DNA methylation is a conserved epigenetic mark in eukaryotes involved in many important biological processes, such as genome integrity, gene imprinting, and gene regulation (1). In genomes, DNA methylation marks can be added through the DNA methylation pathway and can be removed through the DNA demethylation pathway (1). The REPRESSOR OF SILENCING 1 (ROS1)/DEMETER (DME; DEMETER-like, DML) family of DNA demethylases were first reported in plants (2, 3) and now are known to be involved in DNA demethylation in all eukaryotes (1). The bifunctional glycosylase/lyase activities of the ROS1 family of enzymes could initiate DNA demethylation through removal of methylcytosine from the DNA backbone, leaving a single-nucleotide gap that is filled with an unmethylated cytosine through a base excision repair (BER) mechanism (1). In both plants and mammals, the active DNA demethylation pathway depends on the BER mechanism, although different enzymes initiate the pathway in the two systems (1). In recent years, several additional enzymes and regulators involved in the active DNA demethylation pathway have been identified and studied in plants (1). In contrast to the extensive mechanistic understanding of DNA demethylation pathways, however, the developmental function of DNA demethylation factors is largely unknown in both model plants and crops. This knowledge gap is now bridged by the findings reported in PNAS by Liu et al. (4), who show that weak mutations of the OsROS1 gene in rice lead to an increased number of aleurone cell layers through DNA hypermethylation and repression of two putative transcription factor genes, RISBZ1 and RPBF, that increase aleurone layer cells when their expression levels are reduced (5). The OsROS1 mutant contains more lipids, proteins, vitamins, minerals, and dietary fibers than the wild type and could be useful for nutrient improvement in rice breeding (Fig. 1).
Fig. 1.
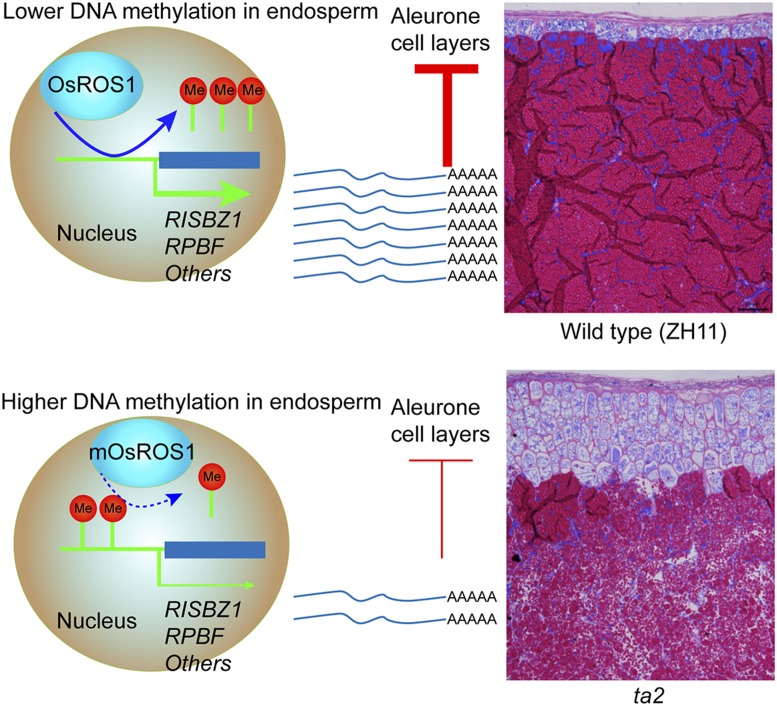
The proposed model for OsROS1 in mediating aleurone development in rice. OsROS1 modulates the expression of genes such as RISBZ1 and RPBF by DNA demethylation in the endosperm of rice. In the wild type, the higher expression of RISBZ1 and RPBF genes is maintained by OsROS1 to ensure the formation of one aleurone cell layer. In ta2 mutants, the attenuated function of mOsROS1 led to increased DNA methylation in the promoter regions, thus repressing the expression of genes such as RISBZ1 and RPBF, which releases their inhibition on the aleurone cell-layer formation. Thus, the ta2 mutants have more aleurone cell layers and contain more nutrients than the wild type (ZH11).
DNA demethylation in mammals is required for many developmental processes, including embryo development and cell differentiation (6). However, in plants, only a few developmental functions have been reported for DNA demethylases before this study. In Arabidopsis, AtDME is highly expressed in central cells of the embryo sac and contributes to gene imprinting (2, 7), whereas AtROS1 is expressed in somatic cells and controls stomatal development through demethylation of EPIDERMAL PATTERNING FACTOR 2 promoter (8). Other roles of DNA demethylases include nodule differentiation and nitrogen fixation in Medicago (9), pericarp ripening and global loss of DNA demethylation (10, 11) due to SlDML2 in tomato, DNA demethylation and bud break (12) due to PtaDML10 in poplar, and, in rice, involvement of OsROS1 in male and female gametogenesis (13).
The study of Liu et al. (4) involved a large-scale screening of rice mutants for aleurone defects using a half-seed assay (5) and it led to the discovery of mutants with a thickened aleurone (ta) phenotype. This study focused primarily on the ta2-1 allele (5) and the finding that the phenotype was caused by a dominant negative effect of altered OsROS1 transcripts (mOsROS1). Consistent with this interpretation, the overexpression of mOsROS1 in wild-type rice was sufficient to increase aleurone layers. Additional OsROS1 weak alleles (named ta2-2 to ta2-6) from a TILLING (5) screen have mutations in the conserved glycosylase domain (ta2-2 and ta2-3) or in nonconserved regions of OsROS1 (ta2-4 to ta2-6). Interestingly, mutant alleles of ta2-4 to ta2-6 showed no visible defects in yield-related traits, but the dominant negative aleurone effect was observed for ta2-3 and weakly for ta2-4, ta2-5, and ta2-6 alleles. The mechanism underlying the dominant negative effect in ta2 mutants is still not known.
To investigate how OsROS1 contributes to aleurone formation, the authors compared DNA methylomes of the wild-type and ta2-1 endosperms. Their study suggests that OsROS1 may regulate DNA demethylation at the promoter regions of RISBZ1 and RPBF, two previously known regulators of aleurone layer formation (5). Compared with the wild type, there was increased DNA methylation and decreased gene expression in ta2-1 for these two genes. They also identified 15,147 hypermethylated genomic regions, and there is a possibility that many other genes could be targeted by OsROS1 and contribute to aleurone formation in rice (Fig. 1). In the future, it will be interesting to investigate additional genes that are hypermethylated and, thus, potentially regulated by OsROS1 in ta2-1 mutants.
In summary, this study reveals a function of DNA demethylase in aleurone formation in rice grains and broadens our understanding of the DNA demethylation pathway in plant development, especially in cereal crops. It also provides a strategy for enhancing the nutritional value of rice, because it is the aleurone layer that stores proteins, lipids, vitamins, and minerals and is the most nutritious part of cereal grains (14). OsROS1 could be modified in different rice varieties using the CRISPR-Cas9 technology (15), especially brown rice varieties in which, unlike in white rice, the aleurone, together with the pericarp and seed coat, is not removed during processing. Gene editing could also be used to find out whether the DNA demethylase-mediated aleurone formation is conserved in maize and other cereals, most of which have one single-cell layered aleurone.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11327 in issue 44 of volume 115.
References
- 1.Zhang H, Lang Z, Zhu JK. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 2.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains. Proc Natl Acad Sci USA. 2018;115:11327–11332. doi: 10.1073/pnas.1806304115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 2009;59:908–920. doi: 10.1111/j.1365-313X.2009.03925.x. [DOI] [PubMed] [Google Scholar]
- 6.Clark AT. DNA methylation remodeling in vitro and in vivo. Curr Opin Genet Dev. 2015;34:82–87. doi: 10.1016/j.gde.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 8.Yamamuro C, et al. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satgé C, et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat Plants. 2016;2:16166. doi: 10.1038/nplants.2016.166. [DOI] [PubMed] [Google Scholar]
- 10.Lang Z, et al. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci USA. 2017;114:E4511–E4519. doi: 10.1073/pnas.1705233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R, et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci USA. 2015;112:10804–10809. doi: 10.1073/pnas.1503362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conde D, et al. Chilling-responsive DEMETER-LIKE DNA demethylase mediates in poplar bud break. Plant Cell Environ. 2017;40:2236–2249. doi: 10.1111/pce.13019. [DOI] [PubMed] [Google Scholar]
- 13.Ono A, et al. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012;71:564–574. doi: 10.1111/j.1365-313X.2012.05009.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Liu J, Li D, Liu CM. Rice caryopsis development II: Dynamic changes in the endosperm. J Integr Plant Biol. 2016;58:786–798. doi: 10.1111/jipb.12488. [DOI] [PubMed] [Google Scholar]
- 15.Mao Y, Botella JR, Zhu JK. Heritability of targeted gene modifications induced by plant-optimized CRISPR systems. Cell Mol Life Sci. 2017;74:1075–1093. doi: 10.1007/s00018-016-2380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]