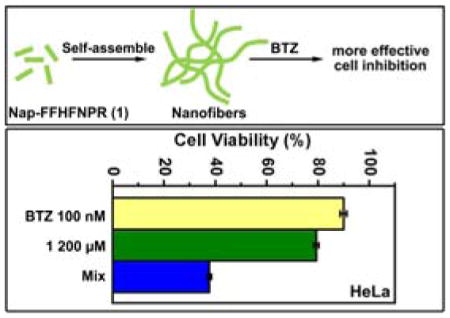
Abstract
Nanofibers of short peptides are emerging as a promising type of agents for inhibiting cancer cells. But the proteolysis of peptides decreases the anticancer efficacy of the peptide nanofibers. Here we show that decreasing the activity of proteasomes enhance the activity of peptide nanofibers for inhibiting cancer cells. Based on the structure of galactin-3, we designed a heptapeptide, which self-assembles to form nanofibers. The nanofibers of the heptapeptide exhibit moderate cytotoxicity to three representative cancer cell lines (HeLa, MCF-7, and HepG2), largely due to the proteolysis of the peptides. Using a clinically approved proteasome inhibitor, bortezomib, to treat the cancer cells significantly decreases the proteolysis of the peptides and enhances the activity of the peptide nanofibers for inhibiting the cancer cells. This work illustrates a promising approach for enhancing the anticancer efficacy of peptide nanofibers by modulating intracellular protein degradation machinery, as well as provides insights for understanding the cytotoxicity of aberrant protein or peptide aggregates in complicated cellular environment.
Keywords: anticancer, nanofibers, self-assemble, peptide, proteolysis
Graphical Abstract
Here we report the design and synthesis of a heptapeptide (1) based on the structure of galactin-3, which can self-assemble into nanofibers. While the proteolysis of the peptides leads to a moderate cytotoxicity of the molecule on cancer cells, co-incubation of a proteasome inhibitor BTZ with the peptide significantly decreases the proteolysis of the peptide and enhances the activity of the peptide nanofibers for inhibiting the cancer cells.
As the consequence of reaching thermodynamic equilibrium, self-assembly autonomously reorganizes components to form ordered structures. When small organic molecules self-assemble in solutions, the assemblies of the molecules usually build up the three dimensional networks to encapsulate water to result in a hydrogel.[1] These small molecules, consequently, are classified as hydrogelators. Share common features such as π-π interactions, hydrogen bonding, hydrophobic interactions, charge interactions, and other non-covalent interactions, the hydrogelators self-assemble and usually form nanofibers. These nanofibers, having the similar morphology as endogenous nanofibers that are normal (e.g., actin filaments and microtubules) or aberrant (e.g., amyloid) cellular structures, are emerging as a new class of molecular entities for controlling the fate of cells.[2]
Among the small molecules being explored for self-assembly, one attracting class of candidates is peptides. Peptide based hydrogelators,[3] possessing inherent biological activities and structural diversity, are accessible in large quantity. Inspired by proteins that self-assemble into nanofibers to serve as critical components of cellular processes, research focusing on the self-assembly of peptides is attracting increasing attentions from scientists.[3a, 3d, 3g, 3h, 3k, 4] The supramolecular assemblies of peptides are potential candidates for biological applications such as 3D cell culture,[5] wound healing[6] and tissue engineering.[7] Studies on the cytotoxicity of nanofibers formed by peptides reveals that nanofibers are able to inhibit cancer cells selectively,[8] by interacting with the cancer cells intracellularly or pericellularly. In addition, the self-assembly of a peptide amphiphile, which contained a cytotoxic peptide fragment, efficiently delivers the cytotoxic peptide to cancer cells and inhibit the cancer cells.[9] Recently, Yang et al.[10] and Wells et al.[11] reported nanofibers formed by the self-assembly of small molecules to inhibit cancer cells. Our group also has been exploring the anticancer properties of nanofibers on cancer cells and reported that the supramolecular assemblies of small molecules behaved differently from the molecule monomers and the assemblies can interact with cells and kill cancer cells.[2d, 8] While nanofibers of peptides are becoming an active research frontier for developing anticancer agents, one of the less explored aspects is how the stability of peptides in biological environment and the proteolysis of these peptides affect the activities of the peptide nanofibers against cancer cells.
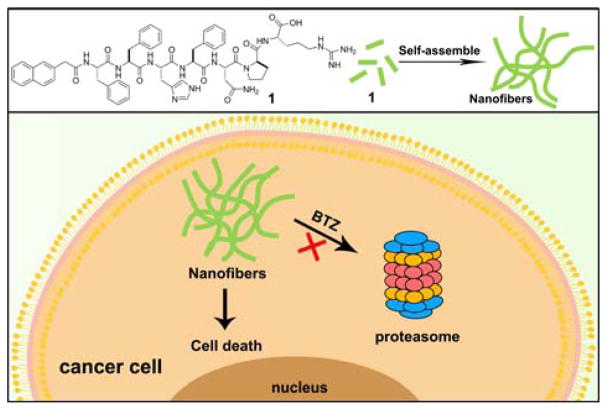
In this study, encouraged by the previous studies on the inhibition of cancer cells by peptide nanofibers, we design and synthesize a peptide (Nap-Phe-Phe-His-Phe-Asn-Pro-Arg, 1 (Scheme 1)), and examine how reducing proteolysis affects the activity of the peptide nanofibers of 1 for inhibiting cancer cells. We choose to follow the structure of galectin-3[12] because we want to increase the interaction of nanofibers and cell surface heparan sulfate for cellular uptake.[13] Since Nap-Phe-Phe is an excellent motif to enable self-assembly and hydrogelation,[14] we connect the peptide fragment His-Phe-Asn-Pro-Arg from galectin-3 to Nap-Phe-Phe for making the hydrogelator 1. The hydrogelator 1 self-assembles into nanofibers. The existence of nanofibers in cancer cells would induce cell death. Cell assays show that the co-incubation 1 with bortezomib (BTZ), a proteasome inhibitor,[15] significantly increases the overall cytotoxicity of 1 in cancer cells. The analysis of cell lysates in the presence of BTZ indicates that the inhibition of proteasome, indeed, reduces the degradation of the peptides, supporting that more nanofibers exist in the cancer cell to lead to higher cytotoxicity. This work illustrates a promising approach for enhancing the anticancer efficacy of peptide nanofibers by modulating intracellular protein degradation machinery, as well as provides insights for understanding the cytotoxicity of aberrant protein or peptide aggregates in complicated cellular environment.
Scheme 1.
Chemical structure of a hydrogelator and the proposed mechanism for its nanofibers to inhibit cancer cells.
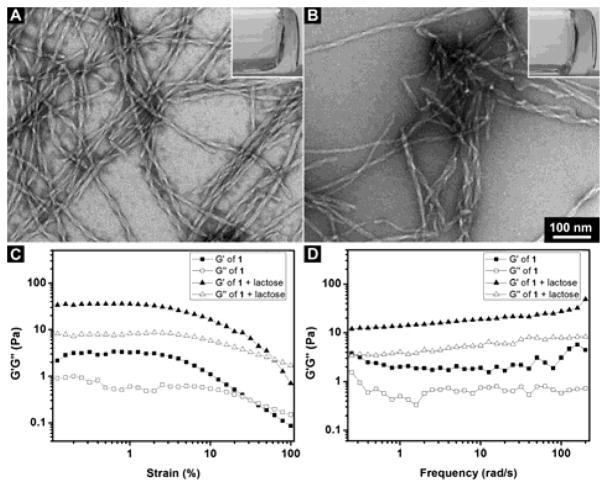
After making the hydrogelator 1 by solid phase peptide synthesis (SPPS),[16] we purify the molecule by high-performance liquid chromatography (HPLC), with the overall yield of 1 about 80%. Molecules are characterized by 1H-NMR (Figure S1–S3). Stable hydrogel forms when 1 (0.4 wt%) dissolves in water at pH7.4. As shown in Figure 1A, transmission electron microscopy (TEM) image of the hydrogel formed by 1 contains uniform and twisted nanofibers with a diameter at 10 ± 2 nm and a well-defined pitch at 52 ± 2 nm. We also test the hydrogelation of 1 at 0.4 wt% together with 1 equivalent of lactose. As shown in Figure 1, a more stable hydrogel is formed when adding 1 eq. of lactose. While the TEM image shows that there are nanofibers with a diameter at 10 ± 2 nm existed in the hydrogel of 1 and lactose, the lengths of pitch of the nanofibers become less uniform. This result indicates that 1 is able to interact with lactose, albeit moderately.
Figure 1.
(A, B) TEM images of the hydrogels (inset: optical images) formed by (A) 0.4 wt% 1; (B) 0.4 wt% 1 and 1 eq. lactose in water at pH7.4 (Scale bar = 100 nm); (C, D) The (C) strain and (D) frequency dependence of dynamic storage modulus G′ and loss modulus G″ of the gels formed by 1 at 0.4 wt% or the gel formed by 1 at 0.4 wt% and 1 eq. lactose at pH7.4 in water.
Rheological test of the hydrogels further elucidates the viscoelastic properties of the hydrogels formed by the heptapeptide. Figure 1C and 1D show the strain and frequency dependence of storage modulus (G′) and loss modulus (G″) of the hydrogels formed by 1 alone or in combination with 1 equivalent of lactose. The G′ values of both the hydrogels are larger than their G″ value; the G′ and G″ of both the hydrogels are nearly independent of frequency. These features confirm the viscoelastic features of the hydrogels. Hydrogel formed by 1 together with lactose shows a higher G′ than that of the hydrogel of 1, with the maximum storage modulus G′ at 36 Pa and indicating that the hydrogel is stronger than the hydrogel formed by hydrogelator 1 alone. The value of critical strain of the hydrogel formed by 1 is at 3.3 % and that of hydrogel formed by the mixture of 1 and lactose at 13.6 %, indicating that the gel formed by the mixture is more resilient than the hydrogel formed by 1 alone. Since part of the sequence of peptide 1 is originated from the saccharide binding domain (FHFNPR) of galactin-3,[17] this peptide should have certain binding affinity to carbohydrate such as lactose. So we prepare the hydrogel of 1 with 1 eq. of lactose to verify the interactions between 1 and lactose. The slight increase of the strength of the hydrogel agrees with the interaction between lactose and 1. These rheological results indicate that the addition of lactose affects the viscoelasticity of the hydrogel and the self-assembling ability of the 1. Such an observation also supports that there are interactions between the nanofibers of 1 and lactose and agrees with that the sequence in molecule 1 originates from the carbohydrate interacting motif in galactin-3.
After the gelation test of the hydrogelator, we test the cytotoxicity of 1 against HeLa (cervical cancer), MCF-7 (breast cancer), and HepG2 (liver cancer) cells. While 1 exhibits the highest cytotoxicity on MCF-7, with the IC50 at 51 μM (48 h), its cytotoxicity against HepG2 is lower (IC50 = 233 μM) than against MCF-7 cells (Figure S4), and 1 shows the lowest cytotoxicity against HeLa cells (IC50 > 500 μM). We then co-incubated 1 with BTZ[18] to treat the aforementioned three cancer cell lines. The results (Figure S5) shows that the addition of BTZ significantly increases the cytotoxicity of 1 against the cancer cells, decreasing the IC50 of 1 against MCF-7, HepG2, and HeLa cells to 38, 56, and 80 μM, respectively. As shown by the summarized cytotoxicity of BTZ, 1 alone, or the combination of 1 and BTZ against the cancer cells (Figure 2A), BTZ alone at 100 nM barely shows cytotoxicity on the cancer cells, 1 at 200 μM exhibits mild cytotoxicity against HeLa cells, with the cell viability of 79 %, and the combination of 1 (200 μM) and BTZ (100 nM) decreases the viability of the HeLa cells to 38 %. Similarly, the cell viability of MCF-7 cells is 43 % after being treated by 1 at 200 μM (TEM image containing nanofibers shown in Figure S6), and the co-incubation of 1 and BTZ decreases the cell viability of MCF-7 cells to 29 %. The combination also decreases the cell viability of HepG2 cells from 57 % (treated by 1 alone) to 34 %. Comparing that it significant increases the cytotoxicity of 1 to HeLa cells, BTZ only moderately increases the cytotoxicity of 1 against MCF-7 and HepG2 cells.
Figure 2.
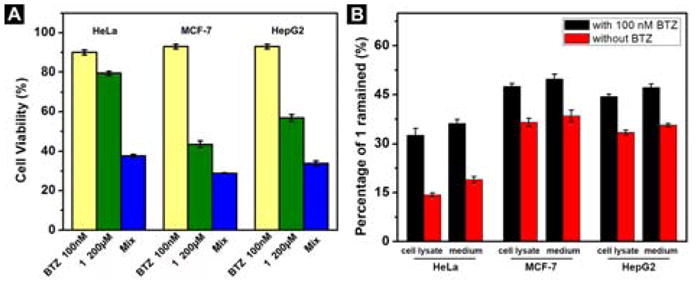
(A) Cell viability of HeLa, MCF-7 and HepG2 cells incubated with 1 (200 μM), BTZ (100 nM) or the mixture of 1 and BTZ for 48 h. (B) Percentage of 1 remained in HeLa cell or in culture medium; MCF-7 cell or in culture medium; HepG2 cell or in culture medium after being treated with 200 μM of 1 alone or together with 100 nM BTZ for 1 h.
To verify whether the decrease of proteolysis results in enhanced cytotoxicity of 1 against the cancer cells, we examine the amount of 1 remained in the cell lysates or in culture medium after the cancer cells are incubated with 1 alone or the mixture of 1 and BTZ for 1 h (Figure 2B). Without the addition of BTZ, most of the hydrogelator 1 inside the cells degrades within 1 hour, with the remaining 1 to be 14 %, 33%, and 39% in HeLa, HepG2, and MCF-7 cells, respectively. This result explains the higher cytotoxicity of 1 on MCF-7 and HepG2 cells since more molecules of 1 present in those two cancer cell lines. The addition of BTZ together with 1 increases the percentages of remaining 1 to be 33%, 44%, and 50% in HeLa, HepG2, and MCF-7 cells, respectively. This result explains the significant increase of cytotoxicity of 1 by combining it with BTZ against HeLa cells. That is, there are more molecules of 1 remaining after the co-incubation of BTZ, which leads to more nanofibers, and thus resulting in higher cytotoxicity.
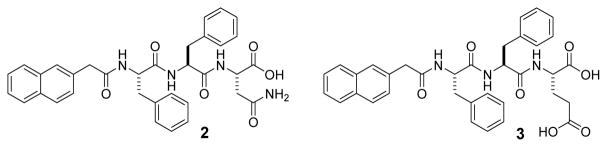
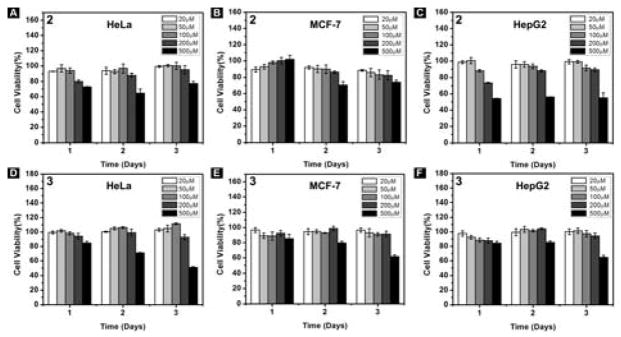
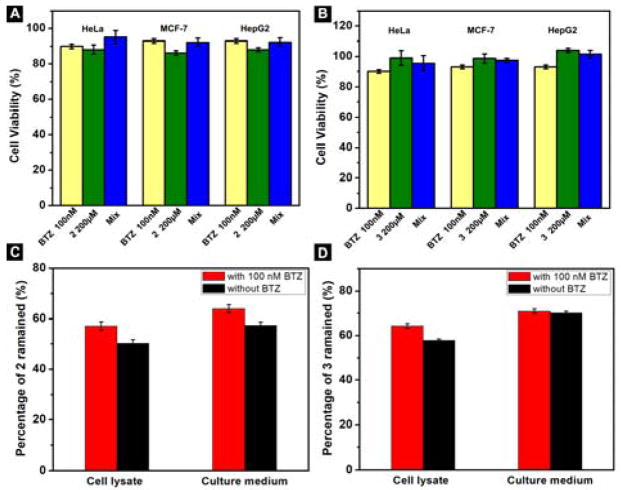
To elucidate the reason that the hydrogelator 1 shows cytotoxicity to cancer cells and exhibits higher cytotoxicity in the presence of BTZ, we synthesize two control molecules: 2 (Nap-Phe-Phe-Asn) and 3 (Nap-Phe-Phe-Glu), as shown in Scheme 2. Containing arsparagine (N) and the self-assembly motif,[19] 2 is a part of 1; and 3 contains a negative charged residue (glutamic acid, E) and the self-assembly motif. 2 exhibits little cytotoxicity against HeLa, MCF-7 and HepG2 cells, except starting to inhibit the cells at 500 μM (Figure 3). Molecule 3 hardly exhibits cytotoxicity against the three cancer cell lines (Figure 3). TEM images of 1, 2, and 3 at 500 μM indicate that the three molecules can form nanofibers at this concentration, which explains the cytotoxicity to cancer cells at 500 μM (Figure S7). These results suggest that the interaction of the assemblies of 1 with cell surface glycans is important because such interactions likely facilitate the nanofibers enter the cells.
Scheme 2.
Chemical structures of molecule 2 and 3.
Figure 3.
(A–C) Cell viability of (A) HeLa, (B) MCF-7, (C) HepG2 cells incubated with 2; (D–F) Cell viability of (D) HeLa, (E) MCF-7, (F) HepG2 cells incubated with 3 for 3 days.
We also co-incubate the molecule 2 and 3 with BTZ to treat the cancer cells, which barely increases the cytotoxicity of 2 and 3 (Figures 4A, 4B, S8, and S9). We incubate the HeLa cells with 2 (or 3) alone or in combination with BTZ for 1 h and examine the molecules remained in cell lysate or culture medium (Figure 4C, 4D), which indicates that both 2 and 3 degrade slower than 1. Intracellular concentrations of 1, 2, and 3 (Table 1) after treating HeLa cells with 1, 2, and 3 (200 μM, 1 h) show that 1 has a much higher intracellular concentration than the incubation concentration, but 2 (or 3) shows much lower intracellular concentrations. This result agrees with that (i) 1 is able to interact with glycans on cells for cellular uptake and (ii) 1 likely self-assembles inside the cells to reduce the exocytosis of the peptides.
Figure 4.
(A, B) Cell viability of HeLa, MCF-7 and HepG2 cells incubated with (A) 2 (200 μM), BTZ (100 nM) or the mixture of 2 and BTZ; (B) 3 (200 μM), BTZ (100 nM) or the mixture of 3 and BTZ for 48 h; (C) Percentage of 2 remained in HeLa cell or in culture medium after being treated with 200 μM of 2 alone or together with 100 nM BTZ for 1 h; (D) Percentage of 3 remained in HeLa cell or in culture medium after being treated with 200 μM of 3 alone or together with 100 nM BTZ for 1 h.
Table 1.
Intracellular concentrations of the hydrogelators in HeLa cells.
| Intracellular concentration (μM) [a] | Average | st.dev |
|---|---|---|
| 1 | 296.8 | 3.61 |
| 2 | 23.5 | 0.82 |
| 3 | 36.4 | 1.01 |
The cell lysates of HeLa cells are collected after 1h incubation with 200 μM of hydrogelators at 37 °C.
In conclusion, here we present a promising approach to enhance the anticancer activity of nanofibers formed by peptide by co-incubating the peptides with a proteasome inhibitor, BTZ. The overall cytotoxicity of peptide 1 is significantly increased after adding BTZ in the cell culture, since BTZ inhibits proteasome and increases the amount of nanofibers inside cells. The results suggest that the cellular uptake and intracellular fate of the peptides are important factors for designing peptide nanofibers for inhibiting cancer cells. Comparing to the use of D-amino acid to increase the long-term bio-stability of peptide materials,[20] this approach should be a useful alternative because L-peptides are more likely to exhibit higher affinity to cellular targets than D-peptides do, which may improve the specificity of the nanofibers against cancer cells. Moreover, these results imply that increasing proteolysis may be an effective way to reduce the cytotoxicity of the aggregates of aberrant proteins.
Experimental Section
Materials
The reagents such as N, N-diisopropylethylamine (DIPEA), O-benzotriazole-N,N,N′,N′-tetramethyluronium-hexafluorophosphate (HBTU), were purchased from ACROS Organics USA. All other amino acid derivatives including Fmoc-Phe-OH, Fmoc-His(Trt)-OH, Fmoc-Asn(Trt)-OH, Fmoc-Pro-OH, Fmoc-Arg(pbf)-OH were purchased from GL Biochem (Shanghai) Ltd. The synthesis of hydrogelator 1 is based on solid phase peptide synthesis (SPPS) and has been reported previously. The crude compound that acquired from SPPS is purified by HPLC and the overall yield of the synthesis is about 80%. 1H-NMR and LC-MS spectra are used to characterize the product.
Instrumentation and characterization
We used Waters Delta600 HPLC system, which equipped with an XTerra C18 RP column and an in-line diode array UV detector to purify the product. LC-MS machine we used was a Waters Acquity Ultra Performance LC with Waters MICROMASS detector. Hydrogen nuclear magnetic resonance (NMR) spectra were recorded on a Varian Unity Inova 400 with DMSO as solvent. We used Morgagni 268 transmission electron microscope to take Transmission electron microscope (TEM) images. MTT assay for cell cytotoxicity was test on DTX880 Multimode Detector.
Supplementary Material
Acknowledgments
This work was partially supported by NIH (CA142746 and AI130560) and NSF (MRSEC-1420382). We thank Brandeis EM and Optical Imaging facilities for TEM.
Footnotes
Supporting information for this article is available on the WWW
References
- 1.a) Estroff LA, Hamilton AD. Chem Rev. 2004;104:1201–1217. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]; b) Yang Z, Liang G, Xu B. Acc Chem Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]; c) Du XW, Zhou J, Shi JF, Xu B. Chem Rev. 2015;115:13165–13307. doi: 10.1021/acs.chemrev.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Pires RA, Abul-Haija YM, Costa DS, Novoa-Carballal R, Reis RL, Ulijn RV, Pashkuleva I. J Am Chem Soc. 2015;137:576–579. doi: 10.1021/ja5111893. [DOI] [PubMed] [Google Scholar]; b) Tanaka A, Fukuoka Y, Morimoto Y, Honjo T, Koda D, Goto M, Maruyama T. J Am Chem Soc. 2015;137:770–775. doi: 10.1021/ja510156v. [DOI] [PubMed] [Google Scholar]; c) Li J, Kuang Y, Shi JF, Zhou J, Medina JE, Zhou R, Yuan D, Yang CH, Wang HM, Yang ZM, Liu JF, Dinulescu DM, Xu B. Angew Chem Int Ed. 2015;54:13307–13311. doi: 10.1002/anie.201507157. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kuang Y, Xu B. Angew Chem Int Ed. 2013;52:6944–6948. doi: 10.1002/anie.201302658. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Hill ZB, Pollock SB, Zhuang M, Wells JA. J Am Chem Soc. 2016;138:13123–13126. doi: 10.1021/jacs.6b06828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Lock LL, Reyes CD, Zhang PC, Cui HG. J Am Chem Soc. 2016;138:3533–3540. doi: 10.1021/jacs.6b00073. [DOI] [PubMed] [Google Scholar]; b) Lock LL, Lo YG, Mao XP, Chen HW, Staedtke V, Bai RY, Ma W, Lin R, Li Y, Liu GS, Cui HG. Acs Nano. 2017;11:797–805. doi: 10.1021/acsnano.6b07196. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Luo ZC, Wu QJ, Yang CB, Wang HM, He T, Wang YZ, Wang ZY, Chen H, Li XY, Gong CY, Yang ZM. Adv Mater. 2017:29. [Google Scholar]; d) Zheng WT, Gao J, Song LJ, Chen CY, Guan D, Wang ZH, Li ZB, Kong DL, Yang ZM. J Am Chem Soc. 2013;135:266–271. doi: 10.1021/ja308690y. [DOI] [PubMed] [Google Scholar]; e) Du XW, Zhou J, Xu B. Chem Asian J. 2014;9:1446–1472. doi: 10.1002/asia.201301693. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Hai ZJ, Li JD, Wu JJ, Xu JC, Liang GL. J Am Chem Soc. 2017;139:1041–1044. doi: 10.1021/jacs.6b11041. [DOI] [PubMed] [Google Scholar]; g) Liu S, Tang AM, Xie ML, Zhao YD, Jiang J, Liang GL. Angew Chem Int Ed. 2015;54:3639–3642. doi: 10.1002/anie.201409952. [DOI] [PubMed] [Google Scholar]; h) Weingarten AS, Kazantsev RV, Palmer LC, Fairfield DJ, Koltonow AR, Stupp SI. J Am Chem Soc. 2015;137:15241–15246. doi: 10.1021/jacs.5b10027. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) McClendon MT, Stupp SI. Biomaterials. 2012;33:5713–5722. doi: 10.1016/j.biomaterials.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Truong WT, Su YY, Meijer JT, Thordarson P, Braet F. Chem Asian J. 2011;6:30–42. doi: 10.1002/asia.201000592. [DOI] [PubMed] [Google Scholar]; k) Shigemitsu H, Hamachi I. Chem Asian J. 2015;10:2026–2038. doi: 10.1002/asia.201500563. [DOI] [PubMed] [Google Scholar]; l) Roy R, Deb J, Jana SS, Dastidar P. Chem Asian J. 2014;9:3196–3206. doi: 10.1002/asia.201402672. [DOI] [PubMed] [Google Scholar]; m) Fang WW, Zhang Y, Wu JJ, Liu C, Zhu HB, Tu T. Chem Asian J. 2018;13:712–729. doi: 10.1002/asia.201800017. [DOI] [PubMed] [Google Scholar]; n) Xu AP, Yang PP, Yang C, Gao YJ, Zhao XX, Luo Q, Li XD, Li LZ, Wang L, Wang H. Nanoscale. 2016;8:14078–14083. doi: 10.1039/c6nr03580a. [DOI] [PubMed] [Google Scholar]; o) Qiao ZY, Hou CY, Zhang D, Liu Y, Lin YX, An HW, Li XJ, Wang H. J Mater Chem B. 2015;3:2943–2953. doi: 10.1039/c4tb02144d. [DOI] [PubMed] [Google Scholar]; p) Zhou J, Li J, Du XW, Xu B. Biomaterials. 2017;129:1–27. doi: 10.1016/j.biomaterials.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Ardoña HAM, Draper ER, Citossi F, Wallace M, Serpell LC, Adams DJ, Tovar JD. J Am Chem Soc. 2017;139:8685–8692. doi: 10.1021/jacs.7b04006. [DOI] [PubMed] [Google Scholar]; b) Liu J, Xu FY, Sun ZL, Pan Y, Tian J, Lin HC, Li XM. Soft Matter. 2016;12:141–148. doi: 10.1039/c5sm02111a. [DOI] [PubMed] [Google Scholar]; c) Priftis D, Leon L, Song ZY, Perry SL, Margossian KO, Tropnikova A, Cheng JJ, Tirrell M. Angew Chem Int Ed. 2015;54:11128–11132. doi: 10.1002/anie.201504861. [DOI] [PubMed] [Google Scholar]; d) Parveen R, Sravanthi B, Dastidar P. Chem Asian J. 2017;12:792–803. doi: 10.1002/asia.201700049. [DOI] [PubMed] [Google Scholar]; e) Chung EJ, Tirrell M. Adv Healthc Mater. 2015;4:2408–2422. doi: 10.1002/adhm.201500126. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Hudalla GA, Sun T, Gasiorowski JZ, Han HF, Tian YF, Chong AS, Collier JH. Nat Mater. 2014;13:829–836. doi: 10.1038/nmat3998. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Wen Y, Waltman A, Han HF, Collier JH. Acs Nano. 2016;10:9274–9286. doi: 10.1021/acsnano.6b03409. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Hamley IW, Dehsorkhi A, Castelletto V. Langmuir. 2013;29:5050–5059. doi: 10.1021/la400163q. [DOI] [PubMed] [Google Scholar]; i) Sawada H, Yamanaka M. Chem Asian J. 2018;13:929–933. doi: 10.1002/asia.201800217. [DOI] [PubMed] [Google Scholar]; j) Roy R, Adalder TK, Dastidar P. Chem Asian J. 2018;13:552–559. doi: 10.1002/asia.201701632. [DOI] [PubMed] [Google Scholar]; k) Laishram R, Maitra U. Chem Asian J. 2017;12:1267–1271. doi: 10.1002/asia.201700289. [DOI] [PubMed] [Google Scholar]; l) Zhang L, Jin Q, Liu M. Chem Asian J. 2016;11:2642–2649. doi: 10.1002/asia.201600441. [DOI] [PubMed] [Google Scholar]; m) Mukherjee S, Kar T, Kumar Das P. Chem Asian J. 2014;9:2798–2805. doi: 10.1002/asia.201402358. [DOI] [PubMed] [Google Scholar]
- 5.a) Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Xu B. J Mater Chem. 2007;17:2385–2393. [Google Scholar]
- 7.Langer R, Vacanti JP. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 8.Kuang Y, Long MJC, Zhou J, Shi JF, Gao Y, Xu C, Hedstrom L, Xu B. J Biol Chem. 2014;289:29208–29218. doi: 10.1074/jbc.M114.600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Standley SM, Toft DJ, Cheng H, Soukasene S, Chen J, Raja SM, Band V, Band H, Cryns VL, Stupp SI. Cancer Res. 2010;70:3020–3026. doi: 10.1158/0008-5472.CAN-09-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang HM, Wei J, Yang CB, Zhao HY, Li DX, Yin ZN, Yang ZM. Biomaterials. 2012;33:5848–5853. doi: 10.1016/j.biomaterials.2012.04.047. [DOI] [PubMed] [Google Scholar]
- 11.Zorn JA, Wille H, Wolan DW, Wells JA. J Am Chem Soc. 2011;133:19630–19633. doi: 10.1021/ja208350u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su JY, Zhang T, Wang PQ, Liu FJ, Tai GH, Zhou YF. Acta Biochim Biophys Sin. 2015;47:192–198. doi: 10.1093/abbs/gmu132. [DOI] [PubMed] [Google Scholar]
- 13.a) Christianson HC, Belting M. Matrix Biol. 2014;35:51–55. doi: 10.1016/j.matbio.2013.10.004. [DOI] [PubMed] [Google Scholar]; b) Ji ZS, Brecht WJ, Miranda RD, Hussain MM, Innerarity TL, Mahley RW. J Biol Chem. 1993;268:10160–10167. [PubMed] [Google Scholar]
- 14.Zhang Y, Kuang Y, Gao Y, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Curr Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Accardi F, Toscani D, Bolzoni M, Palma BD, Aversa F, Giuliani N. BioMed Res Int. 2015 doi: 10.1155/2015/172458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan WC, White PD. Fmoc solid phase peptide synthesis: A Practical Approach. Oxford University Press Inc; New York: 2000. [Google Scholar]
- 17.Atmanene C, Ronin C, Teletchea S, Gautier FM, Djedaini-Pilard F, Ciesielski F, Vivat V, Grandjean C. Biochem Biophys Res Commun. 2017;489:281–286. doi: 10.1016/j.bbrc.2017.05.150. [DOI] [PubMed] [Google Scholar]
- 18.a) Adams J, Kauffman M. Cancer Invest. 2004;22:304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]; b) Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Kuang Y, Gao YA, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Li JY, Gao Y, Kuang Y, Shi JF, Du XW, Zhou J, Wang HM, Yang ZM, Xu B. J Am Chem Soc. 2013;135:9907–9914. doi: 10.1021/ja404215g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhou J, Du XW, Li J, Yamagata N, Xu B. J Am Chem Soc. 2015;137:10040–10043. doi: 10.1021/jacs.5b06181. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhou J, Du X, Chen X, Wang J, Zhou N, Wu D, Xu B. J Am Chem Soc. 2018;140:2301–2308. doi: 10.1021/jacs.7b12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.