Significance
There is an epigenetic requirement for de novo kinetochore assembly in budding yeast. Perturbations that delay S phase suppress assembly defects. We propose a role for replication fork progression in allowing DNA to adopt configurations conducive for centromere function. The function of replication fork stalling upon encounters with DNA damage or other blockades is to allow time for thermal fluctuations of the DNA chain to explore numerous configurations. Biasing thermodynamics provides a mechanism to facilitate macromolecular assembly, DNA repair, and other nucleic acid transactions at the replication fork. These loop configurations are essential for sister centromere separation and kinetochore assembly in the absence of the COMA complex.
Keywords: kinetochore, centromere, COMA
Abstract
De novo kinetochore assembly, but not template-directed assembly, is dependent on COMA, the kinetochore complex engaged in cohesin recruitment. The slowing of replication fork progression by treatment with phleomycin (PHL), hydroxyurea, or deletion of the replication fork protection protein Csm3 can activate de novo kinetochore assembly in COMA mutants. Centromere DNA looping at the site of de novo kinetochore assembly can be detected shortly after exposure to PHL. Using simulations to explore the thermodynamics of DNA loops, we propose that loop formation is disfavored during bidirectional replication fork migration. One function of replication fork stalling upon encounters with DNA damage or other blockades may be to allow time for thermal fluctuations of the DNA chain to explore numerous configurations. Biasing thermodynamics provides a mechanism to facilitate macromolecular assembly, DNA repair, and other nucleic acid transactions at the replication fork. These loop configurations are essential for sister centromere separation and kinetochore assembly in the absence of the COMA complex.
De novo establishment of centromeres is a critical function for kinetochore assembly and ensuring the fidelity of chromosome segregation (1). Centromeres can be lost from chromosomes through a variety of mechanisms, including chromosome breakage, translocation, and deletion (2–4). Cells have the ability to build neocentromeres from new sites in the genome. Analysis of the preferred sites for neocentromere formation indicates that the chromatin environment, in addition to DNA sequence, factors into where centromeres are built (5). Once the centromere specific histone H3 variant CENP-A is deposited, the centromere can be propagated in successive generations independent of the particular organization of pericentric heterochromatin (6).
De novo kinetochore assembly has been studied in budding yeast through the use of a conditionally functional centromere (GALCEN). Induction of transcription on galactose inactivates the adjacent centromere, while repression on glucose is permissive for centromere function and kinetochore formation (7). Chromosomes with two centromeres undergo a breakage–fusion–bridge cycle that persists until stable monocentric derivative chromosomes are generated (2, 4, 8). Dicentric chromosomes with one of the two centromeres as GALCEN can be stably maintained in the population by growth on galactose. Upon GALCEN centromere activation (+glucose), chromosome breakage ensues (2, 9, 10). Several components of the constitutive centromere associated protein network (CCAN in mammals, COMA in yeast) (11–15) have been identified that are required for de novo kinetochore formation at GALCEN (Chl4, Mcm21, Iml3/Mcm19; ref. 10). Chl4 (CENP-N) and Iml3/Mcm19 interact and function with COMA (16). Chl4/CENP-N has been identified as a CENP-A reader involved in CENP-A assembly and structural transitions (17, 18). Ctf19, the receptor for cohesin (19), Chl4, Iml3/Mcm19, and Mcm21 are all required for the enrichment of cohesin in the pericentromere [Ctf19 (20), Chl4 (21), Iml3/Mcm19 (21), and Mcm21 (22)].
Cohesin is loaded at centromeres in late G1 and early S phase where it functions to compact DNA loops between sister centromeres and maintain kinetochore clustering (23–27). Cohesin is enriched about 3× in the 30- to 50-kb pericentric region relative to the chromosome arm (28–30). The pericentric cohesin links adjacent DNA loops into a bottle-brush configuration that functions to stiffen the chromatin axis between sister centromeres (23). The stiffening between sister centromeres provides a means to facilitate sister kinetochore biorientation.
Delaying the rate or duration of DNA replication has been reported to rescue cohesion defects in the pericentromere in select COMA mutants (21). Replication delay allows the cell to restore cohesion in a mechanism independent of kinetochore-mediated cohesin enrichment (21). Thus, replication intermediates may provide key structural templates within the pericentromere that compensate for the reduction of cohesin. This finding highlights the potential role of DNA topology or other structural modalities such as looping in centromere function.
In this report we demonstrate that de novo kinetochore assembly can be restored through exposure to low levels of DNA damaging agents and in mutants exhibiting delays in DNA replication or nucleosome assembly. Direct measure of DNA looping together with simulations of chain motion reveal how a kinetic delay allows time for thermal DNA fluctuations to separate sister centromeres and bias conformation toward kinetochore assembly.
Results
COMA Mutants Are Defective in de Novo Centromere Assembly.
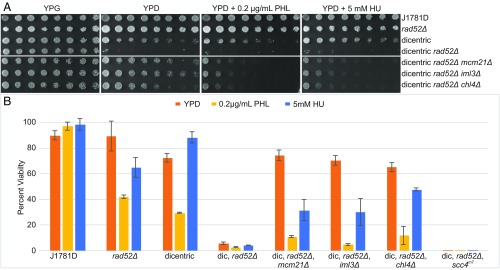
We used a highly sensitive dicentric chromosome breakage assay to quantitate de novo kinetochore assembly. Rad52 is the major component required for double-strand DNA break repair (31). Activation of the GALCEN centromere in rad52Δ mutants with a single chromosome containing GALCEN at HIS4 and endogenous CEN3 leads to lethal chromosome breakage events (Fig. 1A, dicentric rad52Δ, YPG vs. YPD). The ability of chl4Δ mutants to suppress centromere activation and kinetochore assembly (10) can be seen in the increased viability of rad52Δ, chl4Δ mutants with a dicentric chromosome on glucose (Fig. 1A, YPG vs. YPD). Dicentric rad52Δ, chl4Δ mutants exhibit growth out to an approximately eightfold dilution (87.5% viability, Fig. 1B) and 65% viability in single colony assays (Fig. 1B). Other members of the COMA complex including Iml3/Mcm19 and Mcm21 are also required for de novo kinetochore assembly (mcm21Δ and iml3Δ, Fig. 1A, rows 5 and 6; YPG vs. YPD) (10).
Fig. 1.
Viability assay for cells containing an active dicentric chromosome. (A) Serial dilutions of WT and various COMA mutants were pronged on galactose (YPG), glucose (YPD), glucose + phleomycin (PHL), and glucose + hydroxyurea (HU) plates. Strains with a dicentric chromosome are indicated. Dilutions are twofold from Left to Right. Prong plates were repeated in triplicate with identical results. Individual plates and treatments are separated by spaces. (B) Quantitative analysis of cell viability expressed as a percent of single colony growth on galactose (inactive GALCEN). WT cells (J1781D) are not sensitive to low levels of PHL or HU. rad52Δ mutants exhibit reduced viability in the presence of PHL and HU. WT cells containing a dicentric chromosome exhibit reduced viability on YPD due to chromosome breakage. WT cells containing the dicentric chromosome are sensitized to PHL but not HU. Dicentric, rad52Δ viability is dramatically reduced on YPD, PHL, and HU. Dicentric, rad52Δ with mcm21Δ, iml3Δ, or chl4Δ exhibit increased viability on YPD relative to dicentric, rad52Δ cells, indicating suppression of dicentric breakage in these COMA mutants. Dicentric, rad52Δ with mcm21Δ, iml3Δ, or chl4Δ exhibit reduced viability relative to YPD in the presence of PHL and HU, indicative of chromosome breakage. Error bars are SEM. Student’s t-test values can be found in SI Appendix, Table S1. Dilution plating numbers can be found in SI Appendix, Table S2.
To distinguish whether suppression of dicentric chromosome breakage was dependent on cohesin loading, we utilized a mutation in the cohesin loading protein, Scc4 (32). Scc4 is required for cohesin loading, but is not physically part of the kinetochore (33). Reduction of pericentric cohesin in an scc4m7 mutant does not prevent de novo kinetochore assembly (Fig. 1B, inviability of dicentric rad52Δ, scc4m7). The assay also discriminates functional subdomains within the COMA complex as deletion of the cohesin receptor, Ctf19 (19) does not prevent de novo kinetochore assembly (SI Appendix, Fig. S1, ctf19Δ, rad52Δ dicentric cells). Thus, loss of members of the COMA complex, and not reduction of pericentric cohesin, accounts for the defect in de novo kinetochore assembly.
Growth on Sublethal Levels of Phleomycin or Hydroxyurea Bypasses Defects in de Novo Kinetochore Assembly in a Subset of COMA Mutants.
The cohesion defect in chl4Δ and iml3Δ can be rescued through replication delays invoked by low concentrations of hydroxyurea (HU) (21). To examine whether similar treatments might rescue the kinetochore assembly defect, we treated COMA, rad52Δ mutants containing the conditionally dicentric chromosome with sublethal concentrations of phleomycin (PHL) or HU. In the presence of 0.2 µg/mL PHL or 5 mM HU, rad52Δ mutants were viable (Fig. 1A, row 2 YPG, YPD, YPD + PHL, YPD + HU). Quantitation from single colony plating of rad52Δ mutants revealed one break/every two cells (∼50% viability) on 0.2 µg/mL PHL, and one break/every three cells (66% viability) on 5 mM HU (Fig. 1B). Wild-type (WT) cells containing the dicentric chromosome (Fig. 1A, row 3 dicentric) exhibit a similar spectrum of sensitivity to PHL and HU as rad52Δ mutants on glucose (Fig. 1A, compare rows 2 and 3 YPD, +PHL, +HU). PHL treatment may be sensitizing WT cells to the dicentric chromosome, evidenced by their reduction in viability relative to YPD. In contrast, chl4Δ, rad52Δ mutants containing the conditional dicentric chromosome are highly sensitized to PHL and HU relative to YPD alone (Fig. 1A, row 7 YPD, +PHL, +HU). Two other COMA components, iml3Δ and mcm21Δ, exhibit decreased viability on glucose in the presence of PHL and HU (mcm21Δ, iml3Δ/mcm19Δ, Fig. 1B, rows 5 and 6, YPD vs. YPD + PHL, +HU).
To address whether drug treatments extend the cell cycle, thereby bypassing the defect in de novo kinetochore assembly, we measured the growth rates and distribution of cell cycle phases in various conditions [phleomycin 0.2 µg/mL, HU 5 mM, and benomyl (ben) 0. 5 µg/mL, SI Appendix, Fig. S2]. Exposure to 0.5 µg/mL benomyl does not activate the GALCEN centromere in mcm21Δ, iml3Δ, or chl4Δ mutant cells (SI Appendix, Fig. S3A). As shown in SI Appendix, Fig. S2, there are statistical differences in growth rate and distribution of growth phase in the various strains or treatments. However, there are no delays in S phase (as evidenced by reduced unbudded or small budded cells) in WT dicentric ± PHL (columns 1 and 2) or chl4Δ ± PHL (columns 5 and 6). There are a comparable number of small budded cells (22–24%) among chl4Δ untreated, chl4Δ + PHL and chl4Δ + ben. At these concentrations of PHL or HU, there is no effect on cell growth in inactive GALCEN dicentric COMA mutants (SI Appendix, Fig. S3C) or monocentric single mutants (SI Appendix, Fig. S3D).
The statistical differences in the growth curves (SI Appendix, Fig. S2B) do not correlate with centromere activation. The slowest growing cells were WT dicentrics treated with HU or ben. HU treatment activates the GALCEN centromere (SI Appendix, Fig. S3B, 5 and 10 mM HU) while benomyl does not. The fastest growing cells were chl4Δ dicentrics on glucose (centromere inactive), chl4Δ dicentric + PHL (cen active), and WT dicentric glu (cen active). There is no systematic delay in cell cycle progression or growth that accounts for the ability to activate the GALCEN centromere in COMA mutants.
Generation of Monocentric Derivatives Following Replication Delay.
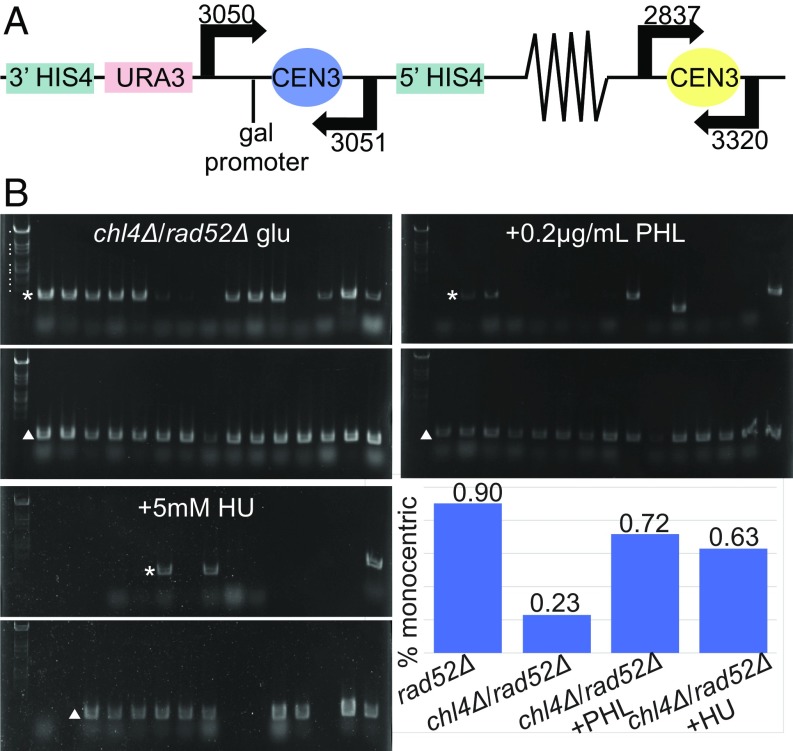
The fraction of viable cells remaining after activation of the conditional centromere in a rad52Δ mutant will become monocentric through nonhomologous recombination (9). To examine the consequences of drug treatment on the physical stability of the dicentric chromosome, we performed PCR analysis on DNA extracted from wild-type and mutant cells grown on glucose in the presence or absence of drug treatment. In viable cells following dicentric chromosome activation, >90% of cells have lost either GALCEN3 or CEN3 (Fig. 2B) (9). In rad52Δ, chl4Δ mutants only 7/30 cells have lost either of the two centromeres (23% chl4Δ/rad52Δ, Fig. 2B, chart). Upon exposure to HU or PHL, there is an increase in the number monocentric derivatives in rad52Δ, chl4Δ mutants, predominately retaining the endogenous CEN3 (Fig. 2B, from 7/30 in glucose to 18/25 PHL and 19/30 HU). The physical analysis confirms and extends the viability assay, demonstrating centromere activation and kinetochore assembly upon exposure to PHL. Exposure to sublethal concentrations of PHL or HU allow chl4Δ, iml3Δ, and mcm21Δ mutants to assemble kinetochores at the GALCEN3 locus.
Fig. 2.
Physical analysis of dicentric chromosome III. (A) Schematic of dicentric chromosome showing locations of GALCEN3 at the HIS4 gene and relevant primers used in PCR assays. (B) DNA was isolated from single colonies of dicentric chl4Δ, rad52Δ cells grown on YPD, YPD + PHL (0.2 µg/mL), and YPD + HU (5 mM). PCR products for GALCEN3 (primers 3050, 3051; 865-bp product indicated with asterisk *) and CEN3 (primers 2837, 3320; 625-bp product indicated with Δ) were subjected to gel electrophoresis. Individual gels were cropped and separated by spaces. On glucose (YPD), only 7/30 chl4Δ, rad52Δ colonies contained chromosome III rearrangements (lacking one of the two centromeres). On PHL, 18/25 were rearranged and on HU, 19/30 were rearranged. In a rad52Δ strain containing the dicentric chromosome, 28/31 colonies exhibited rearrangements. Standard marker sizes in descending order are as follows: 7.8 kb, 4.3 kb, 3.7 kb, 3.3 kb, 2.3 kb, 2.0 kb, 1.8 kb, 1.5 kb, 1.4 kb, and 1.1 kb. Standard marker sizes are identical throughout.
An alternative pathway to monocentric derivative chromosomes is through homologous recombination within the conditional dicentric chromosome in radiation resistant cells (SI Appendix, Fig. S4A) (2). In wild-type cells containing the dicentric chromosome, ∼70% of cells harbor the monocentric derivative chromosome generated by GALCEN3–CEN3 recombination (WT dicentric, SI Appendix, Fig. S4C). In iml3Δ mutants containing the dicentric chromosome, the suppression of dicentric breakage is evident by the lack of monocentric derivatives (SI Appendix, Fig. S4 B and C). There is elevated centromere rearrangement product upon exposure to PHL vs. no drug (∼20% CEN rearrangement product/LEU2, SI Appendix, Fig. S4 B and C).
Deletion of Csm3 Rescues de Novo Kinetochore Assembly.
Cohesion can be reestablished in iml3Δ and chl4Δ COMA mutants without additional cohesin through a kinetic mechanism that depends on replication fork delay (21). The notion that the timing of replication contributes to the cohesive mechanism comes from analysis of Csm3. The Csm3/Tof1 complex travels with the replication fork and is required for fork rotation and stalling. Csm3 is also required for the replication fork to reach its maximal velocity (34). Csm3, like COMA, is required for cohesin enrichment in the pericentromere (21). However, unlike COMA mutants, replication fork delay through HU treatment does not suppress the cohesion defect in csm3Δ mutants (21).
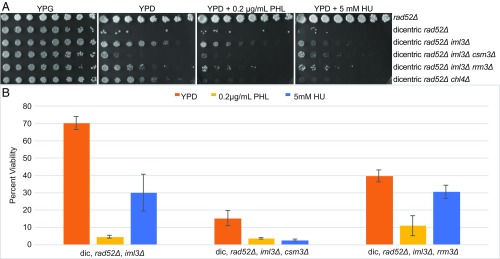
To test whether Csm3 participates in de novo kinetochore assembly, we deleted CSM3 from iml3Δ mutants containing the conditional dicentric chromosome. The fraction of iml3Δ, rad52Δ, csm3Δ viable cells on glucose dropped to 10% compared with 70% viability for the double mutant (iml3Δ, rad52Δ) (Fig. 3A, row 4). Mutants in replication fork passage rescue de novo kinetochore assembly in iml3Δ mutants as evidenced by dicentric chromosome breakage and cell inviability in the absence of DNA repair. Further treatment with HU or PHL only marginally enhances the csm3Δ rescue phenotype (Fig. 3B). This result, together with the rescue of de novo kinetochore assembly in chl4Δ, iml3Δ, and mcm21Δ mutants upon HU or PHL treatment (Fig. 1), indicates that delays in replication fork progression may provide a kinetic mechanism for promoting de novo kinetochore assembly in the absence of the COMA complex.
Fig. 3.
Rescue of de novo kinetochore assembly in COMA mutants through csm3Δ and rrm3Δ. (A) Serial dilutions of rad52Δ, dicentric rad52Δ, and dicentric rad52Δ iml3Δ mutants with csm3Δ or rrm3Δ. Dilutions (twofold) from Left to Right, pronged on galactose (YPG), glucose (YPD), glucose + phleomycin (PHL), and glucose + hydroxyurea (HU) plates. Deletion of the replication fork protection protein Csm3 rescues de novo kinetochore assembly in iml3Δ mutants. Deletion of the Rrm3 helicase has a similar but less dramatic effect. Prong plates were repeated in triplicate with identical results. (B) Quantitative analysis of cell viability expressed as a percent of single colony growth on galactose (inactive GALCEN). Error bars are SEM. Student’s t-test values can be found in SI Appendix, Table S1. Dilution plating numbers can be found in SI Appendix, Table S2.
If the rate of fork progression contributes to de novo kinetochore assembly, then deletion of the helicase that removes roadblocks might also suppress the kinetochore assembly deficiency in iml3Δ mutants. To test this hypothesis, we deleted the RRM3 helicase in iml3Δ, rad52Δ dicentric strains. As predicted, the triple mutant was more sensitive to glucose (40% viable) than the double mutant (70%) (Fig. 3A, rows 3 and 5, and Fig. 3B). Thus, slowing the fork down due to loss of the fork complex, csm3Δ, or the helicase, rrm3Δ, confer varying degrees of de novo assembly function to iml3Δ mutants.
Deletion of Cac1 (of the Chromatin Assembly Complex, CAF1) Suppresses the Defect in de Novo Kinetochore Assembly.
An alternative mechanism for promoting de novo kinetochore assembly could be complexes that modulate histone incorporation. To test the role of replication coupled nucleosome assembly on de novo kinetochore assembly, we constructed chl4Δ, cac1Δ dicentrics (SI Appendix, Fig. S5A). Colonies were heterogeneous on glucose, indicative of dicentric breakage. In cac1Δ mutants, the viability was 67%, comparable to the viability of WT cells containing a dicentric chromosome. To quantitate the extent of breakage, we determined the viability of chl4Δ, cac1Δ, rad52Δ, mutants containing the dicentric chromosome. The viability of the triple mutants dropped to less than 20%, indicative of GALCEN activation on glucose and chromosome breakage. Thus, loss of the chromatin assembly factor (Cac1) bypasses defects in de novo kinetochore assembly. We further examined these cells for monocentric derivative chromosomes and found the diagnostic rearrangement product in 10/10 cells (SI Appendix, Fig. S5B). Loss of Cac1 allows kinetochore assembly as evidenced by dicentric chromosome breakage.
Chromosome Conformation at the Centromere.
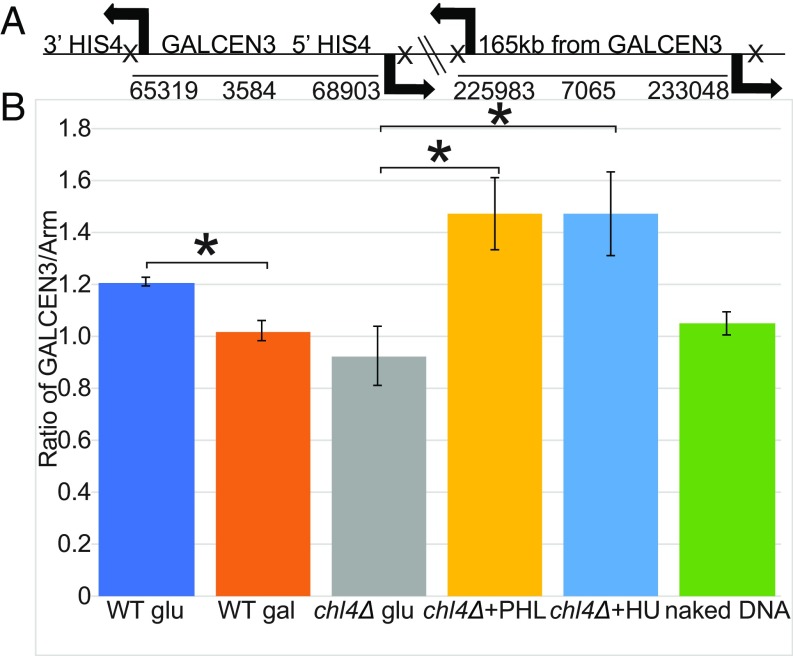
We had previously shown that upon kinetochore formation, a DNA loop with the endogenous centromere at its apex is formed (30). To examine DNA loops at the newly assembled centromere, we utilized an inverse-PCR strategy to capture chromosome conformation (3C) (35). Inverse primer pairs are shown in Fig. 4A for GALCEN3 at the HIS4 locus and a distal arm locus 165 kb from GALCEN3. Chromatin was fixed, digested with NsiI, and ligated under dilute conditions to minimize intermolecular reactions. Each primer lies between 250 and 450 bp from the NsiI restriction enzyme site resulting in PCR products of ∼600 bp when the sites are in ligation proximity. For quantitation of random association due to thermal motion, ligation was performed on DNA isolated from cells in the absence of cross-linking (naked DNA). The ratio of GALCEN3 to the distal arm locus is comparable in cells grown on galactose and uncross-linked, naked DNA samples. There is no tendency for DNA loops at either of these positions in the absence of centromere function. To examine the conformation of functional GALCEN3, cells were switched from galactose to glucose for 3 h. There is a 20% increase in ligation efficiency of loops at GALCEN3 in cells containing the dicentric chromosome (Fig. 4B, WT, glu, 1.2).
Fig. 4.
Loop formation at GALCEN3 is stimulated upon exposure to PHL. The schematic (A) shows the position of oligonucleotide primers on chromosome III (arrows) relative to GALCEN3 at the HIS4 locus (Left) and a distal arm marker, 165 kb from the GALCEN3 (Right). Each pair of oligonucleotides (GALCEN3 vs. arm chromatin) extend away from each other on the linear chromosome. NsiI sites are indicated by X downstream of each oligonucleotide primer. In the linear chromosomal configuration, these oligonucleotides will not amplify DNA in the 3C assay. If there is intramolecular looping, the oligonucleotide pairs will amplify DNA. (B) The products from PCR reactions following cross-linking, restriction digestion, and ligation were quantified as described in Materials and Methods. Ratio of GALCEN3/Arm for WT, chl4Δ cells (glu, 0.2 µg/mL PHL, 10 mM HU), and naked DNA containing the conditionally functional dicentric chromosome are plotted. Error bars are SEM. An asterisk designates a Student’s t-test P value of <0.05. Student’s t-test P value for WT glu vs. WT gal is 0.011; chl4Δ + PHL vs. chl4Δ glu is 0.032; chl4Δ + HU vs. chl4Δ glu is 0.049. WT glu is not statistically different from chl4Δ + PHL, Student’s t-test P value = 0.168. Each sample was prepared once and PCR was repeated. WT glu n = 3, WT gal n = 3, chl4Δ glu n = 3, chl4Δ + PHL n = 4, chl4Δ + HU n = 4, naked DNA n = 6.
To examine the conformation of inactive GALCEN3 in chl4Δ mutants, cells were grown on glucose. There is no increase in ligation efficiency at GALCEN3 in chl4Δ mutants relative to GALCEN3 in WT cells grown on galactose (Fig. 4B, chl4Δ glu vs. WT gal). Any remaining structural components at the inactive kinetochore in chl4∆ mutants on glucose (36) are unable to convert centromere DNA into a looped organization.
To test whether there is a conformational change in the presence of PHL or HU as predicted from the restoration of kinetochore function, cells were grown on glucose with PHL or HU for 3 h. There is a 40% increase in contact frequency 3.5 kb surrounding GALCEN3 upon drug treatment (Fig. 4B). Licensing kinetochore assembly at a de novo centromere is accompanied by organizing the centromere into a loop.
Discussion
The specification of centromere function varies from sequence based in budding yeast to epigenetic inheritance in mammals and a variety of other species. The finding that sequence identity was not sufficient in COMA mutants in budding yeast to build a kinetochore (10, 37) raised the prospect that there are nonsequence-specific requirements even in the simplest point centromere organisms. Centromeres are found at the apex of a DNA loop (30, 38) that accounts in large part for the Rabl organization of chromosomes in budding yeast. An unanswered question is whether the centromere DNA loop is a consequence of kinetochore assembly, or rather that centromere DNA looping is required for timely recognition by the sequence-specific DNA binding protein complex (Cbf3). We have found that pausing replication via DNA damage (39) or limiting nucleotide diphosphates (40) rescues COMA mutants defective in de novo kinetochore assembly. We propose that the function of replication fork stalling upon encounters with DNA damage or other blockades is to allow time for thermal fluctuations of the DNA chain to explore numerous configurations. Biasing thermodynamics provides a mechanism to facilitate macromolecular assembly, DNA repair, and other nucleic acid transactions at the replication fork. In the case of centromere, these loop configurations are essential for kinetochore assembly.
Kinetochore assembly following DNA replication requires the complete replacement of the centromere-specific histone Cse4 (41, 42) and several inner kinetochore components (Dsn1, part of the MIND complex and Spc105) (43). In addition, there is a pause in replication fork progression through the centromere in wild-type cells (44). Whether the increase in duration of replication intermediates is conducive for the unique mode of Cse4 deposition to both strands or for coordinating Cse4 deposition with kinetochore assembly is not known. From the perspective of tension or DNA chain strain, it is important to consider the force generated by DNA polymerase. Replication is force dependent, proceeding to a stall force of 34–45 pN (45, 46), three to four times greater than the force observed at the kinetochore microtubule plus ends (46). We propose that fork stalling, or other dynamical features of the replisome, provides a mechanism to reduce the force exerted by replication and to allow the DNA chains time to explore numerous configurational states favoring kinetochore assembly. The pause in fork progression also prevents the dissipation of positive supercoils. Positive supercoils that precede the fork dissipate into precatenanes behind the fork if the fork is able to swivel. By blocking fork swiveling (Csm3), newly replicated centromeres remain separated, a configuration that may predispose centromeres for kinetochore assembly (47). Additionally, low doses of the topoisomerase II inhibitor etoposide, rescue de novo kinetochore assembly in chl4Δ, mcm21Δ, and iml3Δ mutants (SI Appendix, Fig. S7).
Csm3 is also required for the replication fork to reach its peak velocity (34). The mechanics of replication are in kinetic competition with sister centromere separation and loop formation. Slowing down the rate of replication gives a kinetic advantage for kinetochore components to interact with emerging sister centromere strands to build the kinetochore. This model predicts a dependency on DNA length, such that there is a sufficient amount of DNA to get the two sister centromere DNA loops apart. The instability of small circular minichromosomes relative to endogenous chromosomes has been attributed to their small size (48), but could also reflect the decreased efficiency of kinetochore assembly on such a small template.
DNA motion and the dynamics of biochemical transactions must be integrated into mechanistic models for DNA processes such as repair and transcription (49). To explore the mechanistic basis of replication fork passage we turned to a polymer dynamics simulator (ChromoShake) designed to explore thermodynamic configurations (50). The model uses a series of beads linked by springs and hinges, subjected to Brownian motion (50). Each bead in the simulation represents 10 nm or about 30 bp of b-form DNA. The model successfully captures the behavior of the centromere in budding yeast mitosis (50). We simulated a replication bubble by duplicating a 350-nm region of polymer bead-spring chains within a 1-μm bead-spring chain (SI Appendix, Fig. S5D). To simulate tension on the DNA template we increased the mass of the end beads 100× over the mass of the internal beads. The mass increase of the end bead reflects the estimated drag force from the chromosome arms (SI Appendix, Fig. S6). Tension on the chain from DNA polymerase (SI Appendix, Fig. S5D, pinned) restricts motion and separation of the replicated centromeres (green beads). The distance between the two replication forks along the DNA axis remains larger than the distance between separated sister centromeres (ratio of centromere separation/fork separation = ∼0.5, SI Appendix, Fig. S5D, pinned). If the replicated strands were free to adopt a random coil, the ratio between centromere separation and fork separation should approach 1. To model a pause in replication, we reduced the mass of the end beads to that of the chain beads. As shown in snapshots from the model (SI Appendix, Fig. S5D, unpinned) there is an increase in propensity to form centromere loops, as the ratio of centromere-to-fork separation increased to ∼0.75 (SI Appendix, Fig. S5D, unpinned). Simulations of chromatin compaction by bridging every seventh bead (sevenfold nucleosome compaction) give similar increases in the ratio of centromere separation to fork separation (SI Appendix, Fig. S5D, compacted). In this way, the initial mode of centromere separation is derived from the thermodynamics of the chains and their behavior relative to the replication machinery. We hypothesize that a kinetic delay gives the chains freedom to adopt a looped configuration more favorable for kinetochore formation.
Formation of a centromere DNA loop is likely the physical catalyst for kinetochore assembly. Based on the studies herein, we conclude that the COMA complex is integral to mechanisms required for loop formation of the centromere. Based on the hypothesis that the kinetics of chain motion is a key feature in assembly, we propose a function for COMA in remodeling the centromere into an assembly-conducive conformation. One possibility is that COMA is the centromere–CTCF factor, analogous to the mechanism by which CTCF leads to increased cohesin concentration at the base of loops (51). Interestingly, the centromere DNA binding factor Cbf3 bends centromere DNA as probed with atomic force microscopy (52). Centromere bending may reflect the cumulative function of inner kinetochore complexes that act synergistically to mold the centromere into a kinetochore competent precursor.
Materials and Methods
Yeast cells were cultured and imaged under conditions as described in ref. 25. Simulations were performed in ChromoShake (50), using the geometries of bead spring polymers as shown in SI Appendix, Fig. S5D. In the “pinned” simulation, the end beads remained two orders of magnitude larger vs. the “unpinned” simulation, the mass of the two end beads was reduced to that of a standard bead. Chromosome conformation (3C) was performed as outlined in ref. 30.
Supplementary Material
Acknowledgments
We thank S. Hinshaw and S. Harrison for generously supplying the scc4m7 mutant. This work was supported by NIH Grants R37GM32238 (to K.B.) and T32CA201159–01 (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806791115/-/DCSupplemental.
References
- 1.Scott KC, Sullivan BA. Neocentromeres: A place for everything and everything in its place. Trends Genet. 2014;30:66–74. doi: 10.1016/j.tig.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brock JA, Bloom K. A chromosome breakage assay to monitor mitotic forces in budding yeast. J Cell Sci. 1994;107:891–902. doi: 10.1242/jcs.107.4.891. [DOI] [PubMed] [Google Scholar]
- 3.Leibowitz ML, Zhang CZ, Pellman D. Chromothripsis: A new mechanism for rapid karyotype evolution. Annu Rev Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- 4.McClintock B. Induction of instability at selected loci in maize. Genetics. 1953;38:579–599. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrey EJ, Heun P. Artificial chromosomes and strategies to initiate epigenetic centromere establishment. Prog Mol Subcell Biol. 2017;56:193–212. doi: 10.1007/978-3-319-58592-5_8. [DOI] [PubMed] [Google Scholar]
- 6.Stankovic A, Jansen LET. Quantitative microscopy reveals centromeric chromatin stability, size, and cell cycle mechanisms to maintain centromere homeostasis. Prog Mol Subcell Biol. 2017;56:139–162. doi: 10.1007/978-3-319-58592-5_6. [DOI] [PubMed] [Google Scholar]
- 7.Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mythreye K, Bloom KS. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Pekgöz Altunkaya G, et al. CCAN assembly configures composite binding interfaces to promote cross-linking of Ndc80 complexes at the kinetochore. Curr Biol. 2016;26:2370–2378. doi: 10.1016/j.cub.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol. 2015;25:671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schleiffer A, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal M, Mehta G, Ghosh SK. Role of Ctf3 and COMA subcomplexes in meiosis: Implication in maintaining Cse4 at the centromere and numeric spindle poles. Biochim Biophys Acta. 2015;1853:671–684. doi: 10.1016/j.bbamcr.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 17.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J, et al. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev. 2015;29:1058–1073. doi: 10.1101/gad.259432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinshaw SM, Makrantoni V, Harrison SC, Marston AL. The kinetochore receptor for the cohesin loading complex. Cell. 2017;171:72–84.e13. doi: 10.1016/j.cell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernius J, Marston AL. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009;5:e1000629. doi: 10.1371/journal.pgen.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrimore J, et al. DNA loops generate intracentromere tension in mitosis. J Cell Biol. 2015;210:553–564. doi: 10.1083/jcb.201502046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens AD, et al. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J Cell Biol. 2013;200:757–772. doi: 10.1083/jcb.201208163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens AD, et al. The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol Biol Cell. 2013;24:3909–3919. doi: 10.1091/mbc.E13-06-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens AD, et al. Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J Cell Biol. 2013;203:407–416. doi: 10.1083/jcb.201307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- 30.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resnick MA, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 32.Hinshaw SM, Makrantoni V, Kerr A, Marston AL, Harrison SC. Structural evidence for Scc4-dependent localization of cohesin loading. eLife. 2015;4:e06057. doi: 10.7554/eLife.06057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeeles JTP, Janska A, Early A, Diffley JFX. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell. 2017;65:105–116. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 36.Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durand-Dubief M, et al. SWI/SNF-like chromatin remodeling factor Fun30 supports point centromere function in S. cerevisiae. PLoS Genet. 2012;8:e1002974. doi: 10.1371/journal.pgen.1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Koç A, Wheeler LJ, Mathews CK, Merrill GF. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem. 2004;279:223–230. doi: 10.1074/jbc.M303952200. [DOI] [PubMed] [Google Scholar]
- 41.Pearson CG, et al. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 42.Wisniewski J, et al. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife. 2014;3:e02203. doi: 10.7554/eLife.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenfeder SA, Newlon CS. Replication forks pause at yeast centromeres. Mol Cell Biol. 1992;12:4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 47.Schvartzman JB, Martínez-Robles ML, Hernández P, Krimer DB. The benefit of DNA supercoiling during replication. Biochem Soc Trans. 2013;41:646–651. doi: 10.1042/BST20120281. [DOI] [PubMed] [Google Scholar]
- 48.Murray AW, Schultes NP, Szostak JW. Chromosome length controls mitotic chromosome segregation in yeast. Cell. 1986;45:529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- 49.Graham JE, Marians KJ, Kowalczykowski SC. Independent and stochastic action of DNA polymerases in the replisome. Cell. 2017;169:1201–1213.e17. doi: 10.1016/j.cell.2017.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrimore J, et al. ChromoShake: A chromosome dynamics simulator reveals that chromatin loops stiffen centromeric chromatin. Mol Biol Cell. 2016;27:153–166. doi: 10.1091/mbc.E15-08-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fudenberg G, et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pietrasanta LI, et al. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.