Abstract
Calreticulin (CALR) is a Ca2+ binding multifunctional protein that mostly resides in the endoplasmic reticulum (ER) and plays a number of important roles in various physiological and pathological processes. Although the major functions ascribed to CALR are controlling the Ca2+ homeostasis in ER and acting as a lectin-like ER chaperon for many glycoproteins, this moonlighting protein can be found in various cellular compartments where it has many non-ER functions. To shed more light on the mechanisms underlying polyfunctionality of this moonlighting protein that can be found in different cellular compartments and that possesses a wide spectrum of unrelated biological activities, being able to interact with Ca2+ (and potentially other metal ions), RNA, oligosaccharides, and numerous proteins, we used a set of experimental and computational tools to evaluate the intrinsic disorder status of CALR and the role of calcium binding on structural properties and conformational stability of the full-length CALR and its isolated P- and C-domains.
Keywords: Calreticulin, intrinsically disordered protein, moonlighting protein, chaperone, protein-protein interaction, posttranslational modifications
1. INTRODUCTION
Calreticulin (CALR) is a multifunctional protein that participates in various physiological and pathological processes. Being preferentially resided in the endoplasmic reticulum (ER), CALR is mostly known for its roles in Ca2+ homeostasis (in ER lumen, more than 50% of Ca2+ associates with CALR) (Nakamura et al., 2001) and acting as a lectin-like ER chaperon for many glycoproteins (Labriola, Cazzulo, & Parodi, 1999; Spiro, Zhu, Bhoyroo, & Soling, 1996; Vassilakos, Michalak, Lehrman, & Williams, 1998; Zapun et al., 1998). CALR binds monoglucosylated oligosaccharides (Helenius & Aebi, 2004; Helenius, Trombetta, Hebert, & Simons, 1997), as well as interacts with misfolded proteins that can be either glycosylated or non-glycosylated (Lum et al., 2016; Saito, Ihara, Leach, Cohen-Doyle, & Williams, 1999). As a matter of fact, together with another lectin chaperone (or carbohydrate-binding chaperone), calnexin (CNX), CALR is known to interact with practically all glycoproteins in the ER (Ellgaard & Frickel, 2003). In response to Ca2+, CALR might escort numerous client proteins to their active sites, thereby regulating their functions (Fadel et al., 1999; Jiang, Dey, & Matsunami, 2014). Besides being highly enriched within the ER lumen, CALR is present in other cellular compartments, where it has multiple non-ER functions (Gold et al., 2010). For example, in the cytoplasm, CALR shuttles other proteins between Golgi and endoplasmic reticulum (Fadel et al., 1999; D. B. Williams, 2006). It can also be present in the nucleus, where CALR mediates the export of all nuclear receptors, including the glucocorticoid receptor (GRα) (Dedhar et al., 1994; Holaska et al., 2001). CALR can translocate to the surface of cells undergoing immunogenic cell death (ICD), or immunogenic apoptosis, acting as a part of a damage-associated molecular pattern (DAMP) involved in the phagocytic response (Chao et al., 2010; Gardai et al., 2005). In fact, CALR expression is abnormally high on the cell-surface of a wide variety of cancer cells, including hematological malignancy (Wemeau et al., 2010; Zamanian et al., 2016; Zamanian, Veerakumarasivam, Abdullah, & Rosli, 2013). It has been suggested that, by antagonizing the “eat-me” signal provided by CD47, this abnormal cell-surface expression of CALR may inhibit clearance of cancer cells by the immune system, favoring disease progression (Chao et al., 2010; Obeid et al., 2007). Since there is no transmembrane domain in CALR, this protein is anchored to the membrane via binding of its C-terminal domain to specific adaptors, such as CD59 in neutrophils (Ghiran, Klickstein, & Nicholson-Weller, 2003). On the plasma membrane, CALR acts as receptor for orphan ligands, such as thrombospondin (Goicoechea, Pallero, Eggleton, Michalak, & Murphy-Ullrich, 2002). Furthermore, CALR is involved in cell adhesion by playing a role in regulation of expression of several genes related to focal contacts (Fadel et al., 1999; Villagomez et al., 2009) or via direct interaction with the cytoplasmic KXGFFKR motif of the integrin α-subunit (Coppolino & Dedhar, 1999; Dedhar et al., 1994; Rojiani, Finlay, Gray, & Dedhar, 1991). Finally, CALR was shown to be an RNA-binding protein that regulates mRNA stability (Lu, Weng, & Lee, 2015; Mueller et al., 2008; Nickenig et al., 2002; Totary-Jain et al., 2005) and can be found in stress granules (SGs) in the cytoplasm (Carpio, Lopez Sambrooks, Durand, & Hallak, 2010).
Therefore, these and many other studies clearly show that CALR is a multifunctional moonlighting protein. As with many other moonlighting proteins, the ability of CALR to have a highly diversified functional repertoire is related to the tightly controlled distribution of this protein within the various cellular compartments that allows CALR to act as the right man that appears in the right place at a right time. Furthermore, it is recognized also that the polyfunctionality of CALR can be further controlled and regulated by various posttranslational modifications (PTMs). For example, CALR phosphorylation is essential for mRNA binding (Mueller et al., 2008), whereas CALR association with SGs, being promoted by the decrease in intracellular Ca2+, is arginylation-dependent (Carpio et al., 2010).
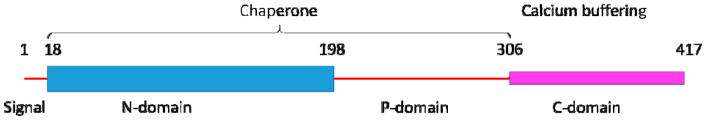
What makes CALR a moonlighting protein? Human CALR is synthesized in a form of a precursor containing cleavable N-terminal signal peptide (residues 1-17) needed for targeting this protein to the ER lumen, where it is accumulated due to the presence of a C-terminal KDEL ER-retention motif (Afshar, Black, & Paschal, 2005). Structurally and functionally, mature human CALR (residues 18-417; UniProt ID: P27797) can be divided into three domains, a globular N-terminal domain (N-domain, N-CARL, residues 18-197), an extended proline-rich P-domain (residues 198-308), and an acidic C-terminal domain (C-domain, C-CARL, residues 309-417) (Chouquet et al., 2011; Michalak, Groenendyk, Szabo, Gold, & Opas, 2009). The chaperone function of this protein requires N-CALR and P-CALR that bind other proteins. Furthermore, this chaperone module is helped by C-CALR that in addition to binding other proteins contains the KDEL motif required for translocation of the protein and its complexes across membranes. Furthermore, since C-CALR contains a large number of negatively charged amino acids, this domain is involved in high-capacity Ca2+ storage (see Figure 1 that shows linear representation of the CALR and indicates domains with their boundaries used in this study).
Figure 1.
Schematic representation of human CALR where positions of the domains with their boundaries used in this study are shown and some basic functions ascribed to different domains are shown.
It became clear at the turn of the 20th century that many biologically active proteins lack unique 3D structures under physiological conditions and instead exist as highly dynamic conformational ensembles consisting of a multitude of interconverting structures (Dunker et al., 2001; Habchi, Tompa, Longhi, & Uversky, 2014; Oldfield & Dunker, 2014; Tompa, 2002, 2012; Uversky, 2013a, 2013c, 2016a; Uversky & Dunker, 2010; Uversky, Gillespie, & Fink, 2000; van der Lee et al., 2014; Wright & Dyson, 1999). This phenomenon is known as protein intrinsic disorder, and it defines high structural heterogeneity of proteins, where either entire protein or any part of it can be disordered to different degree. The inability of intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs) to (completely) fold and form globular well-defined structures with hydrophobic cores, as seen in “normal” ordered proteins, is determined by the presence of specific differences in the amino acid sequences of ordered proteins and IDPs. Since the ability of a polypeptide chain to collapse into compact globule in aqueous media is determined by an interplay between its hydrophobicity and the overall net charge, combination of low overall hydrophobicity and high net charge defines the existence of proteins with extended disorder (Uversky et al., 2000). Statistically, amino acid sequence of IDPs and IDPRs are characterized by the enrichment in the so-called disorder-promoting residues (Arg, Gln, Glu, Gly, Lys, Ser, and Pro) and the depletion in the order-promoting residues (Asn, Cys, Ile, Leu, Phe, Trp, Tyr, and Val) (Dunker et al., 2001; Radivojac et al., 2007; P. Romero et al., 2001; Vacic, Uversky, Dunker, & Lonardi, 2007; R. M. Williams et al., 2001).
According to several bioinformatics studies, IDPs and hybrid proteins possessing ordered domains and IDPRs constitute significant fraction of all proteomes, with the per-proteome amounts of differently disordered proteins increasing with the increase in the organism complexity (Dunker, Obradovic, Romero, Garner, & Brown, 2000; Z. Peng et al., 2015; Walsh et al., 2015; Ward, Sodhi, McGuffin, Buxton, & Jones, 2004; Xue, Dunker, & Uversky, 2012). According to the Protein Data Bank (PDB) analyses, many proteins with known 3-D structures commonly contain IDPRs of different length (DeForte & Uversky, 2016b; Le Gall, Romero, Cortese, Uversky, & Dunker, 2007). Furthermore, even enzymes, which have long been considered an exception to the rule of the prevalence of protein intrinsic disorder due to the structural requirements for catalysis, were recently shown to abundantly contain functional IDPRs (DeForte & Uversky, 2016a, 2017). High natural abundance of IDPs/IDPRs is determined by their functional multifariousness (Dunker, Brown, Lawson, Iakoucheva, & Obradovic, 2002; Dunker, Brown, & Obradovic, 2002; Dunker et al., 2001; Habchi et al., 2014; Uversky et al., 2000; van der Lee et al., 2014), especially in biological processes related to the control and regulation of various signaling pathways (Dunker, Cortese, Romero, Iakoucheva, & Uversky, 2005; Iakoucheva, Brown, Lawson, Obradovic, & Dunker, 2002; Leonova & Galzitskaya, 2015; Oldfield et al., 2005). IDPs/IDPRs are tightly controlled themselves by different means, such as alternative splicing (Buljan et al., 2013; Buljan et al., 2012; P. R. Romero et al., 2006) and numerous enzymatically-catalyzed posttranslational modifications (Iakoucheva et al., 2004; Lenton et al., 2017; Pejaver et al., 2014; Reddy et al., 2017). Intrinsic disorder is crucial for functions of ‘moonlighting’ proteins (Tompa, Szasz, & Buday, 2005), control of generation and disintegration of various cellular membrane-less organelles (Uversky, 2016b, 2017; Uversky, Kuznetsova, Turoverov, & Zaslavsky, 2015), programmed cell death (Z. Peng, Xue, Kurgan, & Uversky, 2013), innate immunity (Xue & Uversky, 2014), antimicrobial peptides of the NK-lysin family (Yacoub, Al-Maghrabi, Ahmed, & Uversky, 2017), βγ-crystallin Hahella chejuensis (Gao, Yang, Zhang, Su, & Huang, 2017), transcription regulation (Fuxreiter et al., 2008; Liu et al., 2006; Toth-Petroczy et al., 2008), regulation of kinase activity (Kathiriya et al., 2014), induction of pluripotent stem cells (Xue, Oldfield, Van, Dunker, & Uversky, 2012), assembly and functionality of various protein complexes (Fuxreiter et al., 2014), and conditional or transient disorder (Jakob, Kriwacki, & Uversky, 2014; Uversky, 2015). This list is far from being complete, and it is believed now that the biological activities of IDPs/IDPRs are complementary to functions of ordered proteins and domains (Dunker et al., 2001; Radivojac et al., 2007; Uversky, 2002). Finally, pathogenesis of different human diseases, such as cancer, cardiovascular disease, amyloidoses and neurodegeneration, is commonly associated with dysfunctions of IDPs/IDPRs (Uversky et al., 2014; Uversky, Oldfield, & Dunker, 2008).
Curiously, several specific structural features and important functions were assigned to the P- (residues 198–308 of the unprocessed human protein, UniProt ID: P27797) and C-terminal domains (residues 309–417 of the unprocessed human protein, UniProt ID: P27797) of CALR. For example, SAXS analysis of the monomeric CALR in solution showed that the P-domain (or P-arm) is characterized by high flexibility, protrudes from the extended globular structure formed by N- and C-domains, and might adopt a spiral-like conformation (Norgaard Toft et al., 2008), whereas molecular dynamics simulations revealed that the CALR P-domain is characterized by high conformational flexibility and that interactions between CALR and its binding partners promotes transition of the P-domain into “open” conformation (Yan, Murphy-Ullrich, & Song, 2010). Subsequent studies indicated that CALR differently interacts with glycosylated and non-glycosylated target proteins, binding of which display distinct kinetic profiles because they induce “open” and “closed” conformations of P-domain, respectively (Wijeyesakere, Rizvi, & Raghavan, 2013). Recently, the presence of such a transition from “open” to “closed” conformation that regulates the accessibility of the dual specificity substrate-binding site, which is able to interact with both carbohydrates and/or proteins, was demonstrated during the structural characterization of CALR isolated from two distinct parasites, Trypanosoma cruzi and Entamoeba histolytica (Moreau et al., 2016). It was shown that the deletion of the acidic tail (residues 359-417) stimulates the ability of CALR to interact with polypeptide substrates and enhance chaperone activity of this protein (Rizvi, Mancino, Thammavongsa, Cantley, & Raghavan, 2004). Application of chemical cross-linking, mass spectrometry, bioinformatics analysis, and computational modelling revealed that Ca2+ binding is accompanied by the structural rearrangement of CALR, where the position of flexible P-domain is dependent on the concentration of Ca2+, part of the disordered acidic C-terminal tail is stabilized by Ca2+, and where Ca2+ induces interactions between the P-loop and the acidic C-terminal tail of CALR (Boelt et al., 2016).
In this article, we used a set of spectroscopic techniques to analyze the effect of calcium on structural properties and conformational stability of the recombinant human CALR and its P- and C-domains. We also utilized a wide spectrum of computational tools to look at the peculiarities of distribution of intrinsic disorder predisposition within the amino acid sequence of this protein and to find if intrinsic disorder may be related to its functional multifariousness.
2. MATERIALS AND METHODS
2.1. Materials
The full-length recombinant CALR and its isolated P- and C-domains (residues 198–308 and 309–417 of the unprocessed human protein, UniProt ID: P27797, respectively) were a kind gift of Prof. Marlene Bouvier (School of Pharmacy, University of Connecticut, Storrs, CT 06269 and Boston Biomedical Research Institute, Watertown, MA 02472). The E. coli strain BNN103 based on a glutathione S-transferase (GST) fusion protein system using the pGEX-3X plasmid (Pharmacia) was utilized for the CALR expression (Bouvier & Stafford, 2000). The expression plasmids for human CALR and its domains have been described previously (Bouvier & Stafford, 2000; Tan, Chen, Li, Mabuchi, & Bouvier, 2006). A glutathione-Sepharose 4B affinity column was used for the purification of the crude GST-CRT fusion protein at 4°C. The eluted protein was digested with Factor Xa. After the proteolytic digestion of the GST tag, the N-terminus of the resulting CALR protein contained additional GIPG residues. Digested GST tag was removed by direct application of the proteolytic mixture onto the glutathione-Sepharose 4B affinity column. Final stage of the protein purification was achieved by applying the CALR containing flow-through of the glutathione-Sepharose 4B affinity column to the Uno Q-6 (Bio-Rad) FPLC ion-exchange column. Purified protein was analyzed by the N-terminal amino acid sequencing and MALDI mass spectrometry to validate the Factor Xa cleavage and to evaluated the protein molecular mass (Bouvier & Stafford, 2000; Tan et al., 2006).
2.2. Computational evaluation of the intrinsic disorder predisposition of human CALR
2.2.1. Evaluation of the per-residue disorder propensity
The intrinsic disorder predisposition of human CALR (UniProt ID: P27797) was evaluated by four algorithms from the PONDR family, PONDR-FIT, PONDR® VLXT, PONDR® VSL2, and PONDR® VL3 (X. Li, Romero, Rani, Dunker, & Obradovic, 1999; Obradovic, Peng, Vucetic, Radivojac, & Dunker, 2005; K. Peng, Radivojac, Vucetic, Dunker, & Obradovic, 2006; K. Peng et al., 2005; P. Romero et al., 2001; Xue, Dunbrack, Williams, Dunker, & Uversky, 2010), as well as by the IUPred web server with its two versions for predicting long and short disordered regions (Dosztanyi, Csizmok, Tompa, & Simon, 2005). PONDR® VSL2 is one of the most accurate stand-alone disorder predictors (Obradovic et al., 2005), PONDR® VL3 possesses a high accuracy for finding long disordered regions (Obradovic et al., 2003), PONDR® VLXT is not the most accurate predictor, but has a high sensitivity to local sequence peculiarities which are often associated with disorder-based interaction sites (Dunker et al., 2001), PONDR-FIT is a metapredictor that is moderately more accurate than each of the component predictors and that is one of the most accurate disorder predictors (Xue et al., 2010), whereas IUPred utilizes the pair-wise energy estimation approach for finding intrinsically disordered residues and regions. The consensus disorder propensity of human CALR was evaluated by averaging disorder profiles of individual predictors. Use of consensus for evaluation of intrinsic disorder is motivated by empirical observations that this approach usually increases the predictive performance compared to the use of a single predictor (Fan & Kurgan, 2014; Z. Peng & Kurgan, 2012; Walsh et al., 2015).
2.2.2. MobiDB-based intrinsic disorder analysis
We further characterized the overall disorder status of human CALR using the MobiDB database (http://mobidb.bio.unipd.it/) (Di Domenico, Walsh, Martin, & Tosatto, 2012; Potenza, Domenico, Walsh, & Tosatto, 2015), that generates consensus disorder scores by combining the outputs of ten predictors, such as two versions of DisEMBL (Linding, Jensen, et al., 2003), two versions of ESpritz (Walsh, Martin, Di Domenico, & Tosatto, 2012), GlobPlot (Linding, Russell, Neduva, & Gibson, 2003), JRONN (Yang, Thomson, McNeil, & Esnouf, 2005), PONDR® VSL2B (Obradovic et al., 2005; K. Peng et al., 2006), and two versions of IUPred (Dosztanyi et al., 2005). MobiDB also has manually curated annotations related to protein function and structure derived from UniProt (Apweiler et al., 2004) and DisProt (Sickmeier et al., 2007), as well as from Pfam (Finn et al., 2014) and PDB (Berman et al., 2000).
2.2.3. Analysis of the CALR functional disorder by D2P2
The D2P2 database (http://d2p2.pro/) (Oates et al., 2013) was used for the complementary evaluation of intrinsic disorder propensity of human CALR. Some important disorder-related functional information was retrieved from this database too. D2P2 is a database of predicted disorder for a large library of proteins from completely sequenced genomes (Oates et al., 2013) that uses outputs of IUPred (Dosztanyi et al., 2005), PONDR® VLXT (P. Romero et al., 2001), PrDOS (Ishida & Kinoshita, 2007), PONDR® VSL2B (Obradovic et al., 2005; K. Peng et al., 2006), PV2 (Oates et al., 2013), and ESpritz (Walsh et al., 2012). The database is further supplemented by data concerning location of various functional domains, sites of curated posttranslational modifications, and predicted disorder-based protein binding sites.
2.2.4. Prediction of potential disorder-based binding sites by ANCHOR and MoRFpred
The presence of the potential disorder-based protein binding sites in human CALR was evaluated by the ANCHOR algorithm [58,59]. This algorithm utilizes the pair-wise energy estimation approach originally used by IUPred [41,60]. This approach is based on the hypothesis that long disordered regions might include localized potential binding sites, which are not capable of folding on their own due to not being able to form enough favorable intrachain interactions, but can obtain the energy to stabilize via interaction with a globular protein partner [58,59].
In addition to ANCHOR, we utilized a MoRFpred algorithm that was designed to find all types of MoRFs, α-helix, β-strand, coil, and complex using a comprehensive dataset of annotated MoRFs (Disfani et al., 2012). This tool utilizes several sequence-derived hallmarks of potential MoRFs, such as their appearance as dips in the disorder predictions and high hydrophobicity and stability. High accuracy of MoRFpred in comparison with several existing tool for finding disorder-based binding sites in proteins is achieved by combining information about evolutionary profiles with several sequence-derived features, such as B-factors, predicted disorder, selected physiochemical properties of amino acids, and solvent accessibility (Disfani et al., 2012).
2.2.5. Analysis of the CALR interactivity by STRING
Additional functional information for these proteins was retrieved using STRING (Search Tool for the Retrieval of Interacting Genes, http://string-db.org/). STRING generates a network of predicted associations based on predicted and experimentally-validated information on the interaction partners of a protein of interest (Szklarczyk et al., 2011). In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line - the presence of fusion evidence; a purple line - experimental evidence; a blue line – co-occurrence evidence; a light blue line - database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence (Szklarczyk et al., 2011). In our analysis, the most stringent criteria were used for selection of interacting proteins by choosing the highest cut-off of 0.9 as the minimal required confidence level.
2.3. Spectroscopic analysis of the full-length CALR and its isolated P- and C-domains
2.3.1. Circular dichroism measurements
CD spectra were collected on an AVIV-60DS spectropolarimeter (AVIV, Lakewood, N.J.), equipped with a temperature-controlled cell-holder. In the far-UV region CD spectra were recorded in 0.01 cm cell from 250–190 nm with a step size of 0.5 nm, a bandwidth of 1.5 nm, and an averaging time of 5 sec. For all spectra, averages of 5 scans were obtained. CD spectra of the appropriate buffer were recorded and subtracted from the protein spectra. Protein concentrations were 1 mg/mL. The following buffers were used in the CD studies: 2 mM HEPES, 200 mM NaCl, pH 7.0 supplemented with either 2 mM EDTA, 50 μM CaCl2, or 1000 μM CaCl2. The molar ellipticity, θ was calculated as the CD signal (millidegrees) × MW (Da)/[number of residues × CALR concentration (mg/mL) × cell pathlength (mm)].
2.3.2. Fluorescence measurements
Since fluorescence intensity dramatically depends on temperature, the temperature should be kept constant to ensure that the observed effects are associated with protein and not changes in temperature. Therefore, all fluorescence measurements were performed using a FluoroMax-2 fluorescence spectrophotometer (Jobin Yvon-Spex, NJ, USA) equipped with a temperature-controlled cell-holder. All fluorescence measurements were performed at room temperature with the final protein concentrations of 0.01 mg/ml.
Intrinsic fluorescence measurements were conducted using an excitation wavelength of 280 nm and recording the emission spectra from 300 to 420 nm. The following buffers were used in the fluorescence experiments: 2 mM HEPES, 200 mM NaCl, pH 7.0 supplemented with either 2 mM EDTA, 50 μM CaCl2, or 1000 μM CaCl2.
In ANS binding studies, a fresh 10 mM ANS stock solution was prepared in double distilled water. The concentration of ANS was determined by UV absorption at 350 nm using a molar extinction coefficient of 5000. An aliquot of 1 μL of the ANS solution was added to 1 mL of buffer to a final concentration of 10 μM. A blank spectrum without protein was recorded before 10 μL of protein solution was added to the solution to a final protein concentration of 0.01 mg/ml. Fluorescence measurements were performed immediately after the addition of protein to the ANS solution in semimicro quartz cuvettes (Hellma, Germany). Spectra were recorded at ambient temperatures with a 1-cm excitation light path using a FluoroMax-2 spectrofluorometer (Instruments S.A., Inc. Jobin Yvon-Spex, USA). The light source was a 150-W xenon lamp. Emission spectra were recorded from 460 nm to 600 nm with excitation at 350 nm, an increment of 1 nm, an integration time of 1 s, and slits of 5 nm for both excitation and emission. For all spectra, averages of 5 scans were obtained. Data were processed using DataMax software.
2.3.3. Acrylamide Quenching
Acrylamide quenching studies of intrinsic fluorescence were performed by adding aliquots from a stock solution of the quencher into a cuvette containing the protein solution. Fluorescence intensities were corrected for dilution effects. Fluorescence quenching data were analyzed using the general form of the Stern-Volmer equation, taking into account not only the dynamic, but also static quenching (Eftink & Ghiron, 1981):
| (1) |
where Io and I are the fluorescence intensities in the absence and presence of quencher, KSV is the dynamic quenching constant, V is a static quenching constant, and [Q] is the quencher concentration. All related experiments were conducted in triplicates.
2.4. Urea-induced unfolding of the full-length CALR and its P- and C-domains
Conformational stability of various CALR-related constructs was evaluated by measuring the intrinsic fluorescence after incubation of proteins in increasing amounts of urea (from 0 M to 8 M). All samples were incubated for four hours at room temperature to insure complete unfolding equilibrium. The final protein concentration was 0.01 mg/ml. The following buffers were used in the urea-induced unfolding studies: 2 mM HEPES, 200 mM NaCl, pH 7.0 supplemented with either 2 mM EDTA, 50 μM CaCl2, or 1000 μM CaCl2. The fluorescence measurements were performed using a FluoroMax-2 fluorescence spectrophotometer (Jobin Yvon-Spex, NJ, USA) with an excitation wavelength of 280 nm (5 nm band pass) and recording the emission spectra from 300 to 420 nm (5 nm band pass). Assuming a two-state folding mechanism, the fraction of the unfolded conformation in the unfolding of the full-length CALR was obtained using the following equation:
| (2) |
where yf and yu represent the intrinsic fluorescence value of y characteristic of the folded and unfolded conformations, respectively, under the conditions where y is being measured. The values of yf and yu were obtained by linear regression on the data points before and after the unfolding transition, respectively. A sigmoidal curve-fit was subsequently used to determine the midpoints of the unfolding transition (Cm). All related experiments were conducted in triplicates.
3. RESULTS AND DISCUSSION
3.1. Effect of Ca2+ binding on structural properties and conformational stability of CALR and its isolated P- and C-domains
CALR is an endoplasmic reticulum (ER) luminal Ca2+-buffering chaperone that serves as one of the ER Ca2+-binding/storage proteins (Nash, Opas, & Michalak, 1994), plays a role in regulation of the intracellular Ca2+ homoeostasis, and is involved in the control of the ER Ca2+ capacity (Fadel et al., 1999; Gelebart, Opas, & Michalak, 2005; Michalak et al., 2009). Therefore, it is not surprising that structure and function of CALR are known to be controlled by Ca2+ binding (Andrin et al., 1998; Carpio et al., 2010; Conte, Keith, Gutierrez-Gonzalez, Parodi, & Caramelo, 2007; Corbett et al., 1999; Holaska, Black, Rastinejad, & Paschal, 2002; Villamil Giraldo et al., 2010; Wijeyesakere, Gafni, & Raghavan, 2011). It was also emphasized that different domains of CALR may play different roles in the calcium sensitivity of this protein. For example, the CALR globular domain is involved in high affinity Ca2+ binding, whereas its acidic C-tail, with high content of acidic residues (22 glutamates and 12 aspartates are spread over the 50 C-terminal residues of this domain defining its net negative charge of −25), is responsible for the high capacity (~20 mol of Ca2+ per 1 mol of CALR) and low affinity (Kd ~ 2 mM) Ca2+-binding activity of CALR and is needed for the Ca2+-buffering properties of this protein (Baksh & Michalak, 1991). Dynamic light scattering and size exclusion chromatography-based analyses revealed that the full-length CALR undergoes noticeable compaction in the presence of Ca2+ (Villamil Giraldo et al., 2010), and the thermal stability of the full-length protein was increased in the Ca2+-dependent manner (Z. Li, Stafford, & Bouvier, 2001; Wijeyesakere et al., 2011). Here, the thermal denaturation midpoint temperatures (Tm) progressively increased from 40.2 °C to 44.3 °C and 46.4 °C when CaCl2 concentration was increased from 0 to 50 μM and 1000 μM, respectively (Z. Li et al., 2001). Earlier studies also showed that the far-UV CD spectrum of the full-length CALR was characterized by a minimum at 208 nm and a prominent shoulder at 229 nm, which was attributed to the contributions from aromatic side chains (Bouvier & Stafford, 2000). In fact, ~11% of all residues in CALR are aromatic (this protein has 18 phenylalanines, 14 tyrosines, and 11 tryptophans) (Bouvier & Stafford, 2000). The shape of the far-UV CD spectrum of the full-length CALR was characterized by an apparent lack of features typical for the proteins having a high content of well-defined secondary structure such as α-helices and β-sheets (Bouvier & Stafford, 2000) and was not affected by the addition of CaCl2 (Z. Li et al., 2001). Although these experiments uncovered interesting effects of calcium on the CALR structure one can argued that the low NaCl concentrations (<10mM) investigated by Wijeyesakere et al. (Wijeyesakere et al., 2011) may not be predictive of the conformational changes induced by Ca2+ at more “physiological” NaCl concentrations (i.e., 150–200 mM) in the human full-length CALR protein.
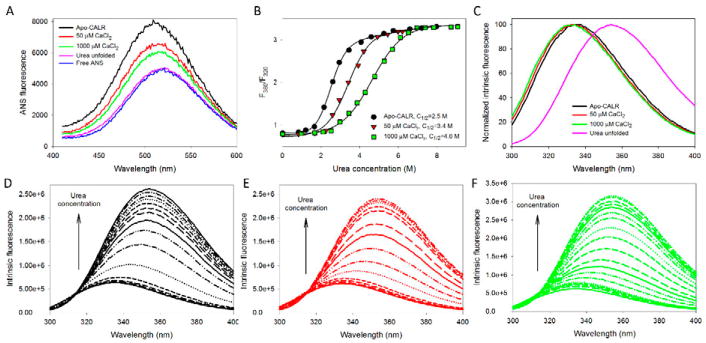
To gain a better understanding of how Ca2+ affects the structure and conformational stability of human CALR, a comprehensive biophysical analysis of the human full-length protein and of its isolated P- and C-domain at 0, 50 and 1000 μM Ca2+ was conducted in solutions containing 200 mM NaCl. In agreement with the results of previous studies, Figure 2A and 2B show that the addition of calcium decreases the overall hydrophobicity of the full-length CALR (measured as changes in the fluorescence parameters of the hydrophobic fluorescent probe ANS, and suggesting the Ca2+-induced increase in the compaction of this protein) and noticeably increases its conformational stability toward the urea-induced unfolding. The presence of some calcium-induced compaction of the full-length CALR is also evidenced by the slight blue shift of its intrinsic fluorescence from 335 nm to 334 nm, and to 333 nm when the CaCl2 concentration was increased from 0 to 50 μM, and to 1000 μM, respectively (see Figure 2C). The presence of the calcium-induced partial compaction is further supported by the reduction in the efficiency of the dynamic fluorescent quenching (as evaluated by the Stern-Volmer constant, KSV), from 4.19±0.08 to 3.91±0.06 and to 3.68±0.08 M−1 for CALR in the presence of 0, 50 and 1000 μM Ca2+ in solutions containing 200 mM NaCl. For comparison, the urea-unfolded CALR in the presence of 10.5 M urea is characterized by the KSV of 19.29 ±0.08 M−1. Furthermore, Figures 2D, 2E, and 2F show that the urea-induced unfolding of the full-length CALR in its apo-form or in the presence of 50 and 1000 μM Ca2+ was accompanied not only by the noticeable red shift of the maximal fluorescence from 333–335 nm in their native states to 353–354 nm in the urea-unfolded states (that reflects a conformational transition from the compact form whose tryptophan residues are mostly located in the hydrophobic environment and protected from interaction with the solvent to the highly expanded state with solvent-accessible tryptophan residues) but also by a dramatic increase in the intrinsic fluorescence intensity, reflecting unfolding-promoted release of some quenching groups from the close vicinity of the tryptophan residues of this protein. We would like to emphasize here that although we came to the conclusions that are similar to those reported in previous studies (Wijeyesakere et al., 2011), the obtained results on the effect of Ca2+ on CALR structure and conformational stability in the presence of 200 mM NaCl are not trivial, since one might expect that protein interactions with Ca2+ (which are obviously electrostatically driven) may be affected by salt.
Figure 2. Analysis of the effect of calcium on structural properties and conformational stability of the full-length human CALR. The following buffers were used in these spectroscopic studies: 2mM HEPES, 200mM NaCl, pH=7.0 supplemented with either 2mM EDTA (Apo), 50μM CaCl2, or 1000μM CaCl2.
A. ANS fluorescence spectra measured for the full-length human CALR in the absence calcium (black line) or in the presence of 50μM CaCl2 (red line), or 1000μM CaCl2 (green line). Spectra of free ANS (blue line) and CALR completely unfolded by 8M urea (pink line) are shown for comparison. In these experiments, protein concentration was 0.01mg/mL, and ANS concentration was kept at the level of 10μM.
B. Urea-induced unfolding of the apo-form of the full-length human CALR (black circles) and the CALR in the presence of 50μM CaCl2 (red triangles), or 1000μM CaCl2 (green squares). Urea-induced unfolding was detected by the characteristic red shift of the intrinsic fluorescence spectrum monitored by the urea-induced changes in the F360/F320 values, where F360 and F320 correspond to the fluorescence intensity at 360 and 320nm, respectively. Experiments were performed in triplicate. Plots represents averaged F360/F320 values. Conformational stability of the full-length protein is progressively increased with the increase in calcium concentration.
C. Normalized intrinsic fluorescence spectra of the full-length CALR in the absence calcium (black line) or in the presence of 50μM CaCl2 (red line), or 1000μM CaCl2 (green line), and CALR completely unfolded by 8M urea (pink line). Spectra were normalized, since there is a dramatic increase in their maximal intensity which makes the visual analysis of their position difficult.
D. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the apo-CALR. Spectra were measured in the presence of 0.0, 0.82, 1.20, 1.65, 2.15, 2.61, 2.94, 3.44, 4.07, 4.59, 5.25, 5.73, 6.59, 7.21, and 7.97 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
E. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the full-length human CALR in the presence of 50μM CaCl2. Spectra were measured in the presence of 0.00, 0.83, 1.68, 2.25, 2.59, 2.94, 3.41, 3.92, 4.21, 4.91, 5.45, 5.98, 6.21, and 6.49 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
F. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the full-length human CALR in the presence of 1000μM CaCl2. Spectra were measured in the presence of 0.00, 0.97, 1.68, 2.10, 2.52, 2.98, 3.44, 3.87, 4.28, 4.78, 5.27, 5.69, 6.10, 6.53, 6.96, 7.39, 7.83, 8.73, and 9.07 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
In the existing structural models, CALR is typically depicted as a rigid body (comprised of the N-CALC and N-terminal part of C-domain) with two flexible arms (P-domain and C-tail) (Chouquet et al., 2011; Michalak et al., 2009; Wijeyesakere et al., 2011). Therefore, technically, analysis of the isolated domains of this protein is justifiable. Furthermore, there is an important question on whether the individual isolated domains of CALR can sense calcium in the presence of 200 mM NaCl. Therefore, to understand if structure and conformational stability of the isolated P-domain and C-domain are impacted by calcium, we analyzed how the addition of different CaCl2 concentrations affects their far-UV CD spectra (Figures 3A and 4A), intrinsic fluorescence (Figures 3B and 4B), and conformational transition curves describing their urea-induced unfolding (Figures 3C and 4C). Figure 3A shows that the far-UV CD spectrum of the isolated P-domain is characterized by a rather unusual shape and is insensitive to the presence or absence of calcium. This shape of the P-domain far-UV CD spectrum, with a strong negative band at ~200 nm, a weak positive band at ~222 nm, and another negative band at ~230 nm, can be attributed to a polyproline II-like conformation that can be present in highly charged polypeptides with high proline content. In fact, this 101 residue-long domain has a net charge of −20 containing 21 aspartates, 13 glutamates, 12 lysines and 2 arginines and 19 prolines account for 17.4% of all its residues.
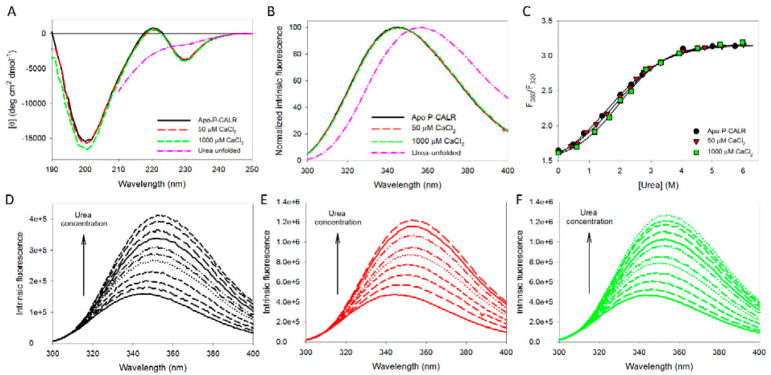
Figure 3. Analysis of the effect of calcium on structural properties and conformational stability of the isolated P-domain of human CALR. The following buffers were used in these spectroscopic studies: 2mM HEPES, 200mM NaCl, pH=7.0 supplemented with either 2mM EDTA (Apo), 50μM CaCl2, or 1000μM CaCl2.
A. Far-UV CD spectra measured for the isolated P-domain of human CALR in the absence calcium (black line) or in the presence of 50μM CaCl2 (red line), or 1000μM CaCl2 (green line). Spectrum of the P-domain of human CALR completely unfolded by 6.0M urea (pink line) is shown for comparison.
B. Normalized intrinsic fluorescence spectra of human P-CALR in the absence calcium (black line) or in the presence of 50μM CaCl2 (red line), or 1000μM CaCl2 (green line), and P-CALR completely unfolded by 6.0M urea (pink line). Spectra were normalized, since there is a dramatic increase in their maximal intensity which makes the visual analysis of their position difficult.
C. Urea-induced unfolding of the apo-form of the P-domain of human CALR (black circles) and the P-domain of human CALR in the presence of 50μM CaCl2 (red triangles), or 1000μM CaCl2 (green squares). Urea-induced unfolding was detected by the characteristic red shift of the intrinsic fluorescence spectrum monitored by the urea-induced changes in the F360/F320 values, where F360 and F320 correspond to the fluorescence intensity at 360 and 320nm, respectively. Experiments were performed in triplicate, and plot represents averaged F360/F320 values. Calcium does not affect the conformational stability of the P-domain.
D. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the apo-P-CALR. Spectra were measured in the presence of 0.00, 0.47, 0.83, 1.42, 2.01, 2.37, 2.72, 3.32, 4.04, 5.01, and 5.74 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
E. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the P-CALR in the presence of 50μM CaCl2. Spectra were measured in the presence of 0.00, 0.41, 0.62, 0.91, 1.23, 1.58, 2.00, 2.32, 2.53, 2.79, 2.99, 3.32, 4.05, 4.76, 5.31, and 6.01 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
F. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the P-CALR in the presence of 1000μM CaCl2. Spectra were measured in the presence of 0.00, 0.59, 1.18, 1.65, 2.01, 2.37, 2.84, 3.32, 3.92, 4.52, 5.25, and 5.98 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
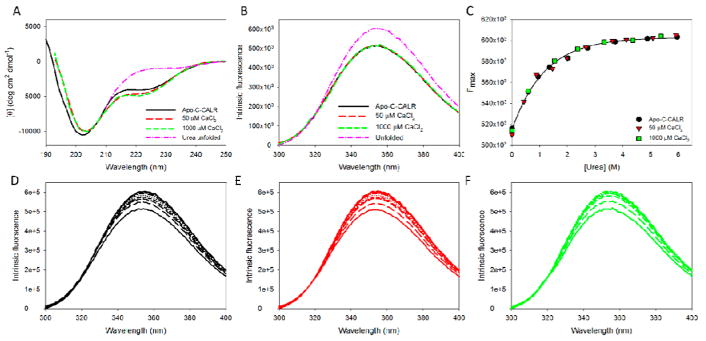
Figure 4. Analysis of the effect of calcium on structural properties and conformational stability of the isolated C-domain of human CALR. The following buffers were used in these spectroscopic studies: 2mM HEPES, 200mM NaCl, pH=7.0 supplemented with either 2mM EDTA (Apo), 50μM CaCl2, or 1000μM CaCl2.
A. Far-UV CD spectra measured for the isolated C-domain of human CALR in the absence calcium (black line) or in the presence of 50μM CaCl2 (red line), or 1000μM CaCl2 (green line). Spectrum of the C-domain of human CALR completely unfolded by 6 M urea (pink line) is shown for comparison.
B. Intrinsic fluorescence spectra of the isolated human C-CALR in the absence calcium (black line) or in the presence of 50μM CaCl2 (red line), or 1000μM CaCl2 (green line), and P-CALR completely unfolded by 6.0M urea (pink line).
C. Urea-induced unfolding of the apo-form of the C-domain of human CALR (black circles) and the C-domain of human CALR in the presence of 50μM CaCl2 (red triangles), or 1000μM CaCl2 (green squares). Since the position of the intrinsic fluorescence spectrum of the isolated C-domain is red-shifted (352 nm) and is affected by the addition of CaCl2 or high urea concentrations, urea-induced unfolding of this domain was detected by the urea-induced increase in the fluorescence intensity. Experiments were performed in triplicate, and plots represents averaged Fmax values. Calcium does not affect the conformational stability of the C-domain.
D. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the isolated apo-C-CALR. Spectra were measured in the presence of 0.00, 0.59, 0.94, 1.35, 2.01, 2.72, 3.72, 4.88, and 5.96 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
E. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the C-CALR in the presence of 50μM CaCl2. Spectra were measured in the presence of 0.00, 0.43. 0.88, 1.46, 1.98, 2.67, 3.62, 4.12, 5.08, and 5.92 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
F. Effect of the increasing urea concentration on the intrinsic fluorescence spectrum of the C-CALR in the presence of 1000μM CaCl2. Spectra were measured in the presence of 0.00, 0.59, 1.54, 2.37, 3.32, 4.36, and 5.37 M urea. Increase in urea concentration causes red shift and increase in fluorescence intensity.
Figure 3B shows that the intrinsic fluorescence of the isolated P-domain both in the absence or presence of CaCl2 was slightly blue-shifted in comparison with that of the urea-unfolded polypeptide (346 nm vs. 352 nm, respectively). This suggested the presence of some protection of tryptophan residues of the isolated P-domain from solvent, which, however, was not affected by the addition of calcium to any significant degree. This conclusion is further supported by the results of the acrylamide-induced fluorescence quenching experiments, which showed that the isolated apo-P-CALR and P-CALR in the presence of 50 and 1000 μM Ca2+ in solutions containing 200 mM NaCl where characterized by the KSV of 5.68±0.08, 5.41±0.07, and 5.29±0.06 M−1, respectively, whereas the urea-unfolded P-CALR had the KSV of 19.93±0.06 M−1.
In comparison with the unfolding curves recorded for the full-length protein (see Figure 3A), unfolding of the isolated P-domain detected by the urea-induced changes in its intrinsic fluorescence is characterized by very low cooperativity, which is typical for the unfolding patterns of mostly disordered proteins (Uversky, 2009). Despite its high negative charge, neither P-domain structure (Figures 3A and 3B) nor conformational stability (Figure 3C) are affected by the addition of CaCl2, indicating the lack of the structure-inducing and/or stabilizing effects of Ca2+ binding for the P-domain of CALR. Similar to the full-length CALR, urea-induced unfolding of the isolated P-CALR was accompanied by both a noticeable red shift of the fluorescence spectrum and a large increase in the intrinsic fluorescence intensity.
Figure 4A shows that in agreement with predictions (see below), the far-UV CD spectrum of the isolated C-domain is characterized by a strong negative band near 203 nm typical for the mostly disordered polypeptides but has a pronounced negative band in the vicinity of 217–227 nm suggesting the presence of some ordered secondary structure. Although the CD spectral properties of this domain are minimally affected by Ca2+, Figure 4A shows the intensity of the negative band near 203 nm decreases and the intensity of the 217–227 shoulder increases in the calcium-dependent manner, suggesting that the structure of the isolated C-domain is sensitive to calcium, shifting from more disordered structure in the absence of Ca2+ to a more ordered form when calcium concentration is raised. This observation is in the line with the previously published study, where a systematic analysis of the various fragments of the C-domain revealed that the addition of calcium mostly affects structure of the C-terminal half of this domain, whereas its N-terminal half was not affected by calcium (Villamil Giraldo et al., 2010). Figure 4B shows that the position of the intrinsic fluorescence spectrum of the isolated C-domain (352 nm) was not noticeably affected by the addition of CaCl2 or high urea concentrations, suggesting high solvent accessibility of its tryptophan residues in all states analyzed in this study. This conclusion is in line with the outputs of the acrylamide-induced fluorescence quenching experiments, where the lack of significant protection of the tryptophan residues in the isolated apo-C-CALR alone or C-CALR in the presence of 50 and 1000 μM Ca2+ or 6 M urea is evidenced by the KSV of 10.5±0.3, 13.2±0.4, 12.6±0.2, and 15.1±0.3 M−1, respectively. However, Figure 4B shows that the addition of urea caused a detectable increase in the intrinsic fluorescence intensity of this domain, and this phenomenon was used to follow urea-induced changes in its structure. Figure 4C represents the corresponding unfolding curve and shows that the urea-induced unfolding of the isolated C-domain is completely non-cooperative and is not affected by calcium. Spectra used for the generation of these unfolding curves are presented in Figures 4D, 4E, and 4F.
Therefore, our analysis revealed that neither structure nor conformational stability of the P-domain were sensitive to calcium. Although highly disordered C-CALR gained a bit more ordered structure at calcium binding, it still remained mostly disordered, and its conformational stability was not affected by calcium. Taken together, our observations indicate that calcium has a noticeable effect on the structure and conformational stability of the full-length protein, making it somewhat more compact and significantly more stable. Since no detectable changes in the structure and conformational stability of the isolated P-domain was induced by calcium, and since the addition of calcium had minimal effects of the intrinsically disordered nature of the isolated C-domain and did not affect its conformational stability, the calcium-induced changes in the behavior of the full-length CALR can be attributed to the calcium-dependent stabilization of an C-embedded structure, where the portion of the C-domain is incorporated into a jelly-roll-like structure formed by its N-domain.
3.2. Peculiarities of structure and intrinsic disorder of human CALR
3.2.1. Peculiarities of the CALR structure
There are three domains in the mature CALR (residues 18-417; UniProt ID: P27797), a globular N-terminal domain (N-domain) followed by an extended proline-rich P-domain and acidic C-terminal domain (C-domain), spanning through residues 18-197, 198-308, and 309-417, respectively (Chouquet et al., 2011; Michalak et al., 2009). Despite serious efforts no 3D structure is available for the full-length protein, suggesting the presence of significant conformational dynamics in some of its parts. In fact, it was emphasized that the flexible nature of the P-domain and the C-terminal tail preclude crystallization of the full-length CALR (Chouquet et al., 2011).
In agreement with this hypothesis, the use of programs for secondary structure prediction revealed that the P-domain is expected to be devoid of regular secondary structure elements mostly due to the high content of proline residues (Ellgaard et al., 2001). The P-domain is characterized by the presence of multiple copies of types 1 and 2 repeat sequences arranged in a specific ‘111222’ pattern (Ellgaard et al., 2001). Figure 5A represents solution NMR structure of the P-domain of the rat CALR (residues 189–288 in mature protein or residues 206-305 in UniProt ID: P18418) and shows that this domain is characterized by an unusual extended hairpin fold with high conformational flexibility. The most stable part of this flexible domain is its central loop (residues 227–247) closed by an anti-parallel β-sheet containing two very short β-strands, residues 224–226 and 248–250 (PDB ID: 1HHN; (Ellgaard et al., 2001)). There are two additional two-stranded antiparallel β-sheets containing pairs of short β-strands, residues 207–209 and 262–264, and 190–192 and 276–278 (Ellgaard et al., 2001). Since remaining parts of this 101 residue-long domain are mostly unstructured, the P-domain is expected to have 1% of α-helical structure (1 helix; 4 residues) and 11% of β-sheet structure (6 strands; 12 residues) (Ellgaard et al., 2001).
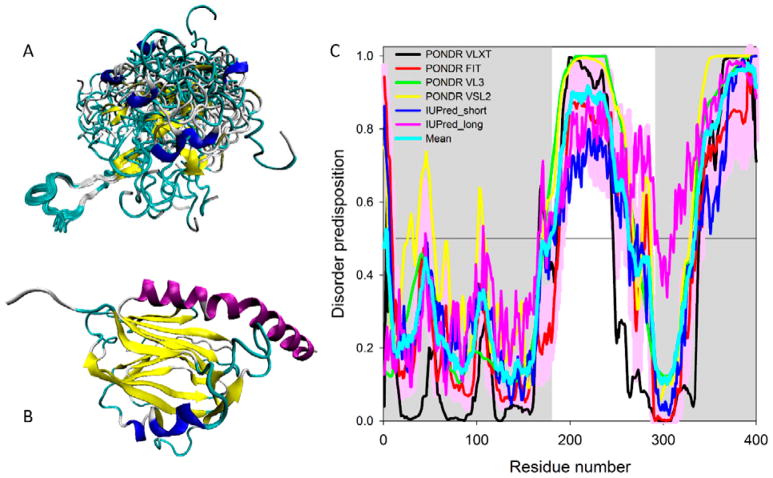
Figure 5. Structural characterization and intrinsic disorder propensity of human CALR.
A. Solution NMR structure of the P-domain of the rat CALR (residues 189–288 in mature protein or residues 206-305 in UniProt ID: P18418) (PDB ID: 1HHN; (Ellgaard et al., 2001)).
B. X-ray crystal structure of the N-domain human of CALR (residues 18-204) and the N-terminal half of the C-domain (residues 302-368) connected by a GSG tripeptide (PDB ID: 3POW; (Chouquet et al., 2011)).
C. Evaluating intrinsic disorder propensity of human CALR (UniProt ID: P27797) by series of per-residue disorder predictors. Disorder profiles generated by PONDR® VLXT, PONDR FIT, PONDR® VL3, PONDR® VSL2, IUPred_short and IUPred_long are shown by black, red, green, yellow, blue, and pink lines, respectively. Light cyan line shows the mean disorder propensity calculated by averaging disorder profiles of individual predictors. Light pink shadow around the PONDR® FIT shows error distribution. In these analyses, the predicted intrinsic disorder scores above 0.5 are considered to correspond to the disordered residues/regions. Positions of the N-domain (residues 1-180 in mature form that correspond to the residues 18-197 in UniProt ID: P27797) and C-domain (residues 293-400 in mature form that correspond to the residues 309-417 of the in UniProt entry P27797) are shown as gray shaded areas.
Also, the X-ray crystal structure was solved for the construct of the human CALR containing the N-domain (residues 18-204) and the N-terminal half of the C-domain (residues 302-368) connected by a GSG tripeptide instead of the extended P-domain (PDB ID: 3POW; (Chouquet et al., 2011)). Figure 5B represents structure of this construct and shows that it is characterized by a jelly-roll fold formed by one convex and one concave anti-parallel β-sheets (Chouquet et al., 2011). The part of the C-domain being integrated into the globular domain, provides two central strands to the β-sandwich structure and adds a long kinked C-terminal α-helix (Chouquet et al., 2011). Based on these observations, a structural model of CALR was proposed, according to which the N-domain and a part of C-domain jointly form a globular structure, from which a hook-like arm of a β-stranded hairpin of P-domain protrudes, with the tip of this arm possessing the binding site for the protein partners (e.g., oxidoreductase ERp57; (Frickel et al., 2002)), whereas the acidic C-terminal domain, also protruding from the globular body, contains a large number of negatively charged amino acids and acts as a high-capacity Ca2+ storage, within the ER lumen (Michalak et al., 2009; Wijeyesakere et al., 2011).
3.2.2. Peculiarities of the intrinsic disorder distribution in CALR
Therefore, currently available structural data suggest that the overall CALR structure can be viewed as a globular body with two flexible and functionally different arms corresponding to the P-domain and C-tail. In agreement with these structural data and model, Figure 5C represents the results of the multiparametric analysis of the intrinsic disorder predisposition of human CALR evaluated by four algorithms from the PONDR family, PONDR-FIT, PONDR® VLXT, PONDR® VSL2, and PONDR® VL3 (X. Li et al., 1999; Obradovic et al., 2005; K. Peng et al., 2006; K. Peng et al., 2005; P. Romero et al., 2001; Xue et al., 2010), as well as by the IUPred web server that predicts probability of a query protein to have short and long IDPRs (Dosztanyi et al., 2005). We also estimated the mean disorder propensity of human CALR which was calculated by averaging disorder profiles of individual predictors. The use of the consensus of multiple predictors for evaluation of intrinsic disorder is motivated by empirical observations that this approach usually increases the predictive performance compared to the use of a single predictor (Fan & Kurgan, 2014; Z. Peng & Kurgan, 2012; Walsh et al., 2015). In fact, 38.25% and 43.65% of the human CALR residues are predicted to be disordered (have disorder scores above the 0.5 threshold) based on the averaging the outputs of the PONDR® VLXT, PONDR® VL3, PONDR® VSL2, PONDR® FIT, IUPred_short, and IUPred_long predictors and by the MobiDB platform that aggregates the outputs from ten disorder predictors (http://mobidb.bio.unipd.it/) (Di Domenico et al., 2012; Potenza et al., 2015), respectively. This clearly places CALR into the category of highly disordered proteins, if the classification of proteins based on their content of predicted disordered residue (CPDR) values is used, where proteins are considered as highly ordered, moderately disordered, or highly disordered if their CPDR<10%, 10% ≤ CPDR<30%, or CPDR≥30%, respectively (Rajagopalan, Mooney, Parekh, Getzenberg, & Kulkarni, 2011)
Figure 5C shows that the N-domain is expected to be mostly ordered, whereas P-domain and the C-terminal tail of C-domain are predicted to be highly disordered. Importantly, the part of the C-domain which was co-crystallized with the N-domain (residues 302-368, see Figure 5B) is predicted to be mostly ordered. Therefore, our computational data where the mostly disordered nature of the C-terminal part of CALR that includes P- and C-domains is shown based on the peculiarities of the amino acid sequence of CALR provides support and explanation for the “two armed body” structural model of this protein.
3.2.3. Potential functional roles of disorder in CALR
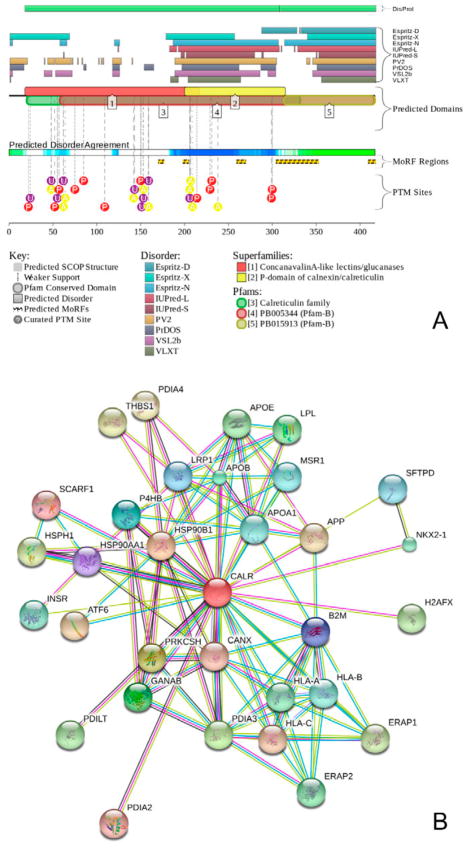
To further illustrate abundance and functionality of intrinsic disorder in human CALR, we used the output of the D2P2 platform (http://d2p2.pro/) (Oates et al., 2013), which shows results of the intrinsic disorder predisposition analysis of a target protein using several common disorder predictors, such as IUPred (Dosztanyi et al., 2005), PONDR® VLXT (P. Romero et al., 2001), PrDOS (Ishida & Kinoshita, 2007), PONDR® VSL2B (Obradovic et al., 2005; K. Peng et al., 2006), PV2 (Oates et al., 2013), and ESpritz (Walsh et al., 2012). In addition, the D2P2 profile of a query protein contains information on the location of various posttranslational modifications and predicted disorder-based protein binding sites. Figure 6A represents the D2P2 profile of human CALR that provides further support to the notion that this protein is expected to be highly disordered. Furthermore, Figure 6A shows that CALR has multiple sites of various posttranslational modifications, such as phosphorylation, acetylation, and glycosylation.
Figure 6. Computational analysis of functional disorder in human CALR.
A. Intrinsic disorder propensity and some important disorder-related functional information generated for human CALR (UniProt ID: P27797) by the D2P2 database (http://d2p2.pro/) (Oates et al., 2013). Outputs of nine disorder predictors are shown by differently colored bars. The green-and-white bar in the middle of the plot shows the predicted disorder agreement between nine predictors, with green parts corresponding to disordered regions by consensus. Yellow bars show the locations of the predicted disorder-based binding sites (molecular recognition features, MoRFs), whereas colored circles at the bottom of the plot show location of various PTMs, such as phosphorylation (red), acetylation (yellow), and ubiquitination (purple).
B. Analysis of the interactivity of human CALR by STRING computational platform that produces the network of predicted associations for a particular group of proteins (Szklarczyk et al., 2011). In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line - the presence of fusion evidence; a purple line - experimental evidence; a blue line – co-occurrence evidence; a light blue line - database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence (Szklarczyk et al., 2011). In our analysis, the most stringent criteria were used for selection of interacting proteins by choosing the highest cut-off of 0.9 as the minimal required confidence level.
Multiple studies revealed that some IDPs/IDPRs are able to undergo at least partial disorder-to-order transitions upon binding, and such binding-induced folding is crucial for recognition, regulation, and signaling functions of these proteins (Dunker et al., 2001; Dyson & Wright, 2002, 2005; Mohan et al., 2006; Oldfield et al., 2005; Z. Peng et al., 2013; Uversky, 2013b; Uversky et al., 2000; Vacic, Oldfield, et al., 2007; Wright & Dyson, 1999). Among these disorder-based functional sites are short order-prone motifs typically located within the long IDPRs. Such motifs are able to undergo disorder-to-order transition as a result of binding to a specific partner. These motifs are known as molecular recognition features (MoRFs) that can be identified computationally (Cheng et al., 2007; Oldfield et al., 2005). In our study, we used a specialized algorithm, ANCHOR, to find potential disorder-based protein binding sites that can fold as a result of interaction with specific partners (Dosztanyi, Meszaros, & Simon, 2009; Meszaros, Simon, & Dosztanyi, 2009). Figure 6A represents results of this analysis and shows that human CALR contains five MoRFs identified by ANCHOR, residues 170-176; 198-204, 259-269, 304-352, 409-417, distributed over the C-terminal part of N-domain, P-domain, and C-domain, suggesting that intrinsic disorder in CALR can be used for protein-protein interactions. This hypothesis is further supported by the results of the pre-CALR analysis by MoRFpred, which is an alternative computational tool for accurate evaluation of disorder-based binding sites undergoing disorder-to-order transition at interaction with specific partners (Disfani et al., 2012). Results of this analysis shows that the pre-CALR contains a set of relatively short regions that have a potential to fold at interaction with binding partners. These regions are residues 1-10, 19-25, 50-55, 76-80, 199-206, 221-225, 236-242, 247-252, 312-314, 332-334, 378-389, and 409-417. Regions in this list shown by bold font either coincide or are contained within the ANCHOR-predicted MoRFs (residues 198-204, 304-352, and 409-417). Curiously, although ANCHOR-predicted MoRFs in regions 170-176 and 259-269 are not identified by MoRFpred, these two regions have noticeably higher average MoRFpred scores when compared to adjacent (in the chain) regions, suggesting that do have increased binding potential. Therefore, ANCHOR and MoRFpred data indicate that CALR can be a highly promiscuous binder that utilizes intrinsic disorder to interact with a broad range of binding partners.
In agreement with this hypothesis, Figure 6B represents the STRING-based interactome of the human CALR and indicates that this protein indeed has several putative binding partners. Note that data shown in this plot were obtained using the most stringent confidence level of 0.9. Furthermore, MobiDB lists 2,791 proteins that interact with human CALR and that are characterized by the levels of intrinsic disorder ranging from 0% to 100% (http://mobidb.bio.unipd.it/entries/P27797). Overall, the vast majority of proteins interacting with CALR are expected to be ordered. In fact, 24, 75, and 2,692 of CALR partners are predicted to be highly or moderately disordered or highly ordered, possessing CPDR≥30%, 10% ≤ CPDR<30%, and CPDR<10%, respectively. Although indicated number of CALR binding partners seems to be very high, one should keep in mind that together with another lectin chaperone, calnexin (CNX), CALR is known to interact with practically all glycoproteins investigated to date (Ellgaard & Frickel, 2003) that makes it indeed a highly promiscuous binder.
4. CONCLUSIONS
Data reported in this study indicate that human CALR contains significant amount of intrinsic disorder mostly concentrated in its P- and C-domains. These observations are in line with the previous notion that the flexible nature of the P-domain and of the C-terminal tail has precluded crystallization of the full-length human CALR so far (Chouquet et al., 2011). Our study represents results of careful structural and conformational analyses of the full-length and the isolated P- and C-domains of human CALR that provide some mechanistic insights on how Ca2+ may affect exposure of both N-terminal and C-terminal halves of CALR (N-CALR and C-CALR, respectively) in vitro and in vivo. In fact, in the existing structural models, CALR is depicted as a rigid body (comprised of the N-CALR and N-terminal part of the C-domain) with two flexible arms (P-domain and C-tail). However, whether the individual isolated domains may sense Ca2+ has not been established as yet. Our analysis revealed that neither structure nor conformational stability of the P-domain were sensitive to Ca2+. Similarly, although the highly disordered C-CALR gained some level of ordered structure after Ca2+-binding, it still remained mostly disordered, and its conformational stability was not affected by Ca2+. These observations indicate that Ca2+ has a noticeable effect on the structure and conformational stability of the full-length protein, making it somewhat more compact and significantly more stable. Since no detectable changes in the structure and conformational stability of the isolated P-domain was induced by Ca2+ and its addition had minimal effects of the intrinsically disordered nature of the isolated C-domain without affecting its conformational stability, the Ca2+-induced changes in the behavior of the full-length CALR can be attributed to Ca2+-dependent stabilization of an C-embedded structure, where the portion of the C-domain is incorporated into a jelly-roll-like structure formed by its N-domain. This close spatial relationship between the two domains make their conformations sensitive to Ca2+ and determines which of the five MoRF motifs of the protein would be exposed and may exert their binding functions.
Acknowledgments
This study was supported by grants from the National Cancer Institute (P01-CA108671) and National Heart, Lung, and Blood Institute (1R01-HL116329) and Associazione Italiana Ricerca Cancro (AIRC 17608). Prof. Marlene Bouvier is gratefully acknowledged for the generous gift of full-length human CALR and its isolated P- and C-domains.
Footnotes
Conflict of interest disclosure: Authors declare no competing financial interest.
Authors’ contribution: VNU performed experiments and analyzed data. ARM and VNU designed research. VNU and ARM analyzed the data and wrote the manuscript. The authors state that the content of this manuscript has not been submitted elsewhere.
References
- Afshar N, Black BE, Paschal BM. Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol Cell Biol. 2005;25(20):8844–8853. doi: 10.1128/MCB.25.20.8844-8853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrin C, Pinkoski MJ, Burns K, Atkinson EA, Krahenbuhl O, Hudig D, Fraser SA, Winkler U, Tschopp J, Opas M, Bleackley RC, Michalak M. Interaction between a Ca2+-binding protein calreticulin and perforin, a component of the cytotoxic T-cell granules. Biochemistry. 1998;37(29):10386–10394. doi: 10.1021/bi980595z. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32(Database issue):D115–119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J Biol Chem. 1991;266(32):21458–21465. [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelt SG, Norn C, Rasmussen MI, Andre I, Ciplys E, Slibinskas R, Houen G, Hojrup P. Mapping the Ca(2+) induced structural change in calreticulin. J Proteomics. 2016;142:138–148. doi: 10.1016/j.jprot.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Bouvier M, Stafford WF. Probing the three-dimensional structure of human calreticulin. Biochemistry. 2000;39(48):14950–14959. doi: 10.1021/bi0019545. [DOI] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Dunker AK, Bateman A, Balaji S, Fuxreiter M, Babu MM. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23(3):443–450. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46(6):871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpio MA, Lopez Sambrooks C, Durand ES, Hallak ME. The arginylation-dependent association of calreticulin with stress granules is regulated by calcium. Biochem J. 2010;429(1):63–72. doi: 10.1042/BJ20091953. [DOI] [PubMed] [Google Scholar]
- Chao MP, Jaiswal S, Weissman-Tsukamoto R, Alizadeh AA, Gentles AJ, Volkmer J, Weiskopf K, Willingham SB, Raveh T, Park CY, Majeti R, Weissman IL. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med. 2010;2(63):63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46(47):13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouquet A, Paidassi H, Ling WL, Frachet P, Houen G, Arlaud GJ, Gaboriaud C. X-ray structure of the human calreticulin globular domain reveals a peptide-binding area and suggests a multi-molecular mechanism. PLoS One. 2011;6(3):e17886. doi: 10.1371/journal.pone.0017886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte IL, Keith N, Gutierrez-Gonzalez C, Parodi AJ, Caramelo JJ. The interplay between calcium and the in vitro lectin and chaperone activities of calreticulin. Biochemistry. 2007;46(15):4671–4680. doi: 10.1021/bi6026456. [DOI] [PubMed] [Google Scholar]
- Coppolino MG, Dedhar S. Ligand-specific, transient interaction between integrins and calreticulin during cell adhesion to extracellular matrix proteins is dependent upon phosphorylation/dephosphorylation events. Biochem J. 1999;340(Pt 1):41–50. doi: 10.1042/0264-6021:3400041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett EF, Oikawa K, Francois P, Tessier DC, Kay C, Bergeron JJ, Thomas DY, Krause KH, Michalak M. Ca2+ regulation of interactions between endoplasmic reticulum chaperones. J Biol Chem. 1999;274(10):6203–6211. doi: 10.1074/jbc.274.10.6203. [DOI] [PubMed] [Google Scholar]
- Dedhar S, Rennie PS, Shago M, Hagesteijn CY, Yang H, Filmus J, Hawley RG, Bruchovsky N, Cheng H, Matusik RJ, et al. Inhibition of nuclear hormone receptor activity by calreticulin. Nature. 1994;367(6462):480–483. doi: 10.1038/367480a0. [DOI] [PubMed] [Google Scholar]
- DeForte S, Uversky VN. Order, Disorder, and Everything in Between. Molecules. 2016a;21(8) doi: 10.3390/molecules21081090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForte S, Uversky VN. Resolving the ambiguity: Making sense of intrinsic disorder when PDB structures disagree. Protein Sci. 2016b;25(3):676–688. doi: 10.1002/pro.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForte S, Uversky VN. Not an exception to the rule: the functional significance of intrinsically disordered protein regions in enzymes. Mol Biosyst. 2017;13(3):463–469. doi: 10.1039/c6mb00741d. [DOI] [PubMed] [Google Scholar]
- Di Domenico T, Walsh I, Martin AJ, Tosatto SC. MobiDB: a comprehensive database of intrinsic protein disorder annotations. Bioinformatics. 2012;28(15):2080–2081. doi: 10.1093/bioinformatics/bts327. [DOI] [PubMed] [Google Scholar]
- Disfani FM, Hsu WL, Mizianty MJ, Oldfield CJ, Xue B, Dunker AK, Uversky VN, Kurgan L. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012;28(12):i75–83. doi: 10.1093/bioinformatics/bts209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25(20):2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Eftink MR, Ghiron CA. Fluorescence quenching studies with proteins. Anal Biochem. 1981;114(2):199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Frickel EM. Calnexin, calreticulin, and ERp57: teammates in glycoprotein folding. Cell Biochem Biophys. 2003;39(3):223–247. doi: 10.1385/CBB:39:3:223. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Riek R, Herrmann T, Guntert P, Braun D, Helenius A, Wuthrich K. NMR structure of the calreticulin P-domain. Proc Natl Acad Sci U S A. 2001;98(6):3133–3138. doi: 10.1073/pnas.051630098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel MP, Dziak E, Lo CM, Ferrier J, Mesaeli N, Michalak M, Opas M. Calreticulin affects focal contact-dependent but not close contact-dependent cell-substratum adhesion. J Biol Chem. 1999;274(21):15085–15094. doi: 10.1074/jbc.274.21.15085. [DOI] [PubMed] [Google Scholar]
- Fan X, Kurgan L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J Biomol Struct Dyn. 2014;32(3):448–464. doi: 10.1080/07391102.2013.775969. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci U S A. 2002;99(4):1954–1959. doi: 10.1073/pnas.042699099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4(12):728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M, Toth-Petroczy A, Kraut DA, Matouschek A, Lim RY, Xue B, Kurgan L, Uversky VN. Disordered proteinaceous machines. Chem Rev. 2014;114(13):6806–6843. doi: 10.1021/cr4007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Yang F, Zhang L, Su Z, Huang Y. Exploring the sequence-structure-function relationship for the intrinsically disordered betagamma-crystallin Hahellin. J Biomol Struct Dyn. 2017:1–11. doi: 10.1080/07391102.2017.1316519. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123(2):321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Gelebart P, Opas M, Michalak M. Calreticulin, a Ca2+-binding chaperone of the endoplasmic reticulum. Int J Biochem Cell Biol. 2005;37(2):260–266. doi: 10.1016/j.biocel.2004.02.030. [DOI] [PubMed] [Google Scholar]
- Ghiran I, Klickstein LB, Nicholson-Weller A. Calreticulin is at the surface of circulating neutrophils and uses CD59 as an adaptor molecule. J Biol Chem. 2003;278(23):21024–21031. doi: 10.1074/jbc.M302306200. [DOI] [PubMed] [Google Scholar]
- Goicoechea S, Pallero MA, Eggleton P, Michalak M, Murphy-Ullrich JE. The anti-adhesive activity of thrombospondin is mediated by the N-terminal domain of cell surface calreticulin. J Biol Chem. 2002;277(40):37219–37228. doi: 10.1074/jbc.M202200200. [DOI] [PubMed] [Google Scholar]
- Gold LI, Eggleton P, Sweetwyne MT, Van Duyn LB, Greives MR, Naylor SM, Michalak M, Murphy-Ullrich JE. Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J. 2010;24(3):665–683. doi: 10.1096/fj.09-145482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Helenius A, Trombetta ES, Hebert DN, Simons JF. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7(5):193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Black BE, Love DC, Hanover JA, Leszyk J, Paschal BM. Calreticulin is a receptor for nuclear export. J Cell Biol. 2001;152(1):127–140. doi: 10.1083/jcb.152.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Black BE, Rastinejad F, Paschal BM. Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol. 2002;22(17):6286–6297. doi: 10.1128/MCB.22.17.6286-6297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35(Web Server issue):W460–464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Kriwacki R, Uversky VN. Conditionally and transiently disordered proteins: awakening cryptic disorder to regulate protein function. Chem Rev. 2014;114(13):6779–6805. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Dey S, Matsunami H. Calreticulin: roles in cell-surface protein expression. Membranes (Basel) 2014;4(3):630–641. doi: 10.3390/membranes4030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiriya JJ, Pathak RR, Clayman E, Xue B, Uversky VN, Dave V. Presence and utility of intrinsically disordered regions in kinases. Mol Biosyst. 2014;10(11):2876–2888. doi: 10.1039/c4mb00224e. [DOI] [PubMed] [Google Scholar]
- Labriola C, Cazzulo JJ, Parodi AJ. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Mol Biol Cell. 1999;10(5):1381–1394. doi: 10.1091/mbc.10.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24(4):325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- Lenton S, Grimaldo M, Roosen-Runge F, Schreiber F, Nylander T, Clegg R, Holt C, Hartlein M, Garcia Sakai V, Seydel T, Marujo Teixeira SC. Effect of Phosphorylation on a Human-like Osteopontin Peptide. Biophys J. 2017;112(8):1586–1596. doi: 10.1016/j.bpj.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova EI, Galzitskaya OV. Cell communication using intrinsically disordered proteins: what can syndecans say? J Biomol Struct Dyn. 2015;33(5):1037–1050. doi: 10.1080/07391102.2014.926256. [DOI] [PubMed] [Google Scholar]
- Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- Li Z, Stafford WF, Bouvier M. The metal ion binding properties of calreticulin modulate its conformational flexibility and thermal stability. Biochemistry. 2001;40(37):11193–11201. doi: 10.1021/bi010948l. [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11(11):1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31(13):3701–3708. doi: 10.1093/Nar/Gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YC, Weng WC, Lee H. Functional roles of calreticulin in cancer biology. Biomed Res Int. 2015;2015:526524. doi: 10.1155/2015/526524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Ahmad S, Hong SJ, Chapman DC, Kozlov G, Williams DB. Contributions of the Lectin and Polypeptide Binding Sites of Calreticulin to Its Chaperone Functions in Vitro and in Cells. J Biol Chem. 2016;291(37):19631–19641. doi: 10.1074/jbc.M116.746321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5(5):e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multiprocess calcium-buffering chaperone of the endoplasmic reticulum. Biochem J. 2009;417(3):651–666. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362(5):1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Moreau C, Cioci G, Iannello M, Laffly E, Chouquet A, Ferreira A, Thielens NM, Gaboriaud C. Structures of parasite calreticulins provide insights into their flexibility and dual carbohydrate/peptide-binding properties. IUCrJ. 2016;3(Pt 6):408–419. doi: 10.1107/S2052252516012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CF, Wassmann K, Berger A, Holz S, Wassmann S, Nickenig G. Differential phosphorylation of calreticulin affects AT1 receptor mRNA stability in VSMC. Biochem Biophys Res Commun. 2008;370(4):669–674. doi: 10.1016/j.bbrc.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Zuppini A, Arnaudeau S, Lynch J, Ahsan I, Krause R, Papp S, De Smedt H, Parys JB, Muller-Esterl W, Lew DP, Krause KH, Demaurex N, Opas M, Michalak M. Functional specialization of calreticulin domains. J Cell Biol. 2001;154(5):961–972. doi: 10.1083/jcb.200102073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash PD, Opas M, Michalak M. Calreticulin: not just another calcium-binding protein. Mol Cell Biochem. 1994;135(1):71–78. doi: 10.1007/bf00925962. [DOI] [PubMed] [Google Scholar]
- Nickenig G, Michaelsen F, Muller C, Berger A, Vogel T, Sachinidis A, Vetter H, Bohm M. Destabilization of AT(1) receptor mRNA by calreticulin. Circ Res. 2002;90(1):53–58. doi: 10.1161/hh0102.102503. [DOI] [PubMed] [Google Scholar]
- Norgaard Toft K, Larsen N, Steen Jorgensen F, Hojrup P, Houen G, Vestergaard B. Small angle X-ray scattering study of calreticulin reveals conformational plasticity. Biochim Biophys Acta. 2008;1784(9):1265–1270. doi: 10.1016/j.bbapap.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztanyi Z, Uversky VN, Obradovic Z, Kurgan L, Dunker AK, Gough J. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013;41(Database issue):D508–516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid M, Tesniere A, Ghiringhelli F, Fimia GM, Apetoh L, Perfettini JL, Castedo M, Mignot G, Panaretakis T, Casares N, Metivier D, Larochette N, van Endert P, Ciccosanti F, Piacentini M, Zitvogel L, Kroemer G. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med. 2007;13(1):54–61. doi: 10.1038/nm1523. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(Suppl 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61(Suppl 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44(37):12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- Pejaver V, Hsu WL, Xin F, Dunker AK, Uversky VN, Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23(8):1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3(1):35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- Peng Z, Kurgan L. On the complementarity of the consensus-based disorder prediction. Pac Symp Biocomput. 2012:176–187. doi: 10.1142/9789814366496_0017. [DOI] [PubMed] [Google Scholar]
- Peng Z, Xue B, Kurgan L, Uversky VN. Resilience of death: intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ. 2013;20(9):1257–1267. doi: 10.1038/cdd.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015;72(1):137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza E, Domenico TD, Walsh I, Tosatto SC. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 2015 doi: 10.1093/nar/gku982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem. 2011;112(11):3256–3267. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KD, Malipeddi J, DeForte S, Pejaver V, Radivojac P, Uversky VN, Deschenes RJ. Physicochemical sequence characteristics that influence S-palmitoylation propensity. J Biomol Struct Dyn. 2017:1–14. doi: 10.1080/07391102.2016.1217275. [DOI] [PubMed] [Google Scholar]
- Rizvi SM, Mancino L, Thammavongsa V, Cantley RL, Raghavan M. A polypeptide binding conformation of calreticulin is induced by heat shock, calcium depletion, or by deletion of the C-terminal acidic region. Mol Cell. 2004;15(6):913–923. doi: 10.1016/j.molcel.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Rojiani MV, Finlay BB, Gray V, Dedhar S. In vitro interaction of a polypeptide homologous to human Ro/SS-A antigen (calreticulin) with a highly conserved amino acid sequence in the cytoplasmic domain of integrin alpha subunits. Biochemistry. 1991;30(41):9859–9866. doi: 10.1021/bi00105a008. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Ihara Y, Leach MR, Cohen-Doyle MF, Williams DB. Calreticulin functions in vitro as a molecular chaperone for both glycosylated and non-glycosylated proteins. EMBO J. 1999;18(23):6718–6729. doi: 10.1093/emboj/18.23.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35(Database issue):D786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro RG, Zhu Q, Bhoyroo V, Soling HD. Definition of the lectin-like properties of the molecular chaperone, calreticulin, and demonstration of its copurification with endomannosidase from rat liver Golgi. J Biol Chem. 1996;271(19):11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Chen M, Li Z, Mabuchi K, Bouvier M. The calcium- and zinc-responsive regions of calreticulin reside strictly in the N-/C-domain. Biochim Biophys Acta. 2006;1760(5):745–753. doi: 10.1016/j.bbagen.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37(12):509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30(9):484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Totary-Jain H, Naveh-Many T, Riahi Y, Kaiser N, Eckel J, Sasson S. Calreticulin destabilizes glucose transporter-1 mRNA in vascular endothelial and smooth muscle cells under high-glucose conditions. Circ Res. 2005;97(10):1001–1008. doi: 10.1161/01.RES.0000189260.46084.e5. [DOI] [PubMed] [Google Scholar]
- Toth-Petroczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. Malleable machines in transcription regulation: the mediator complex. PLoS Comput Biol. 2008;4(12):e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28(7–8):305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013a;22(6):693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsic Disorder-based Protein Interactions and their Modulators. Curr Pharm Des. 2013b;19(23):4191–4213. doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013c;1834(5):932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282(7):1182–1189. doi: 10.1111/febs.13202. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Dancing Protein Clouds: The Strange Biology and Chaotic Physics of Intrinsically Disordered Proteins. J Biol Chem. 2016a;291(13):6681–6688. doi: 10.1074/jbc.R115.685859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2016b;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv Colloid Interface Sci. 2017;239:97–114. doi: 10.1016/j.cis.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dave V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem Rev. 2014;114(13):6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.3.co;2-z. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015;589(1):15–22. doi: 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilakos A, Michalak M, Lehrman MA, Williams DB. Oligosaccharide binding characteristics of the molecular chaperones calnexin and calreticulin. Biochemistry. 1998;37(10):3480–3490. doi: 10.1021/bi972465g. [DOI] [PubMed] [Google Scholar]
- Villagomez M, Szabo E, Podcheko A, Feng T, Papp S, Opas M. Calreticulin and focal-contact-dependent adhesion. Biochem Cell Biol. 2009;87(4):545–556. doi: 10.1139/o09-016. [DOI] [PubMed] [Google Scholar]
- Villamil Giraldo AM, Lopez Medus M, Gonzalez Lebrero M, Pagano RS, Labriola CA, Landolfo L, Delfino JM, Parodi AJ, Caramelo JJ. The structure of calreticulin C-terminal domain is modulated by physiological variations of calcium concentration. J Biol Chem. 2010;285(7):4544–4553. doi: 10.1074/jbc.M109.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh I, Giollo M, Di Domenico T, Ferrari C, Zimmermann O, Tosatto SC. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics. 2015;31(2):201–208. doi: 10.1093/bioinformatics/btu625. [DOI] [PubMed] [Google Scholar]
- Walsh I, Martin AJ, Di Domenico T, Tosatto SC. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28(4):503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wemeau M, Kepp O, Tesniere A, Panaretakis T, Flament C, De Botton S, Zitvogel L, Kroemer G, Chaput N. Calreticulin exposure on malignant blasts predicts a cellular anticancer immune response in patients with acute myeloid leukemia. Cell Death Dis. 2010;1:e104. doi: 10.1038/cddis.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeyesakere SJ, Gafni AA, Raghavan M. Calreticulin is a thermostable protein with distinct structural responses to different divalent cation environments. J Biol Chem. 2011;286(11):8771–8785. doi: 10.1074/jbc.M110.169193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeyesakere SJ, Rizvi SM, Raghavan M. Glycan-dependent and -independent interactions contribute to cellular substrate recruitment by calreticulin. J Biol Chem. 2013;288(49):35104–35116. doi: 10.1074/jbc.M113.507921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119(Pt 4):615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- Williams RM, Obradovic Z, Mathura V, Braun W, Garner EC, Young J, Takayama S, Brown CJ, Dunker AK. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput. 2001:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Xue B, Oldfield CJ, Van YY, Dunker AK, Uversky VN. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst. 2012;8(1):134–150. doi: 10.1039/c1mb05163f. [DOI] [PubMed] [Google Scholar]
- Xue B, Uversky VN. Intrinsic disorder in proteins involved in the innate antiviral immunity: another flexible side of a molecular arms race. J Mol Biol. 2014;426(6):1322–1350. doi: 10.1016/j.jmb.2013.10.030. [DOI] [PubMed] [Google Scholar]
- Yacoub HA, Al-Maghrabi OA, Ahmed ES, Uversky VN. Abundance and functional roles of intrinsic disorder in the antimicrobial peptides of the NK-lysin family. J Biomol Struct Dyn. 2017;35(4):836–856. doi: 10.1080/07391102.2016.1164077. [DOI] [PubMed] [Google Scholar]
- Yan Q, Murphy-Ullrich JE, Song Y. Structural insight into the role of thrombospondin-1 binding to calreticulin in calreticulin-induced focal adhesion disassembly. Biochemistry. 2010;49(17):3685–3694. doi: 10.1021/bi902067f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21(16):3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- Zamanian M, Qader Hamadneh LA, Veerakumarasivam A, Abdul Rahman S, Shohaimi S, Rosli R. Calreticulin mediates an invasive breast cancer phenotype through the transcriptional dysregulation of p53 and MAPK pathways. Cancer Cell Int. 2016;16:56. doi: 10.1186/s12935-016-0329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M, Veerakumarasivam A, Abdullah S, Rosli R. Calreticulin and cancer. Pathol Oncol Res. 2013;19(2):149–154. doi: 10.1007/s12253-012-9600-2. [DOI] [PubMed] [Google Scholar]
- Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273(11):6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]