Abstract
Background and Objectives:
Factors contributing to development of gastric cancer are still under investigation. The JC Virus (JCV), as an oncogenic virus, has been indicated to play a possible role in gastric carcinogenesis. Theoretically, tumor antigen (T-Ag), the viral transforming protein, is capable of binding and inactivating tumor suppressor proteins p53 and pRb, there by promoting cancer development although such a role in gastric cancer is still controversial and additional data is needed to reach a definite conclusion. The prevalence of the virus varies in different geographic regions, therefore, we aimed to investigate JCV presence in cancerous gastric tissues of Iranian patients.
Materials and Methods:
Thirty-one paired samples were included in this study (total of 62 samples). T-Ag sequences were investigated using real-time PCR in formalin fixed paraffin embedded (FFPE) tissue samples from the tumor site and relevant adjacent non-cancerous tissues (ANCT). In positive samples, JCV copy number (viral load) was also measured using real-time PCR. To evaluate T-Ag protein expression, immunohistochemistry examination was performed using an anti-T-Ag specific antibody.
Results:
JCV sequences were detected in 17 out of 31 gastric cancer tissue samples (54.84%) and in 10 out of 31 of the non-cancerous adjacent gastric mucosa (32.25%) (Odds ratio of 2.4). Viral load in tumoral and adjacent tissue samples was not statistically different (p=0.88). Immunohistochemical study confirmed presence of JC T-Ag in the nuclear compartment.
Conclusion:
We showed the presence of the JC virus in gastric carcinoma tissue samples in our geographic region. This finding provides supportive data for a possible contribution of JCV in gastric cell transformation to malignancy. However, we highly recommend additional investigations to further explore JC virus and gastric cancer in order to reach a conclusion.
Keywords: Oncogenic virus, Tumor antigen, Tumor suppressor proteins, Real-time PCR
INTRODUCTION
Although the global incidence and mortality rate of gastric cancer has decreased dramatically over the past decades, gastric cancer remains a major public health problem in many parts of the world (1, 2). Gastric carcinogenesis is a multistep and multifactorial process. The modification in function of genes that control epithelial cell growth and differentiation is an important key factor in the progression of gastric cancer (3). Exposure to biological and chemical carcinogens, such as Helicobacter pylori, oncoviruses, nitrosamines and oxidants which lead to DNA damage are the main known risk factors of gastric cancer (4).
In order to identify the predisposing factors contributing to development of cancers, some infectious agents such as Epstein–Barr virus (EBV) and John Cunningham virus (JCV) have been blamed to play a possible role in different stages of carcinogenesis process (5, 6). JCV belongs to the Polyomaviridae family and is a common human infection (7). The JCV genome is 5.13-kb, double-stranded, supercoiled circular DNA. JCV genome consists of early and late coding regions that are separated by a transcription control region (TCR) containing the promoter and enhancer elements for early and late transcription. Among these genes, two oncoproteins, large T antigen (T-Ag) and small t antigen (t-Ag), have been shown to be transforming and oncogenic proteins in experimental systems (8, 9).
Regardless of PML (Progressive Multifocal Leukoencephalopathy), a fatal CNS complication in immunocompromised patients (10–12), infection with the JC virus has been considered subclinical in humans. However, it has been recently proposed that persistent infection with JC virus might be a potential risk factor for human carcinogenesis (6, 13–15). All Polyomaviruses have the ability to encode a version of large tumor antigen (T-Ag) that is a multifunctional protein capable of promoting transformation of cells through several pathways. T-Ag can modulate cellular signaling pathways and thereby induces the host cells to enter the S-phase, considering its function as ATPase, helicase, polymerase and DNA binding capacity; all of which are essential for DNA replication (16, 17).
Also, this protein has the ability to bind and inactivate tumor suppressor proteins p53 and the cell cycle regulator retinoblastoma gene product, pRb, leading to their impaired function (18–23). In addition to T-Ag transforming function, the JC viral genome may integrate into the host genome which in turn may cause genetic instability leading to malignant cells (18–20). Overall, different cellular mechanisms including rapid division, prolonged life span, enhanced production of plasminogen activator, anchorage-dependent growth, and unstable multicentric chromosome have been proposed for JC T-Ag carcinogenicity (24). Nevertheless, the role of JCV in gastric cancer is still controversial. Few reports provided supportive data on such a role though additional data are still needed to reach a definite conclusion. Here we investigated the presence of JCV T-Ag DNA sequences and its expression in gastric cancer and its non-cancerous adjacent tissue in Iranian patients.
MATERIALS AND METHODS
Samples.
Formalin fixed paraffin embedded (FFPE) tissue sample pairs of gastric cancer and adjacent non-cancerous tissue (ANCT) were obtained from the archives of the pathology department of Ghaem University Hospital, Mashhad, Iran. Samples from the past two years were included to obtain better preserved genome. The FFPE samples belonged to 18 men and 13 women diagnosed with gastric carcinoma.
DNA extraction and real-time PCR.
FFPE tissue samples were deparaffinized and then genomic DNA was extracted. Firstly, one ml xylene was added to 25 mg of slice sample in a microcentrifuge tube and incubated for 30 min at 56°C until the paraffin was completely dissolved. The samples were centrifuged at full speed for 5 min and the supernatant was removed. In the next step, one ml ethanol (96–100%) was added to the deparaffinized tissue sample and gently mixed. After centrifuging and removing the supernatant solution, samples were incubated at 37°C for 15∼20 min to evaporate ethanol residue completely.
Then, genomic DNA was extracted from deparaffinized tissues in a clean pre-PCR room using the QIAamp DNA mini kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions with slight modifications. After extraction, DNA was quantitated spectrophotometrically and kept at −20°C until used for real-time PCR assay.
Before real-time PCR, 400 ng of DNA was treated with 0.26 unit of topoisomerase I (BioLabs M0301S) for 45 minutes at 37°C to enhance primer access to supercoiled JC genomic DNA. For detection of JCV T-Ag sequence, SYBR Green Real Time quantitative PCR was applied using specific primers for T-Ag to produce a 171-bp amplicon related to the conserved N-terminal region of JCV T-Ag. PCR primers were designed using Beacon Designer software. Mad-1 reference sequence from National Center for Bio-technology Information (NCBI) reference genome (accession number NC: 001699.1) (www.ncbi.nlm.nih.gov) was used for primer design. The primer set used in this study was specific for the T-Ag of JCV, distinct from the T-Ag sequences of the homologous viruses such as SV40 and BKV. The primer pairs used in this study are shown in Table 1.
Table 1.
Primer sequences for amplification of JC Virus T-Ag.
| Primer name | Primer sequence | Primer position* |
|---|---|---|
| JCV T-Antigen F | 5′-GGA TTA GTG GCA CAG TTA GG -3′ | 4700-4719 |
| JCV T-Antigen R | 5′-CGA AGA CAA GAT GAA GAG AAT G -3′ | 4870-4849 |
Based on JC virus sequence Genbank
The qPCR amplification was performed in a Light Cycler instrument (Roche Applied Science). The PCR reaction mixture volume was 20 μL consisting of 1.5 μL of template DNA, 10 μL of Takara SYBR Premix Ex Taq II, 6.5 μL dH2O and 1 μL (10 pmol) of each primer. The following PCR protocol was used: initial denaturation step (95°C for 7 minutes), amplification and quantification program repeated 35 times (denaturation at 95°C for 15 seconds, followed by annealing at 60°C for 45 seconds and extension 72°C for 60 seconds with a single fluorescence measurement).
To prepare the positive control, the JCV (Mad-1) T-Ag gene was sub-cloned into pBHA plasmid vector and serially diluted to prepare the standard curve to be used for genome quantification calculation. Master Mix without template DNA served as a negative control. Beta-2 microglobulin (β2-MG) DNA was used as internal control of PCR reactions and reagents/instrumentation efficacy. Melting curve analysis was performed to confirm the specificity of reactions.
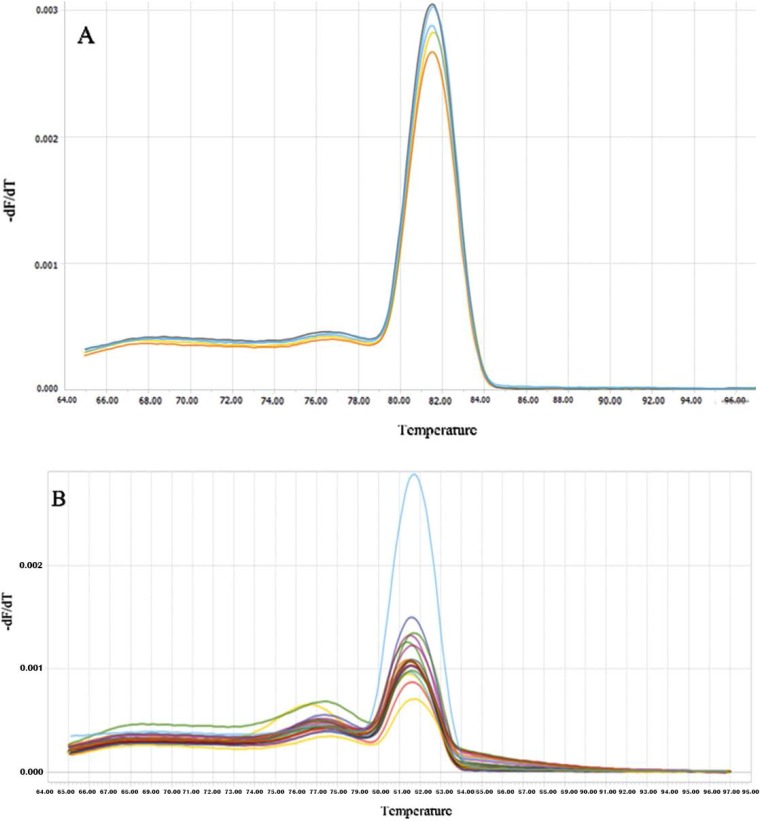
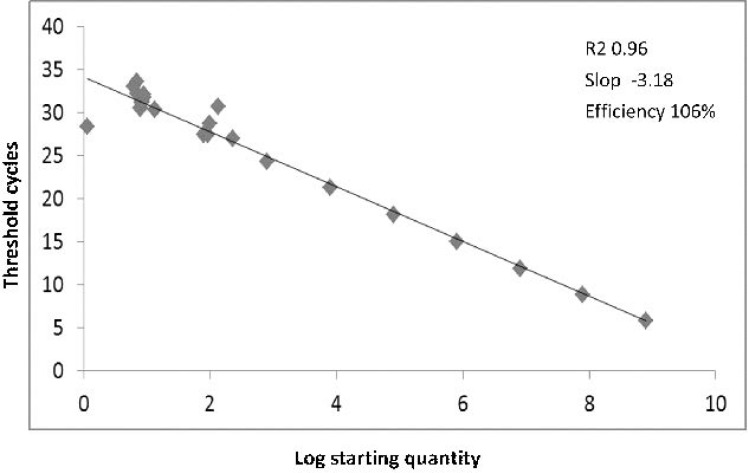
Melt curve analysis was used in order to assess the specificity of qPCR products (Fig. 1). JCV-positive samples were identified by comparison of the test sample melt curves with the melt curves of the positive controls included in each run. The Ct value of the samples were measured and compared to the standard curve to calculate the viral T-Ag copy number (Fig. 2). The number copies was calculated according to the formula described by Rutledge and Stewart (25). Also, the PCR products were electrophoresed to evaluate the amplicon size and later subjected for sequencing. The clean-up process was done by the sequencing company.
Fig. 1.
Melting curve analysis. (A) Amplicons from reference gene. The single peak observed is typically interpreted as representing a pure, single amplicon. (B) Amplicons from samples. The specificity of amplicons was verified by melting curve analysis after 35 cycles and agarose gel electrophoresis. Fluorescence melting peaks for the T-Ag gene, no separate heteroduplex products were apparent. Specific signals had melting temperatures of 81.4 ± 0.05°C.
Fig. 2.
Standard curve analysis. Synthetic DNA standard curves for absolute quantification of JC-T-Ag by real-time PCR. The JCV-Mad-1 T-Ag sequence that cloned in a pBHA plasmid vector was serially diluted (from 100 to 108 copies of positive control DNA) and served as a standard reference. High amplification efficiency has also been shown by a good linear relationship among each concentration.
Immunohistochemical staining for T-Antigen.
Immunohistochemical staining was performed on the FFPE samples in 5 μm thick, mounted on positively charged slides. Briefly, tissue sections were placed in an oven at 65°C to melt the paraffin, followed by deparaffinization with xylene and rehydrated through serially gradient alcohol solutions. The sections subsequently were immersed into 0.01 M sodium citrate buffer (pH 6.0) and were heated intermittently in a microwave oven for 20 minutes to nonenzymatic antigen retrieval. Endogenous peroxidase activity was blocked with endogenous enzyme block solution (Dako Cytomation Inc., Carpinteria, CA) for 10 minutes.
The primary antibody used to detect specific T-Ag protein expression was a mouse monoclonal antibody against SV40 large T-Ag with cross reacting capacity with JCV T-Ag (clone pAb416, 1:100 dilution; Oncogene Research Products, San Diego, CA). Incubation of the primary antibody was performed overnight at 4°C and was followed by incubation in Dako EnVision+system-HRP polymer (Dako Cytomation Inc.) for 30 minutes. Staining was developed by reaction with diaminobenzide chromogen for 5 to 10 minutes. Next, slides were counterstained for 5 minutes with hematoxylin. The brown chromogen complexes were indicative of T-Ag-specific expression in the gastric tissues.
Statistical analysis.
Demographic variables such as age and gender were entered in SPSS program version 21 for analyzing all data. Statistical evaluation was carried out using Wilcoxon Signed Ranks and MacNemar tests to differentiate non-parametric values. P-value less than 0.05 was considered statistically significant.
RESULTS
Viral T-Ag gene detection and JC quantification.
This study was performed on a set of biopsy specimens collected from gastric adenocarcinomas and, in parallel, from the healthy gastric mucosa surrounding the tumor mass from 31 patients including 18 men and 13 women diagnosed with gastric carcinoma and mean age of 64.90 ± 8.87 years.
JCV T-Ag sequences were found in 17 out of 31 (54.84%) neoplastic tissue samples and 10 out of 31 (32.25%) anatomically normal surrounding tissues (MacNemar Test, p=0.54). Only in 5 cases, both cancerous tissue and its adjacent non-cancerous tissue harbored T-Ag sequences. A paired Odds ratio of 2.4 (CI95%=0.84–6.81) was calculated.
Quantitative real-time PCR for T-Ag in paraffin embedded specimens was performed in order to compare viral copy number inside and outside of tumor. Standard curve was constructed using serial dilutions of JCV T-Ag positive control DNA plasmid (corresponding to 100 to 108 copies) (Fig. 2). The number copies was calculated according to the formula described by Rutledge and Stewart (25). The viral DNA loads ranged from 2, 030 to 1.97 × 105 copies/μg DNA (mean ± SD, Median, Interquartile range, 11252±35748, 2250, 0–3770) in tumor tissue, and 389 to 9.42 × 104 copies/μg DNA (mean ± SD; Median, Interquartile range, 7934±18744, 0, 0–6150) in surrounding healthy tissue (Wilcoxon Test, p=0.88).
In order to check the specificity of the reaction, melting curve analysis was performed and a single peak without secondary configurations or nonspecific signals was observed. A peak related to β-2 microglobulin control was observed in all experiments indicating adequate integrity of the extracted DNA and efficacy of reaction.
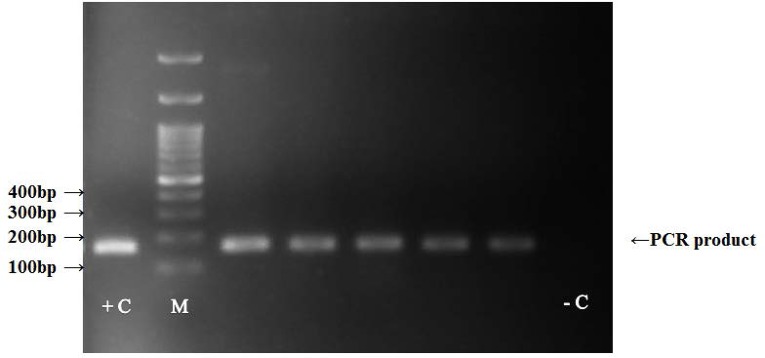
Next, the positive samples of PCR were subjected to gel electrophoresis and a 171 bp bond corresponding to the relevant amplicon size was observed (Fig. 3). Subsequently, T-Ag-positive PCR products were further confirmed by DNA sequencing and in each case, the presence of JCV and its specificity was validated using BLAST (Basic Local Alignment Search Tool) available from NCBI.
Fig. 3.
PCR amplification of T-antigen of JCV in gastric cancer and non-neoplastic tissues. A 171 bp bond corresponding to amplified sequences are shown. (+C; positive control, −C; negative control)
Expression of oncogenic T-Ag protein.
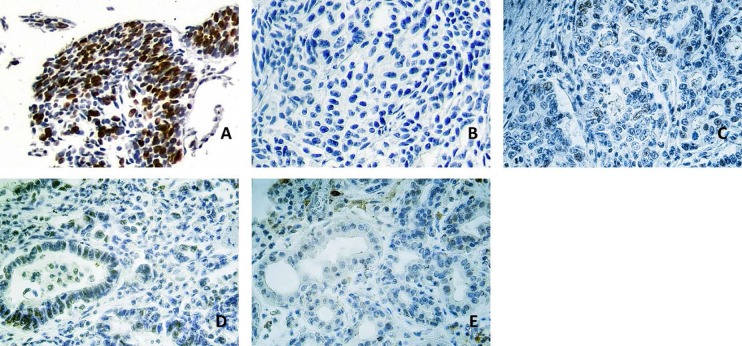
We performed immunohistochemical staining to show T-Ag expression in gastric cancer tissue. Fig. 4 illustrates one representative panel of positive and negative results. In positive samples, the reaction was visualized in the nuclear compartment of the cells (Fig. 4).
Fig. 4.
Panel A: Colon cancer cells infected by JC virus were used as a positive control, of which 20–30% cells were positive. (grade III); Panel B, C, D: These photomicrographs show immunohistochemical staining for JCV T-Ag in gastric tumor tissue samples. B. Tumor sample without expression of T-Ag. C. a weak and focal positive nuclear staining can be observed in the nuclei of tumor cells (shown in brown; grade I) D. positive staining in the nuclei of tumor cells (grade II); Panel E: Positive staining of grade I in the nuclei of endothelial cells of healthy mucosa surrounding a gastric adenocarcinoma. Original magnification ×400.
DISCUSSION
In this study, JCV sequences were found in 54.83% of neoplastic tissue samples and 32.25% of anatomically normal surrounding tissue. These results are in agreement with our previous findings evaluating JCV in colorectal cancer (unpublished data); in addition, these observations are in line with data from other laboratories reporting the presence of JCV DNA sequences in the mucosa of the gastrointestinal tract, like colon cancer tissue, suggesting the possible role of polyomaviruses in development of tumor, mainly due to the presence of potent transforming genes such as large tumor antigen (T-Ag) (6, 14, 15, 26, 27). Analogous to other malignancies, gastric carcinogenesis is a multistage process with a multi-factorial etiology (28).
During the last few decades, there have been several reports emphasizing the association of Polyomavirus with different types of human carcinomas such as brain and colorectal cancers (13, 15, 27, 29, 30). JC virus strains worldwide can be classified into various genotypes based on DNA sequence variations. Although based on the structure of its TCR, JCV can be classified into two forms: the archetype and tandem repeat variants. Among the tandem repeat variants of JCV, the Mad-1 is the exclusive strain selected for replication and persistently infects the gastrointestinal tract. Several studies also have shown different single nucleotide polymorphisms (SNPs) in the T-Ag region of the JC virus (31, 32) though a conserved area of T-Ag sequence was used for primer design in this study.
To the best of our knowledge, this is the first report of JCV DNA in gastric tissues from Iranian patients, a region where gastric cancer is common. It should be noted that except for few reports, little is known about epidemiology of JC infection among the Iranian population (33). In the present study, the sensitivity of the method used was critical; immunohistochemical reactions were less sensitive probably due to T-Ag degradation in formalin-fixed and paraffin-embedded tissue samples. In positive samples, the protein was visualized in nuclei where it mediates its oncogenic role after its expression in the cytoplasm.
T-Ag is a multifunctional protein with the ability to bind and inactivate tumor suppressor proteins including pRb and p53. Different gene-based association studies explored the relationship between single nucleotide polymorphisms (SNPs) of oncogenes and tumor suppressor genes and various cancers such as gastric cancer and colorectal cancer in the Iranian population which should be noted in the interpretation of T-Ag oncogenic function (34, 35). It has been suggested that with these functions, T-Ag can inactivate cell cycle check points and induce the host cells to enter S-phase resulting in unchecking cellular replication (19–21, 23).
Experimentally, it is difficult to detect viral sequences in human tumors and more importantly, it is difficult to reproduce results in different laboratories. In some cases, detection rates for T-Ag in infected tumor cells might be reduced due to a low copy number. Some viruses can induce cellular integration and differentiation process involving genomic rearrangement and loss of the viral genome during the process called “hit-and-run”, which might have pathogenic roles in cancer progression (15). In this case, the target sequences might not be detectable by PCR. Also, a severely undifferentiated cancer tissue may be non-permissive for JCV replication. Technical reasons such as degraded extracted DNA as well as DNA quality could be other explanations for varying rates of JCV detection.
In the present study, the efficacy of our PCR reaction was enhanced by topoisomerase I pretreatment which has been recommended by previous studies in order to unwind supercoiled viral DNA genes resulting in improved access of the primers to their target sequences (36, 37). Investigators previously have used internal controls as a method to monitor the PCR amplification performance including extraction, amplification and detection steps. In this study, β-2 microglobulin (β2-MG) DNA served as a sensitive internal control (38, 39).
Due to high homology between JCVT-Ag with other polyomaviruses, including SV40 and BKV at the nucleotide level, our PCR primers were designed specifically to amplify only JCV T-Ag Mad-1Ref sequences (31). To ensure that our real-time PCR results were accurate; the specificity of T-Ag detection was confirmed by PCR electrophoresis, immunohistochemistry and sequencing. The results showed the presence of the virus in neoplastic tissues and non-cancerous tumor adjacent to the cancerous tissues though the viral load was not significantly different which is consistent with other reports (26, 39). Similar to previous studies in the gastrointestinal tract and in colon cancer, in the present study, we detected Mad-1 or type I-R form of JCV strain in gastric cancer tissue (31).
JCV might not necessarily be the main cause of gastric cancer but it can contribute, to some extent, to development of cancerous cells at one or several stages of tumor progression. To ensure the validity of the results, further molecular, cellular and in vivo investigations are encouraged to evaluate the role of T-Ag as a potential factor involved in pathways leading to the development or progression of cancer. Such studies with large numbers of cases at different stages, including different JCV strains and genotypes will be needed to address the efficacy and possible mechanisms in the field of JCV carcinogenesis.
Some limitations of this study should be noted. First, like many patient-based studies, lack of underlying confounding data should be mentioned. Second, like some other studies, DNA extraction efficacy was determined spectrophotometrically and we did not use a more accurate method of monitoring DNA extraction using extraction of defined number of plasmids. Also, extended sample size could be beneficial. Technically we were unable to include healthy gastric tissue samples. Although the presence of virus in a healthy control sample would not entirely rule out its possible role in tumorigenesis and vice versa, its absence in healthy controls cannot fully confirm the tumorigenicity, however, it could provide supporting data. It should be noted that in the present study, we showed the existence of the virus and we cannot conclude a cause and effect relationship. However, due to tumorigenic features of the virus, the observed association might support a causal relationship. Additional supportive data and multicentral longitudinal cause and effect studies are needed to make a definite conclusion.
To summarize, the current results clearly indicate the presence of JC polyomavirus sequence in human gastric mucosa independent to the copy number. The study is important from the point of view that it was the first attempt at detecting JCV in gastric carcinoma in the Iranian population and provides supportive data for possible role of JC in gastric cancer. Further investigations to explore the extent of its contribution to tumorogenesis is still needed.
ACKNOWLEDGEMENTS
This study was supported by grant No 940386 from Mashhad University of Medical Sciences.
REFERENCES
- 1.Rugge M, Fassan M, Graham DY. Epidemiology of gastric cancer. In: Gastric Cancer. Springer; 2015: 23–34. [Google Scholar]
- 2.Malekzadeh R, Derakhshan MH, Malekzadeh Z. Gastric cancer in Iran: epidemiology and risk factors. Arch Iran Med 2009; 12:576–583. [PubMed] [Google Scholar]
- 3.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—first American cancer society award lecture on cancer epidemiology and prevention. Cancer Res 1992; 52:6735–6740. [PubMed] [Google Scholar]
- 4.Plummer M, Franceschi S, Muñoz N. Epidemiology of gastric cancer. IARC Sci Publ 2004; (157):311–326. [PubMed] [Google Scholar]
- 5.Takano Y, Kato Y, Saegusa M, Mori S, Shiota M, Masuda M, et al. The role of the Epstein-Barr virus in the oncogenesis of EBV (+) gastric carcinomas. Virchows Arch 1999; 434:17–22. [DOI] [PubMed] [Google Scholar]
- 6.Shin SK, Li MS, Fuerst F, Hotchkiss E, Meyer R, Kim T, et al. Oncogenic T-antigen of JC virus is present frequently in human gastric cancers. Cancer 2006; 107:481–488. [DOI] [PubMed] [Google Scholar]
- 7.Khalili K, Del Valle L, Otte J, Weaver M, Gordon J. Human neurotropic polyomavirus, JCV, and its role in carcinogenesis. Oncogene 2003; 22:5181–5191. [DOI] [PubMed] [Google Scholar]
- 8.Haggerty S, Walker D, Frisque R. JC virus-simian virus 40 genomes containing heterologous regulatory signals and chimeric early regions: identification of regions restricting transformation by JC virus. J Virol 1989; 63:2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalili K, Sariyer IK, Safak M. Small tumor antigen of polyomaviruses: role in viral life cycle and cell transformation. J Cell Physiol 2008; 215:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang H, Wang M, Tsai R-T, Lin H-S, Huan J-S, Wang W-C, et al. High incidence of JC viruria in JC-seropositive older individuals. J Neurovirol 2002, 8:447–451. [DOI] [PubMed] [Google Scholar]
- 11.Ling PD, Lednicky JA, Keitel WA, Poston DG, White ZS, Peng R, et al. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14-month longitudinal study. J Infect Dis 2003; 187:1571–1580. [DOI] [PubMed] [Google Scholar]
- 12.Shah KV, Daniel RW, Strickler HD, Goedert JJ. Investigation of human urine for genomic sequences of the primate polyomaviruses simian virus 40, BK virus, and JC virus. J Infect Dis 1997; 176:1618–1621. [DOI] [PubMed] [Google Scholar]
- 13.Del Valle L, Gordon J, Assimakopoulou M, Enam S, Geddes JF, Varakis JN, et al. Detection of JC virus DNA sequences and expression of the viral regulatory protein T-antigen in tumors of the central nervous system. Cancer Res 2001; 61:4287–4293. [PubMed] [Google Scholar]
- 14.Del Valle L, White MK, Enam S, Oviedo SP, Bromer MQ, Thomas RM, et al. Detection of JC virus DNA sequences and expression of viral T antigen and agnoprotein in esophageal carcinoma. Cancer 2005; 103:516–527. [DOI] [PubMed] [Google Scholar]
- 15.Laghi L, Randolph AE, Chauhan D, Marra G, Major EO, Neel JV, et al. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci U S A 1999; 96:7484–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan CS, Tremblay JD, Fewell SW, Lewis JA, Brodsky JL, Pipas JM. Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol Cell Biol 2000; 20:5749–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol 2003; 77:10638–10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dörries K. New aspects in the pathogenesis of polyomavirus-induced disease. Adv Virus Res 1997, 48:205–261. [DOI] [PubMed] [Google Scholar]
- 19.Dyson N, Bernards R, Friend S, Gooding L, Hassell J, Major E, et al. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol 1990; 64:1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bollag B, Chuke W, Frisque RJ. Hybrid genomes of the polyomaviruses JC virus, BK virus, and simian virus 40: identification of sequences important for efficient transformation. J Virol 1989; 63:863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staib C, Pesch J, Gerwig R, Gerber JK, Brehm U, Stangl A, et al. p53 inhibits JC virus DNA replication in vivo and interacts with JC virus large T-antigen. Virology 1996; 219:237–246. [DOI] [PubMed] [Google Scholar]
- 22.Niv Y, Goel A, Boland CR. JC virus and colorectal cancer: a possible trigger in the chromosomal instability pathways. Curr Opin Gastroenterol 2005; 21:85–89. [PubMed] [Google Scholar]
- 23.Nosho K, Shima K, Kure S, Irahara N, Baba Y, Chen L, et al. JC virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia 2009; 11:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neel JV. The Colonel Harlan D. Sanders award address for 1998: JC virus and its possible role in oncogenesis. Am J Med Genet 1999; 83:152–156. [PubMed] [Google Scholar]
- 25.Rutledge RG, Stewart D. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol 2008; 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai Y, Zheng HC, Aziz HOA, Mei H, Kutsuna T, Nakanishi Y, et al. High JC virus load in gastric cancer and adjacent non-cancerous mucosa. Cancer Sci 2007; 98:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricciardiello L, Laghi L, Ramamirtham P, Chang CL, Chang DK, Randolph AE, et al. JC virus DNA sequences are frequently present in the human upper and lower gastrointestinal tract. Gastroenterology 2000; 119:1228–1235. [DOI] [PubMed] [Google Scholar]
- 28.Yustein AS, Harper JC, Petroni GR, Cummings OW, Moskaluk CA, Powell SM. Allelotype of gastric adenocarcinoma. Cancer Res 1999; 59:1437–1441. [PubMed] [Google Scholar]
- 29.Ricciardiello L, Baglioni M, Giovannini C, Pariali M, Cenacchi G, Ripalti A, et al. Induction of chromosomal instability in colonic cells by the human polyomavirus JC virus. Cancer Res 2003; 63:7256–7262. [PubMed] [Google Scholar]
- 30.Martini F, Iaccheri L, Lazzarin L, Carinci P, Corallini A, Gerosa M, et al. SV40 early region and large T antigen in human brain tumors, peripheral blood cells, and sperm fluids from healthy individuals. Cancer Res 1996; 56:4820–4825. [PubMed] [Google Scholar]
- 31.Ricciardiello L, Chang DK, Laghi L, Goel A, Chang CL, Boland CR. Mad-1 is the exclusive JC virus strain present in the human colon, and its transcriptional control region has a deleted 98-base-pair sequence in colon cancer tissues. J Virol 2001; 75:1996–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalkias S, Gorham JM, Mazaika E, Parfenov M, Dang X, DePalma S, et al. ViroFind: A novel target-enrichment deep-sequencing platform reveals a complex JC virus population in the brain of PML patients. PLoS One 2018; 13(1):e0186945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bozorgi SM, Tahaei SME, Mohebbi SR, Sahba N, Damavand B, Romani S, et al. Molecular prevalence of JC virus in Tehran, Iran. Gastroenterol Hepatol Bed Bench 2012; 5:84–89. [PMC free article] [PubMed] [Google Scholar]
- 34.Mojtahedi Z, Haghshenas MR, Hosseini SV, Fattahi MJ, Ghaderi A. p 53 codon 72 polymorphism in stomach and colorectal adenocarcinomas in Iranian patients. Indian J Cancer 2010; 47:31–34. [DOI] [PubMed] [Google Scholar]
- 35.Doosti A, Zamani M, Ghasemi Dehkordi P, Taheri S, Banitalebi-Dehkordi M, Mahmoudzadeh M. Association of the p53 codon 72 polymorphism with colorectal cancer in South West of Iran. Sci Res Essays 2011; 6(15). [Google Scholar]
- 36.Khopde S, Simmons DT. Simian virus 40 DNA replication is dependent on an interaction between topoisomerase I and the C-terminal end of T antigen. J Virol 2008; 82:1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matos A, Duque V, Luxo C. Topoisomerase I improve JC virus DNA detection. Mol Biol 2016; 5:2. [Google Scholar]
- 38.Aberham C, Pendl C, Gross P, Zerlauth G, Gessner M. A quantitative, internally controlled real-time PCR assay for the detection of parvovirus B19 DNA. J Virol Methods 2001; 92:183–191. [DOI] [PubMed] [Google Scholar]
- 39.Mou X, Chen L, Liu F, Lin J, Diao P, Wang H, et al. Prevalence of JC virus in Chinese patients with colorectal cancer. PLoS One 2012; 7(5):e35900. [DOI] [PMC free article] [PubMed] [Google Scholar]