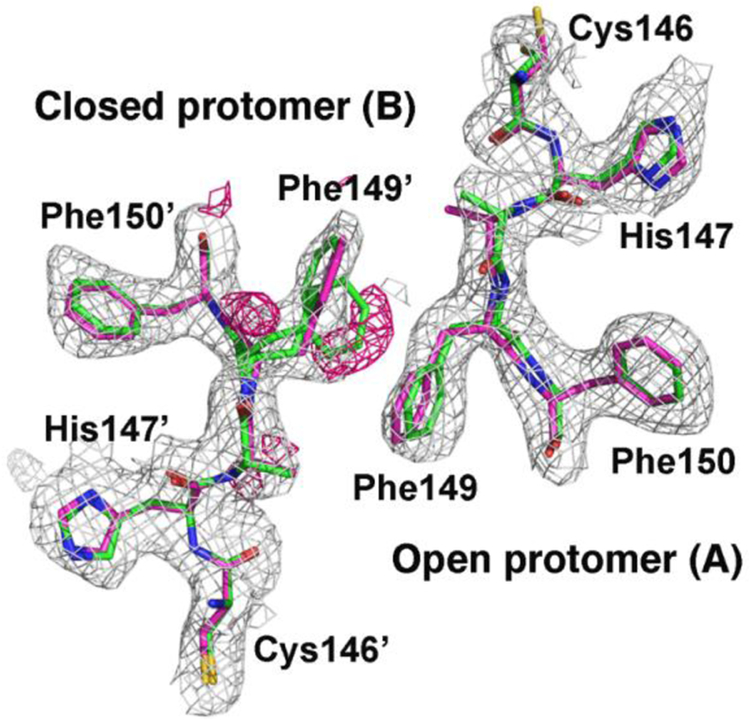
Secondly, there is a minor conformational rearrangement of interface residues Cys146-Phe150 from site A, induced by covalent bond formation between the catalytic Cys and dUMP in site B, which breaks the two-fold symmetry near the catalytic Cys (Figure 5). Cys146 adopts a different rotamer that is not compatible with nucleophilic attack of the catalytic sulfhydryl on C6 of dUMP. Phe149’ from site B is statistically disordered, with two half-occupied side chain rotamers, one of which protrudes into the dimer interface. Statistical disorder of Phe149 is not seen in the wild type EcTS complexes. Thus an increase in conformational entropy near the active site is unique to the asymmetric complex. Similar but more exaggerated structural differences near the active site cysteine were observed in the asymmetric ternary complex of Pneumocystis carinii TS, which had CB3717 bound in only one active site, and the differences were proposed to be responsible for the half-the-sites reactivity of the enzyme27. NMR spectroscopy of EcTS heterodimers with inactivating mutations at one active site revealed that upon binding a single dUMP molecule, subtle structural changes occurred in these same interface residues26. This result suggests the residues may constitute a common pathway for communicating between active sites in EcTS, its C-terminal mutants, and other TS species.
Fig. 5.
Residues 146–150 (chain A, open protomer) and 146’−150’ (chain B, closed protomer) from the I264Am ternary complex dimer interface are shown overlaid with likelihood-weighted 2Fo-Fc (grey contours) and Fo-Fc (pink contours) density maps. The Fo-Fc difference map shows positive peaks corresponding to a second conformation for the side chain Phe149’. Residues are colored by atom type: carbon, green (I264Am) or magenta (wild type); oxygen, red; nitrogen, blue; sulfur, yellow.
An asymmetric interface in fully liganded I264Am may be harnessed to increase product formation by one protomer at the expense of product formation by the other.
Overall the interaction free energy,iG, for dimer formation in the asymmetric structure, estimated from changes in contacts and solvent accessibility using the program PISA28, indicates the interface is ~1.5 kcal/mol more stable than for the symmetric apo or doubly bound enzymes. Interface residues at the phosphate-binding sites contribute a large proportion of the difference in ΔiG between I264Am and wild type EcTS ternary complexes. Thus we propose there is a small penalty for closure of the second site, which would be compensated for by cofactor - protein contacts in the wild type enzyme but not in I264Am, where the C-terminus is absent. Mutation of the key cofactor-binding residue Asp169 in E. coli TS led to a crystal structure of an asymmetric dimer of E. coli D169C-dUMP-CB3717, in which one protomer was in an open conformation with misaligned substrates29. This result corroborates the idea that energy required to close the second protomer in ternary complexes of E. coli TS is sensitively balanced by the interaction energy of the cofactor with the protein.
Alternatively, binding at the incompletely closed site A can be realized as augmenting substrate orientation for hydride transfer at the competent site B. For example, hydrogen bond between Arg126’ Nη1 and His207 Nε2 in protomer B may help to stabilize the closed conformation of the active site and lead to the more favorable ΔiG for this arginine than for Arg126’ at active site A or at the active sites of wild type EcTS complexes (Figure 4b). This interaction, which is not present in I264Am protomer A or the wild type EcTS-dUMP-CB3717 ternary complex, may result from a small shift of His207 in protomer B upon CB3717 binding to protomer A. The asymmetric doubly bound I264Am species represented by our structure must be different in structural detail and/or conformational order than the singly bound intermediate, since the latter species has a less stable reactive conformation, leading to a 4-fold lower rate of dTMP formation and partitioning of intermediate III to HETM-dUMP15.
The ensemble of conformational states in the ternary complex of wild type EcTS with dUMP and cofactor bound to both protomers, is shifted relative to that of the I264Am ternary complex reported here toward a more two-fold symmetric closed state, as seen in crystal structures of analogs of this complex, and this is associated with a ~400-fold higher kcat and 10-fold-lower KmTHF for the wild type enzyme12 compared to I264Am. Nevertheless, the same principle of one active site recruiting binding energy from the second to optimize the tunnel-ready state needed for hydride transfer may pertain. Optimization of one active site at the expense of the second would explain the half-the-sites activity of wild type TS. Since hydride transfer is exquisitely sensitive to ligand alignment as well as protein vibrational modes12, even subtle differences in structure and dynamics between the two protomers could result in large differences in activity.
CONCLUSION
The C-terminal carboxyl of TS has an important role in stabilizing a closed conformation of the enzyme active site during catalysis. When it is deleted in either E. coli or L. casei TS, the closed conformation is destabilized, allowing β-ME to enter the active site and trap 5-methylene-dUMP (reaction intermediate III, Scheme 1) as HETM-dUMP. Our I264Am structure shows that in E. coli TS, the catalytically competent conformation of one active site is stabilized even in the absence of the C-terminus when cofactor is bound in the other active site, allowing the product, dTMP, to form exclusively, at just that site. The asymmetry of the structure suggests that products can be formed at only the site with the catalytically competent conformation. Structural changes at the dimer interface due to closure of the productive active site prevent the ligands in the other site from properly aligning to form exocyclic methylene-substituted dUMP, a common intermediate for formation of both dTMP and HETM-dUMP. At the same time, binding at the non-productive site, can transfer energy to the competent site to augment the probability of reaching a catalytically active state, capable of proton tunneling.
Supplementary Material
Acknowledgments
† This work was supported by National Institutes of Health Grant R01 CA41323 to J. F.-M. and NIH Grants R01 CA63081 and R01 GM024485 to R.M.S.
ABBREVIATIONS AND TEXTUAL FOOTNOTES
- TS
thymidylate synthase
- EcTS
E. coli thymidylate synthase
- dUMP
2’-deoxyuridine 5’-monophosphate
- dTMP
thymidine 5’-monophosphate
- mTHF
5,10-methylene-5,6,7,8-tetrahydrofolate
- THF
5,6,7,8-tetrahydrofolate
- DHF
7,8-dihydrofolate
- DTT
dithiothreitol
- β-ME
β-mercaptoethanol
- HETM-dUMP
5-(2-hydroxyethyl)thiomethyl-dUMP
- CB3717 or CB3
10-propargyl-5,8-dideazafolate
- I264Am
E. coli TS with the C-terminal residue removed
Footnotes
REFERENCES
- [1].Agrawal N, Hong B, Mihai C, and Kohen A (2004) Vibrationally enhanced hydrogen tunneling in the Escherichia coli thymidylate synthase catalyzed reaction, Biochemistry 43, 1998–2006. [DOI] [PubMed] [Google Scholar]
- [2].Hardy LW, Finer-Moore JS, Montfort WR, Jones MO, Santi DV, and Stroud RM (1987) Atomic structure of thymidylate synthase: target for rational drug design, Science 235, 448–455. [DOI] [PubMed] [Google Scholar]
- [3].Maley F, Pedersen-Lane J, and Changchien L (1995) Complete restoration of activity to inactive mutants of Escherichia coli thymidylate synthase: evidence that E. coli thymidylate synthase is a half-the-sites activity enzyme, Biochemistry 34, 1469–1474. [DOI] [PubMed] [Google Scholar]
- [4].Johnson EF, Hinz W, Atreya CE, Maley F, and Anderson KS (2002) Mechanistic characterization of Toxoplasma gondii thymidylate synthase (TS-DHFR)-dihydrofolate reductase. Evidence for a TS intermediate and TS half-sites reactivity, The Journal of biological chemistry 277, 43126–43136. [DOI] [PubMed] [Google Scholar]
- [5].Sapienza PJ, Falk BT, and Lee AL (2015) Bacterial Thymidylate Synthase Binds Two Molecules of Substrate and Cofactor without Cooperativity, Journal of the American Chemical Society 137, 14260–14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sapienza PJ, and Lee AL (2016) Widespread Perturbation of Function, Structure, and Dynamics by a Conservative Single-Atom Substitution in Thymidylate Synthase, Biochemistry 55, 5702–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stroud RM, and Finer-Moore JS (2003) Conformational dynamics along an enzymatic reaction pathway: thymidylate synthase, “the movie”, Biochemistry 42, 239–247. [DOI] [PubMed] [Google Scholar]
- [8].Kholodar SA, and Kohen A (2016) Noncovalent Intermediate of Thymidylate Synthase: Fact or Fiction?, J Am Chem Soc 138, 8056–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matthews DA, Villafranca JE, Janson CA, Smith WW, Welsh K, and Freer S (1990) Stereochemical mechanism of action for thymidylate synthase based on the X-ray structure of the covalent inhibitory ternary complex with 5-fluoro-2’-deoxyuridylate and 5,10-methylenetetrahydrofolate, J Mol Biol 214, 937–948. [DOI] [PubMed] [Google Scholar]
- [10].Hyatt DC, Maley F, and Montfort WR (1997) Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate, Biochemistry 36, 4585–4594. [DOI] [PubMed] [Google Scholar]
- [11].Spencer HT, Villafranca JE, and Appleman JR (1997) Kinetic scheme for thymidylate synthase from Escherichia coli: determination from measurements of ligand binding, primary and secondary isotope effects, and pre-steady-state catalysis, Biochemistry 36, 4212–4222. [DOI] [PubMed] [Google Scholar]
- [12].Wang Z, Abeysinghe T, Finer-Moore JS, Stroud RM, and Kohen A (2012) A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase, J Am Chem Soc 134, 17722–17730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Barrett JE, Maltby DA, Santi DV, and Schultz PG (1998) Trapping of the C5 methylene intermediate in thymidylate synthase, J Am Chem Soc 120, 449–450. [Google Scholar]
- [14].Fritz TA, Liu L, Finer-Moore JS, and Stroud RM (2002) Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access, Biochemistry 41, 7021–7029. [DOI] [PubMed] [Google Scholar]
- [15].Variath P, Liu Y, Lee TT, Stroud RM, and Santi DV (2000) Effects of subunit occupancy on partitioning of an intermediate in thymidylate synthase mutants, Biochemistry 39, 2429–2435. [DOI] [PubMed] [Google Scholar]
- [16].Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode, Macromolecular Crystallography, Pt A 276, 307–326. [DOI] [PubMed] [Google Scholar]
- [17].Navaza J (1994) Amore - an Automated Package for Molecular Replacement, Acta Crystallogr A 50, 157–163. [Google Scholar]
- [18].Stout TJ, Sage CR, and Stroud RM (1998) The additivity of substrate fragments in enzyme-ligand binding, Structure 6, 839–848. [DOI] [PubMed] [Google Scholar]
- [19].Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, and Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination, Acta crystallographica. Section D, Biological crystallography 54, 905–921. [DOI] [PubMed] [Google Scholar]
- [20].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta crystallographica. Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mosca R, Brannetti B, and Schneider TR (2008) Alignment of protein structures in the presence of domain motions, BMC bioinformatics 9, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mosca R, and Schneider TR (2008) RAPIDO: a web server for the alignment of protein structures in the presence of conformational changes, Nucleic acids research 36, W42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Montfort WR, Perry KM, Fauman EB, Finer-Moore JS, Maley GF, Hardy L, Maley F, and Stroud RM (1990) Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate, Biochemistry 29, 6964–6977. [DOI] [PubMed] [Google Scholar]
- [24].Lunin VY, Afonine PV, and Urzhumtsev AG (2002) Likelihood-based refinement. I. Irremovable model errors, Acta Crystallogr A 58, 270–282. [DOI] [PubMed] [Google Scholar]
- [25].Sage CR, Rutenber EE, Stout TJ, and Stroud RM (1996) An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q, Biochemistry 35, 16270–16281. [DOI] [PubMed] [Google Scholar]
- [26].Falk BT, Sapienza PJ, and Lee AL (2016) Chemical shift imprint of intersubunit communication in a symmetric homodimer, Proceedings of the National Academy of Sciences of the United States of America 113, 9533–9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Anderson AC, O’Neil RH, DeLano WL, and Stroud RM (1999) The structural mechanism for half-the-sites reactivity in an enzyme, thymidylate synthase, involves a relay of changes between subunits, Biochemistry 38, 13829–13836. [DOI] [PubMed] [Google Scholar]
- [28].Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- [29].Birdsall DL, Finer-Moore J, and Stroud RM (2003) The only active mutant of thymidylate synthase D169, a residue far from the site of methyl transfer, demonstrates the exquisite nature of enzyme specificity, Protein engineering 16, 229–240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.