Significance
We performed de novo, full-genome sequence analysis of two Populus species, North American quaking and Eurasian trembling aspen, that contain striking levels of genetic variation. Our results showed that positive and negative selection broadly affects patterns of genomic variation, but to varying degrees across coding and noncoding regions. The strength of selection and rates of sequence divergence were strongly related to differences in gene expression and coexpression network connectivity. These results highlight the importance of both positive and negative selection in shaping genome-wide levels of genetic variation in an obligately outcrossing, perennial plant. The resources we present establish aspens as a powerful study system enabling future studies for understanding the genomic determinants of adaptive evolution.
Keywords: genome assembly, natural selection, coexpression, population genetics, Populus
Abstract
The Populus genus is one of the major plant model systems, but genomic resources have thus far primarily been available for poplar species, and primarily Populus trichocarpa (Torr. & Gray), which was the first tree with a whole-genome assembly. To further advance evolutionary and functional genomic analyses in Populus, we produced genome assemblies and population genetics resources of two aspen species, Populus tremula L. and Populus tremuloides Michx. The two aspen species have distributions spanning the Northern Hemisphere, where they are keystone species supporting a wide variety of dependent communities and produce a diverse array of secondary metabolites. Our analyses show that the two aspens share a similar genome structure and a highly conserved gene content with P. trichocarpa but display substantially higher levels of heterozygosity. Based on population resequencing data, we observed widespread positive and negative selection acting on both coding and noncoding regions. Furthermore, patterns of genetic diversity and molecular evolution in aspen are influenced by a number of features, such as expression level, coexpression network connectivity, and regulatory variation. To maximize the community utility of these resources, we have integrated all presented data within the PopGenIE web resource (PopGenIE.org).
The genus Populus comprises ∼30 species, including poplars, cottonwoods, and aspens. Populus has a distribution spanning the Northern Hemisphere, and numerous species and hybrids have been extensively planted globally. Poplars and aspens are pioneer species with among the most rapid growth of any temperate tree species, partly as a result of their characteristic heterophyllous growth. These traits, which are enhanced in interspecific hybrids, render poplars of commercial value with end uses that include biofuel, fiber, timber, bioremediation, and animal feed (1). Poplars are readily amenable to vegetative propagation and, consequently, have been closely associated with agriculture since before the Middle Ages, having been used as windbreaks, to prevent soil erosion and to stabilize river banks. As a result of the relatively small genome (<500 Mbp), suitability for efficient genetic transformation, ease of propagation in tissue culture, and rapid growth, Populus has been firmly established as an important model system for studies of forest tree species with a mature set of genetic and genomic resources (2, 3). Moreover, poplars are “replete with genetic variation at many different levels: among sections within the genus, as well as among species, provenances, populations, individuals, and genes” (4) as a result of their obligate outcrossing (dioecious) nature and airborne dispersal of pollen and seed. These characteristics render Populus an ideal system for advancing our understanding of the transition from juvenile to mature and reproductive phases (5), the genetic architecture underlying natural variation of complex phenotypes, studies of local adaptation (6–9), and studies of the divergence continuum (10, 11). Such studies necessitate available genome assemblies and corresponding genomics and population genetics resources.
Here, we present de novo assembles for two aspen species, Populus tremula and Populus tremuloides. In comparison to the high-quality assembly of the Populus trichocarpa genome, our study provides a starting point for comparative and evolutionary genomics in the field of forest trees. Despite many attributes that differentiate aspens and other poplars, we found that their genomes are remarkably conserved across species. However, the two aspen species display substantially higher levels of heterozygosity both within and between individuals compared with P. trichocarpa. Thus, in addition to examining gene and genome evolution in Populus, the genomes of these species provide an excellent model for studying how evolutionary processes affect patterns of genetic variation across genomes. Access to the dataset of these Populus genome sequences will not only facilitate studies concerning adaptive evolution in natural plant populations but also accelerate the pace at which the untapped reservoir of adaptive genes can be exploited to understand how widespread forest trees may respond to future climate change.
Results and Discussion
Genome Assembly and Annotation.
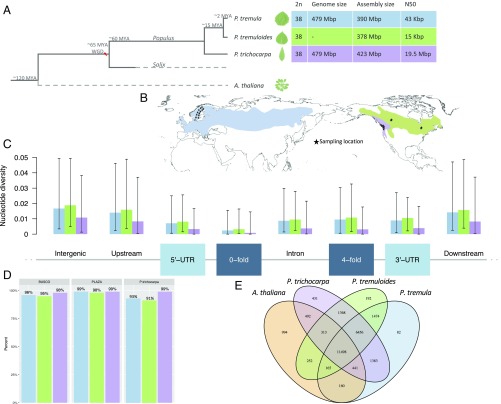
Here, we report the results of the genome assembly, gene, and transposable element (TE) annotation of two aspen species, P. tremula and P. tremuloides (Fig. 1A and SI Appendix for all methods and details); comparative analyses of the two aspens to P. trichocarpa; and resequencing-based analyses of individuals from all three species (Fig. 1B). The Populus section, containing the aspens, diverged from the Tacamachaca section, containing the previously sequenced P. trichocarpa (12), ∼15 Mya (13). Based on read alignments, we estimated that there is ∼2% divergence between P. tremula and P. trichocarpa within coding regions (indicated by alignment mismatches). However, >50% of P. tremula reads could not be aligned to the P. trichocarpa genome, indicating extensive divergence between the two species within noncoding regions (SI Appendix, section 2.5.3). The aspen and P. trichocarpa genomes are of similar size, at about 480 Mbp (SI Appendix, Dataset S2), but a limiting factor for the aspen genome assemblies was the markedly higher nucleotide diversity, as revealed both by the k-mer spectra of unassembled reads and alignment of population resequencing reads (Fig. 1C and SI Appendix, Fig. S2.1). We assembled the P. tremula genome using a hybrid approach that merged 454 and two assemblies of Illumina short-read, paired-end (PE) libraries in a stepwise manner, the result of which was subsequently scaffolded using linking [mate-pair (MP)] libraries (SI Appendix, Fig. S2.3). The P. tremuloides genome was produced using a single assembly of Illumina short-read, PE libraries. Both assemblies contained >100,000 scaffolds and had relatively low contiguity (Table 1). Despite the aspen assemblies being relatively fragmented, 93% of P. trichocarpa (v3.0) genes were represented within the P. tremula assembly (SI Appendix, section 3.3.2), with the BUSCO (14) and PLAZA (15) gene sets further supporting high contiguity and completeness of the gene space in both aspen species (Fig. 1D and SI Appendix, section 3.3). Annotation of the gene space of P. tremula and P. tremuloides identified 35,984 and 36,830 genes, respectively (Table 1 and SI Appendix, section 3.2). Using ab initio and evidence-based methods to identify and annotate repetitive elements in the two aspen genomes (SI Appendix, section 3.1), we found that the majority of elements were long terminal repeat (LTR) TEs from the Ty1-Copia and Ty3-Gypsy families, in addition to a large number of unclassified TEs (Table 1). A phylogenetic analysis of the Ty1-Copia and Ty3-Gypsy families revealed no evidence for any species- or section-specific family members (SI Appendix, Figs. S3.1 and S3.2), demonstrating that the LTR complement predates the divergence of these species. Although long interspersed nuclear elements are rare in all of the genome, two clear examples of postspeciation amplification were identified for P. trichocarpa and, to a more limited extent, for P. tremula (SI Appendix, Fig. S3.3).
Fig. 1.
Genome overview. (A) Simplified phylogram representing estimated divergence times (11, 13) and assembly statistics for the two genome assemblies presented here (P. tremula and P. tremuloides), with P. trichocarpa and A. thaliana indicated for reference. P. trichocarpa assembly statistics are based on Joint Genomes Institute genome release v3.0 of the Joint Genomes Institute. Both genomes were obtained from the Phytozome resource (https://phytozome.jgi.doe.gov/pz/portal.html). (B) Sampling localities (black stars) and distribution of P. tremula, P. tremuloides, and P. trichocarpa. (C) Nucleotide diversity in various genomic contexts calculated from alignments of resequencing data from 24 P. tremula, 22 P. tremuloides, and 24 P. trichocarpa individuals aligned to the corresponding genome assembly. (D) Percentage of genes represented in each Populus genome for BUSCO, PLAZA, and P. trichocarpa v3.0 gene sets. (E) Venn diagram representation of shared and unique gene families among P. tremula, P. tremuloides, P. trichocarpa, and A. thaliana.
Table 1.
Genome assembly, repeat, and gene space annotation summary statistics for P. tremula and P. tremuloides
| Assembly | P. tremula | P. tremuloides |
| No. of scaffolds | 216,318 | 164,504 |
| Total size of scaffolds, bp | 390,124,095 | 377,489,497 |
| No. of scaffolds >500 bp | 57,475 | 59,039 |
| No. of scaffolds >1,000 bp | 31,806 | 39,866 |
| No. of scaffolds >10,000 bp | 5,161 | 10,248 |
| No. of scaffolds >100,000 bp | 687 | 28 |
| N50 bp | 42,844 | 15,222 |
| Repeat annotation (% values) | ||
| Total | 21.54 | 22.09 |
| Ty1-copia | 4 | 4.02 |
| Ty3-gypsy | 7.4 | 7.2 |
| Other-LTR | 0.13 | 0.11 |
| LINEs | 0.38 | 0.35 |
| SINEs | 0.36 | 0.38 |
| DNA | 3.28 | 3.43 |
| NHF | 5.99 | 6.6 |
| Gene annotation (counts) | ||
| High/low-confidence gene loci | 29,252/6,057 | 26,842/8,852 |
| High/low-confidence transcripts | 76,557/8,312 | 34,439/13,899 |
| High/low gene loci expressed | 27,825/4,833 | NA/NA |
LINEs, long interspersed nuclear elements; N50, scaffold length for which at least half of the nucleotides in the assembly belong to scaffolds with the N50 length or longer; NA, not applicable; NHF, no hit found, i.e., elements that are LTR-RTs related but do not have significant similarity with the major families; SINEs, short interspersed nuclear element.
Comparative Genomics Identifies Extensive Conservation of the Populus Gene Space.
On the basis of cross-species gene alignments, 37,238 P. trichocarpa genes were commonly aligned to both aspen species, whereas 1,127 genes were identified as putatively aspen-specific. In addition, 136 P. tremula, 146 P. tremuloides, and 536 P. trichocarpa genes were putatively species-specific (SI Appendix, section 4.2). A gene family clustering analysis placed >90% of genes into ∼22,000 gene families, with 17,954 gene families shared among all three Populus species (Fig. 1E and SI Appendix, section 5.1). We identified 2,246 genes potentially under diversifying selection between aspen and P. trichocarpa [ratio of synonymous to nonsynonymous number of nucleotide substitutions per site (Ka/Ks) > 1], which were enriched for gene ontology (GO) process categories, including “regulation of transcription,” “gene expression,” “biosynthetic process,” and “metabolic process” (SI Appendix, Dataset S1). Among these genes, there were a number of transcription factors from the NAC, MYB, YABBY, bHLH, and WRKY transcription factor families in addition to genes involved in cell wall biogenesis (xyloglucan endotransglucosylase/hydrolase and expansin), abiotic (LEA, heat shock proteins, and RD22) and biotic stress (LRR and disease resistance proteins), cell cycle and developmental regulators [cyclins, epidermal patterning factor, floral time, and homeotic control proteins (CONSTANS-like)], lipid transfer proteins, and an extensive number of proteins of unknown function. We examined the expression characteristics of genes with evidence of selection using population-wide RNA-sequencing data from the Swedish Aspen (SwAsp) collection (16) and the P. tremula tissue expression atlas (17). In the SwAsp gene coexpression network, genes potentially under diversifying selection on the basis of Ka/Ks between the two aspen species had slightly lower within-module (Mann–Whitney test, P = 0.001) and global (Mann–Whitney test, P = 0.01) network connectivity. This suggests that highly connected genes experience lower levels of diversifying selection than genes with fewer connections. The same was also true for genes potentially under diversifying selection on the basis of Ka/Ks from P. trichocarpa, but with a slightly more pronounced effect (Mann–Whitney kdiff_norm P = 0.0006 and kTotal P = 0.03; where kTotal is the total connectivity for a gene within the network and kdiff_norm is the difference between kWithin, which is connectivity of a gene within its assigned module, and kOut, which is the difference between kTotal and kWithin, scaled for module size). This potentially reflects the greater divergence time between P. trichocarpa and P. tremula. Genes potentially under diversifying selection on the basis of Ka/Ks also had significantly higher absolute effect sizes for associated expression quantitative trait loci (eQTLs) (16), both for genes diverged between the two aspen species (Mann–Whitney P < 2.2e-16) and between P. trichocarpa and P. tremula (Mann–Whitney P < 2.2e-16). This is in agreement with the observation that eQTLs were enriched in genes with lower coexpression network connectivity at the periphery of the coexpression network (16). Together, these results indicate that genes with lower network connectivity are more likely to experience diversifying selection, and therefore to contribute to divergence among species driven by expression or regulatory modulation.
We performed comparisons of the genome sequences of the three species using an unsupervised method (18) that combines hidden Markov models and self-organizing maps to segment the individual genomes into unique phylogenetic topologies (SI Appendix, section 5.4). From this, we identified regions exhibiting distinct local topologies congruent with the species taxonomy (SI Appendix, Fig. S5.5) that represent genomic regions of maximal divergence among these species. Functional enrichment testing of genes within these regions identified categories involved in disease resistance (SI Appendix, Table S5.2) for seven of the 12 longest regions, indicating distinct evolutionary pressure on the biotic stress response during the divergence of these species.
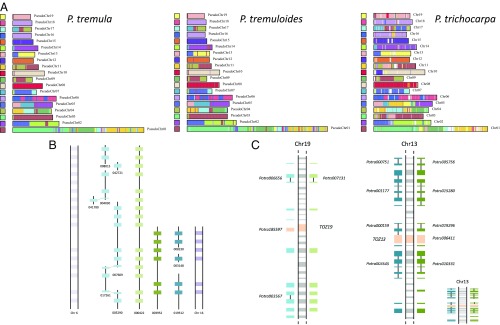
Genetic maps suggest large-scale macrosynteny between aspens and P. trichocarpa (19). By performing whole-genome alignments between P. tremula and P. trichocarpa, we identified commonly retained paralogous regions arising from the Salicaceae whole-genome duplication (WGD) (Fig. 2A), suggesting that the majority of genomic rearrangements following the WGD likely occurred before the split of the two sections. Despite the fragmented nature of the current aspen assemblies, we observed examples of retained local synteny among paralogous regions of the three genomes (Fig. 2B and SI Appendix, section 5.5). Despite the overall similarities of the genomes, aspens are reproductively isolated from all other sections of the Populus genus. One possible contributing factor is the sex determination region, which is located in the pericentromeric region of chromosome 19 (19, 20) in aspens, in contrast to the peritelomeric location in all other poplars (21). Within the aspen sex determination region, the TOZ19 (TORMOZEMBRYO DEFECTIVE) gene has previously been shown to have male-specific expression in P. tremula, with females having a degenerated copy (22, 23). Many of the P. trichocarpa genes in the region of TOZ19 lacked a detectable ortholog in either of our aspen assemblies (Fig. 2C). Assembly of this region was highly fragmented in both aspens, with many scaffolds containing only a single gene or gene fragment. It is important to note that we sequenced a female P. tremula individual; thus, the current assembly lacks the male-specific sequences expected in an XY sex determination system. The region around TOZ19 has unusually low gene density, with the 12 nearest upstream and downstream genes spanning 1.67 Mbp in contrast to 322 Kbp flanking the homologous TOZ13 locus (Fig. 2C), which is also located within the pericentromeric region of chromosome 13 (Fig. 2C). Further research to elucidate the role of this region in sex determination is needed, including male-specific assemblies.
Fig. 2.
Genome synteny. (A) Self-alignments of nucleotide sequences for the P. tremula, P. tremuloides, and P. trichocarpa genomes, showing that paralogous regions from the Salicaceae WGD event are largely retained across the three genomes. Synteny matches following a WGD event are indicated by colored blocks. (B) Schematic representation of scaffold Potra00422, with genes shown as light green boxes, and orthologs in P. tremuloides (light blue) and P. trichocarpa (light purple), illustrating an example of retained local synteny between the three genomes. Paralogs are colored in darker shades of each color (to the right side in the representation). Dotted lines between some of the genes in P. tremuloides indicate that the gene is split across different scaffolds. (C) Schematic representation of the sex-determining region in aspen. (Left) Middle shows TOZ19 and 12 genes upstream and downstream. On both sides, the orthologs (detected using both BLAST and conserved synteny) in P. tremula and P. tremuloides are depicted. (Right) TOZ19 paralog, TOZ13, accompanied by 12 genes on each side. Scaffold IDs are noted for the TOZ19 and TOZ13 genes, as well as in cases of more than one gene per scaffold. Dotted lines indicate paralogs in P. trichocarpa. (Inset, Bottom Right) Region on chromosome 13 drawn to the same scale used for the region on chromosome 19.
Extensive Genetic Variation and Widespread Natural Selection in Aspens.
To better characterize genome-wide patterns of nucleotide diversity, linkage disequilibrium (LD), and recombination, and to further understand mechanisms of adaptive evolution in these species, we analyzed whole-genome resequencing data from 24 P. tremula (11), 22 P. tremuloides (11), and 24 P. trichocarpa (7) individuals (SI Appendix, section 7). We aligned reads from each individual to their respective assembled genome, which enabled us to access a greater proportion of the genomes after filtering (77.6% for P. tremula and 82.1% for P. tremuloides) than alignments to the P. trichocarpa assembly (42.8%; SI Appendix, section 7). We observed that both aspen species harbor substantially higher genome-wide levels of genetic diversity compared with P. trichocarpa (Fig. 1B and SI Appendix, Table S7.2). This can likely be ascribed to different demographic histories and larger distribution ranges that collectively lead to a higher effective population size (Nes) in the two aspen species compared with P. trichocarpa (11, 24). The allele frequency spectrum of polymorphic sites also differed between species, with a more pronounced excess of low-frequency polymorphisms (negative Tajima’s D value) in aspens than in P. trichocarpa (SI Appendix, Table S7.3). This is in accordance with analyses indicating that both aspen species have experienced recent range expansions following the last glaciation (25). Nucleotide diversity showed consistent patterns of variation across coding and noncoding regions in the three species, with diversity being approximately three- to fourfold higher at fourfold synonymous and noncoding sites than at zerofold nonsynonymous sites (Fig. 1B and SI Appendix, Table S7.2). Furthermore, population-scaled recombination rates were inferred to be substantially higher in aspens, particularly in P. tremuloides (SI Appendix, Fig. S7.3), which had the lowest levels of LD (SI Appendix, Fig. S7.2), likely reflecting historical differences in effective population sizes between aspens and P. trichocarpa. Interestingly, genomic regions in close proximity to genes exhibited the highest recombination rates in all three species, (SI Appendix, Fig. S7.3). We also identified insertions and deletions (INDELs) using the same resequencing data (SI Appendix, section 8), finding that >50% of identified INDELs were common to P. tremula and P. tremuloides, suggesting that they occurred after the Populus section split from the Tacamahaca section but before the speciation event that separated P. tremula and P. tremuloides ∼2.3 Mya (11). Short INDELs (<100 bp) were rare in coding regions (1–2%), with a higher proportion of long INDELs (>100 bp) affecting coding regions (15.2%), and, similar to SNPs, there was an excess of low-frequency variants at INDELs, as indicated by negative Tajima’s D values (SI Appendix, Fig. S8.3).
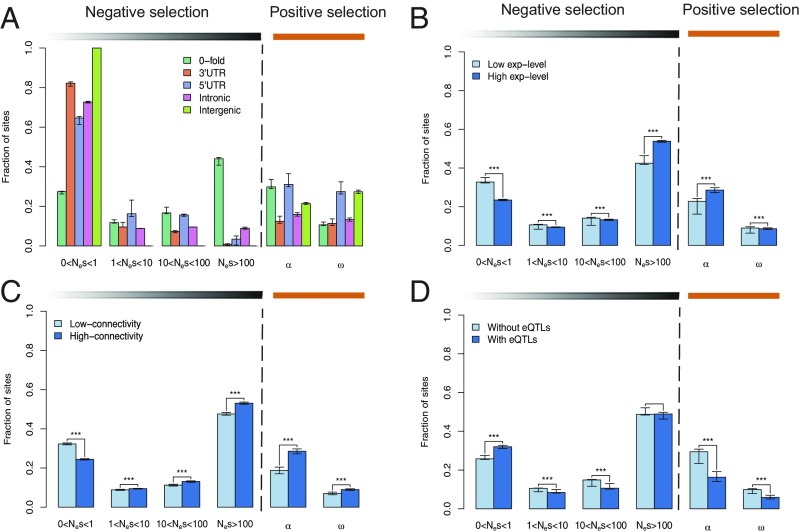
We further used the DFE-alpha approach (26, 27), which jointly infers demographic and selective parameters using polymorphism and divergence data, to quantify the impact of negative and positive selection on sites located in different genomic contexts in the two aspen species (SI Appendix, section 7.2). For both aspen species, we found that negative selection was substantially stronger in coding than noncoding regions, with more than 40% of zerofold nonsynonymous sites being subject to strong negative selection (Nes > 100; Fig. 3A and Table 2). In noncoding regions, 5′ UTRs showed stronger negative selection than other regions (Fig. 3A), with ∼30% of 5′ UTRs under moderate negative selection (1 < Nes < 100) in both aspen species. In addition, we found widespread evidence of positive selection in coding regions, where a high proportion of divergence at zerofold sites has been driven to fixation (α = 30–40%), corroborating a previous study based on far fewer genes (6). Similarly, in both aspen species, there were high proportions of substitutions fixed due to positive selection (α = 30–40%) and higher rates of adaptive substitutions relative to neutral divergence in 5′ UTRs (ω ∼ 30%; Fig. 3A), mirroring results from Drosophila melanogaster (5) and Capsella grandiflora (28). Through analysis of orthologous promoter regions among the three species, we found that genes with highly conserved upstream and downstream regions were enriched for DNA binding activity (SI Appendix, section 4.3), suggesting that these regions are functionally important, and hence provide an explanation for why they are subject to both strong purifying selection and adaptive evolution (Fig. 3A). We caution that estimation of α could be biased if ancient demographic fluctuations are not sufficiently well captured by current polymorphism data (29).
Fig. 3.
Population genetics for P. tremula. (A) Estimates of negative and positive selection on coding and noncoding regions, separated by site type. Error bars represent 95% bootstrap confidence intervals. Estimates of negative and positive selection on zerofold nonsynonymous sites in genes with varying expression level (B), varying connectivity level in coexpression network (C), and with or without eQTLs (D) are shown. Nes categories represent different bins of negative selection strength. α, proportion of divergent sites fixed by positive selection; ω, rate of adaptive substitution relative to neutral divergence. All calculations are based on genomic resequencing of 94 P. tremula individuals with reads aligned to the P. tremula genome assembly. ***P < 0.001.
Table 2.
Estimates of the distribution of fitness effects of new mutations at zerofold nonsynonymous sites, intronic sites, 5′ UTR sites, 3′ UTR sites, and intergenic sites falling in different Nes ranges, and proportion of divergence driven to fixation by positive selection (α) and the rate of adaptive substitution relative to neutral divergence (ω) in P. tremula and P. tremuloides
| Percentage of mutations in Nes range | |||||||
| Species | Category | 0–1 | 1–10 | 10–100 | >100 | α | ω |
| P. tremula | Zerofold | 0.275 (0.263–0.277) | 0.118 (0.115–0.132) | 0.167 (0.163–0.196) | 0.440 (0.408–0.447) | 0.298 (0.289–0.335) | 0.107 (0.103–0.121) |
| 3′ UTR | 0.822 (0.807–0.829) | 0.096 (0.096–0.119) | 0.073 (0.067–0.077) | 0.009 (0.003–0.011) | 0.125 (0.115–0.151) | 0.114 (0.104–0.137) | |
| 5′ UTR | 0.646 (0.614–0.653) | 0.163 (0.148–0.232) | 0.157 (0.150–0.161) | 0.034 (0.005–0.050) | 0.311 (0.299–0.365) | 0.275 (0.263–0.324) | |
| Intronic | 0.727 (0.722–0.731) | 0.088 (0.088–0.089) | 0.096 (0.096–0.096) | 0.089 (0.085–0.095) | 0.159 (0.151–0.167) | 0.133 (0.125–0.141) | |
| Intergenic | 1.000 (1.000–1.000) | 0.000 (0.000–0.000) | 0.000 (0.000–0.000) | 0.000 (0.000–0.000) | 0.215 (0.210–0.220) | 0.273 (0.265–0.282) | |
| P. tremuloides | Zerofold | 0.264 (0.251–0.266) | 0.131 (0.128–0.147) | 0.194 (0.190–0.227) | 0.411 (0.376–0.416) | 0.338 (0.330–0.374) | 0.122 (0.118–0.135) |
| 3′ UTR | 0.853 (0.834–0.858) | 0.095 (0.094–0.120) | 0.051 (0.046–0.056) | 0.001 (0.000–0.002) | 0.094 (0.084–0.132) | 0.086 (0.076–0.120) | |
| 5′ UTR | 0.632 (0.601–0.643) | 0.225 (0.207–0.278) | 0.140 (0.123–0.147) | 0.004 (0.000–0.007) | 0.345 (0.329–0.385) | 0.305 (0.289–0.342) | |
| Intronic | 0.748 (0.713–0.752) | 0.091 (0.090–0.140) | 0.096 (0.095–0.124) | 0.065 (0.023–0.069) | 0.140 (0.133–0.197) | 0.118 (0.111–0.165) | |
| Intergenic | 1.000 (1.000–1.000) | 0.000 (0.000–0.000) | 0.000 (0.000–0.000) | 0.000 (0.000–0.000) | 0.207 (0.200–0.213) | 0.261 (0.250–0.271) | |
Ninety-five percent bootstrap confidence intervals are shown in parentheses.
Gene Expression Characteristics Influence Functional Constraint and Adaptive Evolution.
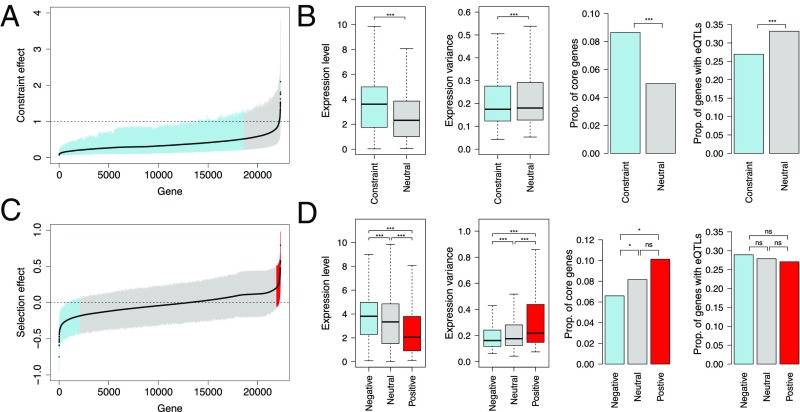
As gene expression and regulation changes have been postulated to be key determinants of rates of adaptive evolution (30, 31), we tested for the relative contributions of negative and positive selection in driving differences in rates of molecular evolution for genes with different expression profiles (SI Appendix, section 7.3). To investigate this, we binned all expressed genes (22,306 genes) from population-wide RNA-sequencing data in winter buds undergoing bud flush of P. tremula (16) into two subsets according to four different features related to gene expression. We used two complementary approaches derived from the McDonald–Kreitman test (32) (SI Appendix, section 7.3) to quantify the relative contributions of negative and positive selection on rates of molecular evolution in genes with different expression profiles. First, we used the DFE-alpha method (26, 27) to estimate the distribution of fitness effects of both coding and noncoding mutations in P. tremula genes (22,306 genes) that have been characterized by various expression features in an earlier eQTL study (16). We also used a generalized linear mixed model (33), which explicitly incorporates the genome-wide effects as fixed effects and individual gene effects as random effects, to separately estimate the constraint effect (f) and selection effect (γ) for genes. We observed that genes with high expression levels and/or high coexpression network connectivity had a significantly higher proportion of zerofold nonsynonymous sites in the strongest category of negative selection compared with genes with low expression and/or low centrality (Fig. 3 B and C). This suggests stronger negative selection acting on nonsynonymous sites in more highly expressed and highly connected genes (16, 34). Additionally, the strength of negative selection acting on these genes was even stronger after controlling for differences in selection on synonymous sites (SI Appendix, section 7.3 and Dataset S2). These patterns were also supported by the regression model, which showed that most expressed genes in the P. tremula genome have evolved under selective constraint (18,612 of 22,306 genes; Fig. 4A), and that genes evolving under selective constraints have significantly higher expression levels and contained a greater fraction of core genes [hubs in coexpression network modules (16)] than genes that are evolving neutrally (Fig. 4B). In comparison to the clear evidence observed at nonsynonymous sites, selection on noncoding sites was not consistent before and after controlling for the synonymous site selection, which could be due, in part, to the general weaker purifying selection on noncoding than coding regions (SI Appendix, section 7.3 and Dataset S2). We thus focused on nonsynonymous sites in the remainder of our analyses.
Fig. 4.
Selection and constraint effects in 22,306 expressed genes that have been characterized by various expression features. (A) Sizes of constraint effect in expressed genes that are sorted in increasing order. The black line follows the average, the vertical line spans the Bayesian credibility intervals, and the dashed line indicates neutrality. Genes with selective constraint effects are marked in blue, and those with neutrality are marked in gray. (B) Characterization of gene expression features [from left to right: expression level, expression variance, proportion (Prop.) of genes that are core genes, Prop. of genes that harbor eQTLs] for genes evolved under selective constraint (blue) and evolved neutrally (gray). (C) Sizes of selective effects in expressed genes that are sorted in increasing order. The black line follows the average, the vertical line spans the Bayesian credibility intervals, and the dashed line indicates neutrality. Genes with negative selection are marked in blue, genes with neutrality are marked in gray, and genes with positive selection are marked in red. (D) Characterization of gene expression features (from left to right: expression level, expression variance, Prop. of genes that are core genes, Prop. of genes that harbor eQTLs) for genes with negative (blue), neutrality (gray), and positive (red) selection. *P < 0.05; ***P < 0.001. ns, not significant.
We found genes with high expression and high connectivity to have significantly higher proportions of adaptive nonsynonymous substitutions (α) than genes with low expression and low connectivity (Fig. 3 B and C). However, we did not observe higher rates of adaptive substitutions (ω) in genes with high expression (Fig. 3B), and there was significantly lower expression among the 395 genes identified as being under positive selection by the regression analysis (Fig. 4 C and D). Together, these results indicate that the higher α we observed for highly expressed genes most likely arises from an increased strength of negative selection acting on genes with high expression that eliminates a substantial fraction of weakly deleterious alleles rather than from an increased rate of fixation of adaptive mutations per se. In contrast to the pattern observed in expression level, genes with high connectivity show higher rates of ω compared with genes with low connectivity (Fig. 3C), and there was also a significantly higher proportion of core genes among the positively selected genes identified by the regression analysis (Fig. 4D). Although the positively selected genes from the regression analysis in P. tremula were enriched for the GO terms “signal transduction,” “cellular response to stimulus,” and “metabolic process” (SI Appendix, Table S7.7), we found that positively selected core genes were highly enriched in nonstructural carbohydrate metabolic and biosynthetic process, biotic stimulus response, and nucleosome and chromatin assembly (SI Appendix, Table S7.8), which may play critical roles in the resilience to stress and life history strategies of long-lived trees (35). Given that core genes are more likely to be required for network integrity and adaptive regulatory evolution than noncore genes (36), the characteristics of both stronger negative and positive selection on core genes may have promoted more rapid and efficient adaptation during the evolutionary history of long-lived forest trees such as aspen.
In addition to expression level and network connectivity, gene expression variation and associated regulatory loci are major contributors to phenotype variation in complex traits, and are thus likely to be subject to selection pressures (37–39). We found that coding sequence evolution in genes with high expression variance among genotypes tended to be under slightly weaker purifying selection and stronger positive selection compared with genes with low expression variance (Fig. 4 B and D and SI Appendix, Dataset S2). Our previous study showed that there is pervasive regulatory variation in P. tremula (16), and we extend this observation here by finding that genes harboring regulatory variation (with identified eQTLs; eGene) have significantly higher proportions of nearly neutral nonsynonymous mutations (Fig. 3D and SI Appendix, Dataset S2), significantly lower proportions of adaptive nonsynonymous substitutions, and lower rates of positive selection than other genes (non-eGene; Fig. 3D). In agreement with a recent study in C. grandiflora (40), our findings support the view that genes with regulatory variation, which we know are less central in the coexpression networks (16, 40), are evolving under weaker negative selection and undergo less adaptive evolution compared with genes without regulation variation. Taken together, our results show there is complex interaction between gene sequence and expression evolution in Populus, and future studies should explore the underlying driving evolutionary forces in greater detail across a wider range of plant species.
Conclusions
Here, and in other recent work, we have demonstrated the power of the aspen genome resources for understanding how natural selection varies among genes differing in gene expression (16) and how effects of linked selection vary across the genome within and among species (25) to help decipher the history of speciation in aspens (11) and identify the genomic basis of local adaptation (8). By developing an extensive genomic resource for aspen, our goal is to enable functional, comparative, and evolutionary genomic analyses in Populus and to extend the utility of the genus as a unique study system in plant and evolutionary biology. Populus has a number of features that makes it novel compared with many other model systems, such as an obligate outcrossing mating system, great longevity, abundant genomic diversity, and species of contrasting effective population size. To facilitate future studies of the many novel aspects of Populus biology, we have integrated all data generated here into the Populus Genome Integrative Explorer (PopGenIE.org) (17) web resource. These data will serve researchers performing evolutionary and comparative analyses as well as functional genomics studies aimed at deciphering genes underlying complex phenotypes through, for instance, aiding with the design of CRISPR guide-RNAs for gene editing.
Materials and Methods
Biological Materials.
P. tremula genome sequencing was performed on DNA extracted from propagated root cuttings of a single wild tree growing on the Umeå University campus (63° 49′17′′N, 20° 18′40′′E). P. tremuloides DNA was extracted from mature, freeze-dried leaves from genotype Dan2-1B7. All DNA was extracted using a DNeasy Plant Mini Kit (Qiagen). Nuclear DNA contents were estimated by propidium iodide flow cytometry.
Sequencing.
P. tremula DNA was sequenced as single-end reads using the Roche 454 platform and as PE and MP read libraries using Illumina short-read platforms. P. tremuloides DNA was sequenced using PE sequencing libraries on the Illumina HiSeq platform. Populus davidiana sequencing data were obtained from the National Institute of Forest Science, Korea. DNA was extracted from mature leaves sampled from mature trees in a common garden experiment in Suwon, Korea. We have deposited all raw sequencing data at the European Nucleotide Archive (ENA) resource (https://www.ebi.ac.uk/ena) as accession no. PRJEB23585, except for P. davidiana sequencing data, which are available from the PopGenIE.org web resource and will be described fully elsewhere.
Assembly.
We used a hierarchical approach to assemble the P. tremula that comprised separate contig assemblies of the 454 and Illumina data that were merged in a stepwise manner, with the final merged assembly subsequently scaffolded using the MP libraries. As we only had PE Illumina data available for P. tremuloides, a single PE assembly was performed. We evaluated gene space coverage of the P. tremula final and substage assemblies by aligning the primary transcripts of the P. trichocarpa reference genome to the various assemblies. We used feature response curves (41) to assess assembly correctness.
Annotation.
To annotate the repetitive fraction of the genome, we searched the P. tremula and P. tremuloides genome assemblies using RepeatScout (42) and retrieved P. trichocarpa repetitive sequences from Repbase (43), merging these to form a repeat library. We then used RepeatMasker (44) to identify repetitive elements in the genome assemblies. To annotate the protein-coding fraction of the genome, we performed gene space annotation in three steps: using MAKER (45) to generate gene annotations, which we iteratively refined using PASA (46) on the basis of four transcriptome datasets before final manual curation. To assess gene space completeness, we used CEGMA (47), BUSCO (14), PLAZA coreGF (15), and alignments of the P. trichocarpa annotation. Cross-species gene alignments were performed using GMAP (48) to identify common and species-specific genes.
Comparative Genomics.
To determine the orthologous relationship between the three Populus species, we collated protein sequences from Arabidopsis thaliana (TAIR10), P. trichocarpa (V3.0), and the two aspen species presented here, and performed an all-against-all BLASTp (49) sequence similarity search, the results of which were used to performed two rounds of clustering using TribeMCL (50) to delineate gene families. We performed multiple sequence alignments of each family and used these to calculate Ks and Ka, and their rate ratio (Ka/Ks). We aligned the aspen and P. trichocarpa genomes using Satsuma (51). To pseudoscaffold the aspen assemblies, we ordered and oriented the scaffolds from each assembly according to synteny to P. trichocarpa using Chromosemble from the Satsuma package. We then generated syntenic self-alignments using Satsuma and visualized these using ChromosomePaint (52).
Population Genetics.
We used whole-genome resequencing data from 24 genotypes of P. tremula, 22 genotypes of P. tremuloides, and 24 genotypes of P. trichocarpa to perform population genetics analyses. We used Trimmomatic (53) to remove adapter sequences and low-quality bases, after which all reads were mapped to their respective genome assembly using bwa-mem (54). SNPs were called using HaplotypeCaller from GATK (55). We used ANGSD (56) to estimate average pairwise nucleotide diversity (Θπ) and Tajima’s D for different genomic contexts (zerofold nonsynonymous, fourfold synonymous, intron, 3′ UTR, 5′ UTR, upstream and downstream regulatory regions, and intergenic sites) over nonoverlapping 1-Kbp windows. To estimate the rate of LD decay, we used PLINK (57) to randomly thin the SNPs and to calculate the squared correlation coefficients (r2) between all pairs of SNPs within a distance of 20 Kbp. Using fourfold synonymous sites as the neutral reference, we estimated the distribution of fitness effects (Nes), the proportion of fixations driven by positive selection (α), and the rate of positive selection (ω) for each category of functional elements using DFE-alpha. To quantify the impact of negative and positive selection on different genomic regions, we used read alignments and SNP calling against the P. trichocarpa reference genome by comparing the site frequency spectra and divergence of different genomic contexts with those for fourfold synonymous sites, which we assumed to be neutral. We binned all expressed genes (22,306 genes) from population-wide RNA-sequencing data in winter buds (16) into two subsets according to four different features related to gene expression to examine the relation of expression to signatures of selection. We ran an unsupervised genome-wide population analysis using Saguaro (18) to segment the genomes into different local topologies based on reads from all populations mapped to P. trichocarpa.
Structural Variants.
We used a combination of Samtools (58) and Varscan (59) to detect short INDELs within mapped reads, Control-FREEC (60) to detect copy number variants based on sequence depth, Breakdancer (61) and Delly (62) to detect structural variants (SVs) based on altered distance between mapped reads from PE data, and Lumpy (63) to integrate multiple SV signals jointly across multiple samples. All methods were run with default parameters. The output from the different methods was combined using an in-house Perl script, and only variants detected by at least two methods were kept for further analyses. Variants were annotated using ANNOVAR (64).
Supplementary Material
Acknowledgments
We thank the Swedish National Genomics Infrastructure hosted at SciLifeLab, the National Bioinformatics Infrastructure Sweden (NBIS), for providing computational assistance and the Uppsala Multidisciplinary Center for Advanced Computational Science for providing computational infrastructure. This work was supported by the Knut and Alice Wallenberg Foundation, the Umeå Plant Science Centre Berzelii Centre, the Stiftelsen för Strategisk Forskning Centre for Plant Developmental Biology, the Kempe Foundation, the Swedish Research Council Vetenskapsrådet, the Research and Development Program for Forestry Technology (Project S111414L070110) provided by Korea Forest Service, and the Stiftelsen Oscar och Lili Lamms Minne (Grant SY2013-0009). J.W. was supported by a scholarship from the Chinese Scholarship Council, B.N. and A.J. were supported by the Knut and Alice Wallenberg Foundation as part of the NBIS at SciLifeLab, P.K.I. was supported by a Young Researcher Award from Umeå University, and N.R.S. was supported by the Trees and Crops for the Future project. Y.V.d.P. acknowledges funding from the European Union Seventh Framework Programme (FP7/2007-2013) under European Research Council Advanced Grant Agreement 322739–DOUBLEUP.
Footnotes
The authors declare no conflict of interest.
Data deposition: All raw sequencing data have been deposited at the European Nucleotide Archive resource, https://www.ebi.ac.uk/ena (accession no. PRJEB23585), except for Populus davidiana sequencing data, which are available from the Populus Genome Integrative Explorer (PopGenIE.org) Lin2018 web resource (accession no. pnas201801437).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801437115/-/DCSupplemental.
References
- 1.Stettler RF, Bradshaw HD, Jr, Heilman P, Hinckley T. Biology of Populus and Its Implications for Management and Conservation. NRC Research Press; Ottawa: 1996. [Google Scholar]
- 2.Wullschleger SD, Weston DJ, DiFazio SP, Tuskan GA. Revisiting the sequencing of the first tree genome: Populus trichocarpa. Tree Physiol. 2013;33:357–364. doi: 10.1093/treephys/tps081. [DOI] [PubMed] [Google Scholar]
- 3.Street NR, Tsai C-J. 2010. Genetics and Genomics of Populus, Plant Genetics and Genomics: Crops and Models, eds Jansson S, Bhalerao R, Groover A (Springer, New York), pp 135–152.
- 4.Stettler RF, Bradshaw HD., Jr . Evolution, genetics, and genetic manipulation. In: Stettler R, Bradshaw HD Jr, Heilman P, Hinckley T, editors. Biology of Populus and Its Implications for Management and Conservation. NRC Research Press; Ottawa: 1996. pp. 1–6. [Google Scholar]
- 5.Wang J-W, et al. miRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol Biol Evol. 2010;27:650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- 7.Evans LM, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat Genet. 2014;46:1089–1096. doi: 10.1038/ng.3075. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. A major locus controls local adaptation and adaptive life history variation in a perennial plant. Genome Biol. 2018;19:72. doi: 10.1186/s13059-018-1444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holliday JA, Zhou L, Bawa R, Zhang M, Oubida RW. Evidence for extensive parallelism but divergent genomic architecture of adaptation along altitudinal and latitudinal gradients in Populus trichocarpa. New Phytol. 2016;209:1240–1251. doi: 10.1111/nph.13643. [DOI] [PubMed] [Google Scholar]
- 10.Stölting KN, et al. Genome-wide patterns of differentiation and spatially varying selection between postglacial recolonization lineages of Populus alba (Salicaceae), a widespread forest tree. New Phytol. 2015;207:723–734. doi: 10.1111/nph.13392. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Street NR, Scofield DG, Ingvarsson PK. Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Mol Biol Evol. 2016;33:1754–1767. doi: 10.1093/molbev/msw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 13.Eckenwalder JJE. Systematics and evolution of Populus. In: Stettler R, Bradshaw HD Jr, Heilman P, Hinckley T, editors. Biology of Populus and Its Implications for Management and Conservation. NRC Research Press; Ottawa: 1996. pp. 7–32. [Google Scholar]
- 14.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 15.Van Bel M, et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158:590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mähler N, et al. Gene co-expression network connectivity is an important determinant of selective constraint. PLoS Genet. 2017;13:e1006402. doi: 10.1371/journal.pgen.1006402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundell D, et al. The plant genome integrative explorer resource:PlantGenIE.org. New Phytol. 2015;208:1149–1156. doi: 10.1111/nph.13557. [DOI] [PubMed] [Google Scholar]
- 18.Zamani N, et al. Unsupervised genome-wide recognition of local relationship patterns. BMC Genomics. 2013;14:347. doi: 10.1186/1471-2164-14-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paolucci I, et al. Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet Genomes. 2010;6:863–875. [Google Scholar]
- 20.Kersten B, Pakull B, Groppe K, Lueneburg J, Fladung M. The sex-linked region in Populus tremuloides Turesson 141 corresponds to a pericentromeric region of about two million base pairs on P. trichocarpa chromosome 19. Plant Biol (Stuttg) 2014;16:411–418. doi: 10.1111/plb.12048. [DOI] [PubMed] [Google Scholar]
- 21.Geraldes A, et al. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus) Mol Ecol. 2015;24:3243–3256. doi: 10.1111/mec.13126. [DOI] [PubMed] [Google Scholar]
- 22.Robinson KM, et al. Populus tremula (European aspen) shows no evidence of sexual dimorphism. BMC Plant Biol. 2014;14:276. doi: 10.1186/s12870-014-0276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pakull B, Kersten B, Lüneburg J, Fladung M. A simple PCR-based marker to determine sex in aspen. Plant Biol (Stuttg) 2015;17:256–261. doi: 10.1111/plb.12217. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L, Bawa R, Holliday JA. Exome resequencing reveals signatures of demographic and adaptive processes across the genome and range of black cottonwood (Populus trichocarpa) Mol Ecol. 2014;23:2486–2499. doi: 10.1111/mec.12752. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Street NR, Scofield DG, Ingvarsson PK. Natural selection and recombination rate variation shape nucleotide polymorphism across the genomes of three related Populus species. Genetics. 2016;202:1185–1200. doi: 10.1534/genetics.115.183152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 28.Williamson RJ, et al. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genet. 2014;10:e1004622. doi: 10.1371/journal.pgen.1004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousselle M, Mollion M, Nabholz B, Bataillon T, Galtier N. Overestimation of the adaptive substitution rate in fluctuating populations. Biol Lett. 2018;14:20180055. doi: 10.1098/rsbl.2018.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 31.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 32.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 33.Eilertson KE, Booth JG, Bustamante CD. SnIPRE: Selection inference using a Poisson random effects model. PLOS Comput Biol. 2012;8:e1002806. doi: 10.1371/journal.pcbi.1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masalia RR, Bewick AJ, Burke JM. Connectivity in gene coexpression networks negatively correlates with rates of molecular evolution in flowering plants. PLoS One. 2017;12:e0182289. doi: 10.1371/journal.pone.0182289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartmann H, Trumbore S. Understanding the roles of nonstructural carbohydrates in forest trees–From what we can measure to what we want to know. New Phytol. 2016;211:386–403. doi: 10.1111/nph.13955. [DOI] [PubMed] [Google Scholar]
- 36.Albert R, Jeong H, Barabási A-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 37.Shaw JR, et al. Natural selection canalizes expression variation of environmentally induced plasticity-enabling genes. Mol Biol Evol. 2014;31:3002–3015. doi: 10.1093/molbev/msu241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci USA. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci USA. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steige KA, Laenen B, Reimegård J, Scofield DG, Slotte T. Genomic analysis reveals major determinants of cis-regulatory variation in Capsella grandiflora. Proc Natl Acad Sci USA. 2017;114:1087–1092. doi: 10.1073/pnas.1612561114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vezzi F, Narzisi G, Mishra B. Feature-by-feature–Evaluating de novo sequence assembly. PLoS One. 2012;7:e31002. doi: 10.1371/journal.pone.0031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saha S, Bridges S, Magbanua ZV, Peterson DG. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008;36:2284–2294. doi: 10.1093/nar/gkn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 44.Smit A, Hubley R, Green P. 1996 RepeatMasker Open-3.0. Available at www.repeatmasker.org. Accessed May 4, 2016.
- 45.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 48.Wu TD, Watanabe CK. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 49.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2008;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grabherr MG, et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010;26:1145–1151. doi: 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sundström G, Zamani N, Grabherr MG, Mauceli E. Whiteboard: A framework for the programmatic visualization of complex biological analyses. Bioinformatics. 2015;31:2054–2055. doi: 10.1093/bioinformatics/btv078. [DOI] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koboldt DC, et al. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boeva V, et al. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rausch T, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.