Significance
Anthrax toxin receptor 1 (ANTXR1), also known as Tumor Endothelial Marker 8, is overexpressed on the surface of tumor cells in over 60% of human cancers. A serious drawback for developing specific ligands for targeted therapy against ANTXR1 is the cross-reactivity with ANTXR2. Recently, ANTXR1 was identified as the high-affinity cellular receptor for Seneca Valley Virus (SVV). SVV has shown promising results as an oncolytic agent in clinical trials, and this discovery offers a powerful biomarker for selecting patient response to treatment. The identification of specific interaction sites between SVV and ANTXR1 lays the foundation to construct potent virus mutants with specific cancer tropism that can escape host antibody response and to expand the development of both antiangiogenic and anticancer antibody therapy.
Keywords: cryo-electron microscopy, picornavirus, virus receptor interaction, Seneca Valley Virus, cancer therapy
Abstract
Recently, the use of oncolytic viruses in cancer therapy has become a realistic therapeutic option. Seneca Valley Virus (SVV) is a newly discovered picornavirus, which has earned a significant reputation as a potent oncolytic agent. Anthrax toxin receptor 1 (ANTXR1), one of the cellular receptors for the protective antigen secreted by Bacillus anthracis, has been identified as the high-affinity cellular receptor for SVV. Here, we report the structure of the SVV-ANTXR1 complex determined by single-particle cryo-electron microscopy analysis at near-atomic resolution. This is an example of a shared receptor structure between a mammalian virus and a bacterial toxin. Our structure shows that ANTXR1 decorates the outer surface of the SVV capsid and interacts with the surface-exposed BC loop and loop II of VP1, “the puff” of VP2 and “the knob” of VP3. Comparison of the receptor-bound capsid structure with the native capsid structure reveals that receptor binding induces minor conformational changes in SVV capsid structure, suggesting the role of ANTXR1 as an attachment receptor. Furthermore, our results demonstrate that the capsid footprint on the receptor is not conserved in anthrax toxin receptor 2 (ANTXR2), thereby providing a molecular mechanism for explaining the exquisite selectivity of SVV for ANTXR1.
Seneca Valley Virus (SVV) is a small (∼30 nm in diameter) icosahedral virus harboring a positive-sense, single-stranded RNA genome of 7.3 kb. The RNA genome encodes seven nonstructural proteins—2A, 2B, 2C, 3A, 3B, 3C, and 3D—and four capsid proteins—VP1, VP2, VP3, and VP4 (1–3). Several studies have highlighted the capacity of SVV to preferentially infect and kill tumor cells with neuroendocrine features without resulting in detrimental off-target effects (4, 5). SVV has been investigated for its antitumor activity in phase I clinical trials against pediatric solid tumors and in phase II clinical trials against small-cell lung cancers (SCLC) (6–8). The efficacy of SVV treatment in oncovirotherapy has been shown to be limited due to the host immune response within 3 wk of viral infusion (7). There is increasing interest in the development of SVV mutants, which can retain their capacity to bind and infect tumor cells while bypassing the host antiviral immune response. Recently published results from our own work and collaborators unveiled anthrax toxin receptor 1 (ANTXR1), a type I transmembrane protein, as the cellular receptor governing specificity for SVV infection (9). In comparison with the second anthrax toxin receptor (ANTXR2), ANTXR1 is weakly expressed in normal tissues but overexpressed in a wide variety of human tumors (10). Little is known about the role of ANTXR1 in the development of normal tissue, and studies of ANTXR1 knockout mice failed to reveal any major phenotypic abnormalities (11). However, tumor growth and tumor blood vessel density were significantly decreased in these mice (12). Moreover, in patients enrolled in a phase I clinical study of SVV in advanced solid tumors with neuroendocrine features, i.v. administration of the oncolytic virus showed high specificity for tumor cells with no evidence of infection and propagation in the surrounding normal tissue (6, 7). In combination these studies provide a valuable strategy for selective targeting of ANTXR1 overexpression in tumor cells with SVV without impact on the adjacent normal tissue. However, the exact locations and the nature of interactions between SVV and ANTXR1 have not been characterized. Cryo-electron microscopy (Cryo-EM) structure of SVV-ANTXR1 from this study identifies surface-exposed loops of VP1, VP2, and VP3 as the sites of receptor attachment. Furthermore, the atomic model of SVV-ANTXR1 complex shows residues participating in the capsid footprint on the receptor are not conserved among ANTXR1 and ANTXR2, thereby providing a plausible explanation for tumor specificity of SVV. Findings from this study, in conjunction with future work on SVV-antibody interaction sites, can provide a promising stage for the development of SVV mutants with improved clinical applications.
Results
Overall Structure of SVV-ANTXR1 Complex.
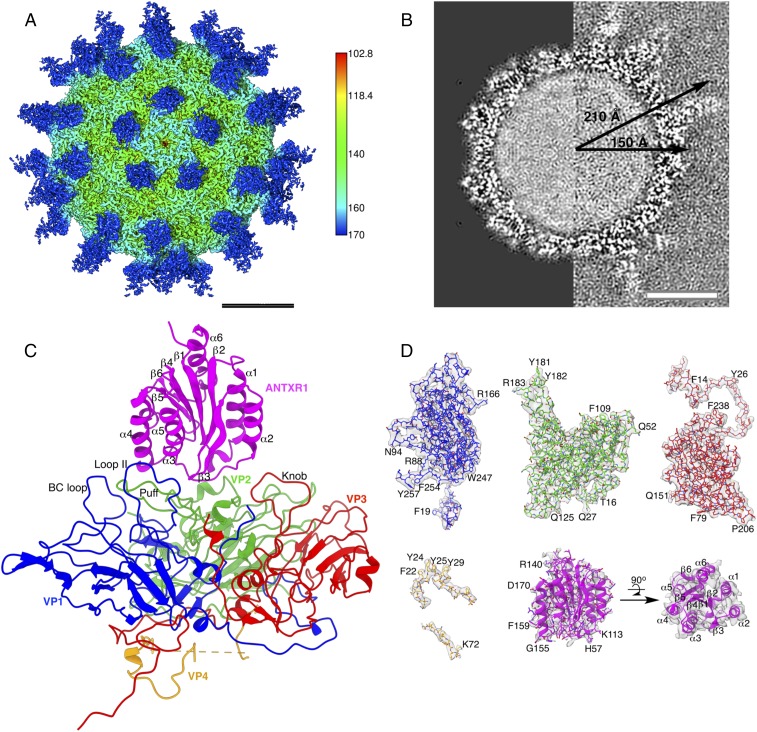
In this study, we used cryo-electron microscopy (cryo-EM) single-particle analysis to study the interaction between SVV and the ANTXR1 receptor. We used a fusion construct of ANTXR1 with the Ig Fc (fragment crystallizable) region, since Fc has previously been reported to be useful in improving the stability and solubility of bound partner protein (13). The structure features a crown-like arrangement of ANTXR1 around the fivefold axis of the capsid (Fig. 1A). The strong electron potential corresponding to the receptor in our 3D reconstruction has a similar overall absolute value as the electron potential inside the capsid (Fig. 1A and Movie S1), suggesting that the equilibrium between the soluble ANTXR1 domain and the SVV-receptor complex in vitro is sufficient to saturate most binding sites located on 60 asymmetric units. The average radius of the receptor-decorated SVV capsid remains unchanged at ∼150 Å as in the native structure, and no rearrangement of the capsid proteins was observed (Fig. 1B). The largest radius of 210 Å spans from the center of the capsid to the Fc region at the tip of the attached ANTXR1 (Fig. 1B). Local resolution estimation of the cryo-EM map indicates 3.1 Å within the capsid structure, while the resolution within the ANTXR1 domain is 4.25–4.75 Å (SI Appendix, Fig. S3 A and B) and within the ANTXR1-linked Fc fragment >5 Å, suggesting some flexibility of this region (SI Appendix, Fig. S1 A and B). Overall, the map features corresponding to the polypeptide chain and most of its side chains were of sufficient quality for atomic model building of capsid proteins VP1–4 and the receptor region of the ANTXR-Fc fusion (Fig. 1 C and D and SI Appendix, Table S1). The model was validated by building two separate atomic models from independent maps as a result of independent refinement and reconstruction (14); its side chain conformations match the known polypeptide sequence in well-resolved areas, and it has been checked for correct stereochemical geometry (SI Appendix, Figs. S2 and S3 and Table S1).
Fig. 1.
Cryo-EM structure of the SVV-ANTXR1 complex. (A) Radially colored iso-potential surface of the reconstructed electron potential map, contoured at 2.4 σ above average, displayed along the icosahedral fivefold axis. ANTXR1 (blue) bound to SVV (green) demonstrates a “crown-like” arrangement around the fivefold axis. (B) Central sections through normalized cryo-EM reconstructions of the undecorated full SVV capsid (left half, EMDB access code 7110) and of the SVV-ANTXR1 complex (right half, present study). The capsid dimension does not change upon receptor binding. The capsid interior appears filled with RNA in both reconstructions. (C) Atomic model in ribbon representation of ANTXR1 complexed to one capsid protomeric unit with VP1, VP2, VP3, VP4, and ANTXR1 colored in blue, green, red, orange, and magenta, respectively. The same color scheme is used throughout the figures. (D) Segmented iso-electron potential surfaces contoured at 2.4 σ of capsid proteins and ANTXR1. Refined side-chain conformations of a fitted atomic model show excellent correspondence with the structure. (Scale bars: 100 Å.)
Interactions Between SVV and ANTXR1.
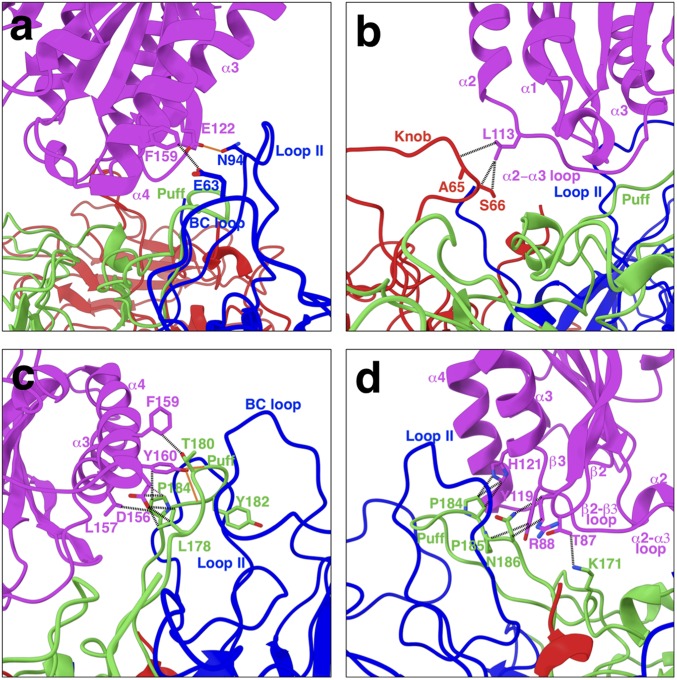
We determined the mechanism of interaction between SVV and ANTXR1 and localized the receptor contact sites to surface-exposed loops on the virus. The BC loop and loop II of VP1, the “puff” of VP2, and the “knob” of VP3 (Fig. 1C) comprise an interface area of 742 Å2 on each ANTXR1-binding site. The involvement of all exterior capsid proteins in receptor attachment is a phenomenon consistent with most species in the Enterovirus genus (15–19). Our refined atomic model of the SVV-ANTXR1 complex suggests that hydrogen-bonding (H-bond) interactions are exclusive to the VP1-ANTXR1 and VP2-ANTXR1 interfaces, whereas a variety of amino acid interactions are shared among all three interfaces. In VP1, the BC loop located at the center of the icosahedral triangle and adjacent loop II establish contacts between helices α4 and α3 of ANTXR1, respectively (Fig. 2A). We detected only one H-bond at the VP1-ANTXR1 interface, formed between N94 on VP1 loop II and E122 on ANTXR1 α3 (Fig. 2A). The opposite end of the receptor is anchored to the capsid near the icosahedral threefold axis by interactions established between the α2–α3 loop of ANTXR1 and the knob of VP3 (Fig. 2B). Interestingly, the majority of the ANXTR1 footprint is located on the puff loop of VP2 elevated between the BC loop and loop II of VP1 (interface area ∼489 Å2) (Fig. 2 C and D). The contact between ANTXR1 and the puff loop is stabilized by H-bonds formed between Y160 on α4 and T180 and Y182 on the puff (Fig. 2C). In addition, the puff loop interacts with helix α3 and with loops α2–α3 and β2–β3 of ANTXR1 (Fig. 2 C and D).
Fig. 2.
Interactions between SVV capsid proteins and ANTXR1. All three proteins located on the exterior of the capsid (VP1-3) are involved in receptor binding. Potential non–H-bond interactions and H-bond interactions are illustrated in black dashed and red solid lines, respectively. (A) Residue E63 on the BC loop of VP1 forms anion-π interactions with F159 on helix ⍺4 of ANTXR1. Our atomic model shows that residue N94 on loop II of VP1 can H-bond with E122 on helix ⍺3 of ANTXR1. (B) Residue L113 on the ⍺2–⍺3 loop of ANTXR1 form potential van der Waals interactions with S66 and the adjacent A65 on the knob of VP3. (C) Residues L157 and F159 on helix ⍺4 of ANTXR1 form van der Waals interactions with P184 and T180 on the puff of VP2, respectively. D156 adjacent to L157 on ANTXR1 participates in establishing van der Waals interactions with L178 and P184 on the puff loop. Potential H-bonds could be formed between Y160 on the ⍺4 helix and T180 and two residues upstream with Y182. Furthermore, Y160 on ⍺4 helix is in contact with P184 on the puff loop via aromatic interactions. (D) H121 on ⍺3 helix and Y119 on the ⍺2–⍺3 loop interact with P184 and P185 by establishing aromatic and van der Waals interactions, respectively. T87 and R88 on the β2–β3 loop are in contact with K171 and N186 on the puff loop via electrostatic interactions.
Changes in Capsid Proteins and the Receptor Post Interaction.
Picornaviruses, especially the members of the Enterovirus genus, undergo significant changes in capsid structure upon receptor binding, which is a prerequisite for the formation of the “A-particle” (16–20). However, the existence of an A-particle has not been reported for SVV or for members of the Aphthovirus genus, such as Foot and Mouth Disease Virus (FMDV). Comparison of the atomic crystal structures of the native SVV capsid and the receptor-decorated capsid from this study shows a root-mean-square deviation (rmsd) of 0.88 in Cα positions (Fig. 3). Although the overall changes are subtle, several regions with significant conformational changes can be distinguished in all four SVV capsid proteins and ANTXR1 (SI Appendix, Fig. S4). These changes are better understood by examining the fit of the X-ray–based model with the refined asymmetric unit coordinates in our cryo-EM map (Fig. 3). In VP1, the major differences are observed in the N-terminal extension located at the interior of the capsid close to the icosahedral fivefold axis (rmsd ∼1.6 Å) (Fig. 3 A, 1). The first four residues are shifted by an average of 2 Å, accounting for the largest deviation in this region. Interestingly, the locally well-resolved map features suggest that this region is not as disordered as in enteroviruses. Minor changes in VP1 can be observed in the GH loop (residues 185–215) located near the icosahedral twofold axis (rmsd 1.1 Å) (Fig. 3 A, 2). Even though the overall conformational changes in the BC loop remain minimal, residues 58–64, which include receptor-binding E63, are shifted by 1.3 Å in their Cα positions, on average (SI Appendix, Fig. S4A). The largest changes within VP2 are observed in the puff and the loop composed of residues 142–151 (Fig. 3 C, 1 and 2). Even though the changes observed in the puff are minimal in general, residues 178–186, which contribute to the bulk of receptor interactions, show considerable shifts in their Cα positions (∼1 Å). A buried region that consists of residues 142–151 undergoes the largest conformational alteration in VP2 (Fig. 3 C, 1) (rmsd ∼1.9 Å). Although this region harbors the integrin-binding motif LDV (21), local conformational changes of this magnitude do not aid in exposing the above-mentioned motif for ANTXR1 interaction. In addition, the VP3 N-terminal extension located near the icosahedral fivefold axis shows considerable conformational changes compared with native structure (rmsd ∼1.3 Å) (Fig. 3 C, 1). Unsurprisingly, the knob of VP3, which establishes van der Waals interactions with ANTXR1, undergoes a major change—in particular in the region composed of residues 62–67 that is shifted closer to the α2–α3 loop of ANTXR1 by an average rmsd of 1.8 Å. G63 in VP4, which has been reported to be in contact with the RNA genome (2), was not well resolved in our cryo-EM reconstruction, suggesting that this residue may become disordered after interaction with the receptor (Fig. 3 D, 1). The loss of order in VP4 G63 is accompanied by changes in RNA appearance in the iso-electron potential map of the SVV-ANTXR1 complex (SI Appendix, Fig. S1C), which appears to be less ordered in comparison with that of mature virions (2). Features of RNA loops were visible only at lower contour levels in our reconstruction. Compared with other capsid proteins, VP4 largely retains its fold (rmsd ∼0.9 Å), except for the missing G63 residue and residues 14–17, 35–38, and 64–65, which shift by 1.3, 1.1, and 1.2 Å, respectively (Fig. 3 D, 1 and 2 and SI Appendix, Fig. S4D).
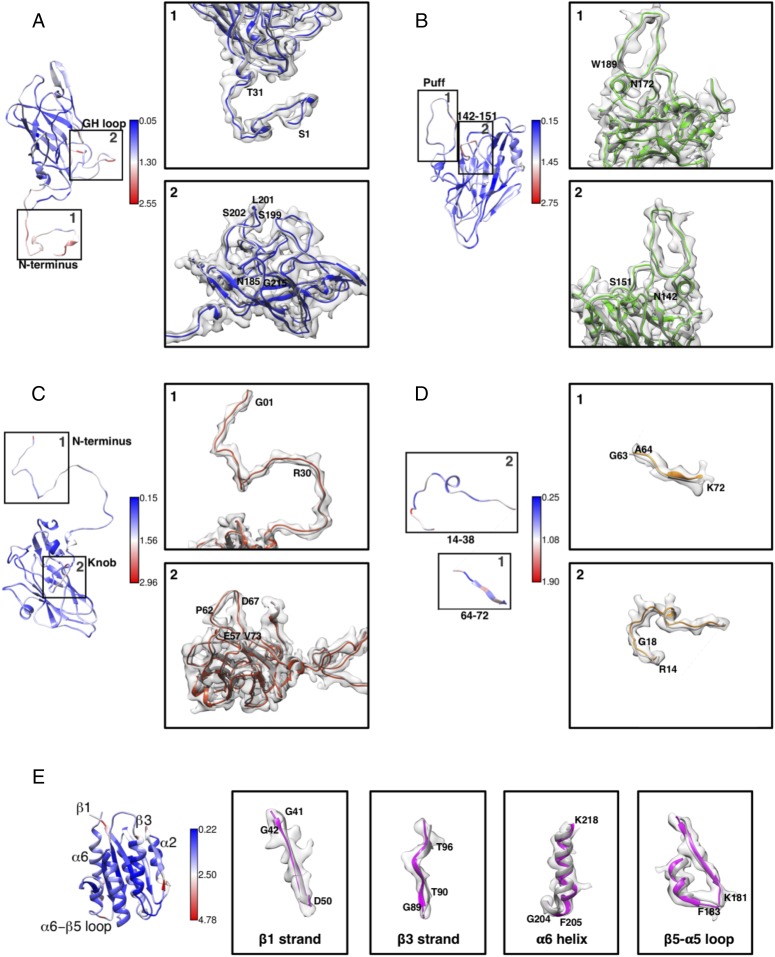
Fig. 3.
Structural changes in capsid proteins and ANTXR1 post receptor binding. Changes in the structure of capsid proteins and ANTXR1 were determined by calculating the shifts in C⍺ positions relative to the initial structure. Gray ribbons represent crystal structures (PDB ID codes 3CJI and 3N2N). Color scales indicate degree of C⍺ rmsd (blue and red colors representing minimal and maximum changes, respectively). (A) N-terminal extension (1) (residues 1–31) and GH loop of VP1 (2) (residues 185–215) deviate most. (B) Puff (residues 172–200) (1) and residues 142–151 (2) on CD loop are two surface-exposed regions on VP2 with the highest conformational changes. (C) In VP3, N-terminal (1) residues (1–30) undergo moderate changes upon receptor binding. The knob (2) (residues 57–73), which is involved in receptor interaction, shows significant conformational changes. (D) VP4 undergoes smaller conformational changes compared with other capsid proteins (1, 2). VP4 is devoid of electron density corresponding to G63 (1), suggesting a disordering of this region upon receptor binding. (E) Changes in ANTXR1 are observed in strands β1 and β3, helix ⍺6, and loop β5–⍺5.
The changes in the ANTXR1 structure upon capsid attachment are more significant (rmsd ∼1.2 Å) (Fig. 3E and SI Appendix, Fig. S4E). Interestingly, these changes are observed in the strands β1 and β3, the helix α6, and the β5–α5 loop, which are not in direct contact with capsid proteins (rmsd of 2.5, 2.5, 1.8, and 2.7 Å, respectively). It is important to note that these individual deviations represent the largest contribution to the overall rmsd in the refined ANTXR1 model. Overall, the changes in SVV capsid proteins and ANTXR1 post interaction remain minimal and do not provide any significant insights into their functional relevance.
Comparison of Capsid Footprint on ANTXR1 with ANTXR2 and Integrins.
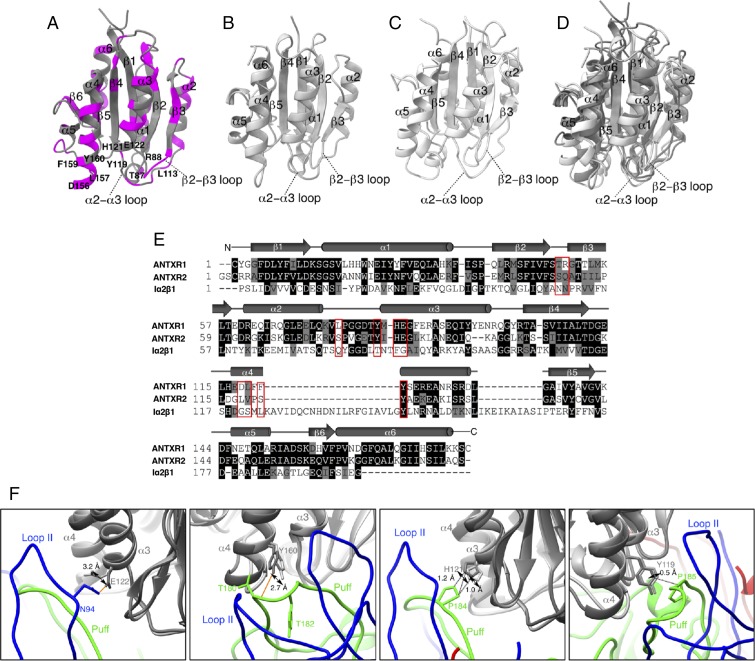
Our structure provides the structural basis to explain the selectivity of SVV to ANTXR1 over ANTXR2 and integrin receptors. ANTXR2, which is abundantly expressed in normal tissues (22), has a 10 times higher affinity for the protective antigen (PA) of anthrax toxin than ANTXR1 (23, 24) and is responsible for its internalization (22, 25, 26). Integrin receptors function as cell-surface receptors for FMDV (27) and adopt an α/β open sheet fold similar to that of ANTXR1 and ANTXR2 (28). When the refined structure of ANTXR1 from our study (Fig. 4A) was superimposed with crystal structures of ANTXR2 [Fig. 4B, Protein Data Bank (PDB) ID code 1SHT] and α2β1 integrin (Fig. 4C, PDB ID code 5HJ2), the three structures agreed very closely in their overall fold (Fig. 4D). However, the sequences of ANTXR1 and α2β1 integrin differ greatly from each other with a sequence homology of only 15%, whereas ANTXR1 and ANTXR2 share a closer sequence homology of 50% (Fig. 4E). When comparing the footprint of ANTXR1 on the SVV capsid to the corresponding regions on ANTXR2, we can distinguish both conserved (Fig. 4A, gray ribbon) and nonconserved residues (Fig. 4A, pink ribbon). Residues 156, 157, 159 on helix α4; T87 and R88 on the β2–β3 loop; and L113 on the α2–α3 loop constitute the nonconserved residues on ANTXR1 that interact with SVV (Fig. 4 A and E). On the other hand, Y160 on helix α4; H121 and E122 on helix α3; and Y119 on the α2–α3 loop fall into the conserved residues that interact with SVV (Fig. 4 A and E). Nevertheless, the side-chain rotamers of E122 and Y160 are visibly shifted in our experimental map of ANTXR1 (Fig. 4F), thereby disfavoring the H-bond formation with N94 (VP1) and T180 (VP2), respectively. Side chains of H121 and Y119 located on helix α3 are in similar locations in ANTXR2 and may therefore interact with SVV as observed at the VP2-ANTXR1 interface. However, these interactions alone may not be sufficient to establish a stable link between ANTXR2 and SVV to facilitate entry into nontumorous host cells.
Fig. 4.
Comparison of SVV-binding footprint on ANTXR1 with corresponding regions on ANTXR2 and α2β1 integrin to understand the specificity of SVV to ANTXR1. Ribbon diagrams illustrate the structures of (A) ANTXR1, (B) ANTXR2 and α2β1, and (C) α2β1 integrin colored in different shades of gray with nonconserved regions of ANTXR1 highlighted in magenta. (D) Superimposed structures of ANTXR1, ANTXR2, and α2β1 integrin. (E) Aligned sequences of ANTXR1, ANTXR2, and α2β1 integrin with capsid-binding sites indicated by red boxes. Residues depicted on black or gray backgrounds represent identical or similar sequences, respectively. Diverging residues are displayed on a white background. (F) Conformational shifts in side chains of conserved residues in ANTXR1 in comparison with ANTXR2. H-bond forming E122 and Y160 residues demonstrates the highest shifts of 3.2 and 2.7 Å in ANTXR2, thereby potentially hindering their capacity to form H-bonds unlike ANTXR1. H121 and Y119 are the only two conserved residues in ANTXR2 that may establish interactions with SVV similar to that of ANTXR1.
Discussion
In summary, our cryo-EM structure allowed us to explain how SVV discriminates between ANTXR1 and ANTXR2. Sequence comparison of ANTXR2 with ANTXR1 bound to SVV revealed that most of the receptor residues contributing to virus-receptor interactions are not conserved among the two receptors. Therefore, this ensures specific targeting of tumors expressing ANTXR1 with SVV, without causing any off-target effect on normal cells that highly express ANTXR2. Interestingly, these residues were found to be among the key amino acids governing the striking difference in PA-binding affinities between ANTXR1 and ANTXR2 in previous studies (29).
Our findings reveal that the affinities of a mammalian virus and a bacterial toxin to the same receptor are differentiated by similar nonconserved regions at their virus-receptor– and toxin-receptor–binding interfaces. Compared with other picornaviruses currently investigated as potential oncolytic agents such as poliovirus (30) and coxsackievirus (31), SVV has the valuable advantage of a high-affinity cellular receptor that is also a widely spread tumor marker. One major challenge in advancing SVV as an oncolytic agent is the response of the patient’s immune response. SVV is not a human pathogen. Therefore, there exists no innate immunity in patients (6, 7). However, after 2–3 weeks following virus administration, the host organism develops immunity, and the virus is cleared from the organism (6, 7). Hence, it would be of major significance for future work to investigate the interactions between SVV and neutralizing antibodies and to identify key residues on the capsid surface involved in these interactions. So far, the only information regarding SVV-antibody interaction stems from the study of genetic variation of stock pathogen SVV strains responsible for neonatal mortality and vesicular lesions in pigs (3). In all of the different SVV strains reported (3), the ANTXR1-binding site is highly conserved, except for the E63T mutation on the BC loop of VP1. Of importance is the ability of these strains to bypass recognition by neutralizing monoclonal antibodies (3, 32). This observation suggests that the ANTXR1-binding site on SVV may partially overlap with the binding epitope of neutralizing antibodies. Future work will involve investigating the interactions between SVV and neutralizing antibodies to identify and mutate residues on the capsid surface that interact with antibodies. Thus, the findings from this study encourage the development of potent SVV mutants in oncovirotherapy capable of evading the host immune response, while retaining their ability to preferentially kill tumors.
Materials and Methods
Virus Production and Purification.
SVV full capsids were purified according to methods adapted from previously published protocols (2). Briefly, SCLC H446wt cells (ATCC, HTB-171) were grown in Roswell Park Memorial Institute medium (RPMI) 1640 (catalog no. 1851354; Gibco) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin–streptomycin to ∼80% confluency in eight T175 flasks. Cells were infected by replacing the existing medium with RPMI medium 1640 containing 2% (vol/vol) FBS and plaque-purified SVV stock at a multiplicity of infection of 1, followed by incubation at 37 °C for 3 d. The detached cells after 3 d were treated with 0.05% (vol/vol) Triton ×100 in RPMI 1640 for 30 min at room temperature to permeabilize cell membranes. Thereafter, the cell suspension was transferred into 250-mL Nalgene bottles and centrifuged at 10,000 × g for 30 min at 4 °C. The resulting supernatant was transferred into six 38.5-mL open-top, polypropylene tubes (catalog no. Z60105SCA; Beckman Coulter) and spun down at 120,000 × g for 1 h in a Beckman Coulter SW32Ti rotor at 4 °C. After discarding the supernatant, the virus pellet was resuspended in CsCl purification buffer (137 mM NaCl, 5 mM KCl, 25 mM Tris base, 0.8 mM NaH2PO4, pH 7.4) overnight at 4 °C. Suspended virus was loaded onto a 1.38-g/mL isopycnic CsCl gradient prepared in a 16.5-mL open-top polypropylene tube (catalog no. Z60303SCA; Beckman Coulter) and centrifuged at 61,580 × g for 18 h in a Beckman Coulter SW 32.1 Ti rotor at 22 °C. Viral bands were collected and dialyzed against PBS buffer overnight at 4 °C. Final virus concentration in dialyzed fraction was measured by a Qubit protein concentration assay kit (catalog no. 1814929; Life Technologies). Samples were stored at −80 °C until used.
SVV-ANTXR1 Interaction.
The interaction between SVV and ANTXR1 was carried out according to previously published protocols (9). In brief, SVV purified from the previous step (0.2 mg/mL) was mixed with recombinant human ANTXR1-Fc (1 mg/mL) (catalog no. 13367-H02H; Sino Biological) in equal volumes. The virus-receptor mixture was incubated at 37 °C for 90 min, followed by another 90 min incubation in an ice bath.
Cryo-EM Sample Preparation and Data Collection.
Cryo-EM specimens were prepared by applying 2.5–4.0 μL of SVV-ANTXR1 mixture from the above step onto glow-discharged QF-1.2/1.3 grids (Quantifoil). Excess liquid was blotted off on a Vitrobot IV (Thermo Fisher Scientific) at 100% humidity for 3 s with blot force 0, immediately followed by plunge-freezing the grid into liquid ethane at liquid nitrogen temperature. Frozen grids were imaged at 73,000× magnification on a Talos Arctica cryo-transmission electron microscope (Thermo Fisher Scientific) operating at 200 kV, with a Falcon III direct electron detector (Thermo Fisher Scientific) in integrating mode at a dose rate of 80 el/pixel/s. Images of grid areas with thin ice were recorded automatically following low-dose procedures with the EPU software (Thermo Fisher Scientific) at a pixel size of 1.42 Å/pixel.
Image Processing and Single-Particle Analysis of the SVV-ANTXR1 Complex.
A total of 39 movie frames (each containing a dose of 1 el/Å2/frame) were aligned, dose-weighted, and summed with MotionCor2 (33), resulting in a total dose of 39 e−/Å2/image. Defocus estimation was performed with Ctffind4 (34). Only micrographs with a good fit correlation to 4.0 Å were used for single-particle analysis of the SVV-ANTXR1 complex. Manual particle selection was carried out in E2BOXER (35), yielding 7,457 single-particle images. Subsequent 2D classification, 3D classification, and 3D refinement were carried out with Relion 1.4 (36). Briefly, picked particles were extracted with a box size of 420 pixels and were subjected to several rounds of reference-free 2D classification. Good class averages were manually identified, and junk particles were omitted. A selection of 6,782 good particles were subjected to reference-based 3D classification by using a low-pass-filtered (50 Å) electron density map of the SVV crystal structure (PDB ID code 3CJI) as the initial 3D model. The resulting experimental 3D model of the SVV-ANTXR1 complex was further refined and postprocessed. Final overall resolution of the reconstruction was estimated as 3.8 Å between independently refined data sets at a Fourier shell correlation of 0.143. A local resolution map was calculated with Resmap (37).
Atomic Model Building and Refinement.
The crystal structures of SVV (PDB ID code 3CJI) and ANTXR1 (PDB ID code 3N2N) were used as starting models to build the atomic model of the SVV-ANTXR1 complex. The aforementioned crystal structures were fitted into the cryo-EM map, first manually and then by using the fitmap function of UCSF Chimera (38). One asymmetric unit and its symmetry-related neighbors were selected and masked, and the surrounding map was excluded. Initial atomic coordinates were subjected to manual rigid-body fitting into the segmented reconstruction with Coot (39). The discrepancies between the atomic model and the cryo-EM map were further minimized by subjecting the atomic model built with Coot to further refinement by Phenix (40) with the phenix.real_space_refine program. Model refinement was carried out for five iterations until satisfactory model refinement statistics were obtained according to MolProbity (41). The model was further validated by building independent atomic models for the two half maps used in the 3D refinement and then comparing these models with one another and with the atomic model built from the full map (14) (SI Appendix, Fig. S2). Chimera and ChimeraX (42) were used as the primary programs for visualization of structures and production of figures. The interactions between SVV and ANTXR1 at the interface (SI Appendix, Fig. S5) were elucidated using Chimera and JsPISA online server (www.ccp4.ac.uk/pisa). Spherical projection of the receptor footprint on SVV was generated using the program RIVEM (43).
Data Availability.
The cryo-EM map of the SVV-ANTXR1 complex was deposited in the EMDataBank (EMDB) database under entry code 7772. Model coordinates were uploaded to the Protein Data Bank with PDB access code 6CX1.
Supplementary Material
Acknowledgments
Electron microscopy work was performed at Otago Micro and Nanoscale Imaging and Okinawa Institute of Science and Technology. M.W. was supported by direct funding from OIST. N.J. was supported by a University of Otago Doctoral Scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the EMDataBank under entry code 7772 and in the Protein Data Bank under entry code 6CX1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810664115/-/DCSupplemental.
References
- 1.Venkataraman S, et al. Structure of Seneca Valley virus-001: An oncolytic picornavirus representing a new genus. Structure. 2008;16:1555–1561. doi: 10.1016/j.str.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss M, et al. Cryo-EM structure of Seneca Valley virus procapsid. J Virol. 2017 doi: 10.1128/JVI.01927-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu W, et al. Genome wide analysis of the evolution of Senecavirus A from swine clinical material and assembly yard environmental samples. PLoS One. 2017;12:e0176964. doi: 10.1371/journal.pone.0176964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, et al. Intravenous injection of oncolytic picornavirus SVV-001 prolongs animal survival in a panel of primary tumor-based orthotopic xenograft mouse models of pediatric glioma. Neuro Oncol. 2013;15:1173–1185. doi: 10.1093/neuonc/not065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy PS, et al. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J Natl Cancer Inst. 2007;99:1623–1633. doi: 10.1093/jnci/djm198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudin CM, et al. Phase I clinical study of Seneca Valley virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res. 2011;17:888–895. doi: 10.1158/1078-0432.CCR-10-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke MJ, et al. Phase I trial of Seneca Valley virus (NTX-010) in children with relapsed/refractory solid tumors: A report of the children’s oncology group. Pediatr Blood Cancer. 2015;62:743–750. doi: 10.1002/pbc.25269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anonymous 2018 Seneca Valley virus-001 after chemotherapy in treating patients with extensive-stage small cell lung cancer. Available at https://clinicaltrials.gov/ct2/show/NCT01017601.
- 9.Miles LA, et al. Anthrax toxin receptor 1 is the cellular receptor for Seneca Valley virus. J Clin Invest. 2017;127:2957–2967. doi: 10.1172/JCI93472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachran C, Leppla SH. Tumor targeting and drug delivery by anthrax toxin. Toxins (Basel) 2016;8:E197. doi: 10.3390/toxins8070197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen M, et al. Host-derived tumor endothelial marker 8 promotes the growth of melanoma. Cancer Res. 2009;69:6021–6026. doi: 10.1158/0008-5472.CAN-09-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhary A, et al. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell. 2012;21:212–226. doi: 10.1016/j.ccr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter PJ. Introduction to current and future protein therapeutics: A protein engineering perspective. Exp Cell Res. 2011;317:1261–1269. doi: 10.1016/j.yexcr.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Hryc CF, et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc Natl Acad Sci USA. 2017;114:3103–3108. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, et al. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat Struct Biol. 2001;8:874–878. doi: 10.1038/nsb1001-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, et al. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol. 2003;77:4827–4835. doi: 10.1128/JVI.77.8.4827-4835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strauss M, et al. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J Virol. 2015;89:4143–4157. doi: 10.1128/JVI.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing L, Casasnovas JM, Cheng RH. Structural analysis of human rhinovirus complexed with ICAM-1 reveals the dynamics of receptor-mediated virus uncoating. J Virol. 2003;77:6101–6107. doi: 10.1128/JVI.77.11.6101-6107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, et al. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci USA. 2008;105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butan C, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol. 2014;88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tselepis VH, Green LJ, Humphries MJ. An RGD to LDV motif conversion within the disintegrin kistrin generates an integrin antagonist that retains potency but exhibits altered receptor specificity. Evidence for a functional equivalence of acidic integrin-binding motifs. J Biol Chem. 1997;272:21341–21348. doi: 10.1074/jbc.272.34.21341. [DOI] [PubMed] [Google Scholar]
- 22.Scobie HM, Rainey GJA, Bradley KA, Young JAT. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scobie HM, et al. A soluble receptor decoy protects rats against anthrax lethal toxin challenge. J Infect Dis. 2005;192:1047–1051. doi: 10.1086/432731. [DOI] [PubMed] [Google Scholar]
- 24.Wigelsworth DJ, et al. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J Biol Chem. 2004;279:23349–23356. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, et al. Capillary morphogenesis protein-2 is the major receptor mediating lethality of anthrax toxin in vivo. Proc Natl Acad Sci USA. 2009;106:12424–12429. doi: 10.1073/pnas.0905409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JAT. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 27.Kotecha A, et al. Rules of engagement between αvβ6 integrin and foot-and-mouth disease virus. Nat Commun. 2017;8:15408. doi: 10.1038/ncomms15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown KL, et al. Salt-bridge modulates differential calcium-mediated ligand binding to integrin α1- and α2-I domains. Sci Rep. 2018;8:2916. doi: 10.1038/s41598-018-21231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu S, et al. The structure of tumor endothelial marker 8 (TEM8) extracellular domain and implications for its receptor function for recognizing anthrax toxin. PLoS One. 2010;5:e11203. doi: 10.1371/journal.pone.0011203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desjardins A, et al. Recurrent glioblastoma treated with recombinant poliovirus. N Engl J Med. 2018;379:150–161. doi: 10.1056/NEJMoa1716435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley S, et al. Applications of coxsackievirus A21 in oncology. Oncolytic Virother. 2014;3:47–55. doi: 10.2147/OV.S56322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M, van Bruggen R, Xu W. Generation and diagnostic application of monoclonal antibodies against Seneca Valley virus. J Vet Diagn Invest. 2012;24:42–50. doi: 10.1177/1040638711426323. [DOI] [PubMed] [Google Scholar]
- 33.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J Struct Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM map of the SVV-ANTXR1 complex was deposited in the EMDataBank (EMDB) database under entry code 7772. Model coordinates were uploaded to the Protein Data Bank with PDB access code 6CX1.