Abstract
A subset of atypical memory B cells accumulates in malaria and several infections, autoimmune disorders and aging in both humans and mice. It has been suggested these cells are exhausted long-lived memory B cells, and their accumulation may contribute to poor acquisition of long-lasting immunity to certain chronic infections, such as malaria and HIV. Here, we generated an immunoglobulin heavy chain knock-in mouse with a BCR that recognizes MSP1 of the rodent malaria parasite, Plasmodium chabaudi. In combination with a mosquito-initiated P. chabaudi infection, we show that Plasmodium-specific atypical memory B cells are short-lived and disappear upon natural resolution of chronic infection. These cells show features of activation, proliferation, DNA replication, and plasmablasts. Our data demonstrate that Plasmodium-specific atypical memory B cells are not a subset of long-lived memory B cells, but rather short-lived activated cells, and part of a physiologic ongoing B-cell response.
Research organism: Mouse
Introduction
Atypical memory B cells (AMB) are an unusual B-cell subset detected in both mouse models and humans in the context of certain infections and autoimmune disorders, including HIV, HCV, tuberculosis, malaria, rheumatoid arthritis and systemic lupus erythematosus, and accumulated with age (Knox et al., 2017b; Naradikian et al., 2016a; Portugal et al., 2017; Rubtsov et al., 2017). In the context of infections, AMB were first described in HIV-viremic subjects, and termed tissue-like memory B cells, due to their similarity to an FCRL4-expressing memory B-cell subset found in human tonsillar tissues (Ehrhardt et al., 2005; Moir et al., 2008). In addition to FCRL4, these cells express relatively high levels of other potentially inhibitory receptors including CD22, CD85j, CD85k, LAIR-1, CD72, and PD-1, and show a profile of trafficking receptors including expression of CD11b, CD11c and CXCR3, consistent with migration to inflamed tissues. They are antigen-experienced class-switched B cells, which lack the expression of CD21 and the hallmark human memory B-cell marker CD27. Further studies demonstrated the expression of the transcription factor T-bet and the cytokine IFNγ by these cells, also characteristic of Th1 CD4+ T cells (Knox et al., 2017b; Obeng-Adjei et al., 2017; Portugal et al., 2017). Due to their poor functional capacity upon in vitro re-stimulation with BCR ligands, AMB were characterized as dysfunctional B cells, and increased frequencies of these cells was proposed to be a consequence of B-cell exhaustion driven by chronic inflammation and stimulation, drawing parallels with T-cell exhaustion during chronic viral infections (Moir et al., 2008; Portugal et al., 2015; Sullivan et al., 2015). It has been hypothesized that expansion of AMB might contribute to the mechanisms driving autoimmune disorders and deficiencies in acquisition of immunity to chronic infections. However, due to lack of good tools and animal models to analyze antigen-specific atypical B cells in greater depth, many of these concepts remain speculative.
Several studies suggest that AMB might contribute to poor acquisition of long-term immunity to Plasmodium infection (Illingworth et al., 2013; Portugal et al., 2015; Sullivan et al., 2015; Sullivan et al., 2016; Weiss et al., 2011; Weiss et al., 2009; Weiss et al., 2010). Indeed, some studies demonstrated that in the absence of constant re-exposure, Plasmodium-specific serum antibody levels rapidly wane, and full protection from clinical symptoms is lost, suggesting that B-cell memory is functionally impaired (Portugal et al., 2013). However, others have reported long-lasting maintenance of Plasmodium-specific antibodies and/or memory B cells in settings of differing malaria endemicity, and similar responses are also observed in mouse malaria models (Dorfman et al., 2005; Ndungu et al., 2009; Ndungu et al., 2013; Ndungu et al., 2012; Wipasa et al., 2010). Moreover, it has been shown that BCRs cloned from P. falciparum-specific AMB from malaria-exposed adults encode P. falciparum-specific IgG antibodies, which could contribute to P. falciparum-specific IgG antibodies in serum (Muellenbeck et al., 2013). These authors proposed that P. falciparum-specific AMB do not prevent, but rather contribute to the control of Plasmodium infection. These apparently contradictory results may reflect the fact that some studies were performed on the general peripheral blood B-cell pool and others focused on Plasmodium-specific B cells. In determining a role for these cells in a chronic infection it would be important to follow antigen-specific responses and to distinguish these from non-specific polyclonal B cell activation.
The study of the development of AMB is challenging and requires suitable mouse models, which allow for identification and isolation of antigen-specific B cells that exist often at very low frequency. Here, we generated a knock-in transgenic mouse with a high frequency of B cells specific to the 21 kDa C-terminal fragment of Plasmodium chabaudi Merozoite Surface Protein 1 (MSP121), to investigate memory B cells generated following mosquito-transmission of the rodent malaria, P. chabaudi. We identified a CD11b+CD11c+FCRL5hi subset of MSP121-specific B cells during the chronic infection with phenotypical and transcriptional features strikingly similar to those of human AMB. These AMB disappeared as the infection progressed, leaving a CD11b—CD11c—FCRL5hi MSP121-specific B-cell compartment with characteristics of long-lived classical memory B cells (Bmem) after the resolution of the infection. These short-lived MSP121-specific AMB were also generated in response to immunization, suggesting they may be a normal but transient component of a B-cell response to antigen. In this chronic P. chabaudi infection, it appears that AMB require ongoing antigenic stimulation driven by the sub-patent infection to persist, and do not represent a true long-lived ‘memory’ B cell subset. Moreover, we show that generation of Plasmodium-specific AMB does not prevent the generation of Plasmodium-specific Bmem, and does not prevent resolution of the infection.
Results
Generation of an immunoglobulin heavy chain knock-in transgenic mouse model to study Plasmodium-specific B cell responses
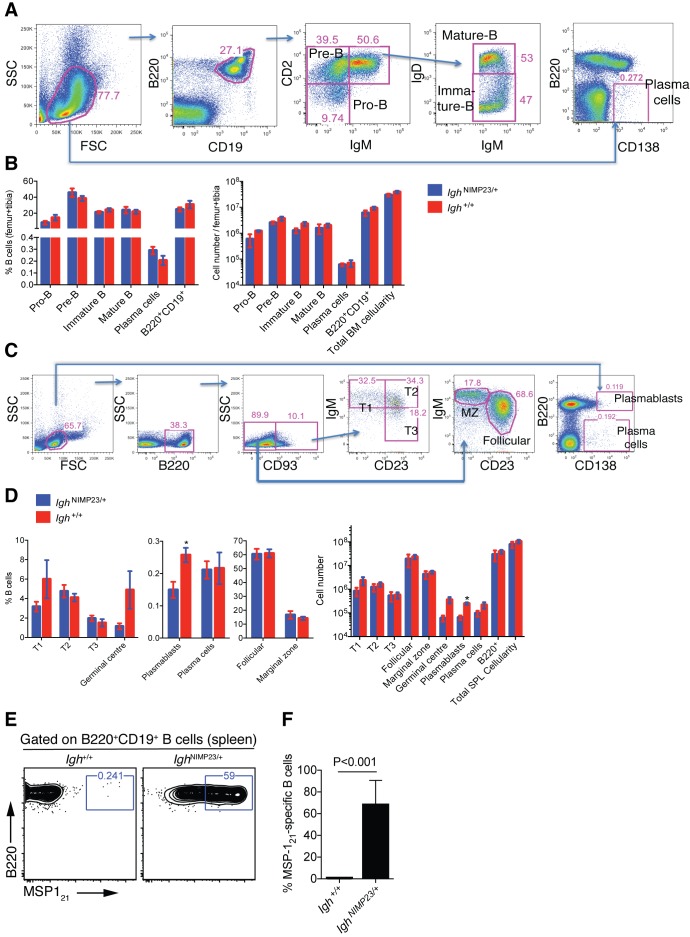
To study Plasmodium-specific B cell responses in a rodent malaria model, we generated an IghNIMP23/+ mouse strain on the C57BL/6J background (Materials and methods and Figure 1—figure supplement 1).
The IghNIMP23/+ mice were healthy, with no unusual behavioral or physical characteristics. There were no alterations in total cellularity, pro-B, pre-B, immature B, mature B, total B220+CD19+ B cells, and plasma cells in the bone marrow (Figure 1A–B), and no alterations in number of T1, T2, T3, follicular, marginal zone, germinal center B cells, plasmablasts, plasma cells, and total cellularity in the spleen of IghNIMP23/+ mice (Figure 1C–D) (Sen et al., 1990; Young et al., 1994). Importantly, The IghNIMP23/+ mice had a greatly increased frequency of B cells specific for MSP121 (approximately 60% of the total B-cell compartment), as demonstrated by flow cytometry analysis of splenocytes with a MSP121 fluorescent probe consisting of biotinylated recombinant MSP121 loaded on streptavidin-PE (Figure 1E–F). Thus, in this model, a recombinant light chain is not required to bring about specificity. This suggest that most endogenous light chains will pair with the NIMP23 heavy chain to generate a BCR with detectable binding to MSP121.
Figure 1. Analysis of total bone marrow and splenic B-cell populations in IghNIMP23/+ and Igh+/+littermates.
(A) Flow cytometry gating strategy to identify different B-cell populations in bone marrow of IghNIMP23/+ mice. Arrows indicate flow of analysis. The same strategy was used for Igh+/+ littermates. (B) Percentages and numbers of different B-cell populations in bone marrow of IghNIMP23/+ and Igh+/+ littermates as defined in (A). (C) Flow cytometry gating strategy to identify different B-cell populations in spleen of IghNIMP23/+ mice. (D) Percentages and numbers of different B-cell populations in spleen of IghNIMP23/+ and Igh+/+ littermates as defined in (C). Data are representative of two independent experiments with four mice per group. (E) Flow cytometry analysis of B cells obtained from spleen of Igh+/+ (left) and IghNIMP23/+ (right) mice stained with anti-B220 and CD19 antibodies in combination with an MSP121 fluorescent probe. The gates show the frequency of B cells specific to MSP121. (F) Frequencies of MSP121-specific splenic B cells in IghNIMP23/+ and wild-type Igh+/+ littermate controls (Mann Whitney U test). Data pooled from two independent experiments with 3–5 mice per group. Mann Whitney U test. *p<0.05. Error bars are SEM.
Figure 1—figure supplement 1. Generation of IghNIMP23/+knock in mice.
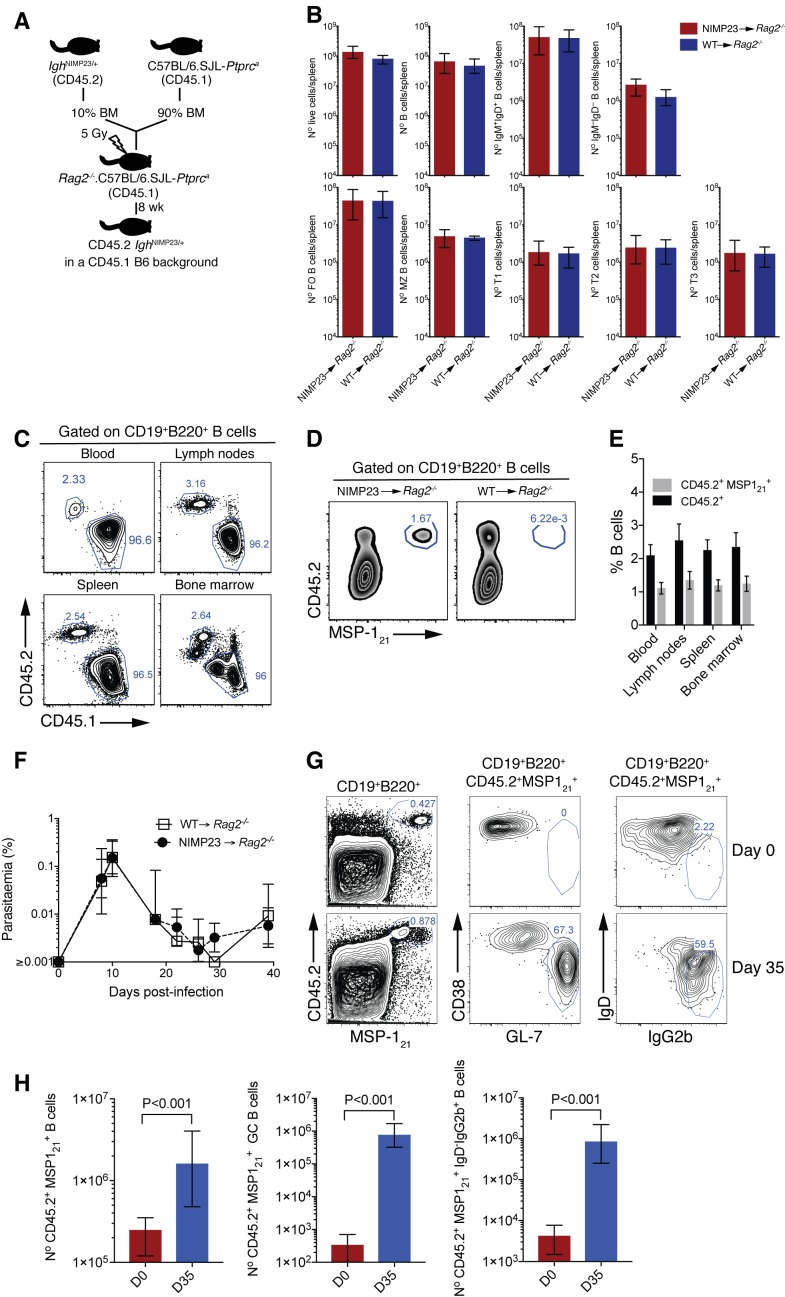
Figure 1—figure supplement 2. Generation of mixed bone marrow chimera model with reduced precursor frequency of IghNIMP23/+ B cells to study MSP121-specific B cell responses during P. chabaudi infection.
Increase in Plasmodium-specific B cells after mosquito transmission of P. chabaudi
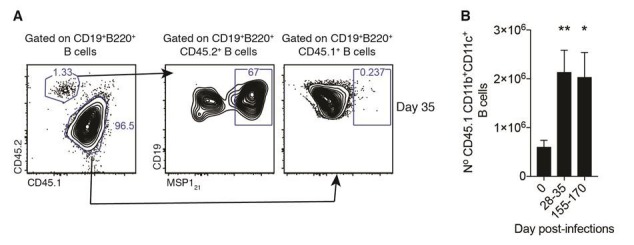
To investigate B cells in P. chabaudi infections, which last several weeks, and to avoid potential problems with activation arising from very high frequencies of MSP1-specific B cells, we reduced the precursor frequency of MSP121-specific B cells to match the natural level expected for antigen-specific B cells more closely, yet still readily detectable by flow cytometry. We generated mixed bone marrow (BM) chimeras by adoptively transferring a mixture of 10% bone marrow from either IghNIMP23/+ or Igh+/+ mice (CD45.2+) together with 90% bone marrow from C57BL/6.SJL-Ptprca mice (CD45.1+) into sub-lethally irradiated Rag2-/-.C57BL/6.SJL-Ptprca mice (CD45.1+) to generate NIMP23→Rag2-/- and WT→Rag2-/- bone marrow chimeric mice respectively (Figure 1—figure supplement 2A–B). In both types of chimeras, 2–3% of the B cells were CD45.2+ and in NIMP23→Rag2-/- mice approximately 1–2% of the B cells were MSP121-specific (Figure 1—figure supplement 2C–E). No MSP121-specific B cells were detected in the control WT→Rag2-/- chimeras (Figure 1—figure supplement 2D).
Infection of C57BL/6J wt mice with P. chabaudi by mosquito bite gives rise to a short (48 hr) pre-erythrocytic infection, followed by an acute blood parasitemia peaking approximately 10d post-transmission. Thereafter, the infection is rapidly controlled, reaching very low parasitemias by 15d post-transmission, with a subsequent prolonged (~90 d), but low-level chronic infection before parasite elimination (Brugat et al., 2017; Spence et al., 2013). NIMP23→Rag2-/- mice infected with P. chabaudi by mosquito bite, showed a similar course of parasitemia to that of control WT→Rag2-/- mice (Figure 1—figure supplement 2F), and C57BL/6J wt mice (Brugat et al., 2017; Spence et al., 2013; Spence et al., 2012). Importantly, the MSP121-specific IghNIMP23/+ B cells (CD45.2+MSP121+) in NIMP23→Rag2-/- chimeras showed a robust response to the infection, as demonstrated by a dramatic increase in the proportions and numbers of GL-7+CD38lo germinal centers (GC) and IgG2b+IgD— class-switched B cells in the spleen at 35 days post-infection (dpi) (Figure 1—figure supplement 2G–H).
Thus, we have generated a mouse model with detectable numbers of functional MSP121-specific B cells capable of responding to P. chabaudi infection.
Generation of Plasmodium-specific AMB after mosquito transmission of P. chabaudi infection
We investigated whether Plasmodium-specific AMB could be identified in mice during a blood-stage P. chabaudi infection. We selected a series of mouse homologues to human cell surface markers described on human AMB (Charles et al., 2011; Kardava et al., 2014; Kardava et al., 2011; Knox et al., 2017a; Li et al., 2016; Moir et al., 2008; Muellenbeck et al., 2013; Portugal et al., 2015; Russell Knode et al., 2017; Sullivan et al., 2015). Human AMB express CD11b, CD11c, Fc receptor-like (FCRL) 3–5, high levels of CD80, low levels of CD21, and are Ig class-switched. Mouse FCRL5 most closely resembles human FCRL3 and is the only mouse FCRL-family member which contains both ITIM and ITAM motifs in its cytoplasmic tail (Davis, 2007; Davis et al., 2004; Won et al., 2006; Zhu et al., 2013). Therefore, our flow cytometry panel for mouse AMB included antibodies against CD11b, CD11c, FCRL5, CD21, IgD, and also CD80 and CD273 which identify mouse B cells that are antigen-experienced and potentially memory cells (Anderson et al., 2007; Tomayko et al., 2010; Zuccarino-Catania et al., 2014).
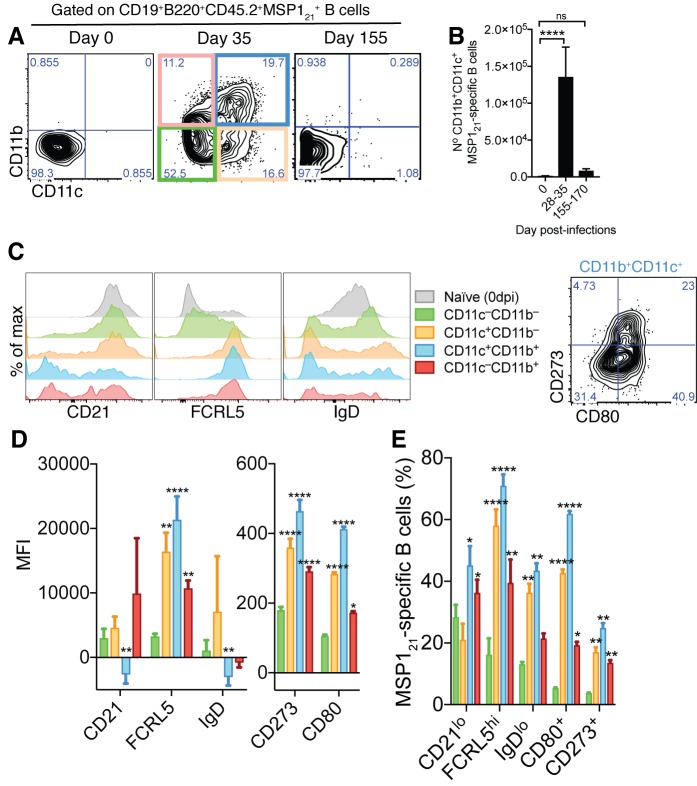
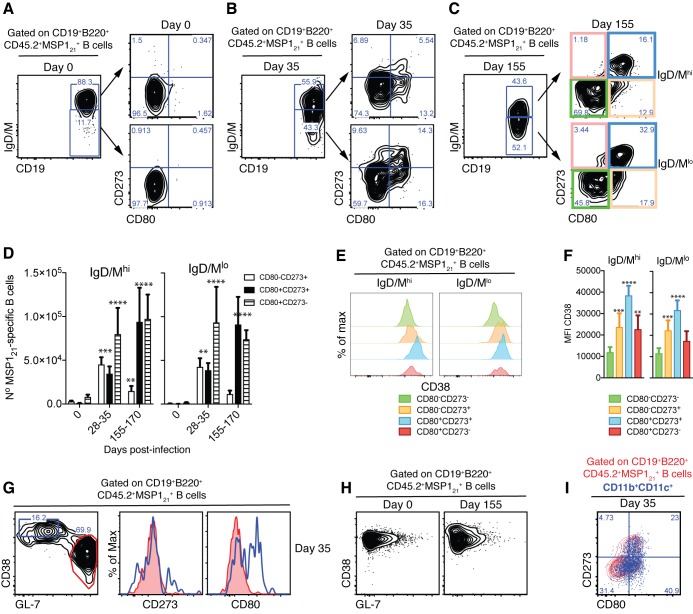
We detected an increased number of cells in a distinct CD11b+CD11c+ MSP121-specific B-cell subset at 28-35dpi, in the chronic phase of P. chabaudi infection (Figure 2A–B). This subset showed several AMB characteristics, including high expression of FCRL5 and low expression of CD21 and IgD (Figure 2C–E). In addition, the CD11b+CD11c+ MSP121-specific B-cell subset was enriched with cells expressing CD80 and CD273 (Figure 2C–E).
Figure 2. Generation of MSP121-specific AMB in response to mosquito transmitted P.chabaudi infection.
(A) Flow cytometry showing differential expression of CD11b and CD11c on splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice before infection (day 0) and at 35 and 155dpi. (B) Numbers of splenic MSP121-specific CD11b+CD11c+ AMB from NIMP23→Rag2-/- during the course of mosquito transmitted P. chabaudi infection. Kruskal-Wallis test vs day 0. ****, p<0.0001 (C) Flow cytometry showing expression of CD21/35, FCRL5, IgD, CD273 and CD80 on different subsets of splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice defined based on CD11b and CD11c expression at 35dpi. (D) Geometric mean fluorescence intensity (MFI) of CD21/35, FCRL5, IgD, CD273 and CD80 expression on different subsets of splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice defined based on CD11b and CD11c expression at 35dpi. (E) Frequencies of CD21/35, FCRL5, IgD, CD273 and CD80 positive cells among different subsets of splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice defined based on CD11b and CD11c expression at 35dpi. Two-way ANOVA vs CD11b-CD11c- subset. *p<0.05; **p<0.01, ***p<0.001; ****p<0.0001. Error bars are SEM. Data pooled from three independent experiments with 3–5 mice per group.
We then explored whether this CD11b+CD11c+ MSP121-specific B cell subset was detected during the memory phase,that is after resolution of the infection. As it takes up to 90 days for a blood-stage P. chabaudi infection to be eliminated from C57BL/6J mice (Achtman et al., 2007; Spence et al., 2013), we measured these responses from 155dpi onwards. Unexpectedly, the numbers of CD11b+CD11c+ MSP121-specific B cells were not significantly higher than background level (Figure 2A–B).
These data demonstrate that a mosquito-borne infection with P. chabaudi generates Plasmodium-specific B cells resembling human AMB. However, these cells do not persist and are not detected above background level after parasite clearance.
Transcriptome analysis confirms the AMB nature of CD11b+CD11c+ MSP121-specific B cells, and reveals a plasmablast-like signature for this subset
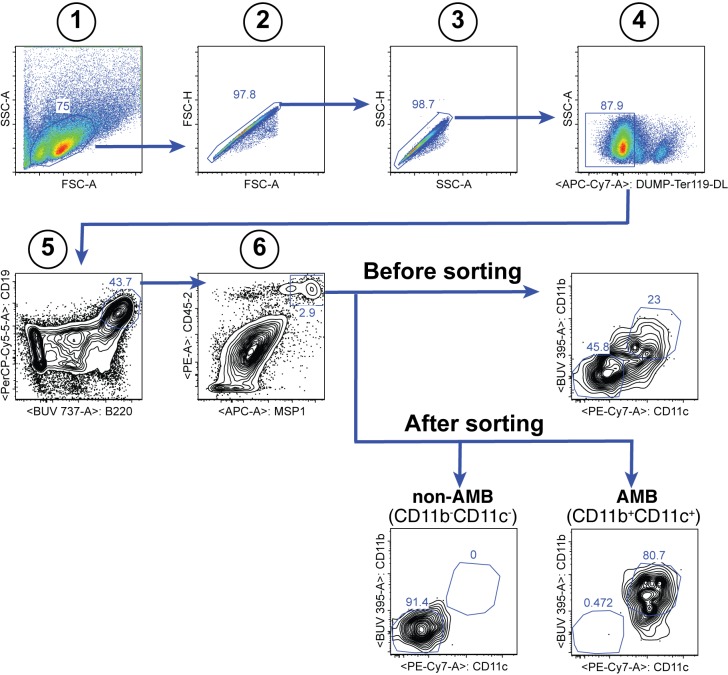
To gain a better understanding of the identity of the CD11b+CD11c+ Plasmodium-specific B cell subset, we isolated both CD11b+CD11c+ and CD11b-CD11c- MSP121-specific B cells from spleens of P. chabaudi-infected NIMP23→Rag2-/- mice (35dpi) (Figure 3—figure supplement 1), and MSP121-specific B cells from the spleen of naïve NIMP23→Rag2-/- mice (Figure 2A), by flow cytometric sorting, and performed an mRNAseq transcriptional analysis on the three populations.
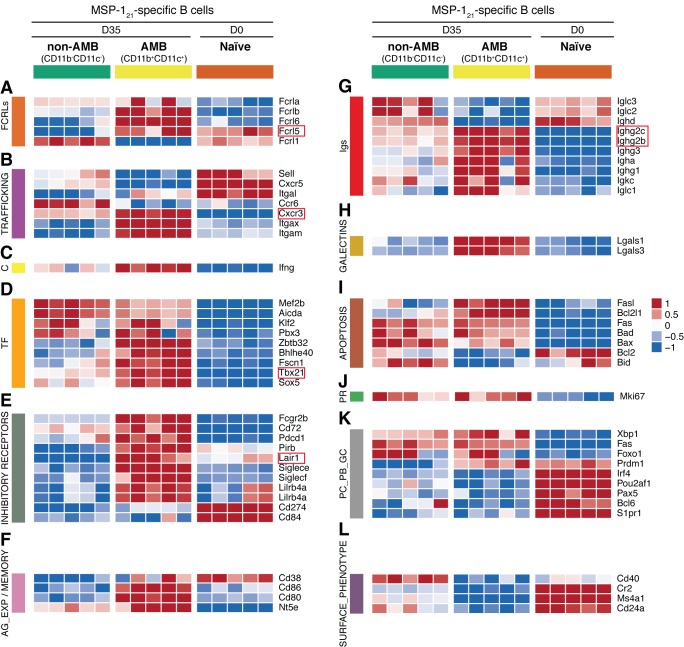
We then selected a large series of key genes previously shown to be either up or downregulated on human AMB (Supplementary file 1), and explored the expression of their mouse homologues on the three different B-cell subsets we sorted at day 35pi. The transcriptome of CD11b+CD11c+ MSP121-specific B cells highly resembled that of human AMB. A series of hallmark genes upregulated in human AMB were also upregulated in CD11b+CD11c+ MSP121-specific B cells, including IgG (Ighg2c and Ighg2b), Cxcr3, Tbx21 (T-bet), Lair1 and Fcrl5 (Figure 3, genes in red boxes, and references in Supplementary file 1). In addition, the MSP121-specific CD11b+CD11c+ B-cell subset showed upregulation of Ifng, Aicda, a large array of inhibitory receptors [including Pd1, Cd72, Cd85k, Fcgr2b (CD32b), Siglece], antigen-experienced/memory markers [Cd80, Cd86, Nt5e (CD73) and high Cd38], and additional class-switched immunoglobulins (i.e. Igha, Ighg1 and Ighg3), all of which have been shown to be upregulated on human AMB (Figure 3C–G, references in Supplementary file 1). These cells also expressed Galectins (Lgals1 and Lgals3), previously implicated in B-cell anergy (Clark et al., 2007) (Figure 3H), and displayed a pro-apoptotic program (e.g. high expression of Fasl, and low expression of Bcl2) (Figure 3I). Interestingly, in agreement with data on human AMB, MSP121-specific CD11b+CD11c+ B cells showed upregulation of Mki67 (Figure 3J), indicative of proliferation, and had characteristics of plasmablasts and/or plasma cells, including upregulation of Cd138, Prdm1 (Blimp1) and Xbp1, and low expression of Cxcr5, Pax5, and Bcl6 (Figure 3B and K). However, these cells showed low expression of Irf4 and S1p1, suggesting that they may be in a pre-plasmablast or pre-migratory plasma-cell stage (Kabashima et al., 2006; Kallies et al., 2007) (Figure 3K). Finally, and similar to human AMB, CD11b+CD11c+ MSP121-specific B cells showed low expression of Cd40, Cr2 (CD21), Ms4a1 (CD20), and Cd24a (Figure 3L).
Figure 3. Transcriptome analysis of sorted splenic MSP121-specific CD11b+CD11c+AMB.
MSP121-specific CD11b+CD11c+ (AMB) and CD11b—CD11c— B cells were flow cytometry sorted from the spleen of NIMP23→Rag2-/- chimeric mice at 35dpi; MSP121-specific B cells were flow cytometry sorted from the spleen of naïve NIMP23→Rag2-/-, and these three B cell populations were submitted to mRNAseq analysis. The heat maps display level of expression of selected individual genes, organized in functional clusters related to (A) Fc receptor like molecules, (B) cell trafficking, (C) cytokines, (D) transcription factors, (E) inhibitory receptors, (F) antigen experience/memory, (G) immunoglobulins, (H) galectins, (I) apoptosis, (J) proliferation, (K) plasma cells/plasmablasts/germinal centers, (L) surface markers. Each column corresponds to data from an individual mouse (n = 5 35 dpi, n = 5 0 dpi).
Figure 3—figure supplement 1. Gating strategy for the sorting of splenic MSP121-specific CD11b+CD11c+ AMB and CD11b—CD11c— B cells at 35dpi.
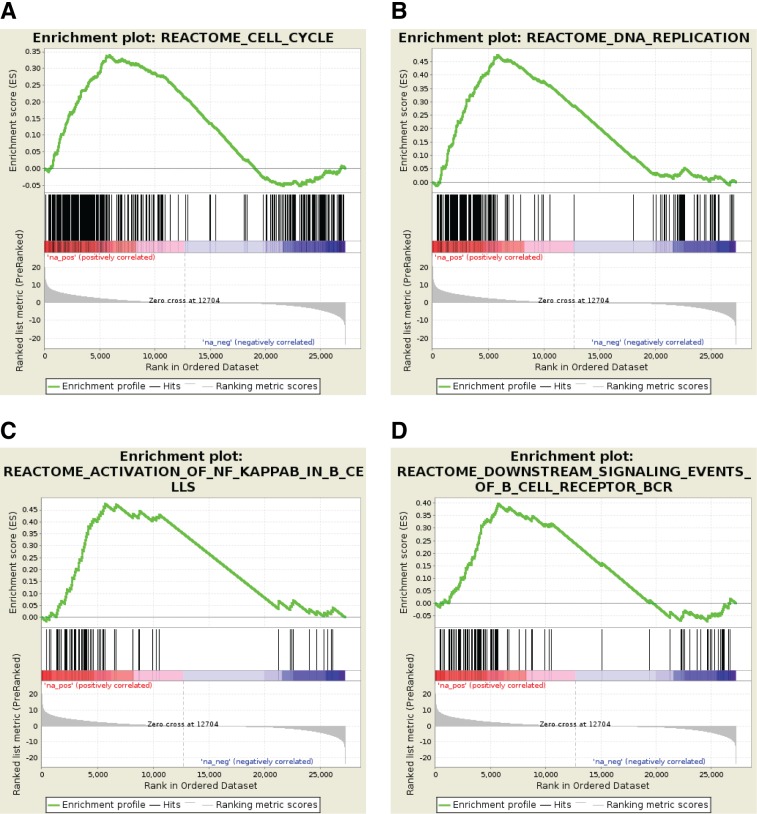
Figure 3—figure supplement 2. Profile of the Running ES Score and positions of GeneSet members on the rank ordered list for selected Reactome pathway gene sets.
We ran a Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) with a gene list ranked according to their differential expression between MSP121-specific CD11b+CD11c+ AMB sorted from infected mice and MSP121-specific B cells sorted from naïve mice, using gene sets a priori obtained from Reactome (Fabregat et al., 2018). Among the gene sets yielding the top 50 significant (fdr <0.001) highest normalized enrichment score (NES) we obtained gene sets corresponding to cell cycle, DNA replication, generation/consumption of energy, regulation of apoptosis, activation of NF-κB on B cells, and downstream signaling events of the BCR (Supplementary file 2 and Figure 3—figure supplement 2). These data further corroborate the activated and proliferative nature of MSP121-specific CD11b+CD11c+ AMB.
Taken together, these data demonstrate that CD11b+CD11c+ MSP121-specific mouse AMB present during the chronic phase of P. chabaudi infection are very similar to human AMB described in several chronic infections. In addition, this B-cell subset shows features of activation, proliferation, DNA replication and plasmablasts, resembling previous observations in human AMB (Muellenbeck et al., 2013).
Generation of Plasmodium-specific AMB in response to immunization
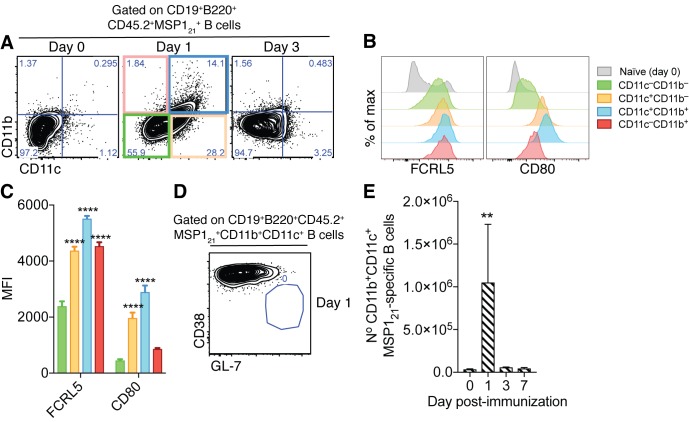
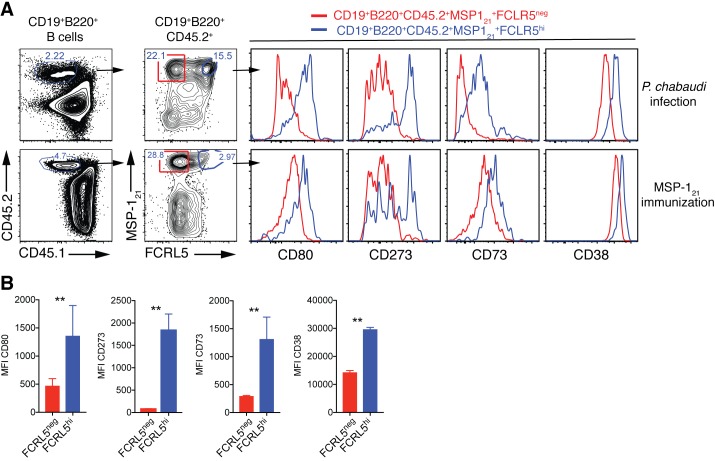
The occurrence of CD11b+CD11c+ AMB might be a consequence of aberrant B-cell activation driven exclusively by certain pathogens. Alternatively, they might be part of a normal B-cell response, which is exacerbated by the persistent nature of certain infections. To test whether CD11b+CD11c+FCRL5+ AMB could be generated in the absence of persistent infection, we immunized mice with MSP121. A previous report had demonstrated the presence of CD11b+CD11c+Tbet+ B-cells 24 hr post-immunization with R848, a TLR7/8 ligand (Rubtsova et al., 2013). Therefore, we immunized IghNIMP23/+ mice with R848 together with the antigen MSP121 and looked for the appearance of MSP121-specific CD11b+CD11c+FCRL5+ atypical B cells. We observed substantial numbers of MSP121-specific CD11b+CD11c+ B cells in the spleens of IghNIMP23/+ mice 24 hr post-immunization (Figure 4A). These cells expressed increased levels of both FCRL5 and CD80 (Figure 4B–C) and did not display GC characteristics (Figure 4D), similar to the MSP121-specific CD11b+CD11c+FCRL5+ atypical B cells generated following Plasmodium infection. The MSP121-specific CD11b+CD11c+ B cells observed after immunization appeared only transiently, as they could no longer be detected at 3 and 7d post-immunization (Figure 4A and E).
Figure 4. Generation of splenic MSP121-specific CD11b+CD11c+AMB in response to immunization.
(A) Flow cytometry showing differential expression of CD11b and CD11c on splenic MSP121-specific B cells from IghNIMP23/+ mice before immunization (day 0) and at days 1 and 3 post-immunization with R848 and MSP121. (B) Flow cytometry showing expression of FCRL5 and CD80 on different subsets of splenic MSP121-specific B cells from IghNIMP23/+ defined based on CD11b and CD11c expression at day one post-immunization and naïve mice. (C) Geometric MFI of FCRL5 and CD80 expression on different subsets of splenic MSP121-specific B cells from IghNIMP23/+ defined based on CD11b and CD11c expression at day one post-immunization. Two-way ANOVA vs CD11b—CD11c— subset. ****p<0.0001. (D) Flow cytometry of CD38 vs GL-7 (GC markers) on CD11b+CD11c+ MSP121-specific B cells from IghNIMP23/+ at day one post-immunization. (E) Numbers of splenic CD11b+CD11c+ MSP121-specific B cells from IghNIMP23/+ during the course of immunization. Kruskal-Wallis test compared to day 0. **p<0.01. Error bars are SEM. Data pooled from three independent experiments with 3–5 mice per group.
These data demonstrate that MSP121-specific CD11b+CD11c+ AMB with no functional characteristics of memory B cells can be generated independently of the infection and the presence of the pathogen, and that they are short-lived cells.
Plasmodium-specific CD80+CD273+ Bmem are generated and persist after resolution of P. chabaudi infection
Identification of mouse Bmem by flow cytometry originally relied on detecting B cells that had undergone Ig class-switching from IgM to IgG, and that did not express GC markers (i.e. IgG+CD38hiGL-7lo) (Lalor et al., 1992; Ridderstad and Tarlinton, 1998). More recently, this set of markers has been extended to include CD80, CD273 (PD-L2) and CD73, with CD273 and CD80 being the most useful to discriminate memory from naïve B cells (Tomayko et al., 2010; Zuccarino-Catania et al., 2014). In combination, these markers allow the identification of different subsets of switched as well as non-class switched (i.e. IgM/D+) Bmem. Therefore, we used cell surface expression of CD80 and CD273 on MSP121-specific B cells to identify Bmem during and after resolution of P. chabaudi infection.
MSP121-specific B cells from spleens of naïve NIMP23→Rag2-/- mixed BM chimeras showed little expression of either CD80 or CD273 (Figure 5A). By contrast, CD80+CD273+, CD80+CD273— and CD80—CD273+ MSP121-specific B cells, both class-switched (IgM/Dlo) and non-class-switched (IgM/Dhi), were readily detected above background levels at 28-35dpi in P. chabaudi-infected NIMP23→Rag2-/- mixed BM chimeras (Figure 5B and D). After resolution of infection (155-170dpi), the numbers of CD80+CD273+ and CD80+CD273— MSP121-specific B cells were either sustained or increased, while the CD80—CD273+ population decreased, compared with 28-35dpi (Figure 5A–D). All three MSP121-specific Bmem subsets showed high CD38 expression (Figure 5E,F). Importantly, the MSP121-specific GC B cells detected at 28-35dpi did not express either CD80 or CD273, further distinguishing GC from memory and atypical memory P. chabaudi-specific B-cell subsets (Figure 5G). No MSP121-specific GC B cells were detected above background level after resolution of the infection (Figure 5H). Finally, and in accordance with Figure 3, approximately 70% of the MSP121-specific CD11b+CD11c+ AMB observed at days 28-35pi expressed either CD80, CD273 or a combination of both (Figure 5I).
Figure 5. Detection of MSP121-specific Bmem after resolution of P.chabaudi infection.
(A), (B) and (C) Flow cytometry showing gating strategy to identify splenic IgM/Dhi and IgM/Dlo CD273+ and/or CD80+ MSP121-specific Bmem in NIMP23→Rag2-/- chimeric mice before infection (day 0), at 35 and 155dpi, respectively. (D) Numbers of splenic IgM/Dhi and IgM/Dlo CD273+ and/or CD80+ MSP121-specific Bmem in NIMP23→Rag2-/- chimeric mice during the course of mosquito transmitted P. chabaudi infection. Two-way ANOVA vs day 0. **p<0.01; ***p<0.001; ****p<0.0001. (E) Flow cytometry showing expression of CD38 on different subsets of splenic IgM/Dhi and IgM/Dlo MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice defined based on CD273 and CD80 expression at 155dpi. (F) Geometric MFI of CD38 expression on different subsets of IgM/Dhi and IgM/Dlo splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice defined based on CD273 and CD80 expression at day 155 post-mosquito transmitted P. chabaudi infection. Two-way ANOVA vs CD273-CD80- subset. **p<0.01; ***p<0.001; ****p<0.0001. Error bars are SEM. (G) Flow cytometry of CD273 and CD80 expression on non-GC (CD38hiGL-7lo, blue) and GC (CD38loGL-7hi, red) splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice at 35dpi. (H) Flow cytometry of CD38 vs GL-7 (GC markers) on splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice at 0 and 155dpi. (I) CD11b+CD11c+ MSP121-specific B cells overlaid on the CD80 vs CD273 plot corresponding to total MSP121-specific B cells. Data pooled from three independent experiments with 3–7 mice per group.
These data show that, in contrast to the transient AMB, splenic CD80+CD273+ and CD80+CD273— class-switched and non-class-switched MSP121-specific Bmem persist after resolution of P. chabaudi infection.
Plasmodium-specific Bmem express high levels of FCRL5
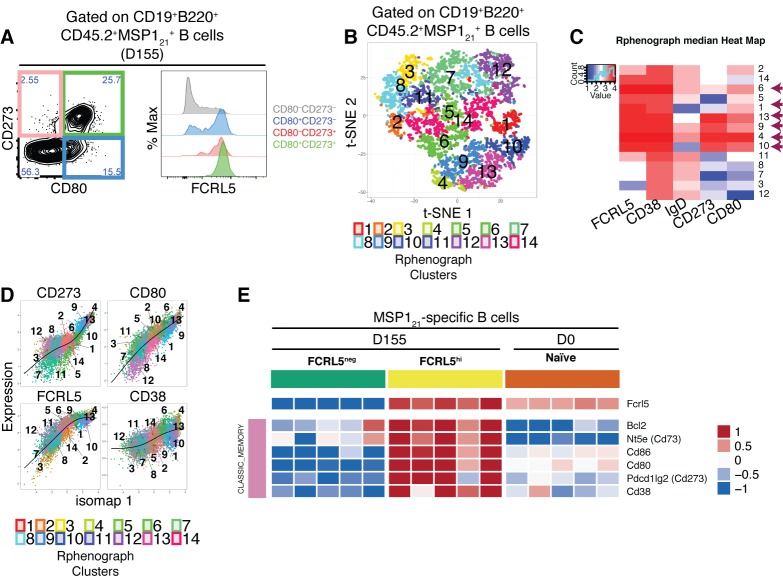
As discussed above, no single marker has been described so far that can identify all mouse Bmem subsets. Surprisingly, we observed that after resolution of infection (155-170dpi), MSP121-specific B cells expressing different combinations of CD80 and CD273 (CD80+CD273+, CD80-CD273+ or CD80+CD273-) all expressed very high levels of FCRL5, in contrast to CD80-CD273- MSP121-specific B cells that express no memory markers at this stage (Figure 6A). This suggests that FCRL5 might be a marker for all Bmem. In order to confirm this, we used unsupervised methods to analyze our multiparameter flow cytometry data. We used PhenoGraph and t-SNE within the Cytofkit package (Materials and methods, Chen et al 2016) to analyze MSP121-specific B cells based on the expression of FCRL5, CD38, IgD, CD80 and CD273 on these cells, as determined by flow cytometry (Figure 6A and B). The analysis identified six clusters of cells with memory characteristics displaying high expression of CD38, CD80 and/or CD273, and variable expression of IgD, all of which expressed high levels of FCRL5 (Figure 6C: clusters identified with purple arrows). We then used Isomap, (Cytofkit package) to infer the relatedness between those cell subsets identified by PhenoGraph. This confirmed high similarities between the cell clusters expressing high levels of FCRL5 with the clusters expressing high levels of the memory markers CD80, CD273 and CD38 (Figure 6D).
Figure 6. FCRL5hi identifies MSP121-specific Bmem after resolution of P.chabaudi infection.
(A) Flow cytometry showing expression of FCRL5 (right) on different subsets of splenic MSP121-specific B cells from NIMP23→Rag2-/- chimeric mice defined based on CD273 and CD80 expression (left) at 155dpi. (B) t-SNE analysis of splenic MSP121-specific B cells based on FCRL5, CD38, IgD, CD273 and CD80 expression measured by flow cytometry (n = 5). Clusters identified by PhenoGraph are colored and numbered. (C) PhenoGraph heat map showing median expression of FCRL5, CD38, IgD, CD273 and CD80 on the different clusters of MSP121-specific B cells. Arrows point at the different clusters displaying a memory B cell phenotype. (D) Expression profiles of FCRL5, CD38, CD273 and CD80 for the different PhenoGraph clusters visualized on the first component of ISOMAP. The regression line estimated using the generalized linear model (GLM) is added for each marker. Data representative of three independent experiments with 4–7 mice per group. (E) Heat map showing expression levels of different genes on splenic FCRL5— and FCRL5hi MSP121-specific B cells sorted at 155dpi, and MSP121-specific B cells sorted before infection (naïve), determined by RNAseq analysis. Each column corresponds to data from an individual mouse (n = 5 155 dpi, n = 5 0 dpi).
Figure 6—figure supplement 1. Gating strategy for the sorting of splenic MSP121-specific FCRL5hi Bmem and FCRL5— B cells at 155dpi.
Figure 6—figure supplement 2. High expression of FCRL5 identifies Bmem.
To confirm the memory identity of MSP121-specific FCRL5hi B-cells detected after resolution of the infection, we isolated MSP121-specific B cells expressing either high levels of FCRL5 or not expressing FCRL5 (i.e. FCRL5hi and FCRL5— MSP121-specific B cells) from the spleen of P. chabaudi-infected NIMP23→Rag2-/- mice (155dpi) (Figure 6—figure supplement 1), and MSP121-specific B cells from the spleen of naïve NIMP23→Rag2-/- mice, by flow cytometric sorting, and performed mRNAseq analysis on these three sorted cell populations (Figure 6E). As expected, the MSP121-specific FCRL5hi B cell subset showed high expression of genes encoding the hallmark memory B cell markers Cd38, Cd80, Cd86, Nt5e (CD73) and Pdcd1lg2 (CD273), when compared with either MSP121-specific FCRL5— B cells sorted at the same time or MSP121-specific B cells sorted from naïve mice (Figure 6E). Moreover, the MSP121-specific FCRL5hi B cells sorted after resolution of the infection upregulated the anti-apoptotic Bcl2 gene, which is an additional hallmark characteristic of memory B cells (Figure 6E). Importantly, FCRL5 also identified CD80+ and CD273+ MSP121-specific Bmem subsets generated following immunization with a model antigen (Figure 6—figure supplement 2).
Thus, after resolution of the infection, high expression of FCRL5 identifies P. chabaudi-specific Bmem.
Plasmodium-specific AMB are a distinct short-lived activated B cell subset
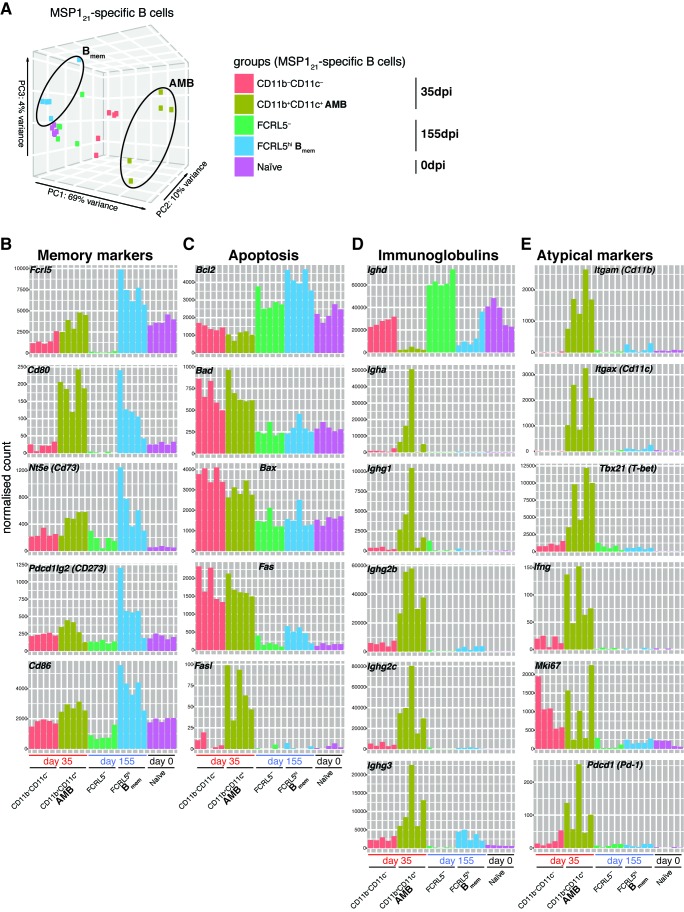
After identifying and sorting MSP121-specific AMB during chronic P. chabaudi infection, and MSP121-specific Bmem after resolution of the infection, we then compared the transcriptome of these two B-cell subsets. Principal component analysis (PCA) demonstrated a strikingly distinct transcriptome of MSP121-specific AMB from that of MSP121-specific Bmem, as well as all other MSP121-specific B-cell subsets sorted in this study (Figure 7A). The MSP121-specific AMB sorted at 35dpi formed a separated cluster at the extreme right of the PC1 axis of the PCA plot, which accounts for the majority of the variance (Figure 7A). All the other subsets [including MSP121-specific CD11b—CD11c— B-cells sorted from the same mice and at the same day post-infection as the MSP121-specific AMB (i.e. 35dpi)] clustered on the left of the PC1 axis, and showed differences mostly along the PC2 axis of the PCA plot, which accounts for only 10% of the variance (Figure 7A). Interestingly, MSP121-specific CD11b—CD11c— and MSP121-specific Bmem clustered on opposite sides of the MSP121-specific naïve B-cell subset along the PC2 axis (Figure 7A), which suggests that the MSP121-specific Bmem more closely resemble MSP121-specific naïve B cells than MSP121-specific CD11b—CD11c— B cells sorted at 35dpi.
Figure 7. MSP121-specific AMB are a distinct short-lived activated B cell subset.
(A) Principal component analysis of RNAseq transcriptome data from splenic MSP121-specific AMB (CD11b+CD11c+, 35dpi), CD11b—CD11c— B cells (35dpi), Bmem (FCRL5hi, 155dpi), FCRL5— B cells (155dpi) and B cells from naïve mice (0dpi). The MSP121-specific AMB and Bmem are contained inside ellipses. (B) (C) (D) (E) Normalized counts corresponding to selected genes representing memory B-cell markers, anti and pro-apoptotic genes, immunoglobulins and atypical memory B-cell markers, respectively, for all five groups described in (A). Each bar represents an individual mouse. Data generated with five mice per group.
MSP121-specific AMB sorted during chronic P. chabaudi infection, and MSP121-specific Bmem sorted after resolution of the infection shared the expression of a series of mouse memory markers, including Cd80, Fcrl5, Nt5e (CD73), and Cd86 (Figure 7B). However, these two subsets showed differences in the expression pattern of anti- and pro-apoptotic genes (Figure 7C). While MSP121-specific Bmem from after infection resolution showed the highest levels of expression of the anti-apoptotic Bcl2 gene, MSP121-specific AMB sorted during chronic P. chabaudi infection showed the lowest levels of expression of this hallmark anti-apoptotic gene (Figure 7C). In contrast to MSP121-specific Bmem, MSP121-specific AMB expressed high levels of the pro-apoptotic genes Bad, Bax, Fas and Fasl (Figure 7C). Interestingly, MSP121-specific AMB expressed very high levels of class-switched immunoglobulins, including Igha, Ighg1, Ighg2b, Ighg2c and Ighg3 (Figure 7D). Finally, MSP121-specific AMB highly expressed Cd11b, Cd11c, Tbx21, Ifng and Pdcd1 (Figure 7E), all hallmarks of human AMB, as well as Mki67, indicative of active cell division, as previously shown in human AMB.
These data demonstrate that AMB and Bmem share the expression of memory markers. However, they show striking differences in the expression of pro- and anti-apoptotic genes, immunoglobulins genes, and cell proliferation genes. The increased expression of Mki67, pro-apoptotic genes and class-switched immunoglobulins in AMB suggests that they resemble activated B cells. By contrast, Bmem express much less Mki67 (similar to naïve cells) and present an anti-apoptotic gene expression pattern, consistent with being long-lived quiescent B cells.
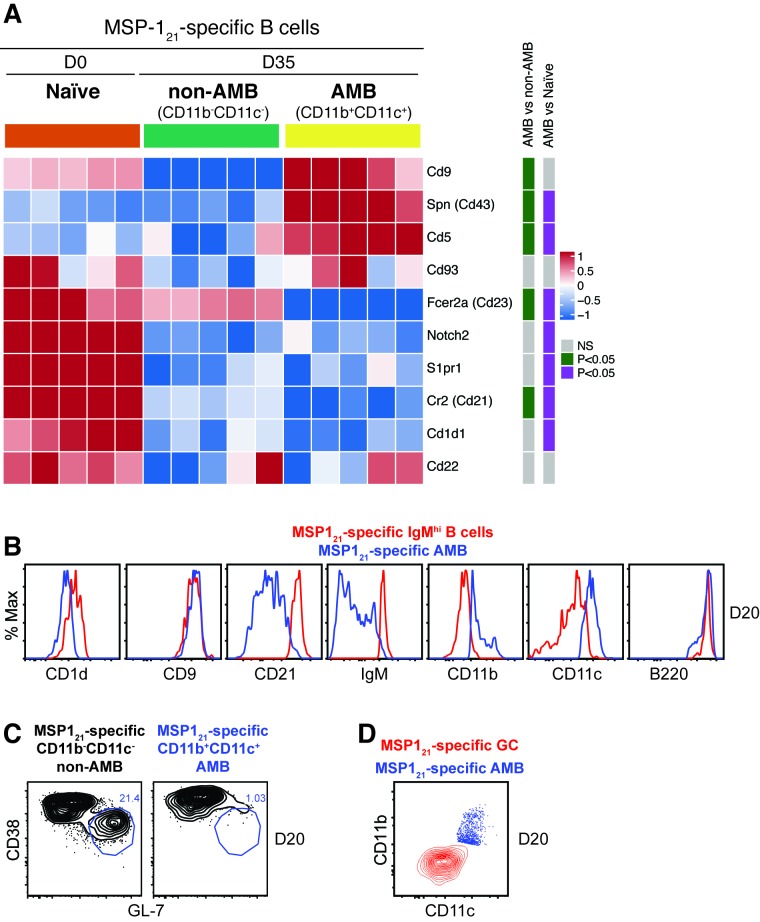
Mouse FCRL5 has been shown to be expressed on marginal zone (MZ) and B1 B cells (Won et al., 2006). Therefore, it is possible that the AMB identified in this study might represent a specific subset of either MZ or B1 B cells. As discussed elsewhere (Baumgarth, 2011; Garraud et al., 2012; Pillai et al., 2005; Zouali, 2011), B1 and MZ B cells are CD1dmid/hi, CD9+, IgMhi and CD23—. B1 are further CD43+, B220lo, and may (B1a) or may not (B1b) express CD5. MZ are further characterised by CD22hi, CD21/CR2hi, and the expression of the lysophospholipid sphingosine-1 phosphate receptor S1P1, and the lineage master regulator Notch2. Indeed, MSP121-specific CD11b+CD11c+ AMB showed high expression of some markers associated with B1 and/or MZ, including CD5, CD9 and CD43. However, these markers have also been shown to be highly expressed by activated B2 B cells and plasma cells (Baumgarth, 2011). Studying all these canonical markers, we found no other similarities between B1/MZ and MSP121-specific CD11b+CD11c+ AMB to suggest a relationship between these subsets (Figure 8A). This was further corroborated by flow cytometry analysis (Figure 8B). Moreover, unlike MZ and B1 B cells, MSP121-specific AMB were class-switched and expressed very high levels of IgG (Figure 7D). To further characterize the identity of MSP121-specific CD11b+CD11c+ AMB, we explored if these cells resemble GC B cells. Flow cytometry analysis demonstrated that MSP121-specific AMB did not display a GC phenotype (i.e. CD38hiGL-7lo) (Figure 8C) while the MSP121-specific CD38loGL-7hi GC B cells showed low expression of both CD11b and CD11c. Thus, MSP121-specific AMB are not GC B cells.
Figure 8. Analysis of MZ, B1 and GC B cell characteristics on MSP121-specific AMB.
(A) MSP121-specific CD11b+CD11c+ (AMB) and CD11b—CD11c— B cells were flow cytometry sorted from the spleen of NIMP23→Rag2-/- chimeric mice at 35dpi; MSP121-specific B cells were flow cytometry sorted from the spleen of naïve NIMP23→Rag2-/-, and these three B cell populations were submitted to mRNAseq analysis. The heat map displays level of expression of selected individual genes known to be up or downregulated on either MZ, B1 B cells or both. Each column represents an individual mouse. (B) Flow cytometry analysis of surface markers of either MZ, B1 B cells or both, on MSP121-specific CD11b+CD11c+ (AMB) (blue) and MSP121-specific IgMhi (red) B cells from the spleen of IghNIMP23/+ mice at 20dpi. (C) Flow cytometry analysis of GC markers on MSP121-specific CD11b—CD11c— (non-AMB, left) and CD11b+CD11c+ (AMB, right) B cells from the spleen of IghNIMP23/+ mice at 20dpi. (D) Flow cytometry analysis showing the expression of CD11b and CD11c on MSP121-specific CD11b+CD11c+ (AMB, blue) compared to GC (CD38loGL-7hi, red) B cells from the spleen of IghNIMP23/+ mice. Data generated with 5–6 mice per group.
All together, these data further show that P. chabaudi-specific AMB represent a distinct subset of short-lived activated B cells.
Discussion
Similar to other chronic infections [e.g. HIV, HCV and Mycobacterium tuberculosis (Knox et al., 2017b; Portugal et al., 2017)], Plasmodium infection, the cause of malaria, leads to an increase in the frequency of AMB [originally termed tissue-like memory B cells (Moir et al., 2008)] in peripheral blood from P. falciparum-exposed subjects (Illingworth et al., 2013; Portugal et al., 2015; Sullivan et al., 2016; Sullivan et al., 2015; Weiss et al., 2011; Weiss et al., 2010; Weiss et al., 2009). However, mouse models to investigate Plasmodium-specific AMB are lacking. Here we have generated an IgH knock-in transgenic mouse strain to study the generation of Plasmodium-specific AMB in a Plasmodium chabaudi infection. We demonstrate the generation of P. chabaudi-specific AMB in response to blood-stage mosquito-transmitted chronic P. chabaudi infection, and show the short-lived nature of these cells. P. chabaudi infections in mice present some outstanding characteristics for the study of AMB; a chronic phase which allows investigation of the impact of constant immune activation driven by persistent subpatent parasitemia, followed by a clearance phase which allows the study of immune responses after the infection is naturally resolved. Thus, it is possible to study both exhaustion driven by chronic immune activation, and memory immune responses which remain after P. chabaudi elimination.
The P. chabaudi-specific AMB we detected during the chronic phase of infection showed strong similarities to human AMB described in chronic malaria. These included being class-switched, and expressing mouse homologues of hallmark human atypical memory B-cell genes such as Itgax (Cd11b), Itgam (Cd11c), Cxcr3, Fcrl5, Tbx21 (T-bet), Ifng, Cd80, Cd86, Aicda, and a series on inhibitory receptors, including Lair1 and Pdcd1 (PD-1). Human AMB have been shown to share several characteristics with plasmablasts (Knox et al., 2017a; Muellenbeck et al., 2013; Obeng-Adjei et al., 2017; Sullivan et al., 2015). Interestingly, we also observed increased expression of several genes associated with plasmablasts in P. chabaudi-specific AMB, including Cd138, Xbp1, Prdm1 (encoding Blimp-1) and Mki67, accompanied by downregulation of Pax5 and Bcl6.
AMB resemble age-associated B cells (ABC) which accumulate with age as well as in autoimmunity, and were also proposed to be a subset of long-lived memory B cells (Naradikian et al., 2016a; Portugal et al., 2017; Rubtsov et al., 2017). Expression of T-bet, CD11c and CXCR3 are shared by AMB, tissue-like memory B cells, and ABC (Knox et al., 2017b; Naradikian et al., 2016a). Moreover, similar to ABC, expansion of human AMB associated with malaria is driven by IFNγ (Obeng-Adjei et al., 2017). IL-21, which is highly expressed by follicular helper T cells in response to Plasmodium infection (Carpio et al., 2015; Obeng-Adjei et al., 2015; Pérez-Mazliah et al., 2015), also directly promotes T-bet expression in B cells in the context of TLR engagement (Naradikian et al., 2016b). Taken together, these data strongly suggest that this is the same T-bet+ B-cell subset, which accumulates with time due to repetitive antigenic exposure. In agreement with previous data (Rubtsova et al., 2013), we show here that immunization with MSP121 and R848, a TLR7/8 ligand, promotes a robust but short-lived CD11b+CD11c+ P. chabaudi-specific AMB response. T-bet+ atypical B cells are critical to eradicate various murine viral infections (Barnett et al., 2016; Rubtsova et al., 2013), and a recent study showed that yellow fever and vaccinia vaccinations of humans stimulated an acute T-bet+ B-cell response and suggested that these T-bet+ B-cell population may function as an early responder during acute viral infections (Knox et al., 2017a). Thus, T-bet+ B cells, even in the context of malaria, are likely to be a normal component of the immune compartment that becomes activated and expands, most probably in response to BCR, endosomal TLR, and IFNγ or IL-21 stimulation. Moreover, a recent study shows a TLR9/IFNγ-dependent activation of autoreactive T-bet+CD11c+ atypical B cells in response to P. yoelii 17XNL infection in mice (Rivera-Correa et al., 2017). However, whether these cells remained part of the long-lived memory B-cell pool after resolution of the infection was not explored.
Here, we show that P. chabaudi-specific AMB are short-lived activated B cells. These cells were absent after resolution of the infection, and immunization with purified antigen and TLR agonists resulted in a transient, yet robust, activation of P. chabaudi-specific AMB which lasted no more than 48 hr. Moreover, the IghNIMP23/+ mouse model allowed us to obtain a deep insight of the transcriptome profile of MSP121-specific AMB during natural infection, and compare it side-by-side with the transcriptome of MSP121-specific naïve B cells. This allowed us to demonstrate their heavily pro-apoptotic and activated transcription profile, further explaining their short-lived nature. P. chabaudi-specific AMB showed very low expression of Bcl2, and high levels of expression of several pro-apoptotic genes including Bad, Bax, Fas and Fasl. In addition, these cells expressed very high levels of class-switched immunoglobulins and genes associated with DNA replication and proliferation.
The association of P. chabaudi-specific AMB with ongoing infection explains several observations in human studies: reduction of HIV plasma viremia by ART resulted in a significant reduction of HIV-specific AMB without altering the frequency of HIV-specific Bmem (de Bree et al., 2017; Kardava et al., 2014); individuals living in high malaria endemicity present higher frequencies of AMB than individuals living in areas with moderate transmission (Illingworth et al., 2013; Sullivan et al., 2015); repetitive Plasmodium episodes result in higher frequencies of AMB (Obeng-Adjei et al., 2017); the percentage of AMB is larger in children with persistent asymptomatic Plasmodium falciparum parasitemia as compared with parasite-free children (Weiss et al., 2009); previously exposed subjects significantly reduce the frequency of AMB following a year of continuous absence of exposure to Plasmodium falciparum infection (Ayieko et al., 2013). These observations all support the view that constant immune activation rather than impaired memory function leads to the accumulation of AMB in malaria.
After resolution of infection, P. chabaudi-specific AMB did not persist, but instead subsets of P. chabaudi-specific Bmem were readily detected. These cells expressed different combinations of previously described mouse B-cell memory markers [i.e. CD80, CD273 and CD73 (Anderson et al., 2007; Tomayko et al., 2010; Zuccarino-Catania et al., 2014)]. P. chabaudi-specific Bmem included both class-switched and non-class-switched cells, which show different responses to a secondary challenge infection (Krishnamurty et al., 2016). Independently of the combination of previously described memory markers expressed on P. chabaudi-specific Bmem, all of these cells displayed very high expression of FCRL5. Previous data showed most prominent expression of FCRL5 in marginal zone B cells, while much less evident in the newly-formed and follicular splenic B cell subpopulations (Davis et al., 2004; Won et al., 2006). In agreement with this, we observed expression of FCRL5 on a subset of splenic P. chabaudi-specific B cells obtained from naïve mice. However, the level of FCRL5 expression on P. chabaudi-specific Bmem detected after resolution of the infection was noticeably higher than that of naïve B cells both at the protein and mRNA levels. In contrast to MSP121-specific AMB, these CD11b—CD11c—FCRL5hi MSP121-specific Bmem showed high expression of the hallmark memory and anti-apoptotic gene Bcl2 (Bhattacharya et al., 2007). Thus, after resolution of the infection, high expression of FCRL5 acted as a universal Bmem marker. Due to its complex dual ITIM/ITAM signaling capacity (Zhu et al., 2013), it is tempting to speculate that FCRL5 might serve as an important signal in the differentiation/maintenance of Bmem.
Tracking the fate of the different MSP121-specific B-cell subsets identified in this work will allow detailing the interplay between them. A model can be proposed in which antigen-specific AMB serve as an intermediate stage of differentiation between naïve and Bmem. Alternatively, antigen-specific AMB might represent early plasmablasts or recent GC emigrates, based on class-switching and the high expression of IgG by these cells. These scenarios are not necessarily antagonist, and might even occur in parallel. Moreover, B1 B cells have been shown to class-switch and contribute to serum IgG1, IgG2a and IgA to influenza (Baumgarth et al., 2005), and IgG-producing B1a B cells have been shown to accumulate in the spleen of a mouse model of systemic lupus erythematosus (Enghard et al., 2010). Therefore, we can’t rule out the possibility that AMB might represent a particular B1 B cell subset that expands in the spleen and blood in response to Plasmodium infection.
Our data suggest that the expansion of AMB in malaria is not a consequence of B-cell exhaustion, but rather a physiologic stage of B-cell activation, and that these cells are sustained in high frequencies by ongoing chronic infections. Thus, Plasmodium-specific AMB are neither ‘memory’, nor ‘atypical’. Importantly, our data demonstrate that robust expansion of Plasmodium-specific AMB does not hinder clearance of the infection, activation of germinal centers, or generation of Plasmodium-specific long-lived quiescent Bmem upon resolution of the infection.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference |
Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) |
B6.SJL-Ptprca Pepcb/BoyJ (B6.CD45.1) |
The Jackson Laboratory |
MGI:4819849 | Bred in the specific pathogen-free facilities of the MRC National Institute for Medical Research and The Francis Crick Institute |
| Genetic reagent (M. musculus) |
Rag2tm1Fwa(Rag2-/-) | The Jackson Laboratory |
MGI:1858556 | Bred in the specific pathogen- free facilities of the MRC National Institute for Medical Research and The Francis Crick Institute |
| Genetic reagent (M. musculus) |
IghNIMP23/+ | This paper | _ | Bred in the specific pathogen-free facilities of the MRC National Institute for Medical Research and The Francis Crick Institute |
| Strain, strain background (Plasmodium chabaudi chabaudi, strain AS) |
P. chabaudi | other | _ | European Malaria Reagent Repository, University of Edinburgh. |
| Strain, strain background (Anopheles stephensi, strain SD500, female) |
mosquitos | PMID: 23217144 | _ | Bred in Jean Langhorne's lab |
| Antibody | Monoclonal Rat Anti-CD11b |
BD Biosciences | 563553 | (dil 1/50) |
| Antibody | Monoclonal Hamster Anti-Mouse CD11c |
BD Biosciences | 561022 | (dil 1/50) |
| Antibody | Monoclonal Rat Anti-Mouse CD138 |
BD Biosciences | 553714 | (dil 1/400) |
| Antibody | Monoclonal Rat Anti-Mouse CD19 |
BD Biosciences | 565076 | (dil 1/200) |
| Antibody | Monoclonal Rat anti-Mouse CD19 |
Biolegend | 115530 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD19 |
Biolegend | 115543 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD1d |
Biolegend | 123510 | (dil 1/100) |
| Antibody | Monoclonal Rat anti-mouse CD2 |
Biolegend | 100112 | (dil 1/100) |
| Antibody | Monoclona Rat Anti-Mouse CD21/35 |
BD Biosciences | 563176 | (dil 1/100) |
| Antibody | Monoclona Rat Anti-Mouse CD21/35 |
BD Biosciences | 553818 | (dil 1/100) |
| Antibody | Monoclonal Rat anti-Mouse CD23 |
eBioscience | 25–0232 | (dil 1/100) |
| Antibody | Monoclonal Rat anti-Mouse CD273 |
BD Biosciences | 564245 | (dil 1/25) |
| Antibody | Armenian Hamster anti-Mouse CD3 |
Biolegend | 100336 | (dil 1/100) |
| Antibody | Monoclonal Rat anti-Mouse CD38 |
Biolegend | 102718 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD38 |
eBioscience | 17–0381 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD38 |
BD Biosciences | 740697 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD4 |
Biolegend | 100414 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD45.1 |
Biolegend | 110706 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD45.1 |
Biolegend | 110728 | (dil 1/400) |
| Antibody | Monoclonal Mouse anti-Mouse CD45.2 |
BD Biosciences | 563685 | (dil 1/50) |
| Antibody | Monoclonal Mouse anti-Mouse CD45.2 |
Biolegend | 109814 | (dil 1/50) |
| Antibody | Monoclonal Mouse anti-Mouse CD45.2 |
Biolegend | 109808 | (dil 1/50) |
| Antibody | Monoclonal Rat anti-Mouse CD45R/B220 |
BD Biosciences | 564449 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD45R/B220 |
Biolegend | 103224 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD45R/B220 |
eBioscience | 25–0452 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD73 |
BD Biosciences | 550741 | (dil 1/100) |
| Antibody | Monoclonal Armenian Hamster anti-Mouse CD80 |
Biolegend | 104729 | (dil 1/25) |
| Antibody | Monoclonal Rat anti-Mouse CD8a |
Biolegend | 100734 | (dil 1/400) |
| Antibody | Monoclonal Rat anti-Mouse CD9 |
BD Biosciences | 558749 | (dil 1/100) |
| Antibody | Monoclonal Rat anti-Mouse CD93 (AA4.1) |
eBioscience | 17–5892 | (dil 1/100) |
| Antibody | FCRL5 | PMID: 17082595 | _ | Produced in Randall Davis' lab (dil 1/400) |
| Antibody | Polyclonal Sheep anti-Mouse FCRL5 |
R and D Systems | FAB6756G | (dil 1/50) |
| Antibody | Monoclonal Rat Anti-Mouse T- and B-Cell Activation Antigen GL7 |
BD Biosciences | 562080 | (dil 1/100) |
| Antibody | Monoclonal Rat Anti-Mouse IgD |
Biolegend | 405725 | (dil 1/400) |
| Antibody | Monoclonal Rat Anti-Mouse IgD |
Biolegend | 405723 | (dil 1/400) |
| Antibody | Monoclonal Rat Anti-Mouse IgD |
Biolegend | 405710 | (dil 1/400) |
| Antibody | Monoclonal Rat Anti-Mouse IgG2b |
Biolegend | 406708 | (dil 1/25) |
| Antibody | Monoclonal Rat Anti-Mouse IgM |
Biolegend | 406512 | (dil 1/100) |
| Recombinant DNA reagent | VDJHNIMP23
anti-MSP121 variable region coding exon containing the Leader-V segment intron from gDNA of the NIMP23 hybridoma |
PMID: 7141700 | _ | Produced in Jean Langhorne's lab |
| Recombinant DNA reagent | C57Bl/6 IgH HEL variable region knock-in construct |
PMID: 12668643 | _ | Donated by Robert Brink of the Garvan Institute of Medical Research, New South Wales, Australia |
| Peptide, recombinant protein | MSP121 | PMID: 11254580 | _ | Produced in Jean Langhorne's lab |
| Commercial assay or kit | EZ-Link Sulfo-NHS-LC-Biotinylation Kit |
Thermo Scientific | 21435 | |
| Commercial assay or kit | RiboPure RNA Purification Kit |
Invitrogen | AM1924 | |
| Commercial assay or kit | Qubit 1X dsDNA HS Assay Kit |
Invitrogen | Q33231 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing |
Takara | 634889 | |
| Commercial assay or kit | Ovation Ultralow Library System V2 |
Nugen | 0344–32 | |
| Commercial assay or kit | LIVE/DEAD Fixable Aqua Dead Cell Stain Kit |
Invitrogen | L34957 | |
| Commercial assay or kit | LIVE/DEAD Fixable Blue Dead Cell Stain Kit |
Invitrogen | L23105 | |
| Chemical compound, drug | Streptavidin-R-Phycoerythrin | Prozyme | PJRS25 | |
| Chemical compound, drug | Streptavidin-Allophycocyanin | Prozyme | PJ27S | |
| Chemical compound, drug | TiterMax Gold Adjuvant |
Merck (formerly Sigma-Aldrich) | T2684-1ML | |
| Chemical compound, drug | R848 (Resiquimod) | Invivogen | tlrl-r848 | |
| Chemical compound, drug | TRI Reagent Solution |
Invitrogen | AM9738 | |
| Software, algorithm | cutadapt v1.9.1 | doi:10.14806/ej.17.1.200 | ||
| Software, algorithm | RSEM v1.2.31 | doi:10.1186/1471-2105-12-323 | ||
| Software, algorithm | STAR v2.5.1b | doi:10.1093/ bioinformatics/bts635 |
||
| Software, algorithm | DESeq2 | doi:10.1186/s13059-014-0550-8 | ||
| Software, algorithm | R v3.4.0 | other | https://www.r-project.org | |
| Software, algorithm | Bioconductor v3.5 |
other |
http://www.
bioconductor.org |
|
| Software, algorithm | Broad's Gene Set Enrichment Analysis (GSEA) |
other | http://software.broadinstitute.org/gsea/index.jsp | |
| Software, algorithm | FlowJo version 9.6 or higher | Tree Star | ||
| Software, algorithm | Cytofkit | doi:10.1371/journa l.pcbi.1005112.s009 |
||
| Software, algorithm | Prism v6 | GraphPad |
Mice
5-12 week-old female mice were used for experiments. C57BL/6J, C57BL/6.SJL-Ptprca (CD45.1 congenic), Rag2-/-.C57BL/6.SJL-Ptprca (CD45.1 congenic) and BALB/c mouse strains were bred in the specific pathogen-free facilities of the MRC National Institute for Medical Research and The Francis Crick Institute, and were housed conventionally with sterile bedding, food and irradiated water. Room temperature was 22°C with a 12 hr light/dark cycle; food and water were provided ad libitum. The study was carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 (Home Office license 80/2538 and 70/8326), was approved by the MRC National Institute for Medical Research Ethical Committee and was approved by The Francis Crick Institute Ethical Committee.
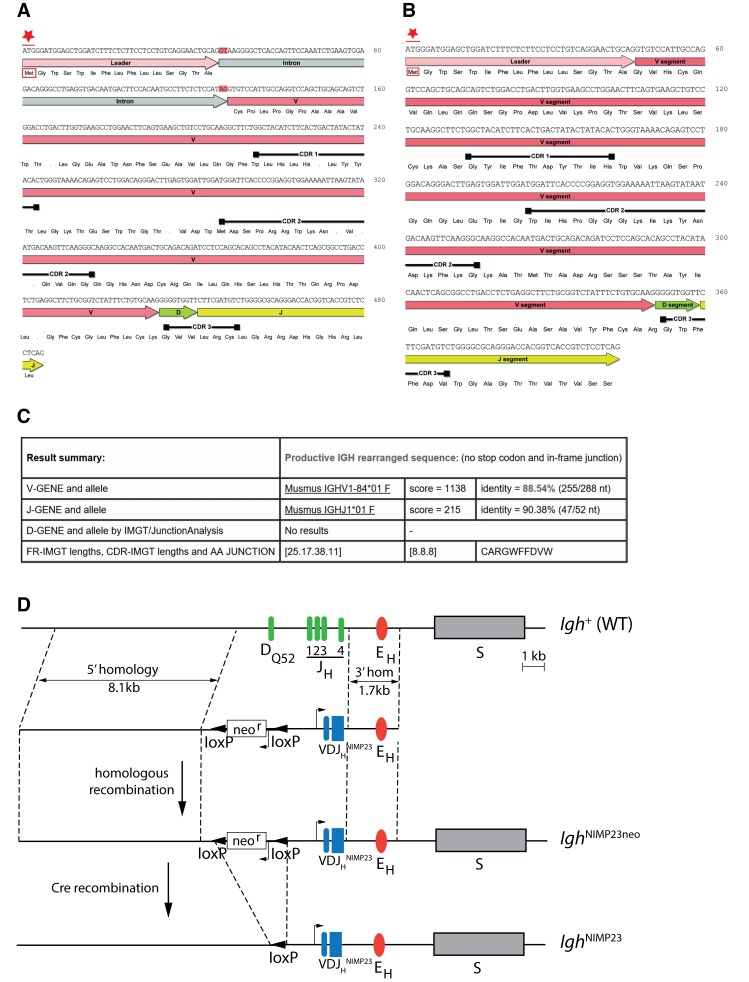
To produce MSP121–specific B cell knock-in mice capable of undergoing class switch recombination on the C57BL/6J genetic background, the VDJHNIMP23 anti-MSP121 variable region coding exon containing the Leader-V segment intron from gDNA of the NIMP23 hybridoma (Boyle et al., 1982) was inserted by homologous recombination into the 5’ end of the endogenous IgH locus (Taki et al., 1993) (Figure 1—figure supplement 1A–C). The VDJHNIMP23 anti-MSP121 variable region coding exon was inserted into a previously described IgH targeting construct, replacing the anti-HEL heavy chain variable region coding exon that was already in it (Phan et al., 2003) (Figure 1—figure supplement 1D). The final targeting construct included a loxP-flanked neomycin resistance cassette in reverse transcriptional orientation to the Igh locus, located immediately 5’ to the rearranged VDJHNIMP23 variable region and its associated promoter (Figure 1—figure supplement 1D). Electroporation of C57BL/6N-derived PRX embryonic stem cells with the targeting construct and selection of homologous recombinant clones was performed using standard techniques by PolyGene AG (Switzerland). One targeted ES clone was used for production of chimeric mice using standard techniques at the Biological Research Facilities of the MRC National Institute for Medical Research, London, UK. Male chimeric mice were crossed to C57BL/6J females and progeny carrying the IghNIMP23neo allele were crossed to PC3Cre mice (O'Gorman et al., 1997) to delete the neor gene in the germline and generate mice carrying the IghNIMP23 allele (Figure 1—figure supplement 1D). The IghNIMP23/+ strain was maintained by backcrossing for at least 10 generations to C57BL/6J mice.
Mixed bone marrow chimeras
Femurs and tibias were excised from female mice and cleaned of flesh using forceps and scalpel, and BM was obtained by flushing out with IMDM supplemented with 2 mM L-glutamine, 0.5 mM sodium pyruvate, 100 U penicillin, 100 mg streptomycin, 6 mM Hepes buffer, and 50 mM 2-ME (Gibco, Invitrogen), using a syringe with a needle. Thereafter, single BM cell suspensions were obtained by mashing through a 70 μm filter mesh, further sieved through 40 μm filter mesh and washed once. Live cells were resuspended in sodium chloride solution 0.9% (Sigma) at 4 × 106 cells/200 μl. Rag2-/-.C57BL/6.SJL-Ptprca mice were sub-lethally irradiated (5Gy) using a [137Cs] source and reconstituted less than 24 hr after irradiation by i.v. injections of a 10% IghNIMP23/+:90% C57BL/6.SJL-Ptprca combination of donor BM cells. Recipient mice were maintained on acidified drinking water and analyzed for reconstitution after 6–8 weeks.
Plasmodium chabaudi infection
Plasmodium chabaudi chabaudi AS was transmitted by Anopheles stephensi mosquitoes, strain SD500, as described elsewhere (Spence et al., 2012). Briefly, C57BL/6J mice were injected i.p. with 105 P. chabaudi-infected red blood cells and used to feed mosquitos two weeks after the injection. Two weeks after mosquito feeding/infection, each experimental mouse was exposed to 20 infected mosquitos for 30 min. Blood parasitemia in infected experimental mice was routinely monitored by thin blood smears.
Immunizations
Mice were immunized i.p. with a combination of 100 μg of MSP121 (Quin and Langhorne, 2001) and 50 μl of Titermax Gold emulsion (Sigma), or a combination of 50 μg of MSP121 and 50 μg of R848 (Invivogen).
Flow cytometry and cell sorting
Spleens, lymph nodes and bone marrows were dissected and single cell suspensions were obtained by mashing the organs through a 70 μm filter mesh in HBSS, 6 mM Hepes buffer (Gibco, Invitrogen). After removal of red blood cells from spleens and bone marrows by treatment with lysing buffer (Sigma), the remaining cells were resuspended in complete Iscove's Modified Dulbecco's Medium [IMDM supplemented with 10% FBS Serum Gold (PAA Laboratories, GE Healthcare), 2 mM L-glutamine, 0.5 mM sodium pyruvate, 100U penicillin, 100 mg streptomycin, 6 mM Hepes buffer, and 50 mM 2-ME (all from Gibco, Invitrogen)] and viable cells were counted using trypan blue (Sigma) exclusion and a hemocytometer. Cells were then resuspended in PBS and incubated with APC- or PE-labelled MSP121 fluorescent probes and/or different combinations of fluorochrome-conjugated antibodies (key resources table), and either acquired after two washes with PBS, or fixed with 2% paraformaldehyde and stored in staining buffer at 4°C until acquisition.
The APC and PE MSP121 fluorescent probes were produced as previously described (Krishnamurty et al., 2016; Taylor et al., 2012). Briefly, purified MSP121 (Quin and Langhorne, 2001) was biotinylated using an EZ-link Sulfo-NHS-LC- Biotinylation kit (Thermo Fisher Scientic) using a 1:1 ratio of biotin to protein, and loaded onto Streptavidin-APC conjugated or Phycolink Streptavidin-R-PE conjugated (ProZyme) in a 6:1 ratio of MSP121:Streptavidin-fluorochrome.
Cell sorting was performed on a MoFlo XDP (Beckman Coulter) or a BD FACSAria Fusion (BD Biosciences) and the target cell populations were directly dispensed into TRIreagent (Ambion) and stored at −80°C until RNA isolation. Purity checks were routinely performed for all assays by sorting aliquots of cells into PBS containing 2% FCS and reacquiring them on the cell sorter.
Dead cells were routinely excluded from the analysis by staining with LIVE/DEAD Fixable Aqua or Blue stain (Invitrogen). Singlets were selected based on FCS-A vs FCS-H and further based on SSC-A vs SSC-H. ‘Fluorescence minus one’ (FMO) controls were routinely used to verify correct compensation and to set the thresholds for positive/negative events. Analysis was performed with FlowJo software version 9.6 or higher (Tree Star).
PhenoGraph and t-distributed stochastic neighbor embedding (t-SNE) were combined to analyze multiparameter flow cytometry data using the Cytofkit package (Chen et al., 2016). t-SNE renders high-dimensional single-cell data based on similarities into only two dimensions, and thus helps visualize multiparameter data (van der Maaten, 2008). PhenoGraph (Levine et al., 2015) allows partitioning of high-dimensional single-cell data into phenotypically coherent subpopulations (i.e. clusters). The relatedness of the cell clusters identified by PhenoGraph was inferred using Isomap (Cytofkit package), in which related clusters/subsets can be visualised close to each other.
RNA isolation, sequencing and data analysis
Total RNA from 1−5 × 104 cells sorted into TRIreagent (Ambion) was isolated using the Ribopure kit (Ambion). Concentration of purified RNA was determined by Qubit fluorometric quantitation using the HS assay kit (ThermoFisher Scientific), and the quality analyzed with a 2100 Bioanalyzer (Agilent). Samples with a RIN score above 8.50 were used for the next steps. cDNA was generated from total RNA with the SMART-Seq v4 Ultra Low Input RNA Kit (Takara Bio USA). Next-generation sequencing libraries were produced with the Ovation Ultralow System V2 (Nugen), and run as PE100 on a HiSeq 4000 sequencer (Illumina). GEO accession: GSE115155.
For bioinformatics analysis, paired-end sequence reads were adapter and quality trimmed using cutadapt v1.9.1 (Martin, 2011) with the following non-default settings: ‘-a AGATCGGAAGAGC -A AGATCGGAAGAGC --minimum-length 30 -q 20,20’. Gene-level abundance estimates were generated from the trimmed reads using RSEM v1.2.31 (Li and Dewey, 2011) running STAR v2.5.1b (Dobin et al., 2013) with default settings, aligned against the Mus musculus Ensembl release 89 transcriptome (mm10). All further analysis was conducted using the DESeq2 (Love et al., 2014) package from Bioconductor v3.5 run in R v3.4.0. The expected counts were imported and rounded to integers to generate a counts matrix. Differential expression between phenotype groups was assessed using the DESeq function with default settings. In the case of comparisons of different MSP121-specific B cell subsets obtained from the same experimental mouse, an additional mouse factor was added to the design formula to accommodate the paired nature of the data. Significance was thresholded using an FDR ≤ 0.01. PCA analysis was conducted using DESeq's plot PCA function with the regularized log (rlog) transformed count data. Heat maps were generated using the regularized log (rlog) transformed count data, scaled per gene using a z-score. Mouse homologues to genes previously associated with human AMB were selected (Supplementary file 1), and those showing significant differential expression on MSP121-specific AMB were used to produce separate heat maps split by functional annotation (Figure 3). The GSEA pre-ranked function from the Broad's Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) suite was used to assess significant enrichment of MSigDB's C2 Reactome gene sets associated with differential expression between cell types. The function was run using a list of genes ranked for differential expression using DESeq2's Wald test statistic with default settings except for: collapse dataset to gene = false enrichment statistic = classic
Statistical analysis
Statistical analysis was performed using Mann Whitney U test, Kruskal-Wallis test followed by Dunn's multiple comparisons test, or Two-Way ANOVA followed by Dunnett's multiple comparisons test on Prism software version 6 (GraphPad). p<0.05 was accepted as a statistically significant difference.
Acknowledgements
We are grateful to Graham Preece, Philip Hobson and the Flow Cytometry Facility, Jackie Holland and the Biological Research Facility, Richard Mitter and the Bioinformatics Facility, and the Advanced Sequencing Facility at the Francis Crick Institute and the former MRC National Institute for Medical Research, London, UK. We thank Robert Brink, Garvan Institute of Medical Research, New South Wales, Australia, for donating the the C57Bl/6 IgH HEL variable region knock-in construct; Marion Pepper, University of Washington, for her support with the preparation of the MSP121 tetramers; and Dinis Calado and Matthew Lewis, Francis Crick Institute, for their critical reading of the manuscript.
This work was supported by the Francis Crick Institute which receives its core funding from the UK Medical Research Council (FC001101), Cancer Research UK (FC001101) and the Wellcome Trust (FC001101); by the Wellcome Trust (grant reference WT101777MA). Irene Tumwine is the recipient of a Francis Crick PhD studentship.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Jean Langhorne, Email: Jean.Langhorne@crick.ac.uk.
Facundo D Batista, Ragon Institute of MGH, MIT and Harvard, United States.
Michel C Nussenzweig, The Rockefeller University, United States.
Funding Information
This paper was supported by the following grants:
National Institutes of Health AI110553 to Randall S Davis.
Medical Research Council FC001101 and FC001194 to Victor LJ Tybulewicz.
Cancer Research UK FC001101 and FC001194 to Victor LJ Tybulewicz.
Wellcome Trust FC001101 and FC001194 to Victor LJ Tybulewicz.
Medical Research Council FC001101 to Jean Langhorne.
Wellcome Trust FC001101 to Jean Langhorne.
Cancer Research UK FC001101 to Jean Langhorne.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing—original draft.
Conceptualization, Formal analysis, Validation, Investigation, Methodology, Writing—review and editing.
Conceptualization, Formal analysis, Supervision, Investigation, Methodology, Writing—review and editing.
Investigation, Writing—review and editing.
Investigation, Writing—review and editing.
Investigation, Writing—review and editing.
Resources, Writing—review and editing.
Conceptualization, Supervision, Methodology, Writing—review and editing.
Conceptualization, Supervision, Validation, Methodology, Funding acquisition, Writing—review and editing.
Conceptualization, Supervision, Funding acquisition, Validation, Methodology, Project administration, Writing—review and editing.
Ethics
Animal experimentation: The study was carried out in accordance with the UK Animals (Scientific Procedures) Act 1986, Home Office license 80/2538 and 70/8326. Was approved by the MRC National Institute for Medical Research Ethical Committee, The Francis Crick Institute Ethical Committee, and further approved by the Home Office of the UK upon granting of the HO license.
Additional files
Data availability
RNAseq transcriptome data have been deposited in GEO under accession code GSE115155.
The following dataset was generated:
Pérez-Mazliah D, Gardner PJ, Schweighoffer E, McLaughlin S, Hosking C, Tumwine I, Davis RS, Potocnik A, Tybulewicz V, Langhorne J. 2018. Plasmodium-specific atypical memory B cells are short-lived activated B cells. NCBI Gene Expression Omnibus. GSE115155
References
- Achtman AH, Stephens R, Cadman ET, Harrison V, Langhorne J. Malaria-specific antibody responses and parasite persistence after infection of mice with Plasmodium chabaudi chabaudi. Parasite Immunology. 2007;29:435–444. doi: 10.1111/j.1365-3024.2007.00960.x. [DOI] [PubMed] [Google Scholar]
- Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. The Journal of Experimental Medicine. 2007;204:2103–2114. doi: 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayieko C, Maue AC, Jura WG, Noland GS, Ayodo G, Rochford R, John CC. Changes in B Cell Populations and Merozoite Surface Protein-1-Specific Memory B Cell Responses after Prolonged Absence of Detectable P. falciparum Infection. PLoS ONE. 2013;8:e67230. doi: 10.1371/journal.pone.0067230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett BE, Staupe RP, Odorizzi PM, Palko O, Tomov VT, Mahan AE, Gunn B, Chen D, Paley MA, Alter G, Reiner SL, Lauer GM, Teijaro JR, Wherry EJ. Cutting Edge: B Cell-Intrinsic T-bet Expression Is Required To Control Chronic Viral Infection. The Journal of Immunology. 2016;197:1017–1022. doi: 10.4049/jimmunol.1500368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N, Tung JW, Herzenberg LA. Inherent specificities in natural antibodies: a key to immune defense against pathogen invasion. Springer Seminars in Immunopathology. 2005;26:347–362. doi: 10.1007/s00281-004-0182-2. [DOI] [PubMed] [Google Scholar]
- Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nature Reviews Immunology. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. The Journal of Immunology. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle DB, Newbold CI, Smith CC, Brown KN. Monoclonal antibodies that protect in vivo against Plasmodium chabaudi recognize a 250,000-dalton parasite polypeptide. Infection and Immunity. 1982;38:94–102. doi: 10.1128/iai.38.1.94-102.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugat T, Reid AJ, Lin J, Cunningham D, Tumwine I, Kushinga G, McLaughlin S, Spence P, Böhme U, Sanders M, Conteh S, Bushell E, Metcalf T, Billker O, Duffy PE, Newbold C, Berriman M, Langhorne J. Antibody-independent mechanisms regulate the establishment of chronic Plasmodium infection. Nature Microbiology. 2017;2:16276. doi: 10.1038/nmicrobiol.2016.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpio VH, Opata MM, Montañez ME, Banerjee PP, Dent AL, Stephens R. IFN-γ and IL-21 Double Producing T Cells Are Bcl6-Independent and Survive into the Memory Phase in Plasmodium chabaudi Infection. PloS ONE. 2015;10:e0144654. doi: 10.1371/journal.pone.0144654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles ED, Brunetti C, Marukian S, Ritola KD, Talal AH, Marks K, Jacobson IM, Rice CM, Dustin LB. Clonal B cells in patients with hepatitis C virus-associated mixed cryoglobulinemia contain an expanded anergic CD21low B-cell subset. Blood. 2011;117:5425–5437. doi: 10.1182/blood-2010-10-312942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS Computational Biology. 2016;12:e1005112. doi: 10.1371/journal.pcbi.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Chen S, Zhang H, Brady GF, Ungewitter EK, Bradley JK, Sackey FN, Foster MH. Multifunctional regulators of cell growth are differentially expressed in anergic murine B cells. Molecular Immunology. 2007;44:1274–1285. doi: 10.1016/j.molimm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Davis RS, Stephan RP, Chen CC, Dennis G, Cooper MD. Differential B cell expression of mouse Fc receptor homologs. International Immunology. 2004;16:1343–1353. doi: 10.1093/intimm/dxh137. [DOI] [PubMed] [Google Scholar]
- Davis RS. Fc receptor-like molecules. Annual Review of Immunology. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- de Bree GJ, Wheatley AK, Lynch RM, Prabhakaran M, Grijsen ML, Prins JM, Schmidt SD, Koup RA, Mascola JR, McDermott AB. Longitudinal dynamics of the HIV-specific B cell response during intermittent treatment of primary HIV infection. PloS ONE. 2017;12:e0173577. doi: 10.1371/journal.pone.0173577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman JR, Bejon P, Ndungu FM, Langhorne J, Kortok MM, Lowe BS, Mwangi TW, Williams TN, Marsh K. B cell memory to 3 Plasmodium falciparum blood-stage antigens in a malaria-endemic area. The Journal of Infectious Diseases. 2005;191:1623–1630. doi: 10.1086/429671. [DOI] [PubMed] [Google Scholar]
- Ehrhardt GR, Davis RS, Hsu JT, Leu CM, Ehrhardt A, Cooper MD. The inhibitory potential of Fc receptor homolog 4 on memory B cells. PNAS. 2003;100:13489–13494. doi: 10.1073/pnas.1935944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt GR, Hsu JT, Gartland L, Leu CM, Zhang S, Davis RS, Cooper MD. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. The Journal of Experimental Medicine. 2005;202:783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enghard P, Humrich JY, Chu VT, Grussie E, Hiepe F, Burmester GR, Radbruch A, Berek C, Riemekasten G. Class switching and consecutive loss of dsDNA-reactive B1a B cells from the peritoneal cavity during murine lupus development. European Journal of Immunology. 2010;40:1809–1818. doi: 10.1002/eji.200940050. [DOI] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P. The Reactome Pathway Knowledgebase. Nucleic Acids Research. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraud O, Borhis G, Badr G, Degrelle S, Pozzetto B, Cognasse F, Richard Y. Revisiting the B-cell compartment in mouse and humans: more than one B-cell subset exists in the marginal zone and beyond. BMC Immunology. 2012;13:63. doi: 10.1186/1471-2172-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth J, Butler NS, Roetynck S, Mwacharo J, Pierce SK, Bejon P, Crompton PD, Marsh K, Ndungu FM. Chronic exposure to Plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. The Journal of Immunology. 2013;190:1038–1047. doi: 10.4049/jimmunol.1202438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G, Wu TT. Kabat Database and its applications: future directions. Nucleic Acids Research. 2001;29:205–206. doi: 10.1093/nar/29.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Haynes NM, Xu Y, Nutt SL, Allende ML, Proia RL, Cyster JG. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. The Journal of Experimental Medicine. 2006;203:2683–2690. doi: 10.1084/jem.20061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM, Nutt SL. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–566. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Kardava L, Moir S, Wang W, Ho J, Buckner CM, Posada JG, O'Shea MA, Roby G, Chen J, Sohn HW, Chun TW, Pierce SK, Fauci AS. Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. Journal of Clinical Investigation. 2011;121:2614–2624. doi: 10.1172/JCI45685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardava L, Moir S, Shah N, Wang W, Wilson R, Buckner CM, Santich BH, Kim LJ, Spurlin EE, Nelson AK, Wheatley AK, Harvey CJ, McDermott AB, Wucherpfennig KW, Chun TW, Tsang JS, Li Y, Fauci AS. Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. Journal of Clinical Investigation. 2014;124:3252–3262. doi: 10.1172/JCI74351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JJ, Buggert M, Kardava L, Seaton KE, Eller MA, Canaday DH, Robb ML, Ostrowski MA, Deeks SG, Slifka MK, Tomaras GD, Moir S, Moody MA, Betts MR. T-bet+ B cells are induced by human viral infections and dominate the HIV gp140 response. JCI Insight. 2017a;2 doi: 10.1172/jci.insight.92943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JJ, Kaplan DE, Betts MR. T-bet-expressing B cells during HIV and HCV infections. Cellular Immunology. 2017b;321:26–34. doi: 10.1016/j.cellimm.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurty AT, Thouvenel CD, Portugal S, Keitany GJ, Kim KS, Holder A, Crompton PD, Rawlings DJ, Pepper M. Somatically Hypermutated Plasmodium-Specific IgM. Immunity. 2016:402–414. doi: 10.1016/j.immuni.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalor PA, Nossal GJ, Sanderson RD, McHeyzer-Williams MG. Functional and molecular characterization of single, (4-hydroxy-3-nitrophenyl)acetyl (NP)-specific, IgG1+ B cells from antibody-secreting and memory B cell pathways in the C57BL/6 immune response to NP. European Journal of Immunology. 1992;22:3001–3011. doi: 10.1002/eji.1830221136. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el-AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe'er D, Nolan GP. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Borrego F, Nagata S, Tolnay M. Fc receptor-like 5 expression distinguishes two distinct subsets of human circulating tissue-like memory b cells. The Journal of Immunology. 2016;196:4064–4074. doi: 10.4049/jimmunol.1501027. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:31. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O'Shea MA, Roby G, Kottilil S, Arthos J, Proschan MA, Chun TW, Fauci AS. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. The Journal of Experimental Medicine. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellenbeck MF, Ueberheide B, Amulic B, Epp A, Fenyo D, Busse CE, Esen M, Theisen M, Mordmüller B, Wardemann H. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. The Journal of Experimental Medicine. 2013;210:389–399. doi: 10.1084/jem.20121970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naradikian MS, Hao Y, Cancro MP. Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunological Reviews. 2016a;269:118–129. doi: 10.1111/imr.12380. [DOI] [PubMed] [Google Scholar]
- Naradikian MS, Myles A, Beiting DP, Roberts KJ, Dawson L, Herati RS, Bengsch B, Linderman SL, Stelekati E, Spolski R, Wherry EJ, Hunter C, Hensley SE, Leonard WJ, Cancro MP. Cutting Edge: IL-4, IL-21, and IFN-γ Interact To Govern T-bet and CD11c Expression in TLR-Activated B Cells. The Journal of Immunology. 2016b;197:1023–1028. doi: 10.4049/jimmunol.1600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndungu FM, Cadman ET, Coulcher J, Nduati E, Couper E, Macdonald DW, Ng D, Langhorne J. Functional memory B cells and long-lived plasma cells are generated after a single Plasmodium chabaudi infection in mice. PLoS Pathogens. 2009;5:e1000690. doi: 10.1371/journal.ppat.1000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndungu FM, Olotu A, Mwacharo J, Nyonda M, Apfeld J, Mramba LK, Fegan GW, Bejon P, Marsh K. Memory B cells are a more reliable archive for historical antimalarial responses than plasma antibodies in no-longer exposed children. PNAS. 2012;109:8247–8252. doi: 10.1073/pnas.1200472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndungu FM, Lundblom K, Rono J, Illingworth J, Eriksson S, Färnert A. Long-lived Plasmodium falciparum specific memory B cells in naturally exposed Swedish travelers. European Journal of Immunology. 2013;43:2919–2929. doi: 10.1002/eji.201343630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. PNAS. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng-Adjei N, Portugal S, Tran TM, Yazew TB, Skinner J, Li S, Jain A, Felgner PL, Doumbo OK, Kayentao K, Ongoiba A, Traore B, Crompton PD, Yeste A, Mascanfroni ID, Nadeau M, Burns EJ, Tukpah A-M, Santiago A, Wu C, Patel B, Kumar D, Quintana FJ. Circulating th1-cell-type tfh cells that exhibit impaired b cell help are preferentially activated during acute malaria in children. Cell Reports. 2015;13:425–439. doi: 10.1016/j.celrep.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng-Adjei N, Portugal S, Holla P, Li S, Sohn H, Ambegaonkar A, Skinner J, Bowyer G, Doumbo OK, Traore B, Pierce SK, Crompton PD. Malaria-induced interferon-γ drives the expansion of Tbethi atypical memory B cells. PLoS Pathogens. 2017;13:e1006576. doi: 10.1371/journal.ppat.1006576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mazliah D, Ng DH, Freitas do Rosário AP, McLaughlin S, Mastelic-Gavillet B, Sodenkamp J, Kushinga G, Langhorne J. Disruption of IL-21 signaling affects T cell-B cell interactions and abrogates protective humoral immunity to malaria. PLoS Pathogens. 2015;11:e1004715. doi: 10.1371/journal.ppat.1004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Amesbury M, Gardam S, Crosbie J, Hasbold J, Hodgkin PD, Basten A, Brink R. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. The Journal of Experimental Medicine. 2003;197:845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annual Review of Immunology. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- Portugal S, Pierce SK, Crompton PD. Young lives lost as B cells falter: what we are learning about antibody responses in malaria. The Journal of Immunology. 2013;190:3039–3046. doi: 10.4049/jimmunol.1203067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal S, Tipton CM, Sohn H, Kone Y, Wang J, Li S, Skinner J, Virtaneva K, Sturdevant DE, Porcella SF, Doumbo OK, Doumbo S, Kayentao K, Ongoiba A, Traore B, Sanz I, Pierce SK, Crompton PD. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. eLife. 2015;4:e07218. doi: 10.7554/eLife.07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal S, Obeng-Adjei N, Moir S, Crompton PD, Pierce SK. Atypical memory B cells in human chronic infectious diseases: An interim report. Cellular Immunology. 2017;321:18–25. doi: 10.1016/j.cellimm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quin SJ, Langhorne J. Different regions of the malaria merozoite surface protein 1 of Plasmodium chabaudi elicit distinct T-cell and antibody isotype responses. Infection and Immunity. 2001;69:2245–2251. doi: 10.1128/IAI.69.4.2245-2251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderstad A, Tarlinton DM. Kinetics of establishing the memory B cell population as revealed by CD38 expression. Journal of Immunology. 1998;160:4688–4695. [PubMed] [Google Scholar]
- Rivera-Correa J, Guthmiller JJ, Vijay R, Fernandez-Arias C, Pardo-Ruge MA, Gonzalez S, Butler NS, Rodriguez A. Plasmodium DNA-mediated TLR9 activation of T-bet+ B cells contributes to autoimmune anaemia during malaria. Nature Communications. 2017;8:1282. doi: 10.1038/s41467-017-01476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov AV, Marrack P, Rubtsova K. T-bet expressing B cells - Novel target for autoimmune therapies? Cellular Immunology. 2017;321:35–39. doi: 10.1016/j.cellimm.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Rubtsova K, Rubtsov AV, van Dyk LF, Kappler JW, Marrack P. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. PNAS. 2013;110:E3216–E3224. doi: 10.1073/pnas.1312348110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell Knode LM, Naradikian MS, Myles A, Scholz JL, Hao Y, Liu D, Ford ML, Tobias JW, Cancro MP, Gearhart PJ. Age-associated b cells express a diverse repertoire of vh and vκ genes with somatic hypermutation. The Journal of Immunology. 2017;198:1921–1927. doi: 10.4049/jimmunol.1601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen J, Rosenberg N, Burakoff SJ. Expression and ontogeny of CD2 on murine B cells. Journal of Immunology. 1990;144:2925–2930. doi: 10.1002/eji.1830190722. [DOI] [PubMed] [Google Scholar]
- Spence PJ, Jarra W, Lévy P, Nahrendorf W, Langhorne J. Mosquito transmission of the rodent malaria parasite Plasmodium chabaudi. Malaria Journal. 2012;11:407. doi: 10.1186/1475-2875-11-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence PJ, Jarra W, Lévy P, Reid AJ, Chappell L, Brugat T, Sanders M, Berriman M, Langhorne J. Vector transmission regulates immune control of Plasmodium virulence. Nature. 2013;498:228–231. doi: 10.1038/nature12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RT, Kim CC, Fontana MF, Feeney ME, Jagannathan P, Boyle MJ, Drakeley CJ, Ssewanyana I, Nankya F, Mayanja-Kizza H, Dorsey G, Greenhouse B. FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Plasmodium falciparum Exposure. PLoS Pathogens. 2015;11:e1004894. doi: 10.1371/journal.ppat.1004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RT, Ssewanyana I, Wamala S, Nankya F, Jagannathan P, Tappero JW, Mayanja-Kizza H, Muhindo MK, Arinaitwe E, Kamya M, Dorsey G, Feeney ME, Riley EM, Drakeley CJ, Greenhouse B. B cell sub-types following acute malaria and associations with clinical immunity. Malaria Journal. 2016;15 doi: 10.1186/s12936-016-1190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki S, Meiering M, Rajewsky K. Targeted insertion of a variable region gene into the immunoglobulin heavy chain locus. Science. 1993;262:1268–1271. doi: 10.1126/science.8235657. [DOI] [PubMed] [Google Scholar]
- Taylor JJ, Martinez RJ, Titcombe PJ, Barsness LO, Thomas SR, Zhang N, Katzman SD, Jenkins MK, Mueller DL, Martinez RJ, Mueller DL. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. The Journal of experimental medicine. 2012;209:2065–2077. doi: 10.1084/jem.20112272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomayko MM, Steinel NC, Anderson SM, Shlomchik MJ. Cutting edge: Hierarchy of maturity of murine memory B cell subsets. The Journal of Immunology. 2010;185:7146–7150. doi: 10.4049/jimmunol.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- Weiss GE, Crompton PD, Li S, Walsh LA, Moir S, Traore B, Kayentao K, Ongoiba A, Doumbo OK, Pierce SK. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. The Journal of Immunology. 2009;183:2176–2182. doi: 10.4049/jimmunol.0901297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GE, Traore B, Kayentao K, Ongoiba A, Doumbo S, Doumtabe D, Kone Y, Dia S, Guindo A, Traore A, Huang CY, Miura K, Mircetic M, Li S, Baughman A, Narum DL, Miller LH, Doumbo OK, Pierce SK, Crompton PD. The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. PLoS Pathogens. 2010;6:e1000912. doi: 10.1371/journal.ppat.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GE, Clark EH, Li S, Traore B, Kayentao K, Ongoiba A, Hernandez JN, Doumbo OK, Pierce SK, Branch OH, Crompton PD. A positive correlation between atypical memory B cells and Plasmodium falciparum transmission intensity in cross-sectional studies in Peru and Mali. PLoS ONE. 2011;6:e15983. doi: 10.1371/journal.pone.0015983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipasa J, Suphavilai C, Okell LC, Cook J, Corran PH, Thaikla K, Liewsaree W, Riley EM, Hafalla JC. Long-lived antibody and B Cell memory responses to the human malaria parasites, Plasmodium falciparum and Plasmodium vivax. PLoS Pathogens. 2010;6:e1000770. doi: 10.1371/journal.ppat.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won WJ, Foote JB, Odom MR, Pan J, Kearney JF, Davis RS. Fc receptor homolog 3 is a novel immunoregulatory marker of marginal zone and B1 B cells. The Journal of Immunology. 2006;177:6815–6823. doi: 10.4049/jimmunol.177.10.6815. [DOI] [PubMed] [Google Scholar]
- Young F, Ardman B, Shinkai Y, Lansford R, Blackwell TK, Mendelsohn M, Rolink A, Melchers F, Alt FW. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes & Development. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Li R, Li H, Zhou T, Davis RS. FCRL5 exerts binary and compartment-specific influence on innate-like B-cell receptor signaling. PNAS. 2013;110:E1282–E1290. doi: 10.1073/pnas.1215156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouali M. Marginal zone B-cells. A Gatekeeper of Innate Immunity. 2011;2 doi: 10.3389/fimmu.2011.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nature Immunology. 2014;15:631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]