Summary
Class switch recombination (CSR) is a DNA recombination reaction that diversifies the effector component of antibody responses. CSR is initiated by activation-induced cytidine deaminase (AID), which targets transcriptionally active immunoglobulin heavy chain (Igh) switch donor and acceptor DNA. The 3′ Igh super-enhancer, 3′ regulatory region (3′RR), is essential for acceptor region transcription, but how this function is regulated is unknown. Here, we identify the chromatin reader ZMYND8 as an essential regulator of the 3′RR. In B cells, ZMYND8 binds promoters and super-enhancers, including the Igh enhancers. ZMYND8 controls the 3′RR activity by modulating the enhancer transcriptional status. In its absence, there is increased 3′RR polymerase loading and decreased acceptor region transcription and CSR. In addition to CSR, ZMYND8 deficiency impairs somatic hypermutation (SHM) of Igh, which is also dependent on the 3′RR. Thus, ZMYND8 controls Igh diversification in mature B lymphocytes by regulating the activity of the 3′ Igh super-enhancer.
Keywords: class switch recombination, germline transcription, Igh super-enhancer, 3′ regulatory region, somatic hypermutation, ZMYND8
Graphical Abstract
Highlights
-
•
ZMYND8 is required for GLT of acceptor S regions and Class Switch Recombination
-
•
ZMYND8 supports efficient somatic hypermutation of the Igh variable regions
-
•
ZMYND8 binds B cell super-enhancers, including the 3′ Igh enhancer
-
•
ZMYND8 modulates the transcriptional status and activity of the 3′ Igh enhancer
Antibody diversity is essential for the establishment of an effective immune response. Delgado-Benito and Rosen et al. report that diversification of the immunoglobulin heavy chain (Igh) gene in mature B lymphocytes is regulated by the chromatin reader ZMYND8, which binds to and modulates the activity of the 3′ Igh locus super-enhancer.
Introduction
Class switch recombination (CSR) is a B-lymphocyte-specific somatic recombination reaction that replaces the Cμ constant region of the immunoglobulin heavy chain (Igh) locus with one of the downstream constant genes (Chaudhuri and Alt, 2004, Pavri and Nussenzweig, 2011). As a consequence, B cells switch from expressing antibody molecules of the immunoglobulin M (IgM) class to IgG, IgE, or IgA, which harbor the same antigen specificity but a different effector function. This process is essential for the establishment of an effective immune response as evidenced by primary human immunodeficiency syndromes that are associated with defects in CSR (Durandy et al., 2013).
The Igh locus spans over 250 kb, and comprises a rearranged variable, diversity, and joining (VDJ) exon, encoding the variable portion of the antibody molecule, followed by exons encoding constant (C) regions (Cμ, Cγ3, Cγ1, Cγ2b, Cγ2a, Cε, and Cα in mice), each preceded by highly repetitive stretches of DNA, known as switch (S) regions. The Igh locus contains two important transcriptional enhancers, Eμ and 3′ regulatory region (3′RR) that are essential for B cell development and function. In addition to supporting Igh expression, Eμ is required for efficient V(D)J recombination and early B cell development (Banerji et al., 1983, Gillies et al., 1983, Marquet et al., 2014, Perlot et al., 2005). The 3′RR is essential for late B cell differentiation, when it regulates antibody gene diversification by CSR and somatic hypermutation (SHM) in mature B cells (Cogné et al., 1994, Manis et al., 1998, Pinaud et al., 2001, Rouaud et al., 2013, Saintamand et al., 2015a, Vincent-Fabert et al., 2010).
CSR is mediated by activation-induced deaminase (AID), which targets cytosine residues within the S regions of activated B cells (Muramatsu et al., 2000, Revy et al., 2000). The lesions induced by AID initiate a cascade of enzymatic reactions resulting in the formation of DNA double-strand breaks (DSBs) (Boboila et al., 2012). Paired breaks at donor and acceptor S regions are then repaired by components of the nonhomologous end-joining (NHEJ) pathway that includes Ku70/80, DNA ligase IV, 53BP1, and its downstream interactor Rif1, thus resulting in deletion of the intervening sequence and expression of the newly switched heavy chain (Boboila et al., 2012, Chapman et al., 2013, Di Virgilio et al., 2013, Escribano-Díaz et al., 2013).
AID targeting is dependent on transcription across the S regions (germline transcription [GLT]), which exposes single-stranded DNA that is the substrate for this enzyme (Chaudhuri et al., 2003, Dickerson et al., 2003, Ramiro et al., 2003). GLT is initiated at a promoter coupled with an I (intervening) exon located upstream of each S region and terminates downstream of the corresponding CH gene (Gauchat et al., 1990, Lebman et al., 1990, Lennon and Perry, 1985, Lutzker and Alt, 1988, Radcliffe et al., 1990, Rothman et al., 1990a, Rothman et al., 1990b). Whereas transcription of the donor Sμ region is constitutive in naive B cells, GLT of acceptor regions is induced in a cytokine-dependent manner, which targets CSR to different isotypes (Berton et al., 1989, Collins and Dunnick, 1993, Esser and Radbruch, 1989, Gauchat et al., 1990, Lebman et al., 1990, Lutzker et al., 1988, Rothman et al., 1988, Severinson et al., 1990, Shockett and Stavnezer, 1991, Stavnezer et al., 1985, Stavnezer et al., 1988). The 3′RR acts as a major regulator of this process (Birshtein, 2014, Pinaud et al., 2011). The 3′RR is located downstream of Cα and contains four lymphoid-specific transcriptional enhancers (DNase 1 hypersensitive sites hs3a, hs1,2, hs3b, and hs4) in mice (Giannini et al., 1993, Lieberson et al., 1991, Madisen and Groudine, 1994, Matthias and Baltimore, 1993, Michaelson et al., 1995, Pettersson et al., 1990). hs1,2 is at the center of a ∼25-kb palindrome delimited by two inverted copies of the hs3 enhancers (hs3a and hs3b), with the distal hs4 module lying outside and downstream of the palindrome (Birshtein, 2014, Pinaud et al., 2011). Both hs core enhancers and surrounding sequences have proven to be crucial to promote CSR by regulating GLT and accessibility of the S regions (Cogné et al., 1994, Garot et al., 2016, Le Noir et al., 2017, Manis et al., 1998, Pinaud et al., 2001, Saintamand et al., 2015b, Vincent-Fabert et al., 2010). However, the mechanism by which the activity of the 3′RR is regulated has yet to be defined precisely.
Here, we identified zinc finger MYND-type containing 8 (ZMYND8) protein as a factor required for physiological levels of CSR. ZMYND8 is dispensable for repair of CSR breaks but acts upstream DSB formation by controlling 3′RR activity.
Results
The Chromatin Reader ZMYND8 Is Required for CSR
To investigate the regulation of antibody diversification by CSR, we determined the protein interactome of the CSR and DSB repair factor Rif1 in switching B lymphocytes. To this end, we applied the proteomics-based technique isotopic differentiation of interactions as random or targeted (I-DIRT) (Tackett et al., 2005) to B lymphocytes stimulated to undergo CSR (Figure S1). We employed primary cultures of splenocytes isolated from Rif1FH/FH mice (Cornacchia et al., 2012), which express physiological levels of a knockin 1×Flag-2×hemagglutinin (HA)-tagged version of Rif1 that supports normal levels of CSR (Figures S1A–S1C). As expected, the two top hits were the Rif1 bait and its phospho-dependent interactor 53BP1 (Figures S1D and S1E). Several proteins exhibited I-DIRT ratios that were multiple SDs above the mean, suggesting that they are specifically associated with Rif1. Among these were a number of transcriptional regulators, including MGA, Zfp592, BACH2, TCEB1, and ZMYND8 (Figures S1D and S1E).
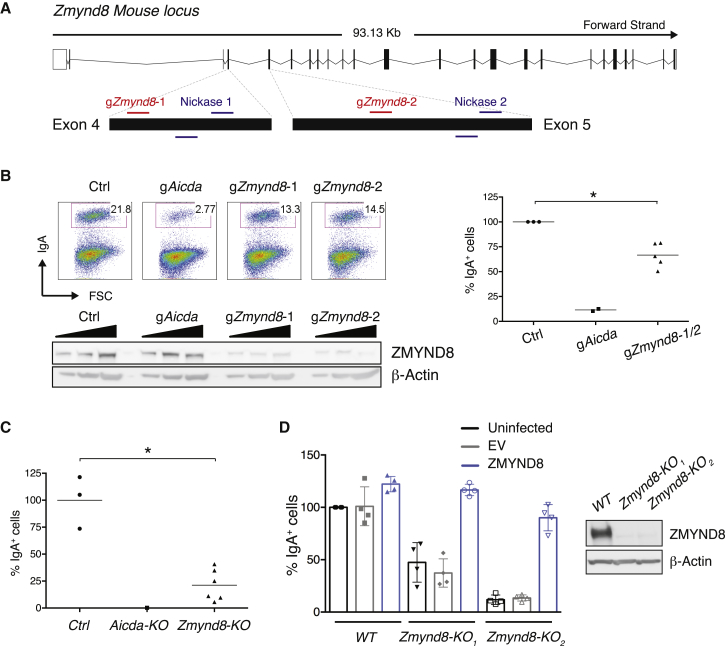
To identify bona fide CSR factors, candidates with high I-DIRT ratio were assessed by somatic targeting using CRISPR-Cas9 in the B cell lymphoma line CH12 (Nakamura et al., 1996; Figures S1F and S1G). Upon cytokine stimulation, CH12 cells express AID and undergo class switching to IgA with high efficiency. Moreover, ablation of factors known to be essential for CSR, such as AID, 53BP1, and Rif1, reduces CSR in CH12 cells (Figure S1G). Among the candidates tested, ZMYND8 showed the greatest impairment in CSR (Figures 1A, 1B, and S1G). In agreement with the targeting results in bulk cultures, ZMYND8-deficient CH12 clones exhibited reduced levels of CSR compared to controls (Figure 1C). Furthermore, complementation of Zmynd8−/− CH12 cell lines with full-length ZMYND8 fully rescued CSR, thus confirming that the defect is specifically caused by ZMYND8 deficiency (Figure 1D).
Figure 1.
ZMYND8 Ablation in CH12 Impairs CSR
(A) Scheme of Zmynd8 genomic locus and location of guide RNAs (gRNAs) used in this study.
(B) (Left top) Representative flow cytometry plots measuring CSR to IgA in activated Cas9/gZmynd8-nucleofected CH12 cells. (Left bottom) Western blot analysis of whole-cell extracts from nucleofected CH12 cultures is shown. (Right) Summary dot plot indicating CSR as a percentage of the control (Ctrl) value within the same experiment is shown. The graph summarizes three independent experiments. Aicda, AID-encoding gene.
(C) Graph depicting CSR to IgA in activated CH12 clonal derivatives. Each cell line was normalized to the mean value of the control clones, which was set to 100%. Graph is representative of at least two independent experiments.
(D) (Left) Summary dot plot for four independent experiments measuring CSR to IgA 48 hr after activation of two Zmynd8−/− CH12 cell lines (KO1 and KO2) reconstituted with either empty vector (EV) or full-length ZMYND8 (top legend). CSR is expressed as a percentage of the uninfected WT within the same experiment. (Right) Representative WB analysis of the CH12 cell lines is shown.
Significance in (B) and (C) was calculated with the Mann-Whitney U test. Error bars in (D) represent SD. ∗p ≤ 0.05. See also Figure S1.
To validate the I-DIRT result and confirm that ZMYND8 is a Rif1 interactor, we performed reciprocal co-immunoprecipitation experiments in primary B cells. We found that ZMYND8 was efficiently co-immunoprecipitated with Rif1 and vice versa (Figures S1H and S1I). Ionizing irradiation (IR)-induced DNA damage resulted in the association between Rif1 and 53BP1 in a manner dependent on the DSB repair kinase ataxia-telangiectasia mutated (ATM) (Figure S1H; Di Virgilio et al., 2013). In contrast, ZMYND8 co-immunoprecipitated with Rif1 irrespective of DNA damage or ATM activity (Figures S1H and S1I). We concluded that ZMYND8 interacts with Rif1 in a DNA-damage-independent manner.
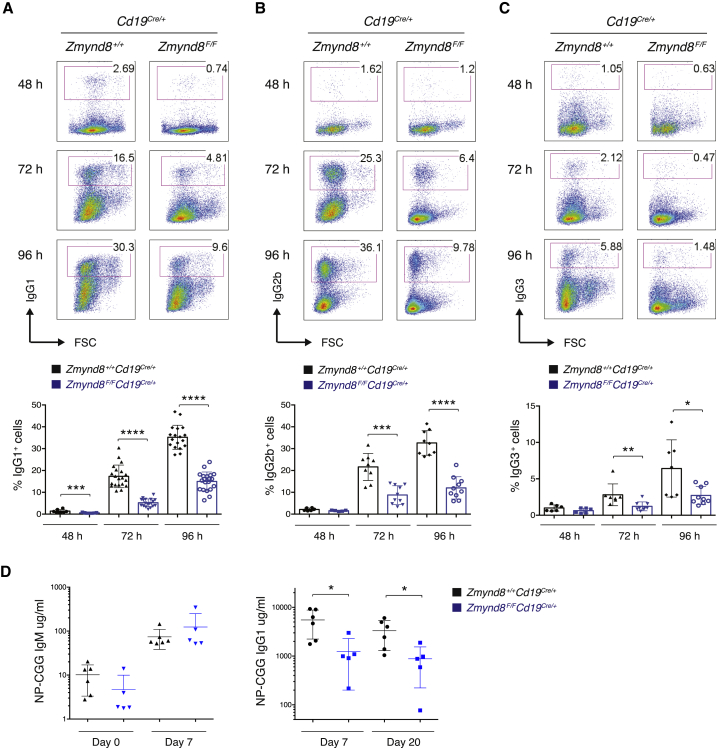
To confirm the findings obtained in CH12 cells, we specifically deleted ZMYND8 in B cells by combining a Zmynd8F conditional allele with Cd19-Cre, which drives Cre expression at early stages of B cell differentiation (Rickert et al., 1997; Figure S2A). B cells from Zmynd8F/FCd19Cre/+ mice developed normally (Figures S2B and S2C), thus indicating that ZMYND8 is dispensable for V(D)J recombination and B cell development. To assess whether ZMYND8 is required for CSR in primary B cells, we activated splenocytes with appropriate stimuli to induce switching to IgG1, IgG2b, and IgG3. ZMYND8 protein was readily detectable in resting B cells, and expression levels did not change following activation (Figure S2D). Mature ZMYND8-deficient B cells showed a severe defect in CSR to all tested isotypes (Figures 2A–2C). To assess whether the defect we observed in primary B cell cultures reflected a reduced capability to mount a proper immune response in vivo, we immunized Cd19Cre/+ and Zmynd8F/FCd19Cre/+ mice with the T-cell-dependent antigen 4-Hydroxy-3-nitrophenylacetyl hapten conjugated to Chicken Gamma Globulin (NP-CGG). The generation of NP-specific IgG1 antibodies was reduced nearly 5-fold in ZMYND8-deficient mice compared to control group (Figure 2D). We conclude that ZMYND8 is essential for physiologic levels of CSR in primary B lymphocytes.
Figure 2.
ZMYND8 Is Required for CSR In Vivo
(A–C) (Top) Representative flow cytometry plots measuring CSR to IgG1 (A), IgG2b (B), and IgG3 (C) in activated splenocytes. (Bottom) Summary dot plot for at least 5 mice per time point per genotype is shown.
(D) NP-specific IgM and IgG1 antibody titers in serum collected before (day 0) and at day 7 and 20 after immunization with NP-CGG. NP-IgG1 antibodies were undetectable at day 0.
Significance in (A)–(D) was calculated with the Mann-Whitney U test. Error bars in (A)–(D) represent SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S2.
ZMYND8 Is Dispensable for DSB Repair
CSR is dependent on the detection, signaling, and repair of AID-induced DSBs (Boboila et al., 2012, Pavri and Nussenzweig, 2011). To determine whether loss of ZMYND8 alters any of these processes, we produced and analyzed ZMYND8-deficient mouse embryonic fibroblasts (MEFs) cell lines (Figures S3A and S3B). DSB signaling was measured by assaying for formation of IR-induced foci (IRIF) of γH2AX, 53BP1, and Rif1. In all cases, formation of DNA damage foci was normal in ZMYND8-deficient cell lines (Figure S3C). Thus, ZMYND8 is not required for DSB detection and early IR-induced signaling in this model system.
In addition, Zmynd8−/− MEFs and Zmynd8−/− CH12 cell lines showed no measurable defect in survival after IR (Figures S4A–S4C). Given reports that implicated ZMYND8 in homologous recombination (HR)-dependent repair of DSBs within transcriptionally active loci (Gong et al., 2015, Gong et al., 2017, Kloet et al., 2015, Spruijt et al., 2016, Xia et al., 2017), we assessed cell survival and genomic instability in response to treatment with PARP inhibitor (PARPi), which promotes DNA-replication-associated damage that is physiologically repaired by HR. ZMYND8-deficient MEFs displayed no increase in sensitivity to PARPi (Figures S4D and S4E). Furthermore, DNA end protection from resection and repair of CRISPR-Cas9-induced DSBs were normal in Zmynd8−/− CH12 cells (Figures S4F and S4G). Altogether, these findings indicate that ZMYND8 is dispensable for DSB signaling and repair in MEFs and CH12 cells.
To determine whether ZMYND8 contributes to repair of AID-induced breaks, we analyzed residual Sμ-Sγ1 junctions cloned from Zmynd8F/FCd19Cre/+ B cells stimulated with lipopolysaccharide (LPS) and interleukin-4 (IL-4). We found that the frequency and length of microhomology at the junctions were similar in Cd19Cre/+ and Zmynd8F/FCd19Cre/+ mice, thus indicating that ZMYND8 deficiency does not bias the end-joining repair of AID-induced Igh breaks toward the use of microhomology (Table S1). We concluded that ZMYND8 does not influence the choice between NHEJ and alternative end-joining (A-EJ) of S regions DSBs.
ZMYND8 Promotes Germline Transcription of Acceptor S Regions
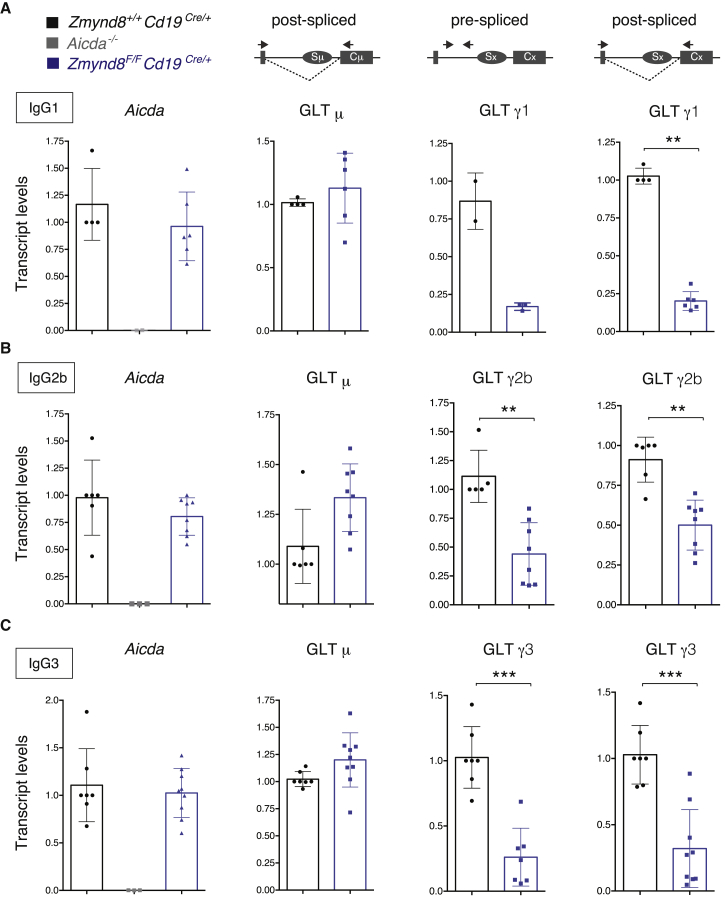
The level of CSR in activated B cells is directly dependent on the level of AID expression (Dorsett et al., 2008, Takizawa et al., 2008, Teng et al., 2008). To determine whether decreased CSR in Zmynd8F/FCd19Cre/+ splenocytes is due to altered AID expression, we measured AID mRNA by qPCR assays. AID expression in ZMYND8-deficient splenocytes was indistinguishable from controls (Figures 3A–3C). Moreover, a general survey of the transcriptome of Zmynd8−/− and control CH12 cell lines by RNA sequencing (RNA-seq) (Figures S5A and S5B; Tables S2 and S3) showed no downregulation of the expression of known CSR factors (Figure S5C). We concluded that ZMYND8 deletion does not alter CSR by interfering with the expression of AID or other known essential regulators of CSR.
Figure 3.
ZMYND8 Deficiency Causes Defective Germline Transcription of Acceptor S Regions
qPCR analysis for Aicda mRNA, Igμ, and Igγ1 (A), Igγ2b (B), and Igγ3 (C) GLT levels in B cells activated to undergo CSR to the corresponding isotypes. The schematic representations indicate the location of primers employed to analyze pre- and post-spliced germline transcripts. One Cd19Cre/+ mouse within each experiment was assigned an arbitrary value of 1 (error bars represent SD). Significance was calculated with the Mann-Whitney U test. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001. See also Figure S5.
In addition to AID expression, CSR is dependent on normal cell cycle progression. To determine whether ZMYND8 deficiency is required for cell division in B cells, we analyzed the proliferation of activated Zmynd8F/FCd19Cre/+ splenocytes by cell tracking dye dilution. The proliferation profile of ZMYND8-deficient B cells was indistinguishable from their wild-type (WT) counterpart at all time points after activation (Figures S6A–S6C), whereas CSR levels were reduced irrespective of the cell cycle division stage (Figure S6D). Furthermore, Cd19Cre/+ and Zmynd8F/FCd19Cre/+ splenocytes exhibited similar growth curves following activation (Figure S6E). These findings indicate that ZMYND8 deficiency does not alter cell proliferation in B cells.
AID targeting is dependent on noncoding transcription (GLT) across the S regions (Chaudhuri and Alt, 2004). To examine the possibility that ZMYND8 promotes S region transcription, we measured GLT levels by qPCR at both donor (Sμ) and acceptor switch regions in ZMYND8-deficient splenocytes stimulated to undergo CSR to different isotypes. Sμ GLT was not affected by ZMYND8 depletion irrespective of the stimulation conditions (Figures 3A–3C), and we did not detect any significant decrease in the frequency of AID-induced mutations at 5′ Sμ in ZMYND8-deficient splenocytes (Figures S6F and S6G). In contrast, both pre- and post-spliced acceptor GLTs were reduced in Zmynd8F/FCd19Cre/+ cells (Figures 3A–3C), indicating the ZMYND8 deficiency specifically interferes with the induction of acceptor S region GLT. We next employed global nuclear run-on sequencing (GRO-seq) (Core et al., 2008) to visualize nascent RNA transcription at the Igh locus of B cells under stimulating conditions that induce GLTμ and ε and CSR to IgG1 and IgE (Figures S5D and S5E). In agreement with the GLT qPCR results, ZMYND8-deficient cells exhibited lower levels of nascent RNA at Sγ1 and Sε, whereas transcription at donor Sμ was unaffected (Figures S5D and S5E). We concluded that ZMYND8 is required to promote transcription of S acceptor regions and that the CSR defect of ZMYND8-deficient B cells is due to inefficient accessibility of these regions.
ZMYND8 Binds the Igh Enhancers and Represses Transcription at 3′RR
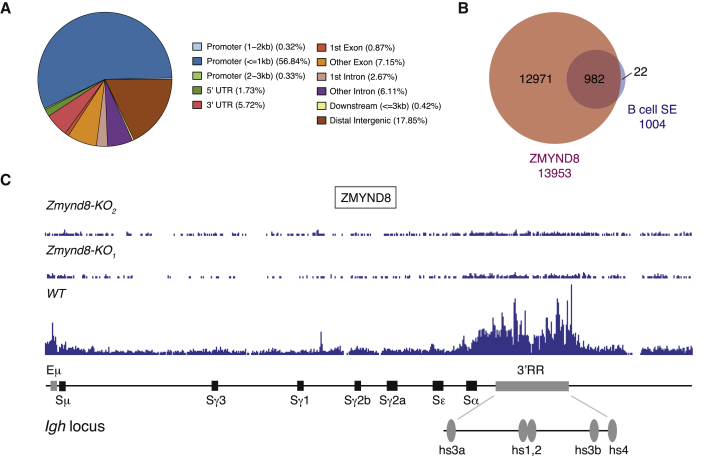
ZMYND8 is a histone mark reader that associates with promoters and enhancers in several cell types and mediates either transcriptional activation or, more frequently, repression (Adhikary et al., 2016, Basu et al., 2017a, Basu et al., 2017b, Li et al., 2016, Malovannaya et al., 2011, Poleshko et al., 2010, Savitsky et al., 2016, Shen et al., 2016, Spruijt et al., 2016, Zeng et al., 2010). To examine the mechanism responsible for defective germline Igh transcription in Zmynd8−/− B cells, we monitored ZMYND8 association with chromatin in CH12 by chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq). The vast majority of ZMYND8 peaks colocalized with promoters and distal intergenic regions (Figure 4A). To specifically assess whether ZMYND8 binds to super-enhancers in B cells, we defined the pool of B cell super-enhancers (Qian et al., 2014) that overlapped with ZMYND8 peaks. Remarkably, ZMYND8 bound to nearly all of these regulatory elements (Figure 4B). Altogether, these findings indicate that ZMYND8 preferentially associates with promoters and super-enhancers in B cells.
Figure 4.
ZMYND8 Binds Promoters and Super-Enhancers in B Cells
(A) Genomic distribution of ZMYND8 ChIP-seq peaks in CH12 cells.
(B) Venn diagram of the overlap between ZMYND8 peaks and B cells super-enhancers. SE, super-enhancer.
(C) ZMYND8 occupancy at the Igh locus in WT and Zmynd8−/− CH12 cell lines (KO1 and KO2). A schematic representation of the murine Igh locus showing location of enhancers (in gray) and S regions (in black) is represented below.
Data in (A)–(C) are representative of two independent experiments.
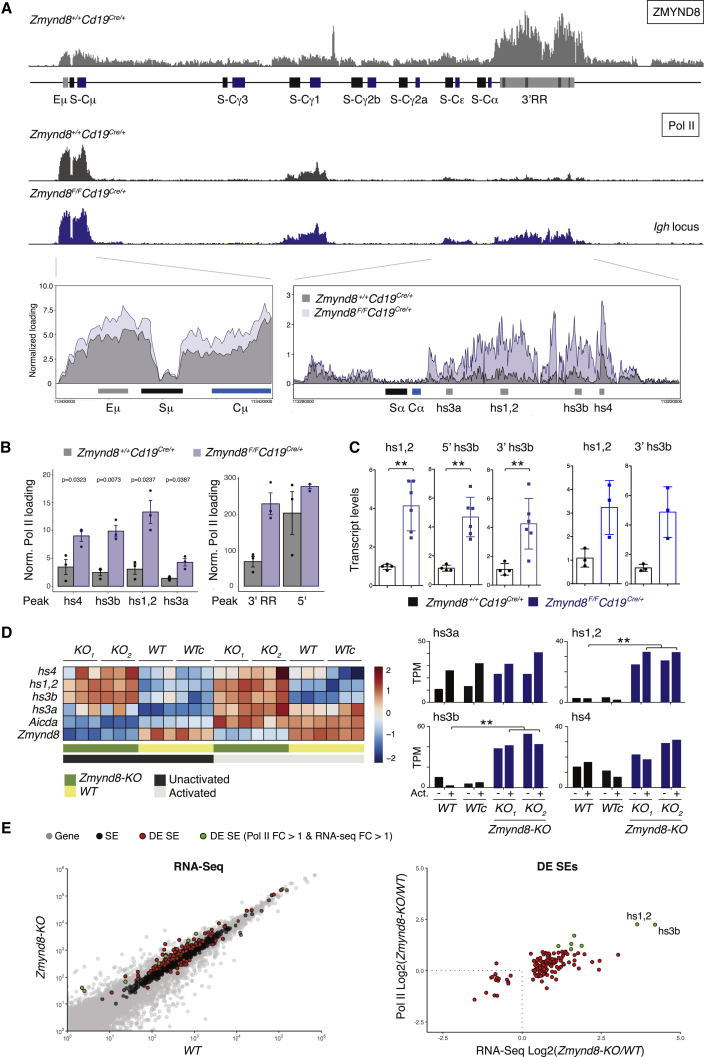
The Igh locus contains two enhancer elements, Eμ and 3′RR, that are located at 5′ of Sμ and 3′ of Cα, respectively (Birshtein, 2014, Perlot and Alt, 2008). The 3′RR is a prototypical super-enhancer (Pinaud et al., 2011, Whyte et al., 2013), which is required for GLT induction and CSR in mature B lymphocytes and, to a lesser extent, in CH12 cells (Issaoui et al., 2018, Kim et al., 2016). As expected from the genome-wide distribution (Figures 4A and 4B), ZMYND8 associates with the Igh locus in activated CH12 and B cells and it binds the 5′ Eμ and 3′RR enhancers (Figures 4C and 5A).
Figure 5.
ZMYND8 Represses 3′RR Enhancer Transcription
(A) (Top) ZMYND8 and Pol II loading at the Igh locus in splenocytes stimulated for 72 hr with LPS and IL-4. (Bottom) Pol II ChIP-seq tracks overlay at regions encompassing Eμ and 3′RR enhancers.
(B) Pol II loading quantification at Igh enhancers. Graph summarizes ChIP-seq data from three mice per genotype (error bars represent SD). Significance was calculated with Welch 2 sample unpaired t test.
(C) qPCR analysis for hs1,2 and hs3b eRNA levels in B cells 48 hr after stimulation with LPS and IL-4 (left) or LPS only (right). Two sets of primers amplifying the 5′ (5′ hs3b) and 3′ (3′ hs3b) regions of hs3b were employed. The data summarize 4–6 mice per genotype (error bars represent SD). One Cd19Cre/+ mouse within each experiment was assigned an arbitrary value of 1.0. Significance was calculated with the Mann-Whitney U test.
(D) (Left) Heatmap showing Igh 3′RR enhancers differential transcript expression as determined by RNA-seq in controls (WT bulk and clonal derivative WTc) and two independent Zmynd8−/− CH12 clones (KO1 and KO2). Expression counts are row-normalized by Z score. Three independent RNA-seq replicates per sample are shown. (Right) Bar graphs depicting relative transcript levels at hs4, hs1,2, hs3b, and hs3a are shown. The adjusted p values were calculated with the Wald test and corrected for multiple testing with the Benjamini-Hochberg method.
(E) (Left) Graph depicting ZMYND8-bound B cell super-enhancers that are differentially expressed in Zmynd8−/− versus WT CH12 as measured by RNA-seq. DE SE, differentially expressed SE. (Right) Plot of fold change of Pol II loading on the differentially expressed super-enhancers defined in the left panel is shown.
Loss of ZMYND8 results in enhancer overactivation as seen by increased enhancer RNA (eRNA) transcription (Shen et al., 2016). To determine whether ZMYND8 occupancy at Igh enhancers alters their activity, we monitored RNA polymerase II (Pol II) occupancy at the Igh locus in ZMYND8-deficient cells by ChIP-seq. Pol II loading was significantly increased at the 3′RR core enhancers hs1,2 and hs3b in ZMYND8-deficient splenocytes and CH12 cells (Figures 5A, 5B, S7A, and S7B). In contrast, no significant increase above controls was observed in Rif1−/− cells (Figures S7A and S7B). In agreement with increased Pol II loading, transcription of hs1,2 and hs3b eRNAs was increased in the absence of ZMYND8 both at the nascent (GRO-seq analysis; Figure S5D) and steady-state levels (Figures 5C and 5D). These findings suggest that ZMYND8 modulates 3′RR function by suppressing its transcriptional activity.
To determine whether ZMYND8 controls the transcriptional status of B cell super-enhancers other than the Ighs, we analyzed the transcription profiles of ZMYND8-bound super-enhancers (Figure 4B) in the WT versus Zmynd8-KO cells. In total, 130 out of the 982 ZMYND8-occupied B cell super-enhancers were differentially expressed based on RNA-seq (Figure 5E; Table S4; log2FoldChange > 1; red and green dots). Of those, 90 displayed increased Pol II loading in the Zmynd8-KO cells when compared to WT (Figure 5E; log2FoldChange > 0). Seven super-enhancers exhibited the most dramatic increase in Pol II loading in the absence of ZMYND8 (Figure 5E; log2FoldChange > 1), with the 3′RR core enhancers hs1,2 and hs3b showing the highest enrichment. We concluded that ZMYND8 deficiency correlates with increased transcription of B cell super-enhancers.
ZMYND8 Is Required for Efficient SHM of the Igh Variable Region
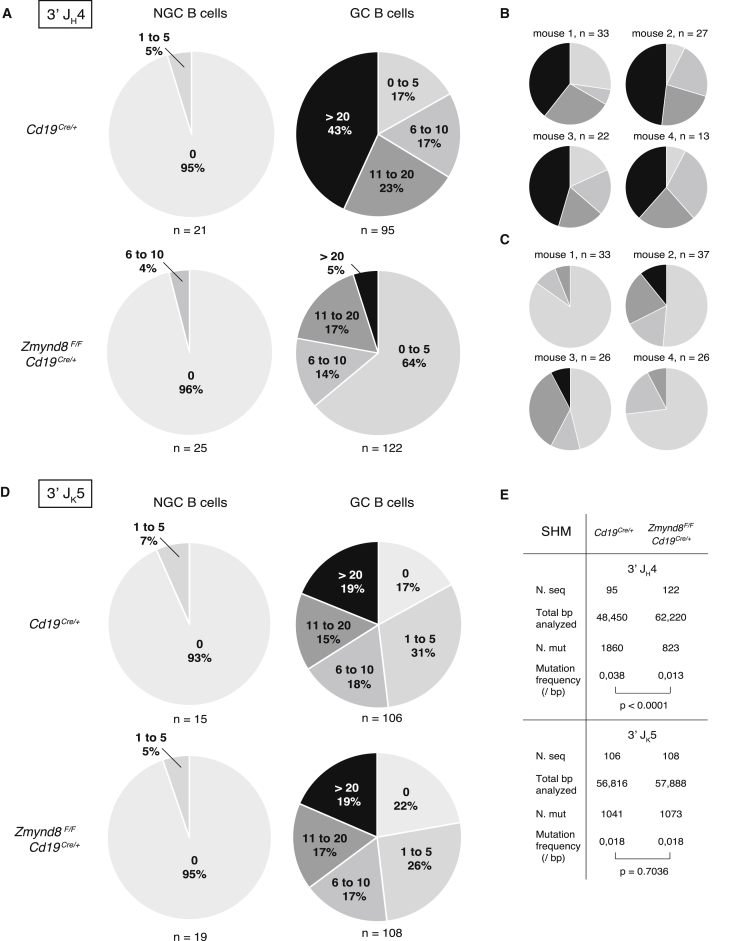
The 3′RR is also essential for somatic hypermutation of Igh variable region genes (Rouaud et al., 2013). To determine whether ZMYND8 is required for SHM of the Igh variable regions, we sequenced the intronic region downstream the JH4 element of germinal center (GC) B cells from Peyer’s patches of aged mice (Jolly et al., 1997). As expected, GC B cells from control mice displayed a high mutation frequency (Figures 6A, 6B, and 6E), which reflects the chronic nature of the B cell stimulation in Peyer’s patches (González-Fernández et al., 1994). Nearly all sequences contained mutations (97%; 92/95), and a considerable portion of clones was heavily mutated (Figures 6A, 6B, and 6E). In contrast, GC B cells from Zmynd8F/FCD19Cre/+ mice accumulated mutations at a significantly lower rate (Figures 6A, 6C, and 6E), and the proportion of mutated sequences was decreased (77%; 94/122). Furthermore, when comparing the number of mutations per sequence, Zmynd8F/FCD19Cre/+ mice exhibited a skewed distribution with a considerable increase in the number of sequences bearing few mutations and the nearly complete absence of highly mutated clones (Figures 6A and 6C). We concluded that ZMYND8 ablation in mature B cells impairs SHM of the Igh locus.
Figure 6.
ZMYND8 Is Required for Efficient SHM of the Heavy, but Not Light, Ig Chain
(A and D) Cumulative pie charts showing percentage of sequences bearing indicated mutations in 3′ JH4 (A) and 3′ Jk5 (D) regions cloned from sorted Peyer’s patches B cells of aged unimmunized mice (4 mice per group). Total number of analyzed sequences is indicated below each pie chart. Mutations were quantified over 510 bp downstream JH4 gene segment and 536 bp downstream JK5 gene segment. GC, germinal center; NGC, non-germinal center.
(B and C) Distribution of mutations per cloned sequences in individual Cd19Cre/+ (B) and Zmynd8F/FCd19Cre/+ (C) mice.
(E) Summary table listing number of analyzed sequences and total length, number of mutations, and mutation frequency at JH4 and JK5 introns in germinal center B cells from (A) and (D). Significance was calculated with the Mann-Whitney U test.
To assess whether ZMYND8 deficiency causes a general defect in somatic hypermutation, we analyzed SHM of the Igk locus, which is not controlled by the 3′RR. To do so, we monitored the mutation frequency immediately downstream JK5 gene segment (Rouaud et al., 2013). We found a similar number of mutated sequences in ZMYND8-deficient (78%; 84/108) and control mice (83%; 88/106). Furthermore, both the distribution of mutations per sequence and mutation frequency were indistinguishable between the two groups (Figures 6D and 6E). Thus, ZMYND8 ablation specifically impairs the SHM process in the heavy-chain locus without affecting the k light-chain locus. Altogether, these findings indicate that, under physiologic conditions, ZMYND8 is an essential regulator of the 3′ Igh super-enhancer.
Discussion
Despite initially identifying ZMYND8 as an interactor of the DNA repair factor Rif1, we were unable to document a significant effect of ZMYND8 in DSB repair in ZMYND8-deficient CH12 or MEF cells, irrespective of the source of DNA damage. This observation is consistent with the finding that the interaction between Rif1 and ZMYND8 in B cells undergoing CSR is not influenced by DNA damage or the ATM kinase. Pol II loading at the 3′RR was not altered in Rif1-deficient cells, thus indicating that, in contrast to ZMYND8, Rif1 does not act as a regulator of the 3′RR. Altogether, these results raise the possibility that the interaction between Rif1 and ZMYND8 might be accidental. However, we cannot exclude the intriguing possibility that Rif1 and ZMYND8 also contribute to CSR or other cellular functions by a common mechanism that is independent of their respective roles in DNA end protection and 3′RR regulation.
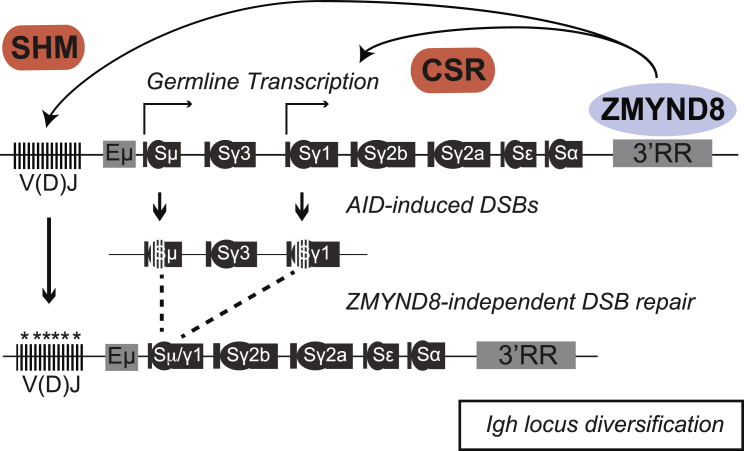
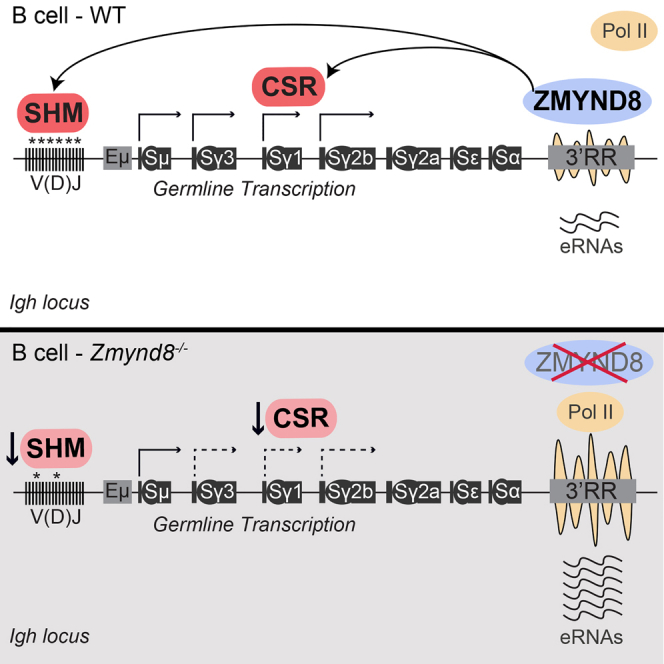
Mutation of the 3′RR in mice showed that it is necessary to support CSR and SHM (Cogné et al., 1994, Garot et al., 2016, Le Noir et al., 2017, Manis et al., 1998, Pinaud et al., 2001, Rouaud et al., 2013, Saintamand et al., 2015b, Saintamand et al., 2016, Vincent-Fabert et al., 2010). More specifically, the 3′RR regulates the early steps of CSR, including donor and acceptor GLT transcription, Pol II pausing, AID targeting, and DSB formation; however, it is dispensable for resolution of CSR breaks and the DSB repair pathway choice between classical and alternative NHEJ. We have found that deletion of ZMYND8 increases 3′RR-transcriptional activity and phenocopies key aspects of the 3′RR deficiency. Consistent with this idea, ZMYND8 is necessary to support GLT of acceptor S regions and CSR. Furthermore, ZMYND8 deficiency impairs SHM specifically at the Igh locus. Given ZMYND8 association with 3′RR and its ability to repress enhancer transcription, these findings suggest that ZMYND8 is a key regulator of the 3′RR activity in mature B cells (Figure 7).
Figure 7.
Model for ZMYND8 Function in Igh Gene Diversification
ZMYND8 controls both CSR and Igh SHM via its ability to regulate the activity of the 3′ Igh super-enhancer. GTL for S-Cγ1 is representative of acceptor S region induction.
The spectrum of phenotypes exhibited by ZMYND8-deficient B cells and cells lacking the full 3′RR is not entirely overlapping. ZMYND8 acts on the accessibility of the acceptor S region rather than the donor Sμ region. In this regard, the phenotype of Zmynd8F/FCd19Cre/+ cells is more reminiscent of B cells harboring deletions or replacements of some portions of the 3′RR (Δhs1,2 or Δ(hs3b-hs4); Cogné et al., 1994, Pinaud et al., 2001), which reduced GLT of several acceptor regions while leaving donor GLTμ unaffected. However, whereas these partial deletions within the 3′RR did not significantly affect γ1 GLT and switching to IgG1 (Cogné et al., 1994, Manis et al., 1998, Pinaud et al., 2001), ZMYND8 deficiency considerably reduced IgG1 CSR, albeit to a lesser extent than the full 3′RR deletion (Saintamand et al., 2015b, Vincent-Fabert et al., 2010). Given the intrinsic differences and degrees of phenotype penetrance exhibited by various deletions of internal portions of the 3′RR (Bébin et al., 2010, Cogné et al., 1994, Garot et al., 2016, Le Noir et al., 2017, Manis et al., 1998, Pinaud et al., 2001, Saintamand et al., 2016, Vincent-Fabert et al., 2009), and the fact that ZMYND8 is a trans-acting factor that appears to differentially affect 3′RR modules, it is not at all surprising that ZMYND8 deficiency does not entirely phenocopy the full 3′RR deletion.
In plasma cells and resting B cells, the 3′RR contacts regions surrounding the 5′ Eμ enhancer (VDJ-Eμ), and upon activation, specific S regions are recruited into the VDJ-3′RR loop in a cytokine-dependent manner (Ju et al., 2007, Wuerffel et al., 2007). This topological organization is thought to facilitate both GLT expression and synapsis of recombining S regions. 4C analysis with a bait within the 3′RR did not show differences in the profile of long-range chromatin interactions within the Igh locus, suggesting that ZMYND8 binding to the Igh super-enhancers might not contribute to the establishment or maintenance of this architectural structure (data not shown).
Instead, the data support the possibility that ZMYND8 suppression of RNA Pol II loading on the 3′RR enhancer favors GLT transcription by removing local competition for Igh transcription factors. In support of this model, replacement of individual (hs1,2 or hs3a) or paired (hs3b and hs4) core enhancers with an actively transcribed neomycin gene cassette resulted in a more severe defect in acceptor GLT expression and CSR than deletion of the same modules (Cogné et al., 1994, Manis et al., 1998, Pinaud et al., 2001). Thus, deregulated transcription within the 3′RR interferes with its activity and disrupts the physiological regulation of GLT and CSR. Our data identify ZMYND8 as a key regulator of 3′RR transcriptional activity under physiologic circumstances.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-CD40 Clone 1C10 | BioLegend | Cat# 102810; RRID: AB_629065 |
| Rat anti-CD180 (RP/14) | BD Biosciences | Cat# 552128; RRID: AB_394343 |
| Mouse anti-Flag M2 | Sigma-Aldrich | Cat# F3165; RRID: AB_259529 |
| Mouse anti-Flag M2 (HRP conjugated) | Sigma-Aldrich | Cat# A8592; RRID: AB_439702 |
| Rabbit anti-Rif1 | Di Virgilio et al., 2013 | N/A |
| Rabbit anti-ZMYND8 | Sigma-Aldrich | Cat# HPA020949; RRID: AB_1857223 |
| Mouse anti-γH2AX (Ser139) Clone JBW301 | Millipore/Merck | Cat# 05-636; RRID: AB_309864 |
| Mouse anti-β Actin Clone AC-15 | Sigma-Aldrich | Cat# A5441; RRID: AB_476744 |
| Mouse anti-53BP1 | Bethyl | Cat# A300-272A; RRID: AB_185520 |
| Mouse anti-53BP1 Clone BP18 | Millipore/Merck | Cat# 05-725; RRID: AB_309939 |
| Mouse anti-RNA polymerase II CTD repeat YSPTSPS (phospho S5) Clone 4H8 | Abcam | Cat# ab5408; RRID: AB_304868 |
| Rat anti-CD21/CD35-FITC Clone 7G6 | BD Biosciences | Cat# 561769; RRID: AB_10924591 |
| Rat anti-IgD-FITC Clone 11-26c.2a | BD Biosciences | Cat# 562022; RRID: AB_10894208 |
| Rat anti-IgM-PE Clone R6-60.2 | BD Biosciences | Cat# 553409; RRID: AB_394845 |
| Rat anti-IgM-FITC Clone II/41 | BD Biosciences | Cat# 553437; RRID: AB_394857 |
| Rat anti-CD43-PE Clone S7 | BD Biosciences | Cat# 561857; RRID: AB_10926206 |
| Rat anti-CD23-PE Clone B3B4 | BioLegend | Cat# 101607; RRID: AB_312832 |
| Rat anti-CD3-PE Clone 17A2 | BioLegend | Cat# 100206; RRID: AB_312663 |
| Rat anti-CD19-APC Clone 6D5 | BioLegend | Cat# 115512; RRID: AB_313647 |
| TruStain fcX Rat anti-CD16/32 Clone 93 | BioLegend | Cat# 101320; RRID: AB_1574975 |
| Rat anti-B220/CD45R-FITC Clone RA3-6B2 | BioLegend | Cat# 103205; RRID: AB_312990 |
| Rat anti-CD38-Alexa700 Clone 90 | Invitrogen | Cat# 56-0381-82; RRID: AB_657740 |
| Hamster anti-CD95/Fas-PE Clone Jo2 | BD Biosciences | Cat# 561985; RRID: AB_10895586 |
| Rat anti-IgG2b-PE Clone RMG2b-1 | BioLegend | Cat# 406707; RRID: AB_2563380 |
| Goat anti-IgA-PE | Southern Biotech | Cat# 1040-09 |
| Rat anti-IgG1-APC Clone X56 | BD Biosciences | Cat# 550874; RRID: AB_398470 |
| Rat anti-IgG3-BIOT Clone R40-82 | BD Biosciences | Cat# 553401; RRID: AB_394838 |
| Streptavidin-APC | BD Biosciences | Cat# 554067; RRID: AB_10050396 |
| Streptavidin-AP Conjugate | Sigma-Aldrich | Cat# 11089161001 |
| Goat anti-Lamda-UNLB | Southern Biotech | Cat# 1060-01 |
| Goat anti-Kappa-UNLB | Southern Biotech | Cat# 1050-01; RRID: AB_2737431 |
| Mouse IgM-UNLB Clone 11E10 | Southern Biotech | Cat# 0101-01; RRID: AB_2629437 |
| Mouse IgG1-UNLB Clone 15H6 | Southern Biotech | Cat# 0102-01 |
| Goat anti-IgM, Human ads-BIOT | Southern Biotech | Cat# 1020-08; RRID: AB_2737441 |
| Goat anti-IgG1, Human ads-BIOT | Southern Biotech | Cat# 1070-08 |
| Rat anti-CD43 (Ly-48) Microbeads | Miltenyi Biotec | Cat# 130-049-801 |
| Goat anti-rabbit Alexa546 | Invitrogen | Cat# A-11035; RRID: AB_2534093 |
| Goat anti-mouse Alexa488 | Invitrogen | Cat# A-11029; RRID: AB_2534088 |
| Anti-BrdU beads | Santa Cruz Biotech | Cat# sc-32323; RRID: AB_626766 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LPS | Sigma-Aldrich | Cat# L2630 |
| IL-4 (mouse recombinant) | Sigma-Aldrich | Cat# I1020 |
| BAFF (human recombinant) | PeproTech | Cat# 310-13 |
| TGFβ-1 (mouse recombinant) | R&D Systems | Cat# 7666-MB-005/CF |
| Puromycin dihydrochloride | Sigma-Aldrich | Cat# P8833 |
| SILAC-RPMI | Thermo Scientific | Cat# 88421 |
| Dialyzed fetal bovine serum | GIBCO | Cat# 26400-044 |
| 13C6 L-arginine | Cambridge Isotope Laboratories | Cat# CLM-2265-H-PK-1 |
| 13C6 L-lysine | Cambridge Isotope Laboratories | Cat# CLM-2247-H-PK-1 |
| Non-labeled L-arginine | Sigma-Aldrich | Cat# A8094 |
| Non-labeled L-lysine | Sigma-Aldrich | Cat# L8662 |
| Polyvinylpyrrolidone | Sigma-Aldrich | Cat# PVP40 |
| Complete EDTA-free protease inhibitor cocktail | Roche | Cat# 11873580001 |
| Phosphatase inhibitor cocktail tablets | Roche | Cat# 04906837001 |
| Benzonase | Sigma-Aldrich | Cat# E1014 |
| Glutaraldehyde | Sigma-Aldrich | Cat# 340855 |
| 3XFlag peptide | Sigma-Aldrich | Cat# F4799 |
| NuPage LDS Sample buffer | Thermo Fisher Scientific | Cat# NP0008 |
| ATMi KU55933 | Tocris Bioscience | Cat# 3544 |
| Carboxyfluorescein succinimidyl ester (CFSE) | Invitrogen | Cat# 65-0850-84 |
| CellTrace Violet | Thermo Fisher Scientific | Cat# C34557 |
| Alum | Sigma-Aldrich | Cat# 31242 (Discontinued) |
| NP-CGG16 | Biosearch Technologies | Cat# N5055B |
| Paraformaldehyde | Sigma-Aldrich | Cat# P6148 |
| Crystal Violet | Sigma | Cat# C0775 |
| PARPi Olaparib/AZD2281, Ku-0059436 | Selleckchem | Cat# S1060 |
| Pierce 16% Formaldehyde Methanol-free | Thermo Scientific | Cat# 28906 |
| TRIzol | Invitrogen | Cat# 15596018 |
| Colcemid | Sigma-Aldrich | Cat# 10295892001 |
| KaryoMAX Giemsa Stain Solution | GIBCO | Cat# 10092013 |
| Gurr Buffer Tablets | GIBCO | Cat# 10582013 |
| Dynabeads Protein A | Invitrogen | Cat# 10001D |
| Dynabeads M-270 Epoxy | Invitrogen | Cat# 14301 |
| TRIzol LS Reagent | Invitrogen | Cat# 10296028 |
| Critical Commercial Assays | ||
| Neon Transfection System, 100 μL Kit | Invitrogen | Cat# MPK10025 |
| APC BrdU Flow Kit | BD Biosciences | Cat# 552598 |
| AllPrep DNA/RNA Mini Kit. Dan | QIAGEN | Cat# 80204 |
| Ribo-Zero Gold rRNA Removal Kit | Illumina | Cat# MRZG12324 |
| TruSeq Stranded Total RNA Library Prep Kit | Illumina | Cat# 20020597 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| SuperScript VILO cDNA Synthesis Kit | Invitrogen | Cat# 11754250 |
| RapidOut DNA Removal Kit | Thermo Scientific | Cat# K2981 |
| TOPO TA Cloning Kit | Invitrogen | Cat# 450641 |
| Phusion High-Fidelity DNA Polymerase | Thermo Scientific, NEB | Cat# F530L, Cat# M0530L |
| Luna Universal qPCR Mastermix | NEB | Cat# M3003 |
| NucleoSpin DNA Purification Kit | Macherey-Nagel | Cat# 740499 |
| TURBO DNase | Invitrogen | Cat# AM2239 |
| RNA Fragmentation Reagents | Invitrogen | Cat# AM8740 |
| BioSpin P-30 Gel Columns | Bio-Rad | Cat# 7326232 |
| T4 Polynucleotide Kinase | NEB | Cat# M0201L |
| Exonuclease I | NEB | Cat# M0293L |
| ChIP DNA Clean and Concentrator Kit | Zymo Research Corporation | Cat# D5205 |
| RNase H | Thermo Scientific | Cat# 18021071 |
| APE1 | NEB | Cat# M0282L |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE118794 |
| Original scans and IF images | This paper; Mendeley Data | https://doi.org/10.17632/jktyjvy9hg.1 |
| Experimental Models: Cell Lines | ||
| Murine: CH12 (CH12F3) - WT, parental | Nakamura et al., 1996 | N/A |
| Murine: CH12 - WTc, clonal derivative | This paper | N/A |
| Murine: AID−/− CH12 | This paper and Kerafast | Cat# ESP013 |
| Murine: 53bp1−/− CH12 | This paper | N/A |
| Murine: Rif1−/− CH12 | This paper | N/A |
| Murine: Zmynd8-KO1 CH12 | This paper | N/A |
| Murine: Zmynd8-KO2 CH12 | This paper | N/A |
| Murine: Zmynd8-KO3 CH12 | This paper | N/A |
| Murine: iMEFs - WTc, clonal derivative | This paper | N/A |
| Murine: iMEFs - Rc-1 | This paper | N/A |
| Murine: iMEFs - Rc-2 | This paper | N/A |
| Murine: Zmynd8-KO1 iMEFs | This paper | N/A |
| Murine: Zmynd8-KO2 iMEFs | This paper | N/A |
| Murine: Zmynd8-KO3 iMEFs | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL6: WT | The Jackson Laboratory | JAX# 664 |
| Mouse: Rif1FH/FH | Cornacchia et al., 2012 | N/A |
| Mouse: AID−/− | Muramatsu et al., 2000 | N/A |
| Mouse: Zmynd8tm1a | European Mouse Mutant Archive | EMMA# 05720 |
| Mouse: ROSA26Flpo | The Jackson Laboratory | JAX# 007844 |
| Mouse: Cd19Cre/+ | Rickert et al., 1997 | N/A |
| Mouse: Zmynd8F/F | This paper | N/A |
| Mouse: Zmynd8F/FCd19Cre/+ | This paper | N/A |
| Oligonucleotides | ||
| ZMYND8 genotyping primers | See Table S5 | N/A |
| gRNAs for gene editing | See Table S5 | N/A |
| Primers for qPCR | See Table S5 | N/A |
| gRNAs for end resection assay | See Table S5 | N/A |
| Primers for end resection assay | See Table S5 | N/A |
| Primers for Sμ-Sγ1 junctions | See Table S5 | N/A |
| Primers for SHM | See Table S5 | N/A |
| Recombinant DNA | ||
| Plasmid: pMX-ZMYND8-3XFlag | This paper | N/A |
| Plasmid: pX458 (pSpCas9(BB)-2A-GFP (PX458)) | Addgene | Cat# 48138 |
| Plasmid: pX458 expressing Cas9D10A | This paper | N/A |
| Software and Algorithms | ||
| MaxQuant (v.1.2.2.5) | Cox and Mann, 2008 | N/A |
| BWA mem | Li and Durbin, 2009 | N/A |
| HOMER ChIP-Seq program | Heinz et al., 2010 | N/A |
| R and ChIPseeker package | Yu et al., 2015 | N/A |
| kallisto v.0.43.0 | Bray et al., 2016 | N/A |
| R package tximport | Soneson et al., 2015 | N/A |
| DESeq2 | Love et al., 2014 | N/A |
| Bowtie | Langmead et al., 2009 | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michela Di Virgilio (michela.divirgilio@mdc-berlin.de).
Experimental Model and Subject Details
Source of cell lines and mouse models used in the study is reported in the Key Resources Table. All experiments were performed in compliance with EU Directive 2010/63/EU, and in agreement with protocols approved by Landesamt für Gesundheit und Soziales (LAGeSo, Berlin), The Rockefeller University (NY), and the National Institutes of Health (NIH) Institutional Animal Care and Use Committees.
Method Details
Mice
Rif1FH/FH (Cornacchia et al., 2012), Cd19Cre mice (Rickert et al., 1997), and AID(Aicda)−/− (Muramatsu et al., 2000) were previously described. The conditional Zmynd8F allele bears LoxP sites flanking Exon 4 (ENSMUSE00001273891), and was generated by crossing the knockout first allele with conditional potential Zmynd8tm1a (European Mouse Mutant Archive, EMMA #05720) with the ROSA26Flpo deleter strain (The Jackson Laboratory). Germline transmission was confirmed and positive pups were bred with Cd19Cre mice to generate Zmynd8F/FCd19Cre/+ mice. Mice were maintained in a specific pathogen-free (SPF) barrier facility under standardized conditions (20+/−2°C temperature; 55% ± 15% humidity) on a 12 h light/12 h dark cycle. Both male and female mice were used for the experiments. Primers used for genotyping the Zmynd8F allele are listed in Table S5.
Cell Cultures and Retroviral Infection
B lymphocytes were isolated from mouse spleens using anti-CD43 MicroBeads (Miltenyi Biotec) and stimulated to undergo class switching with 25 μg/ml LPS (Sigma-Aldrich) and 5 ng/ml of mouse recombinant IL-4 (Sigma-Aldrich) for CSR to IgG1; 25 μg/ml LPS, 10 ng/ml BAFF (PeproTech) and 2 ng/ml TGFβ (R&D Systems) for CSR to IgG2b; or 25 μg/ml LPS only for CSR to IgG3. Mice 7 to 20 weeks old were used in age-matched groups for splenocytes isolation and CSR assays. CH12 cells (Nakamura et al., 1996) were stimulated to undergo CSR to IgA by treatment with 5-15 μg/mL αCD40 (BioLegend), 5 ng/ml TGFβ and 5 ng/ml of mouse recombinant IL-4 for 48 h. pMX-ZMYND8-3XFlag retroviral vector was generated by cloning the cDNA for murine ZMYND8 into pMX-IRES-GFP with a C-terminal 3XFlag tag. For CH12 infections with pMX-based retroviral vectors, cells were subjected to 2 rounds of infection with amphotrophic retroviral supernatant, with or without selection with 1 μg/ml of Puromycin (Sigma-Aldrich), followed by activation for 48 h before analysis for CSR efficiency.
I-DIRT
B cells from Rif1FH/FH and WT mice were isolated and cultured in SILAC medium composed by RPMI (-Arg, -Lys) medium (Thermo Scientific), L-glutamine, sodium pyruvate, HEPES, 50 μM 2-mercaptoethanol, antibiotic/antimitotic, 10% dialyzed fetal bovine serum (GIBCO), LPS, IL-4 and RP/14 (BD-Biosciences), and supplemented with either 13C6 L-arginine and 13C6 L-lysine (Cambridge Isotope Laboratories) (heavy medium; Rif1FH/FH culture), or non-labeled L-arginine and L-lysine (Sigma-Aldrich) (light medium; WT culture). Cells were cultured in SILAC medium for 96 h to ensure near-complete incorporation of the labeled amino acids. For IR treatment, cells were exposed to an X-rays source at a rate of 278.2 Rads/min for 431 s (20 Grays), followed by recovery at 37°C for 45 min.
Immunoisolation of RIF1FH Complexes
Rif1-containing complexes were extracted in soluble form and immune-isolated after sub-stoichiometric treatment with glutaraldehyde in order to stabilize labile interactions without perturbing the native composition of protein complexes. Specifically, 2.6x109 cells per culture (Rif1FH/FH and WT) were collected by centrifugation, resuspended in 20 mM HEPES containing 1.2% polyvinylpyrrolidone (Sigma-Aldrich), protease and phosphatase inhibitor cocktails (Roche), 0.5 mM DTT, and frozen in liquid nitrogen. Rif1FH/FH and WT frozen cells were mixed in a 1:1 ratio and cryogenically disrupted by wet milling in a Planetary Ball Mill PM 100 (Retsch). The resulting frozen cell grindate was rapidly thawed in extraction buffer (20mM Tris-Cl pH 8, 150 mM NaCl, 0.5% Igepal CA-630, 1.5 mM MgCl2, Benzonase, protease and phosphatase inhibitor) supplemented with 2.5 mM Glutaraldehyde (Sigma-Aldrich), and quenched with 100 mM Tris-CL pH8.0 buffer after 5 min incubation. The lysate was clarified by 10 min centrifugation at 13,000 rpm at 4°C, and immediately incubated with magnetic beads (M-270 epoxy beads, Invitrogen) conjugated with anti-Flag M2 (Sigma-Aldrich) antibody for 1 h at 4°C (Di Virgilio et al., 2013). The bead preparation was then washed in extraction buffer, and RIF1FH bait and associated proteins were eluted twice under native conditions by two rounds of incubation with 2.5 μg/μl 3XFlag peptide (Sigma-Aldrich) for 45 min at 4°C with shaking.
Mass Spectrometric Analysis
Rif1 baits and co-purifying proteins were resolved by 4%–12% Bis-Tris gel and visualized by Coomassie blue staining. The gel was divided into upper and lower parts along the 39kDa molecular weight marker, with only the lower part fixed. The protein-containing upper and lower parts were cut into 5 and 1 regions respectively, and the gel samples were subjected to in-gel tryptic digestion. Peptides were extracted and purified, analyzed by LCMS using a Thermo Q Exactive Plus mass spectrometer, with a Thermo Easy-nLC 1000 HPLC and a Thermo Easy-Spray electrospray source. Identification and quantification of proteins was performed by searching against a mouse protein sequence database with the MaxQuant software (version 1.2.2.5) (Cox and Mann, 2008). Protein H/(H+L) ratios were derived using peptides’ H/L intensity values in MaxQuant output.
CRISPR-Cas9 Gene Targeting
gRNAs for functional screens for loss of CSR function in CH12 were cloned into the U6 cassette of pX458 plasmid (pSpCas9(BB)-2A-GFP, Addgene #48138). For the loss of CSR screen, 3-6 gRNAs per candidate were cloned and tested, either individually or in pools. CH12 cells were transfected with Cas9-gRNAs expressing constructs via electroporation with Neon Transfection System (Thermo Fisher Scientific), sorted for GFP-positive cells after 40 h, and left to recover for 72 h before activation for CSR analysis. CH12 and MEFs clonal derivatives were generated via electroporation with either single gRNA and WT Cas9-based plasmid (pX458) or gRNA pairs and nickase Cas9-based plasmid. In the latter case, tandem U6 cassettes were cloned into a mutated version of pX458 expressing Cas9D10A. Single GFP-positive cells were sorted in 96-well plates and clones were allowed to grow for 12 days (CH12) or 17 days (MEFs) days before expansion. Clones were validated at the level of genomic scar and protein expression. The sequences of the gRNAs employed in these studies are listed in Table S5. The scheme of Zmynd8 genomic locus in Figure 1A is adapted from Ensembl ENSMUST00000109269.7. Controls for gRNA-nucleofected CH12 bulk cultures were cells nucleofected either with empty vector or gRNAs against random sequences not present in mouse genome. Controls for CH12 / MEF clonal derivatives included both WT cultures, WT clonal derivatives, and clones derived from targeting CH12 / MEF with random sequences not present in mouse genome.
Cell Lysates and CoIP Assay
Co-immunoprecipitation analyses were performed as for I-DIRT pull-down with the only exception that protein elution from magnetic beads was performed by incubation with NuPage LDS Sample buffer (Thermo Fisher) supplemented with 50mM DTT for 10 min at 70C. Where indicated, 10 μM ATMi KU55933 (Tocris Bioscience) was added 1h before irradiation. Western blot analysis of protein levels was performed on whole cell lysates prepared by lysis in RIPA buffer supplemented with Complete EDTA free proteinase inhibitor (Roche). The antibodies used for WB analysis are: anti-Rif1 (Di Virgilio et al., 2013), anti-Flag M2 (Sigma-Aldrich), anti-ZMYND8 (Sigma-Aldrich), anti-γH2AX (Millipore), anti-β Actin (Sigma-Aldrich), and anti-53BP1 (Bethyl).
Flow Cytometry
For class switching assays, cell suspensions were stained with fluorochrome-conjugated anti-IgG1, anti-IgG3 (BD-Biosciences), anti-IgG2b (BioLegend), or anti-IgA (Southern Biotech). Samples were acquired on a LSRFortessa cell analyzer (BD-Biosciences). Analysis of B cell development and differentiation was performed using anti-CD21/CD35-FITC, anti-IgD-FITC, anti-IgM-PE, anti-IgM-FITC, anti-CD43-PE (BD-Biosciences) and anti-CD23-PE, anti-CD3-PE and anti-CD19-APC (BioLegend) antibodies. For cell proliferation analysis by cell tracking dye dilution, primary B cells were pulsed with 2 μM carboxyfluorescein succinimidyl ester (CFSE) (Invitrogen) or 5 μM CellTrace Violet (Thermofisher) for 10 min at 37C. CFSE/CellTrace covalently labels intracellular molecules, and each cell division halves the signal intensity. For cell cycle analysis, CH12 cells were collected, fixed, and permeabilized using Fixation/Permeabilization Solution (included in BrdU Flow Kit, BD-Biosciences) according to the manufacturer’s instructions. BrdU pulse and staining was performed by using BrdU Flow Kit (BD-Biosciences) according to the manufacturer’s instructions.
Mice Immunization and Ig Serum Titers
For T cell-dependent immunization, 9 to 18 week old mice were injected intraperitoneally with 100 μg alum (Sigma-Aldrich)-precipitated NP-CGG16 (Biosearch Technologies). Blood was collected before and at day 7 and 20 after immunization. Serum specific IgM and IgG1 antibodies against NP hapten were determined by ELISA. NP-IgM and NP-IgG1 preparations for standard curves were a kind gift from Klaus Rajewsky (MDC).
Quantitative PCR
mRNA levels for AID, pre- and post-spliced germline transcripts, and 3′RR hs eRNA levels were measured as it follows. Total RNA was extracted from splenocytes cultures 48 h after activation using TRIzol (Invitrogen) or RNeasy Mini Kit (QIAGEN) according to manufacturer’s instructions, and retro-transcribed with SuperScript VILO cDNA Synthesis Kit (Invitrogen). When using TRIzol, genomic DNA was removed by RapidOut DNA Removal Kit (Thermo Scientific). Transcripts were amplified using StepOnePlus Real-Time PCR System (Applied Biosystems) with Luna Universal qPCR Mastermix (NEB). Primers used for qPCR (this paper; Muramatsu et al., 2000, Oruc et al., 2007, Xu et al., 2015) are listed in Table S5.
Immunofluorescence
iMEFs were grown on coverslips overnight. Cells were irradiated (10 Gy IR) and allowed to recover for 90 min or 6 h. Upon fixation with 4% paraformaldehyde (Sigma-Aldrich) and permeabilization with 0.5% Triton X-100, cells were stained with anti-γH2AX (Millipore), rabbit anti-Rif1 serum (Di Virgilio et al., 2013), mouse anti-53BP1 (Millipore), mouse anti-Flag M2 (Sigma-Aldrich) or rabbit anti-ZMYND8 (Sigma-Aldrich) antibodies as primary antibodies, and with goat anti-rabbit Alexa546 and goat anti-mouse Alexa488 (Invitrogen) as secondary antibodies. Images were acquired using inverted LSM700 laser scanning confocal microscope (Zeiss).
Clonogenic Assay
For the colony formation assay following IR, iMEFs were plated in 60mm dishes, irradiated after 24 hr with the indicated doses and incubated at 37°C in the presence of 5% CO2. After 14 days, colonies were fixed with 15% acetic acid: methanol (v/v) for 5 min and stained with 0.5% Crystal Violet (Sigma-Aldrich) for 30 min for colony visualization. For the colony formation assay in the presence of PARPi (Olaparib/AZD2281, Selleckchem), 0.1, 1, and 5 μM PARPi was added 24 after seeding and cells allowed to grow for 14 days before fixation, with fresh media and PARPi replenished on day 7.
Metaphase Analysis
iMEFs were treated with 2 μM PARPi for 21 h followed by 1 h incubation at 37°C with Colcemid (Sigma-Aldrich). Metaphase preparation and aberration analysis were performed as it follows. Cell pellets were resuspended in 0.075M KCl at 37°C for 20 min to perform a hypotonic shock, and washed/fixed with methanol/glacial-acetic acid solution. Metaphase spreads were prepared by dropping fixed cells on microscope slides, which were air-dried and placed at 42°C for 1 h. Giemsa staining was performed by using KaryoMAX Giemsa Stain Solution and Gurr Buffer Tablets (GIBCO). Metaphases were acquired with the Automated Metaphase Finder System Metafer4 (MetaSystems).
End Resection Assay
Single guide RNAs targeting two sequences 2276 bp apart within the ROSA26 locus were cloned into pX458 plasmid (gDSB-1/2). CH12 cells were electroporated with a 1:1 mix of gDSB-1 and gDSB-2 constructs using the Neon Transfection System (Thermo Fisher Scientific), and allowed to recover for 24 h before collection. Genomic DNA was extracted according to standard protocols and individual repair junctions were amplified using nested PCR reactions. PCR products were extracted from agarose gel and sequenced. Single guide RNAs and primers used for the end resection assay are listed in Table S5.
CRISPR-Cas9-Induced CSR Assay
Constructs for expression of gRNA-Sμ and gRNA-Sα were generated by cloning single guide RNAs directed to the 5′ Sμ and 3′ Sα regions respectively of the Igh locus (Ramachandran et al., 2016) in tandem U6 cassettes on a modified pX458 plasmid backbone. Control construct was generated by cloning random sequences not targeting the mouse genome in tandem U6 cassettes on same plasmid. CH12 cells were electroporated with the constructs using the Neon Transfection System, and allowed to recover for 12 h before CSR analysis.
MutPE-Seq
Primary cultures of splenocytes were collected at either 72 h or 96 h post-activation. gDNA was extracted with phenol-chlorofom-isoamyl alcohol, and 5′ Sμ amplicon was amplified and prepared for deep-sequencing by two rounds of PCR. The first round of PCR amplification was performed with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) (annealing temperature of 64.9°C, 15,” 20 cycles) and locus-specific primers for 5′ Switch μ. Two/thirds of DNA from the first reaction was used as template for the second reaction (annealing temperature of 64.6°C, 15,” 15 cycles) to introduce sequencing adapters and genotype-specific barcodes. Gel-extracted amplicons were pooled and sequenced via 2 × 300bp Paired-end Read Sequencing on an Illumina MiSeq platform. Mutations present in only one of the paired reads were considered a sequencing artifact and discarded. Primers used for MutPE-Seq (Wang et al., 2017) are listed in Table S5.
ChIP-Seq
ChIP-seq for ZMYND8 and RNA Pol II were performed as it follows. Splenocytes (72 h post-activation) or CH12 cells (48 h post-activation) were fixed with 1% (16% Formaldehyde Methanol-free, Thermo Scientific) at 37°C for 10 min followed by addition of 1/20 volume of 2.5 M glycine (dissolved in PBS pH 7.4) and swirling. Fixed cells were washed with cold PBS,centrifuged and aliquoted. 20 million cells were resuspended in 100 μL of 1% SDS of RIPA buffer supplemented with Complete EDTA free proteinase inhibitor (Roche) and sonication was performed with a Covaris S220 Focused Ultrasonicator at peak value 105, duty factor 5, cycle/burst 200 for 10 min. After sonication, samples were adjusted to 0.1% SDS final concentration by diluting with non-SDS containing RIPA buffer. Chromatin fragments were then pre-cleaned by incubation with Dynabeads Protein A (Invitrogen) with rotation at 4°C for 1 h, and immunoprecipitated with antibody-bound Dynabeads. 10 ug antibodies specific for ZMYND8 (Sigma-Aldrich) or RNA Pol II polymerase (4H8, Abcam) were used for each sample. DNA libraries were prepared by using Illumina compatible adaptors (Bio Scientific) and sequenced on an Illumina NextSeq 500 Sequencer. FASTQ files were aligned against mouse genome (mm10) using BWA mem (Li and Durbin, 2009). Processing and peak-calling of ChIP-seq data were accomplished with HOMER ChIP-Seq program (Heinz et al., 2010). Peak annotation was done using R and ChIPseeker package (Yu et al., 2015). The schematic representation of the murine Igh locus in Figures 4C, 5A, and S7A encompasses nucleotides 113,175,000-113,441,797 on Chr12.
RNA-Seq
Samples used in RNA-seq were cultured in activated and unactivated conditions for 48 h. Cultured cells were collected by centrifugation and RNA was extracted with AllPrep DNA/RNA Mini Kit (QIAGEN). Ribosomal RNA was depleted using Ribo-Zero Gold rRNA Removal Kit (Illumina). Libraries were prepared with TruSeq Stranded Total RNA Library Prep Kit (Illumina). Three biological repeats were performed and run in two lanes on the same flow cells on NextSeq High Output 75 SR (Illumina). For data analysis, reads were pseudo-aligned to an index created from the Ensembl mouse GRCm38.p5 assembly and custom annotations of Igh locus features. Transcript-level abundances were quantified using kallisto v0.43.0 (Bray et al., 2016), and subsequently summarized to the gene-level using the R package tximport (Soneson et al., 2015). Differential gene expression analysis was performed using DESeq2 (Love et al., 2014).
GRO-Seq
The GRO-Seq analysis was performed on splenocytes 24 h after stimulation with LPS and IL-4 as it follows. Global run-on and library preparation for sequencing was performed on nuclei from 10-15 million cells. After run-on reaction, the RNA was extracted with TRIzol LS Reagent (Invitrogen). RNA was treated with TURBO DNase (Invitrogen), fragmented using RNA Fragmentation Reagents (Invitrogen) and purified by running through BioSpin P-30 Gel Columns (Bio-Rad). The 3′ end of the fragmented RNA was dephosphorylated with T4 Polynucleotide Kinase (NEB) followed by heat-inactivation. Dephosphorylation reactions were purified using anti-BrdU beads (Santa Cruz Biotech) and precipitated overnight. Poly(A)-tailing and cDNA synthesis were performed the next day. For reverse transcription, an oligo allowing custom barcoding during final amplification was used (RT-primer).
After cDNA synthesis, Exonuclease I (NEB) was used to catalyze the removal of excess oligo. The DNA–RNA hybrid was purified using ChIP DNA Clean & Concentrator Kit (Zymo Research Corporation), RNase H (Thermo Scientific) treated and circularized. Circularized cDNAs were relinearized using APE1 (NEB) and were amplified for 12–14 cycles with primers oNTI201 and oNTI200. The final product was ran on a gel, purified, and cleaned up as above. DNA libraries were sequenced on a Hi-Seq 2500 for 50 cycle, single end run and the standard pipeline (1.8.2) was used for image analysis and base calling. Reads were aligned to the National Center for Biotechnology Information mouse genome data (July 2007; NCBI37/mm9) by using the alignment software Bowtie (Langmead et al., 2009) with the following options: -S –m 1 –p [num_of_thread] -a–best–starta –n 2 –l [read_length] –trim5 2. These options report reads that align uniquely to the best stratum and allowing up to 2 mismatches after trimming 2 nucleotides from the 5′ end. For visualization, stranded density tracks were generated by using bedtools genomecov program with a normalizing scale factor to calculate rpm separately for each strand. Rpkm value was calculated for comparative analysis and one sample t.test in R package was used for the p values. Primers used for GRO-Seq (Ingolia et al., 2009) are listed in Table S5.
Switch Junction Analysis
Genomic DNA was extracted from splenocytes stimulated for 96 h with LPS and IL-4, and Sμ-Sγ1 junctions were amplified by two rounds of PCR using Phusion High-Fidelity DNA Polymerase (Thermo Scientific). PCR annealing conditions were 72°C, 2′ and 10 cycles for the first round and 72°C, 2′ and 20 cycles for the second round. PCR products were run on agarose gels, and fragments corresponding to 350-1000 bp were extracted with NucleoSpin DNA Purification Kit (Macherey-Nagel), cloned using TOPO-TA cloning kit (Invitrogen) and sequenced using M13 forward and reverse universal primers. Primers used for Switch junction analysis (Di Virgilio et al., 2013) are listed in Table S5.
SHM Analysis
Single-cell suspensions from Peyer’s patches of 30-31 weeks old mice were first incubated with anti-mouse CD16/32 (BioLegend) and then labeled with B220-FITC– (BioLegend), CD19-APC– (BioLegend), CD38-Alexa700– (Thermo Scientific), and CD95/Fas-PE– (BD-Biosciences) conjugated antibodies. Non-germinal center (CD38+ Fas−) and germinal center B cells (CD38− Fas+) were sorted on an Aria BD sorter. Genomic DNA was extracted and the 5′ portions of JH4 (Igh) and JK5 (Igk) introns were amplified by PCR using Phusion High-Fidelity DNA Polymerase (Thermo Scientific). The 800 bp JH4 and 700 bp JK5 PCR products were gel extracted, cloned into a pCR2.1 vector using the TOPO TA Cloning Kit (Invitrogen) and sequenced. Mutations were quantified over 510 bp downstream JH4 and 536 bp downstream JK5 gene segments. Primers used for SHM analysis (Rouaud et al., 2013, Sander et al., 2015) are listed in Table S5.
Quantification and Statistical Analysis
The statistical significance of differences between groups/datasets was determined by the Mann–Whitney U test for all data presented in this study, with the following exceptions. The significance for the quantification of RNA Pol II ChIP-seq data (Figure 5B) was calculated with the Welch 2 sample unpaired t test. The adjusted p values for the quantification of relative transcript levels at hs4, hs1,2, hs3b, hs3a (Figure 5D) were calculated with the Wald test, and corrected for multiple-testing with the Benjamini-Hochberg method. ∗∗ is q < 1x10ˆ-7 for hs1,2 and q < 1x10ˆ-9 for hs3b. The pvalues for the differential gene analysis of GRO-Seq data (Figure S5E) were calculated with one-sided one sample t.test (mu = 0) on log2FC. Statistical details of experiments can be found in the figure legends.
Data and Software Availability
The accession number for the deep-sequencing data reported in this paper (ChIP-seq, RNA-seq, and GRO-seq) is GEO: GSE118794. Original imaging and western blot data used to generate any of the figure panels have been deposited at Mendeley: https://doi.org/10.17632/jktyjvy9hg.1.
Acknowledgments
We thank S. Buonomo (The University of Edinburgh, Edinburgh) for the Rif1FH/FH mice; S. Balasubramanian, A. Rahjouei, and V. Coralluzzo (MDC, Berlin) for assistance with metaphase spread, initial qPCR analyses, and support with genotyping, respectively; U. Höpken (MDC) for assistance with B cell development analysis and guidelines for immunization protocols; T. Weber and K. Schmidt from K. Rajewsky’s group (MDC) for sharing their expertise for the Igh SHM analysis and Ig levels assessment; C. Birchmeier (MDC) for kindly granting access to the lab LSM700 confocal microscope; the MDC Advanced Light Microscopy (ALM) technology platform for their technical support with the IF assays; the MDC FACS Core Facility and H.P. Rahn for assistance with cell sorting; and N. Zampieri (MDC) for discussion and advices. This work was supported by ERC grant 638897 (M.D.V.), NIH grant AI037526-24 (M.C.N.), and PHS GM103314 and GM109824 grants (B.T.C.). D.B.R. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. M.C.N. is a Howard Hughes Medical Institute (HHMI) Investigator. M.D.V. is a Helmholtz Young Investigators Group leader (Helmholtz Association).
Author Contributions
Conceptualization, V.D.-B., D.B.R., M.C.N., and M.D.V.; Methodology, V.D.-B., D.B.R., R.I.S., and M.D.V.; Investigation, V.D.-B., D.B.R., Q.W., A.G., D.S., W.Z., M.A., L.K., K.-R.K.-K., A.P., M.D., and M.D.V.; Formal Analysis, V.D.-B., D.B.R., J.A.P., T.Y.O., S.J., and M.D.V.; Writing – Original Draft, M.D.V.; Writing – Review & Editing, M.C.N. and M.D.V.; Visualization, M.D.V.; Supervision, R.C., B.T.C., M.C.N., and M.D.V.; Project Administration, M.D.V.
Declaration of Interests
The authors declare no competing interests.
Published: October 4, 2018
Footnotes
Supplemental Information includes seven figures and five tables and can be found with this article online at https://doi.org/10.1016/j.molcel.2018.08.042.
Supplemental Information
References
- Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D.K., Roy S., Das C. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 2016;291:2664–2681. doi: 10.1074/jbc.M115.679985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Basu M., Khan M.W., Chakrabarti P., Das C. Chromatin reader ZMYND8 is a key target of all trans retinoic acid-mediated inhibition of cancer cell proliferation. Biochim. Biophys. Acta. 2017;1860:450–459. doi: 10.1016/j.bbagrm.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Basu M., Sengupta I., Khan M.W., Srivastava D.K., Chakrabarti P., Roy S., Das C. Dual histone reader ZMYND8 inhibits cancer cell invasion by positively regulating epithelial genes. Biochem. J. 2017;474:1919–1934. doi: 10.1042/BCJ20170223. [DOI] [PubMed] [Google Scholar]
- Bébin A.G., Carrion C., Marquet M., Cogné N., Lecardeur S., Cogné M., Pinaud E. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J. Immunol. 2010;184:3710–3717. doi: 10.4049/jimmunol.0901978. [DOI] [PubMed] [Google Scholar]
- Berton M.T., Uhr J.W., Vitetta E.S. Synthesis of germ-line gamma 1 immunoglobulin heavy-chain transcripts in resting B cells: induction by interleukin 4 and inhibition by interferon gamma. Proc. Natl. Acad. Sci. USA. 1989;86:2829–2833. doi: 10.1073/pnas.86.8.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birshtein B.K. Epigenetic regulation of individual modules of the immunoglobulin heavy chain locus 3′ regulatory region. Front. Immunol. 2014;5:163. doi: 10.3389/fimmu.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C., Alt F.W., Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Alt F.W. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Cogné M., Lansford R., Bottaro A., Zhang J., Gorman J., Young F., Cheng H.L., Alt F.W. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Collins J.T., Dunnick W.A. Germline transcripts of the murine immunoglobulin gamma 2a gene: structure and induction by IFN-gamma. Int. Immunol. 1993;5:885–891. doi: 10.1093/intimm/5.8.885. [DOI] [PubMed] [Google Scholar]
- Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D., Dileep V., Quivy J.P., Foti R., Tili F., Santarella-Mellwig R., Antony C., Almouzni G., Gilbert D.M., Buonomo S.B. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012;31:3678–3690. doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., McBride K.M., Jankovic M., Gazumyan A., Thai T.H., Robbiani D.F., Di Virgilio M., Reina San-Martin B., Heidkamp G., Schwickert T.A. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandy A., Kracker S., Fischer A. Primary antibody deficiencies. Nat. Rev. Immunol. 2013;13:519–533. doi: 10.1038/nri3466. [DOI] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Esser C., Radbruch A. Rapid induction of transcription of unrearranged S gamma 1 switch regions in activated murine B cells by interleukin 4. EMBO J. 1989;8:483–488. doi: 10.1002/j.1460-2075.1989.tb03401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garot A., Marquet M., Saintamand A., Bender S., Le Noir S., Rouaud P., Carrion C., Oruc Z., Bébin A.G., Moreau J. Sequential activation and distinct functions for distal and proximal modules within the IgH 3′ regulatory region. Proc. Natl. Acad. Sci. USA. 2016;113:1618–1623. doi: 10.1073/pnas.1514090113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauchat J.F., Lebman D.A., Coffman R.L., Gascan H., de Vries J.E. Structure and expression of germline epsilon transcripts in human B cells induced by interleukin 4 to switch to IgE production. J. Exp. Med. 1990;172:463–473. doi: 10.1084/jem.172.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini S.L., Singh M., Calvo C.F., Ding G., Birshtein B.K. DNA regions flanking the mouse Ig 3′ alpha enhancer are differentially methylated and DNAase I hypersensitive during B cell differentiation. J. Immunol. 1993;150:1772–1780. [PubMed] [Google Scholar]
- Gillies S.D., Morrison S.L., Oi V.T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Clouaire T., Aguirrebengoa M., Legube G., Miller K.M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 2017;216:1959–1974. doi: 10.1083/jcb.201611135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Fernández A., Gilmore D., Milstein C. Age-related decrease in the proportion of germinal center B cells from mouse Peyer’s patches is accompanied by an accumulation of somatic mutations in their immunoglobulin genes. Eur. J. Immunol. 1994;24:2918–2921. doi: 10.1002/eji.1830241151. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaoui H., Ghazzaui N., Saintamand A., Carrion C., Oblet C., Denizot Y. The IgH 3′ regulatory region super-enhancer does not control IgA class switch recombination in the B1 lineage. Cell. Mol. Immunol. 2018;15:289–291. doi: 10.1038/cmi.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C.J., Klix N., Neuberger M.S. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z., Volpi S.A., Hassan R., Martinez N., Giannini S.L., Gold T., Birshtein B.K. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3′ regulatory region. J. Biol. Chem. 2007;282:35169–35178. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- Kim A., Han L., Santiago G.E., Verdun R.E., Yu K. Class-switch recombination in the absence of the IgH 3′ regulatory region. J. Immunol. 2016;197:2930–2935. doi: 10.4049/jimmunol.1600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet S.L., Baymaz H.I., Makowski M., Groenewold V., Jansen P.W., Berendsen M., Niazi H., Kops G.J., Vermeulen M. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 2015;282:1774–1785. doi: 10.1111/febs.12972. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Noir S., Boyer F., Lecardeur S., Brousse M., Oruc Z., Cook-Moreau J., Denizot Y., Cogné M. Functional anatomy of the immunoglobulin heavy chain 3′ super-enhancer needs not only core enhancer elements but also their unique DNA context. Nucleic Acids Res. 2017;45:5829–5837. doi: 10.1093/nar/gkx203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebman D.A., Nomura D.Y., Coffman R.L., Lee F.D. Molecular characterization of germ-line immunoglobulin A transcripts produced during transforming growth factor type beta-induced isotype switching. Proc. Natl. Acad. Sci. USA. 1990;87:3962–3966. doi: 10.1073/pnas.87.10.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G.G., Perry R.P. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature. 1985;318:475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Li Y., Lv J., Zheng X., Wen H., Shen H., Zhu G., Chen T.Y., Dhar S.S., Kan P.Y. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell. 2016;63:470–484. doi: 10.1016/j.molcel.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberson R., Giannini S.L., Birshtein B.K., Eckhardt L.A. An enhancer at the 3′ end of the mouse immunoglobulin heavy chain locus. Nucleic Acids Res. 1991;19:933–937. doi: 10.1093/nar/19.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzker S., Alt F.W. Structure and expression of germ line immunoglobulin gamma 2b transcripts. Mol. Cell. Biol. 1988;8:1849–1852. doi: 10.1128/mcb.8.4.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzker S., Rothman P., Pollock R., Coffman R., Alt F.W. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell. 1988;53:177–184. doi: 10.1016/0092-8674(88)90379-0. [DOI] [PubMed] [Google Scholar]
- Madisen L., Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- Malovannaya A., Lanz R.B., Jung S.Y., Bulynko Y., Le N.T., Chan D.W., Ding C., Shi Y., Yucer N., Krenciute G. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., van der Stoep N., Tian M., Ferrini R., Davidson L., Bottaro A., Alt F.W. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet M., Garot A., Bender S., Carrion C., Rouaud P., Lecardeur S., Denizot Y., Cogné M., Pinaud E. The Eμ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J. Immunol. 2014;193:1171–1183. doi: 10.4049/jimmunol.1302868. [DOI] [PubMed] [Google Scholar]
- Matthias P., Baltimore D. The immunoglobulin heavy chain locus contains another B-cell-specific 3′ enhancer close to the alpha constant region. Mol. Cell. Biol. 1993;13:1547–1553. doi: 10.1128/mcb.13.3.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson J.S., Giannini S.L., Birshtein B.K. Identification of 3′ alpha-hs4, a novel Ig heavy chain enhancer element regulated at multiple stages of B cell differentiation. Nucleic Acids Res. 1995;23:975–981. doi: 10.1093/nar/23.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kondo S., Sugai M., Nazarea M., Imamura S., Honjo T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- Oruc Z., Boumédiène A., Le Bert M., Khamlichi A.A. Replacement of Igamma3 germ-line promoter by Igamma1 inhibits class-switch recombination to IgG3. Proc. Natl. Acad. Sci. USA. 2007;104:20484–20489. doi: 10.1073/pnas.0608364104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Nussenzweig M.C. AID targeting in antibody diversity. Adv. Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Perlot T., Alt F.W. Cis-regulatory elements and epigenetic changes control genomic rearrangements of the IgH locus. Adv. Immunol. 2008;99:1–32. doi: 10.1016/S0065-2776(08)00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T., Alt F.W., Bassing C.H., Suh H., Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson S., Cook G.P., Brüggemann M., Williams G.T., Neuberger M.S. A second B cell-specific enhancer 3′ of the immunoglobulin heavy-chain locus. Nature. 1990;344:165–168. doi: 10.1038/344165a0. [DOI] [PubMed] [Google Scholar]
- Pinaud E., Khamlichi A.A., Le Morvan C., Drouet M., Nalesso V., Le Bert M., Cogné M. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- Pinaud E., Marquet M., Fiancette R., Péron S., Vincent-Fabert C., Denizot Y., Cogné M. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv. Immunol. 2011;110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8. [DOI] [PubMed] [Google Scholar]
- Poleshko A., Einarson M.B., Shalginskikh N., Zhang R., Adams P.D., Skalka A.M., Katz R.A. Identification of a functional network of human epigenetic silencing factors. J. Biol. Chem. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.R., Resch W., Liang G., Tang Z., Mathé E., Benner C. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe G., Lin Y.C., Julius M., Marcu K.B., Stavnezer J. Structure of germ line immunoglobulin alpha heavy-chain RNA and its location on polysomes. Mol. Cell. Biol. 1990;10:382–386. doi: 10.1128/mcb.10.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Haddad D., Li C., Le M.X., Ling A.K., So C.C., Nepal R.M., Gommerman J.L., Yu K., Ketela T. The SAGA deubiquitination module promotes DNA repair and class switch recombination through ATM and DNAPK-mediated γH2AX formation. Cell Rep. 2016;15:1554–1565. doi: 10.1016/j.celrep.2016.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A.R., Stavropoulos P., Jankovic M., Nussenzweig M.C. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman P., Lutzker S., Cook W., Coffman R., Alt F.W. Mitogen plus interleukin 4 induction of C epsilon transcripts in B lymphoid cells. J. Exp. Med. 1988;168:2385–2389. doi: 10.1084/jem.168.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman P., Chen Y.Y., Lutzker S., Li S.C., Stewart V., Coffman R., Alt F.W. Structure and expression of germ line immunoglobulin heavy-chain epsilon transcripts: interleukin-4 plus lipopolysaccharide-directed switching to C epsilon. Mol. Cell. Biol. 1990;10:1672–1679. doi: 10.1128/mcb.10.4.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman P., Lutzker S., Gorham B., Stewart V., Coffman R., Alt F.W. Structure and expression of germline immunoglobulin gamma 3 heavy chain gene transcripts: implications for mitogen and lymphokine directed class-switching. Int. Immunol. 1990;2:621–627. doi: 10.1093/intimm/2.7.621. [DOI] [PubMed] [Google Scholar]
- Rouaud P., Vincent-Fabert C., Saintamand A., Fiancette R., Marquet M., Robert I., Reina-San-Martin B., Pinaud E., Cogné M., Denizot Y. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J. Exp. Med. 2013;210:1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintamand A., Rouaud P., Garot A., Saad F., Carrion C., Oblet C., Cogné M., Pinaud E., Denizot Y. The IgH 3′ regulatory region governs μ chain transcription in mature B lymphocytes and the B cell fate. Oncotarget. 2015;6:4845–4852. doi: 10.18632/oncotarget.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintamand A., Rouaud P., Saad F., Rios G., Cogné M., Denizot Y. Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion. Nat. Commun. 2015;6:7084. doi: 10.1038/ncomms8084. [DOI] [PubMed] [Google Scholar]
- Saintamand A., Vincent-Fabert C., Garot A., Rouaud P., Oruc Z., Magnone V., Cogné M., Denizot Y. Deciphering the importance of the palindromic architecture of the immunoglobulin heavy-chain 3′ regulatory region. Nat. Commun. 2016;7:10730. doi: 10.1038/ncomms10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S., Chu V.T., Yasuda T., Franklin A., Graf R., Calado D.P., Li S., Imami K., Selbach M., Di Virgilio M. PI3 kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity. 2015;43:1075–1086. doi: 10.1016/j.immuni.2015.10.021. [DOI] [PubMed] [Google Scholar]
- Savitsky P., Krojer T., Fujisawa T., Lambert J.P., Picaud S., Wang C.Y., Shanle E.K., Krajewski K., Friedrichsen H., Kanapin A. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 2016;17:2724–2737. doi: 10.1016/j.celrep.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinson E., Fernandez C., Stavnezer J. Induction of germ-line immunoglobulin heavy chain transcripts by mitogens and interleukins prior to switch recombination. Eur. J. Immunol. 1990;20:1079–1084. doi: 10.1002/eji.1830200520. [DOI] [PubMed] [Google Scholar]
- Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016;165:331–342. doi: 10.1016/j.cell.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockett P., Stavnezer J. Effect of cytokines on switching to IgA and alpha germline transcripts in the B lymphoma I.29 mu. Transforming growth factor-beta activates transcription of the unrearranged C alpha gene. J. Immunol. 1991;147:4374–4383. [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt C.G., Luijsterburg M.S., Menafra R., Lindeboom R.G., Jansen P.W., Edupuganti R.R., Baltissen M.P., Wiegant W.W., Voelker-Albert M.C., Matarese F. ZMYND8 co-localizes with NuRD on target genes and regulates poly(ADP-ribose)-dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep. 2016;17:783–798. doi: 10.1016/j.celrep.2016.09.037. [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Sirlin S., Abbott J. Induction of immunoglobulin isotype switching in cultured I.29 B lymphoma cells. Characterization of the accompanying rearrangements of heavy chain genes. J. Exp. Med. 1985;161:577–601. doi: 10.1084/jem.161.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Radcliffe G., Lin Y.C., Nietupski J., Berggren L., Sitia R., Severinson E. Immunoglobulin heavy-chain switching may be directed by prior induction of transcripts from constant-region genes. Proc. Natl. Acad. Sci. USA. 1988;85:7704–7708. doi: 10.1073/pnas.85.20.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett A.J., DeGrasse J.A., Sekedat M.D., Oeffinger M., Rout M.P., Chait B.T. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- Takizawa M., Tolarová H., Li Z., Dubois W., Lim S., Callen E., Franco S., Mosaico M., Feigenbaum L., Alt F.W. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J. Exp. Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G., Hakimpour P., Landgraf P., Rice A., Tuschl T., Casellas R., Papavasiliou F.N. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Fabert C., Truffinet V., Fiancette R., Cogné N., Cogné M., Denizot Y. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J. Immunol. 2009;182:6926–6932. doi: 10.4049/jimmunol.0900214. [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Fiancette R., Pinaud E., Truffinet V., Cogné N., Cogné M., Denizot Y. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- Wang Q., Kieffer-Kwon K.R., Oliveira T.Y., Mayer C.T., Yao K., Pai J., Cao Z., Dose M., Casellas R., Jankovic M. The cell cycle restricts activation-induced cytidine deaminase activity to early G1. J. Exp. Med. 2017;214:49–58. doi: 10.1084/jem.20161649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R., Wang L., Grigera F., Manis J., Selsing E., Perlot T., Alt F.W., Cogne M., Pinaud E., Kenter A.L. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L., Huang W., Bellani M., Seidman M.M., Wu K., Fan D., Nie Y., Cai Y., Zhang Y.W., Yu L.R. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell. 2017;31:653–668.e7. doi: 10.1016/j.ccell.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Chapman J.R., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zeng W., Kong Q., Li C., Mao B. Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochem. Biophys. Res. Commun. 2010;394:1024–1029. doi: 10.1016/j.bbrc.2010.03.115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.