Significance
An aberrant covalent histone modification which drives dysregulation of transcriptional program is related to human diseases such as cancer. Thus, identifying signaling pathways modulating transcription factors and epigenetic enzymes are coming into view as attractive therapeutic targets. In this study, we provide evidence that KDM3A is tyrosine-phosphorylated by JAK2, and tyrosine-phosphorylated KDM3A acts as a coactivator for STAT3, thereby exerting increased cancer cell growth and motility. We propose that JAK2-dependent tyrosine phosphorylation of KDM3A could be a potential therapeutic target for epigenetic control of oncogenic effect governed by JAK2−STAT3 signaling pathway.
Keywords: KDM3A, JAK2, STAT3, histone demethylation, phosphorylation
Abstract
Janus tyrosine kinase 2 (JAK2)−signal transducer and activator of transcription 3 (STAT3) signaling pathway is essential for modulating cellular development, differentiation, and homeostasis. Thus, dysregulation of JAK2−STAT3 signaling pathway is frequently associated with human malignancies. Here, we provide evidence that lysine-specific demethylase 3A (KDM3A) functions as an essential epigenetic enzyme for the activation of JAK2−STAT3 signaling pathway. KDM3A is tyrosine-phosphorylated by JAK2 in the nucleus and functions as a STAT3-dependent transcriptional coactivator. JAK2−KDM3A signaling cascade induced by IL-6 leads to alteration of histone H3K9 methylation as a predominant epigenetic event, thereby providing the functional and mechanistic link between activation of JAK2−STAT3 signaling pathway and its epigenetic control. Together, our findings demonstrate that inhibition of KDM3A phosphorylation could be a potent therapeutic strategy to control oncogenic effect of JAK2−STAT3 signaling pathway.
Epigenetic changes such as histone modification and DNA methylation are actively involved in diverse cellular processes (1–7). Among the histone modifications, lysine methylation is one of the reversible regulations to attach methyl group to or remove it from lysine residue by corresponding histone methyltransferases and histone demethylases. Lysine methylation plays a crucial role in both gene activation and repression for dynamic gene regulation and provides a binding site for chromatin-associated proteins (8–11). Primarily, one of the histone lysine demethylases, KDM3A (also known as JHDM2A and JMJD1A), is a member of Jumonji domain-containing protein that eliminates monomethylation and dimethylation of histone H3 lysine 9 (H3K9) for exerting transcriptional activation function (12). Cofactors such as Fe (II) and α-ketoglutarate are indispensable for KDM3A demethylase activity.
KDM3A has been shown to control the transcriptional activation process in metabolism and spermatogenesis, as proven by phenotypes of Kdm3a knockout (KO) mice having obesity and male infertility (13, 14). KDM3A positively regulates activation of androgen receptor target genes such as PSA and NKX3.1 in prostate cancer (12). Although KDM3A has been shown to possess oncogenic function, it is unclear how transcriptional activation and repression cycles can be orchestrated in cancer progression.
JAK2−STAT3 signaling pathway is involved in a variety of physiological processes, including stem cell homeostasis, cell cycle progression, and apoptosis (15–21). JAK2 is a nonreceptor tyrosine kinase that induces cytoplasmic signaling cascades, while it has been shown that JAK2 is also present in the nucleus of human hematopoietic cells, functioning as the direct kinase of histone H3Y41 (22, 23). Once activated, it activates the transcription factor STAT3. STAT3 is tyrosine-phosphorylated by JAK2 and forms stable homodimers or heterodimers, which leads to local activation of target genes. Aberrant activation of JAK2−STAT3 signaling pathway has been shown to induce abnormal cell proliferation, migration, and antiapoptosis in various types of cancers (24–29). Therefore, inhibition of JAK2−STAT3 signaling axis could be an appropriate approach to attenuate oncogenic properties, and finding specific regulators in JAK2−STAT3 signaling axis would establish a way to tightly control the associated cancers. Given that JAK2 is also present in the nucleus (22, 30), we hypothesized that JAK2 might have other substrates including epigenetic enzymes in the nucleus.
Here, we provide evidence that KDM3A functions as a key epigenetic factor for JAK2−STAT3 activation in cancer cells. KDM3A is directly tyrosine-phosphorylated by JAK2 at 1101 residue, and phosphorylated KDM3A acts as a coactivator for transcriptional activation of STAT3 target genes such as MYC along with decreased H3K9me2 levels, leading to the increased cancer cell proliferation and motility. Altogether, our data reveal that JAK2−KDM3A axis contributes to enhancement of oncogenic effects and suggest a strategy for inhibition of KDM3A phosphorylation in cancer development.
Results
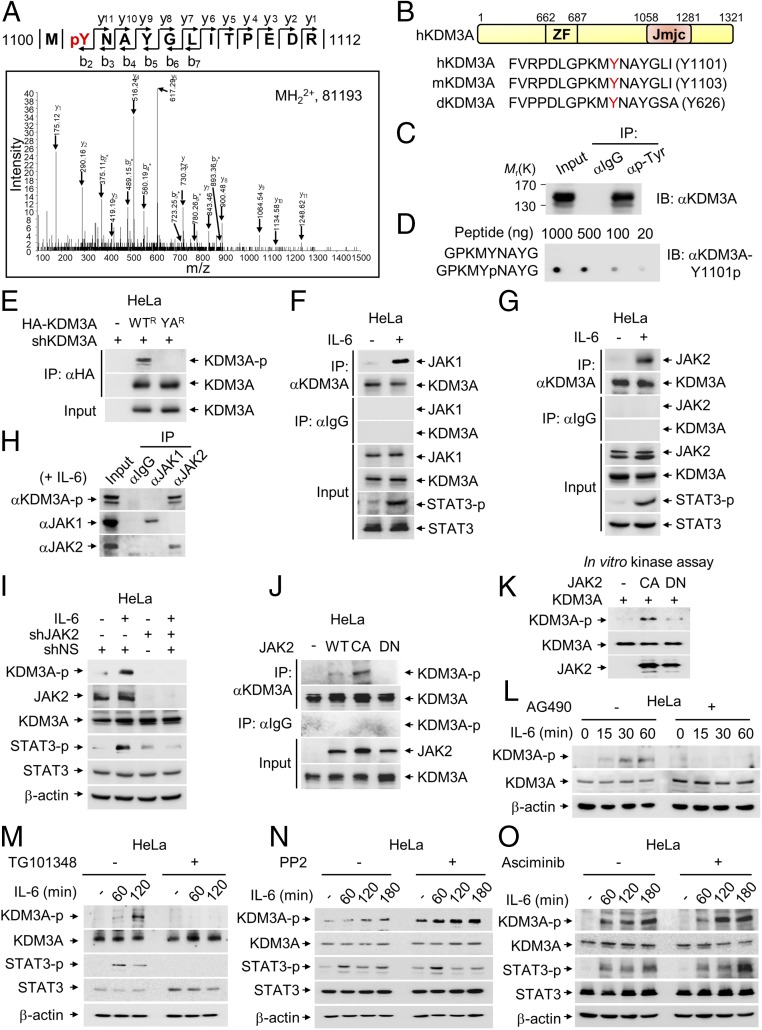
KDM3A Is Tyrosine-Phosphorylated by JAK2 at Tyrosine 1101 Residue.
Liquid chromatography−mass spectrometry/mass spectrometry (LC-MS/MS) analysis showed that KDM3A is phosphorylated at tyrosine 1101 (Y1101) residue (Fig. 1A). Interestingly, the Y1101 residue and the surrounding amino acids of KDM3A are well conserved among species (Fig. 1B). Coimmunoprecipitation (Co-IP) assay using phosphorylated tyrosine-specific antibodies revealed that KDM3A is tyrosine-phosphorylated (Fig. 1C). Therefore, we generated a specific antibody against phosphorylated KDM3A at Y1101 residue (KDM3A-Y1101p) and confirmed no cross-reactivity and its efficacy by immunoblot analysis (Fig. 1D). By utilizing anti−KDM3A-Y1101p antibody, we found that Y1101A (YA) mutant form of KDM3A, in which the 1101st amino acid residue tyrosine is replaced with alanine, abrogated tyrosine phosphorylation of KDM3A (Fig. 1E). These data indicate that KDM3A is tyrosine-phosphorylated at Y1101 residue.
Fig. 1.
JAK2 phosphorylates KDM3A at tyrosine 1101 residue. (A) Identification by LC-MS/MS of a tyrosine phosphorylation site on KDM3A. (B) Schematic of KDM3A showing zinc finger (ZF) and Jumonji C (Jmjc) domains. Sequences around tyrosine residue (Y, marked by red) are conserved among Drosophila (d), mouse (m), and human (h). (C) Protein extracts from HeLa cells were subjected to pull-down with anti−pan-phospho-tyrosine antibody. Phosphorylation of KDM3A was assessed by immunoblot with anti-KDM3A antibody. (D) Dot blotting of unmodified or phosphorylated KDM3A peptides with anti−phospho-KDM3A (KDM3A-Y1101p) antibody. (E) Endogenous KDM3A was knocked down using shRNA, and an shRNA-resistant form of HA-KDM3A WT (WTR) or HA-KDM3A YA mutant (YAR) was reconstituted in HeLa cells. KDM3A was immunoprecipitated using an anti-HA antibody and analyzed by immunoblot with anti−phospho-KDM3A antibody. (F and G) Co-IP assay of KDM3A with (F) JAK1 or (G) JAK2 was conducted in HeLa cells with or without IL-6 treatment (50 ng/mL) for 2 h. (H) Co-IP assay of JAK1 or JAK2 with phosphorylated KDM3A was conducted in HeLa cells treated with IL-6 for 2 h. Phosphorylation level of KDM3A was assessed by immunoblot analysis using anti−phospho-KDM3A antibody. (I) Endogenous JAK2 was knocked down using shRNA, and phosphorylation level of KDM3A was assessed by immunoblot analysis using anti−phospho-KDM3A antibody in HeLa cells with or without IL-6 treatment for 2 h. (J) WT, constitutively active mutant (CA) or dominant negative mutant (DN) of JAK2 was transfected to HeLa cells. Cell lysates were immunoprecipitated with anti-KDM3A antibody followed by immunoblot analysis with anti−phospho-KDM3A antibody to detect phosphorylated KDM3A. (K) In vitro kinase assay with JAK2 CA or DN mutant using recombinant KDM3A proteins. (L–O) HeLa cells, serum-starved for 24 h, were treated with IL-6 (50 ng/mL) for indicated times. Phosphorylation levels of KDM3A were analyzed by immunoblot with anti−phospho-KDM3A antibody in the absence or presence of JAK2 inhibitors (L) AG490 (10 μM) or (M) TG101348 (2.5 μM), (N) an Src family inhibitor PP2 (10 μM), or (O) an Abl inhibitor Asciminib (250 nM).
Next, we decided to search for upstream tyrosine kinases responsible for KDM3A phosphorylation. Given that KDM3A is localized predominantly in the nucleus, we explored tyrosine kinases present in the nucleus. As JAK1, JAK2, Src, and Abl1 are present in the nucleus as well as in the cytoplasm, we hypothesized that they might phosphorylate KDM3A in the nucleus. Co-IP data revealed that KDM3A bound to both JAK1 and JAK2 in the presence of IL-6, which is a well-known upstream physiological inducer for JAK activation and nuclear translocation (Fig. 1 F and G). Next, we examined whether activated JAK1 or JAK2 is responsible for the induction of KDM3A phosphorylation upon IL-6 treatment. We found that JAK2, but not JAK1, increased KDM3A tyrosine phosphorylation following IL-6 treatment (Fig. 1H), indicating that KDM3A tyrosine phosphorylation at Y1101 residue is JAK2-specific. Further, IL-6 treatment induced KDM3A phosphorylation at endogenous expression level as assessed by anti−KDM3A-Y1101p antibody, but JAK2-induced KDM3A phosphorylation upon IL-6 treatment was abrogated by knockdown of JAK2 by shRNA (Fig. 1I).
To examine whether JAK2 enzymatic activity is important for KDM3A phosphorylation, kinase activities of wild type (WT), constitutively active V617F mutant (CA), and dominant negative kinase-inactive mutant (DN) of JAK2 were analyzed in HeLa cells. JAK2 WT and CA mutant, but not DN mutant, exerted kinase activity on KDM3A both in vivo (Fig. 1J) and in vitro (Fig. 1K). Indeed, IL-6−induced KDM3A phosphorylation was almost completely abolished upon treatment with AG490 and TG101348, which are JAK2 inhibitors (Fig. 1 L and M), but not with other inhibitors including PP2, an Src-family inhibitor (Fig. 1N), or Asciminib, an Abl inhibitor (Fig. 1O). Together, these data indicate that KDM3A is tyrosine-phosphorylated by JAK2 in the nucleus both in vivo and in vitro.
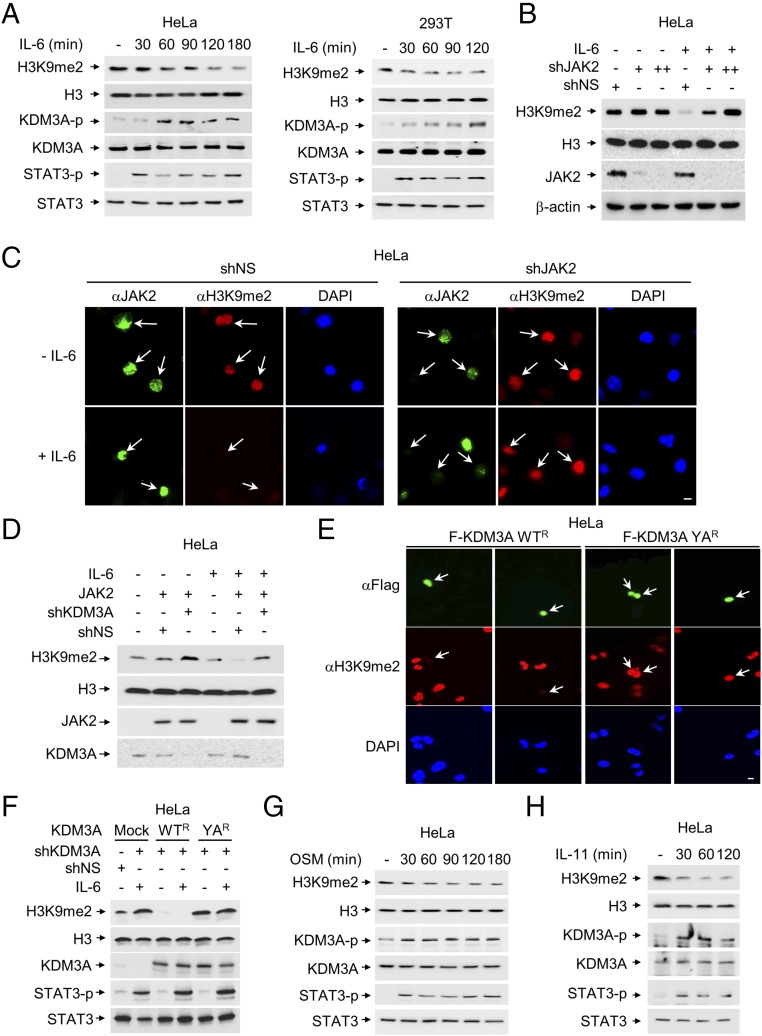
IL-6−Induced KDM3A Tyrosine Phosphorylation Increases Its Demethylase Activity.
Although JAK2 is known to exclude HP-1α from STAT target genes for transcriptional activation in human hematopoietic cells (22, 30, 31), whether JAK2 signaling modulates KDM3A-dependent histone H3K9 demethylation, a transcriptional activation mark, has not been elucidated. Therefore, we tested the possibility that the level of H3K9me2 is changed during IL-6 treatment based on the finding that JAK2-dependent phosphorylation of KDM3A is induced by IL-6 treatment. Indeed, H3K9me2 level was decreased during IL-6 treatment in HeLa and HEK 293T cells, along with increased KDM3A phosphorylation and STAT3 phosphorylation (Fig. 2A). Next, we examined whether IL-6−induced JAK2 is responsible for decreased H3K9me2 level. Indeed, IL-6 treatment decreased H3K9me2 levels, but knockdown of JAK2 by shRNA led to an increased level of H3K9me2 with IL-6 treatment, as confirmed by immunoblot (Fig. 2B) and immunostaining analyses (Fig. 2C).
Fig. 2.
KDM3A phosphorylation by JAK2 increases its demethylase activity. (A) The level of H3K9me2 was analyzed in HeLa and HEK293T cells after IL-6 treatment. (B) The level of H3K9me2 was analyzed after knockdown of JAK2 by shRNA in the absence or presence of IL-6. (C) Representative images of cells knocked down by JAK2 shRNA in the absence or presence of IL-6. The cells were stained with anti-H3K9me2 or anti-JAK2 antibodies. Nuclei were counterstained with DAPI. Arrows indicate cells expressing JAK2 and the level of H3K9me2. (Scale bar, 10 μm.) (D) Demethylation of H3K9me2 was induced by JAK2, and knockdown of KDM3A abolished JAK2-dependent demethylation of H3K9me2 in the presence of IL-6. (E) HeLa cells were knocked down by KDM3A shRNA and reconstituted with shRNA-resistant form of KDM3A WT (WTR) or YA (YAR). Transfected cells were fixed and stained for anti-H3K9me2 and anti-Flag antibodies. The cells were counterstained with DAPI to visualize cell nuclei. Arrows indicate cells expressing KDM3A and the level of H3K9me2. (Scale bar, 10 μm.) (F) Demethylation of H3K9me2 induced by IL-6 is dependent on KDM3A. Protein extracts from HeLa cells were knocked down by KDM3A shRNA and reconstituted with KDM3A WTR or YAR. Transfected cells were collected to determine the H3K9me2 levels in the absence or presence of IL-6 by immunoblot analysis. (G and H) The level of H3K9me2 was analyzed after treatment of (G) OSM (20 ng/mL) or (H) IL-11 (5 ng/mL), which share a gp130-mediated signaling molecule in HeLa cells, serum-starved for 24 h.
We examined whether demethylation of H3K9 mediated by JAK2 is dependent on KDM3A. JAK2-dependent reduction of H3K9me2 levels in the presence of IL-6 was abrogated by knockdown of KDM3A (Fig. 2D). To test whether JAK2-dependent phosphorylation of KDM3A affects its intrinsic histone demethylase activity, we reconstituted an shRNA-resistant form of KDM3A WTR or YAR mutant in KDM3A knockdown cells. Intriguingly, the reconstitution of YA mutant of KDM3A showed significantly reduced enzymatic activity on H3K9me2 compared with WT of KDM3A, as assessed by immunostaining (Fig. 2E). Moreover, when we introduced KDM3A WTR or YAR mutant in KDM3A knockdown cells in the absence or presence of IL-6 treatment, the level of H3K9me2 decreased synergistically with KDM3A WT, but not YA mutant, in the presence of IL-6 (Fig. 2F).
Further, we treated cells with other cytokines to examine whether other cytokines also induce KDM3A phosphorylation along with reduction of H3K9me2 level. Indeed, KDM3A phosphorylation was induced, along with subsequent demethylation of H3K9me2 by Oncostatin M (OSM) and IL-11, other STAT3-activating stimuli (Fig. 2 G and H). Since IL-6, IL-11, and OSM share the gp130-mediated signal-transducing molecules as their receptors (32), they appeared to show similar effects on KDM3A phosphorylation. However, we cannot exclude the possibility that a specific mode of action and downstream target genes could be differentially regulated by each STAT3-activating stimulus. These data indicate that JAK2-dependent tyrosine phosphorylation of KDM3A is responsible for the enhanced enzymatic activity, resulting in the reduction of H3K9me2 levels.
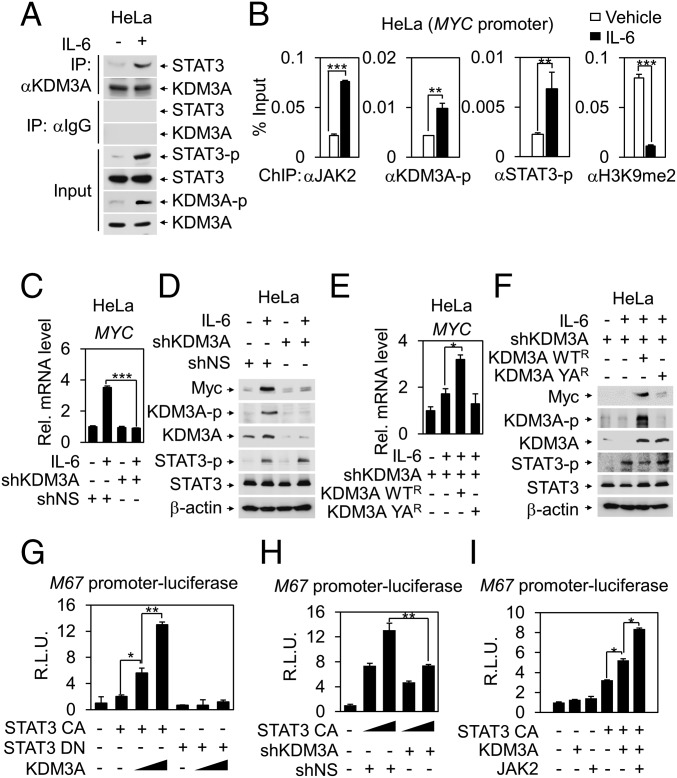
KDM3A Functions as a Transcriptional Coactivator in JAK2−STAT3 Signaling Pathway.
JAK2 interacts with STAT3 and activates STAT3 target genes (24, 33–35). Activated JAK2 induces STAT3 phosphorylation for nuclear translocation and transcriptional activation. Moreover, nuclear JAK2 has been shown to function as an oncogenic factor by acting on other specific substrates such as histones in addition to STAT3 in the nucleus. To test the possibility that tyrosine-phosphorylated KDM3A functions as a transcriptional coactivator for JAK2−STAT3 signaling pathway, we first examined whether KDM3A binds to STAT3. Co-IP assay showed that the binding between KDM3A and STAT3 was significantly enhanced by IL-6 treatment (Fig. 3A).
Fig. 3.
KDM3A functions as a transcriptional coactivator of STAT3 in JAK−STAT signaling pathway. (A) Co-IP assay was performed to detect interaction between STAT3 and KDM3A in HeLa cells with or without IL-6 treatment for 2 h. (B) ChIP assays were performed using anti-JAK2, anti−phospho-STAT3, anti−phospho-KDM3A, and anti-H3K9me2 antibodies on the MYC promoters after IL-6 treatment in HeLa cells. **P < 0.01; ***P < 0.001 (Student’s t test). (C) Quantitative RT-PCR analysis of MYC mRNA levels after knockdown of KDM3A by shRNA in HeLa cells following IL-6 treatment. ***P < 0.001 (Student’s t test). (D) Immunoblot analysis of MYC protein levels after knockdown of KDM3A by shRNA following IL-6 treatment in HeLa cells. (E) MYC mRNA levels were measured by quantitative RT-PCR in HeLa cells after rescuing resistant forms of KDM3A WTR or YAR in shRNA-mediated KDM3A knockdown cells following IL-6 treatment. *P < 0.05 (Student’s t test). (F) Immunoblot analysis in HeLa cells after rescuing resistant forms of KDM3A WTR or YAR in shRNA-mediated KDM3A knockdown cells following IL-6 treatment. (G–I) STAT-responsive M67 promoter luciferase reporter was transfected into HEK293T cells with indicated plasmids. Luciferase reporter activity was measured at 48 h after transfection and normalized by β-galactosidase activity. Values are expressed as mean ± SD for three independent experiments. *P < 0.05; **P < 0.01 (Student’s t test).
To examine whether KDM3A is recruited to the JAK2−STAT3 target promoters via STAT3, we performed chromatin immunoprecipitation (ChIP) assays on MYC promoter containing functional STAT binding sites. ChIP assay showed the IL-6−dependent corecruitment of JAK2, STAT3-p, and KDM3A-p on MYC promoter, concomitant with decreased H3K9me2 levels (Fig. 3B). These results indicate that tyrosine-phosphorylated KDM3A is directly involved in IL-6−induced transcriptional activation of JAK2−STAT3 target genes. In parallel, knockdown of KDM3A by shRNA significantly attenuated IL-6−induced MYC mRNA and protein levels (Fig. 3 C and D). Further, introduction of an shRNA-resistant form of KDM3A WT, but not YA mutant following knockdown of KDM3A, increased STAT3-dependent MYC mRNA and protein levels upon IL-6 treatment (Fig. 3 E and F).
Besides, luciferase assays using M67 promoter-luciferase reporter containing functional STAT3 response elements as readouts showed that KDM3A phosphorylation increased M67 promoter luciferase reporter activity in the presence of the constitutively active form of STAT3 (STAT3 CA), but not the dominant negative form of STAT3 (STAT3 DN), which failed to bind to STAT3 response element (Fig. 3G). STAT3-dependent luciferase reporter activity was attenuated by KDM3A knockdown (Fig. 3H) but elevated by KDM3A overexpression (Fig. 3I). Together, these data indicate that KDM3A function is critical for the IL-6−dependent induction of JAK2−STAT3 target gene expression as a coactivator of STAT3.
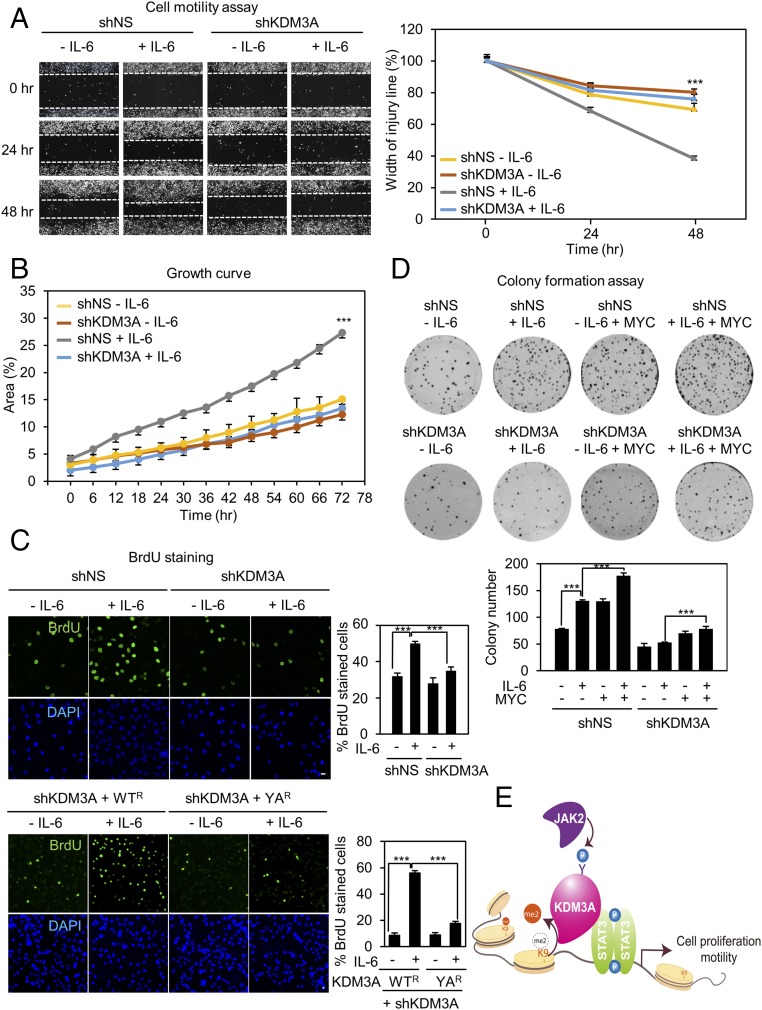
KDM3A Phosphorylation Is Responsible for Increased Cell Proliferation and Motility.
Since dysregulation of JAK2−STAT3 signaling pathway is frequently associated with human malignancies, we examined whether IL-6−induced phosphorylation of KDM3A by JAK2 is involved in modulating cancer cell growth, proliferation, motility, and colony formation. We performed a cell motility assay in the absence or presence of IL-6 that induces KDM3A phosphorylation. IL-6 treatment increased cell motility, and it took almost 48 h for cells to close the gap, whereas cells knocked down by KDM3A shRNA showed significantly reduced cell motility (Fig. 4A). These data indicate that KDM3A is involved in enhancement of cell migration in the presence of IL-6.
Fig. 4.
KDM3A phosphorylation is responsible for increased cell proliferation and motility. (A) (Left) Photomicrographs from the scratch-motility assay of HeLa cells expressing KDM3A shRNA with or without IL-6 treatment. Wound closure was monitored at 24-h intervals for 48 h in HeLa cells. (Right) Cell migration (percent) was quantified by calculating the wound width. ***P < 0.001 (Student’s t test). (B) Proliferation was monitored at 6-h intervals in HeLa cells ectopically expressing either control shRNA or KDM3A shRNA. Proliferation efficiency (percent) was quantified by calculating areas of cell population as shown in the graph. ***P < 0.001 (Student’s t test). (C) Confocal images of cells stained with BrdU. The fraction of increased BrdU-positive (BrdU+) cells after IL-6 treatment decreased following knockdown of KDM3A by shRNA. HeLa cells were knocked down by KDM3A shRNA and reconstituted with an shRNA-resistant form of KDM3A WT (WTR) or YA (YAR). Nuclei were counterstained with DAPI. (Scale bar, 10 μm.) ***P < 0.001 (Student’s t test). (D) (Top) Colony formation assay of HeLa cells transfected with either control shRNA or KDM3A shRNA in combination with MYC with or without IL-6 treatment. Cells were fixed and stained with crystal violet solution. (Bottom) Colony number was quantified as shown in the graph. ***P < 0.001 (Student’s t test). (E) Schematic model depicting JAK2−KDM3A−STAT3 signaling axis. Modulation of H3K9 methylation signature by JAK2-dependent phosphorylation of KDM3A is one of the predominant epigenetic events in transcriptional regulation of STAT3 target genes.
Next, we measured cell proliferation efficiency through growth curve analysis, 5-bromo-2-deoxyuridine (BrdU) staining, and colony formation assay. Results similar to the case of a cell motility assay were acquired. IL-6 treatment increased cell growth and BrdU incorporation, whereas knockdown of KDM3A significantly attenuated them in the presence of IL-6 (Fig. 4 B and C). Reconstitution of KDM3A WTR, but not YAR mutant, in KDM3A knockdown cells restored the increased BrdU incorporation in the presence of IL-6 (Fig. 4C). We performed colony formation assay by introduction of MYC with or without KDM3A knockdown upon IL-6 treatment. Reconstitution of MYC in KDM3A knockdown cells restored the reduced colony-forming ability upon IL-6 treatment (Fig. 4D). Our data indicate that KDM3A-induced MYC expression upon IL-6 treatment is potentially important for increased colony formation leading to increased cancer progression. Together, these data indicate that phosphorylated KDM3A further facilitates cell growth mediated by IL-6, thereby positively regulating cell survival and cancer progression (Fig. 4E).
Discussion
In this study, we provide a functional and mechanistic link between upstream stimuli-triggered intracellular JAK2−STAT3 signaling cascades and downstream epigenetic modification mechanism by KDM3A. We propose that modulation of H3K9 methylation conferred by JAK2-induced tyrosine-phosphorylated KDM3A is one of the predominant epigenetic events leading to cancer cell proliferation and motility. Several studies have demonstrated noncanonical functions of JAK2. The Drosophila JAK overactivation globally disrupts heterochromatin gene silencing, an epigenetic tumor-suppressive mechanism, and HP1 counteracts tumorigenesis induced by JAK−STAT overactivation (22, 36). Therefore, disruption of heterochromatin gene silencing has been shown to be essential for the Drosophila JAK overactivation-induced tumorigenesis. Genetic studies in Drosophila revealed that JAK gain of function suppresses and loss of function enhances heterochromatic gene silencing. In human hematopoietic cells, direct phosphorylation of histone H3Y41 by nuclear JAK2 is followed by a decrease of HP1α binding from chromatin, contributing to the oncogenic function of JAK2, while the displacement of HP1α by JAK2 is likely to be tightly regulated in normal cells (22, 36). Moreover, inhibition of JAK2 activity in human leukemic cells decreases both the expression of the hematopoietic oncogene lmo2 and the phosphorylation of H3Y41 at its promoter.
Our data provide a previously unrecognized JAK2−KDM3A−H3K9me2 signaling axis in the nucleus triggered by IL-6, leading to increased cancer cell motility, proliferation, and growth. JAK2 activation confers KDM3A to function as a coactivator for STAT3 through enhanced binding to STAT3 and increased H3K9 demethylation. Given that JAK2−STAT3 signaling pathway is evolutionarily well conserved in maintaining cellular homeostasis, dysregulation of JAK2−STAT3 signaling pathway may lead to severe consequences, such as cancers. Therefore, identification of signal-dependent modulation of epigenetic enzymes and their dysregulation could provide insights into mechanisms of cancer progression and pave the way for drug development.
There are more than 30 members of the Jumonji-containing KDM family (37). Over the past decade, development of clinical target candidates has not been successful in the case of Jumonji-containing KDMs, whereas four compounds already have been in clinical trials against LSD1. The big hurdle for the development of selective compounds modulating Jumonji-containing KDMs is the high structural similarity and potential functional redundancy among its family members. Since we identify the JAK2−KDM3A−H3K9me2 signaling axis, targeting KDM3A phosphorylation by JAK2 could provide a therapeutic strategy through perturbation of crucial oncogenic signaling cascade.
Materials and Methods
Experimental methods for reporter assay, quantitative RT-PCR, ChIP assays, identification of phosphorylation sites by LC-MS/MS, immunocytochemistry, in vitro cell motility assay, and cell proliferation assay are provided in SI Appendix.
Cell Culture and Reagents.
Cells were cultured at 37 °C in an atmosphere of 5% (vol/vol) CO2. HEK293T and HeLa cells were cultured in DMEM supplemented with 10% FBS (Gibco) with penicillin (100 U/mL) and streptomycin (100 µg/mL). IL-6 (50 ng/mL; R&D systems), OSM (20 ng/mL; R&D systems), IL-11 (5 ng/mL; R&D systems), AG490 (10 μM; Sigma), TG101348 (2.5 μM; Selleckchem), Asciminib (250 nM; Selleckchem), or PP2 (10 μM; Sigma) were treated for indicated times. The following antibodies were used: anti-JAK1 (#3332), anti-JAK2 (#3230), anti−phospho-STAT3 (#9131), and anti-H3K9me2 (#9753, #4658, ab1220) (Cell Signaling Technology, Abcam); anti-STAT3 (sc-8019) (Santa Cruz); anti-KDM3A (nb100-77282) (Novus); anti−phospho-Tyrosine 4G10 (05-1050) (Millipore); anti-HA (MMS-101R) (Covance); and anti-FLAG (F3165) (Sigma). Specific anti−phospho-KDM3A antibody was generated by Peptron (Korea) using immunized rabbits. Synthetic phosphopeptide (KMYNApYGLI) was used as the immunogen.
Statistical Analysis.
Statistical differences between the test and control samples were determined by a Student’s t test. Bars in graphs indicate mean ± SEM. Graphpad Prism was used for all analyses.
Supplementary Material
Acknowledgments
We thank Profs. W.-J. Lee and J. Chung and Drs. G. Lee and S. H. Kim (Seoul National University) for valuable help and discussion. This work was supported by Creative Research Initiative Program NRF-2017R1A3B1023387 (to S.H.B.), the Bio & Medical Technology Development Program NRF-2018M3A9E2023523 (to S.H.B.), the Science Research Center Program NRF-2016R1A5A1010764 (to S.H.B.), and the Basic Science Research Program NRF-2014R1A6A3A04057910 (to H.K.) from the National Research Foundation grant funded by the Korean government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805662115/-/DCSupplemental.
References
- 1.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos EI, Stafford JM, Reinberg D. Epigenetic inheritance: Histone bookmarks across generations. Trends Cell Biol. 2014;24:664–674. doi: 10.1016/j.tcb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Jimenez CP, Sandoval J. Epigenetic crosstalk: A molecular language in human metabolic disorders. Front Biosci (Schol Ed) 2015;7:46–57. doi: 10.2741/S424. [DOI] [PubMed] [Google Scholar]
- 6.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 8.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosammaparast N, Shi Y. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 12.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 13.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 14.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of JHDM2A in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 16.Aaronson DS, Horvath CM. A road map for those who don’t know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 17.Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parganas E, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths DS, et al. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol. 2011;13:13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson MA, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zouein FA, Duhé RJ, Booz GW. JAKs go nuclear: Emerging role of nuclear JAK1 and JAK2 in gene expression and cell growth. Growth Factors. 2011;29:245–252. doi: 10.3109/08977194.2011.614949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marotta LL, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colomiere M, et al. Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br J Cancer. 2009;100:134–144. doi: 10.1038/sj.bjc.6604794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshikawa H, et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 27.Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30:1005–1014. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Jove R. The STATs of cancer–New molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 30.Dawson MA, et al. Three distinct patterns of histone H3Y41 phosphorylation mark active genes. Cell Rep. 2012;2:470–477. doi: 10.1016/j.celrep.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bravo J, Heath JK. Receptor recognition by gp130 cytokines. EMBO J. 2000;19:2399–2411. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 34.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: The JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 36.Shi S, et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maes T, Carceller E, Salas J, Ortega A, Buesa C. Advances in the development of histone lysine demethylase inhibitors. Curr Opin Pharmacol. 2015;23:52–60. doi: 10.1016/j.coph.2015.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.