Significance
Cell-fate determination and cellular behavior in plants rely mainly on positional information and intercellular communication. A plethora of cues are perceived by surface receptors and integrated into an adequate cellular output. Here, we show that the small receptor-like protein RLP44 acts as an intermediary to connect the receptors for two well-known signaling molecules, brassinosteroid and phytosulfokine, to control cell fate in the root vasculature. Furthermore, we show that the brassinosteroid receptor has functions that are independent from the responses to its hormone ligands and reveal that phytosulfokine signaling promotes procambial cell identity. These results provide a mechanistic framework for the integration of multiple signaling pathways at the plasma membrane by shifting associations of receptors in multiprotein complexes.
Keywords: cell fate, plant development, xylem, brassinosteroids, phytosulfokine
Abstract
Multicellularity arose independently in plants and animals, but invariably requires a robust determination and maintenance of cell fate that is adaptive to the environment. This is exemplified by the highly specialized water- and nutrient-conducting cells of the plant vasculature, the organization of which is already prepatterned close to the stem-cell niche, but can be modified according to extrinsic cues. Here, we show that the hormone receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) is required for root vascular cell-fate maintenance, as BRI1 mutants show ectopic xylem in procambial position. However, this phenotype seems unrelated to canonical brassinosteroid signaling outputs. Instead, BRI1 is required for the expression and function of its interacting partner RECEPTOR-LIKE PROTEIN 44 (RLP44), which, in turn, associates with the receptor for the peptide hormone phytosulfokine (PSK). We show that PSK signaling is required for the maintenance of procambial cell identity and quantitatively controlled by RLP44, which promotes complex formation between the PSK receptor and its coreceptor. Mimicking the loss of RLP44, PSK-related mutants show ectopic xylem in the position of the procambium, whereas rlp44 is rescued by exogenous PSK. Based on these findings, we propose that RLP44 controls cell fate by connecting BRI1 and PSK signaling, providing a mechanistic framework for the dynamic balancing of signaling mediated by the plethora of plant receptor-like kinases at the plasma membrane.
A key function of signaling networks in multicellular organisms is to ensure robust determination and maintenance of cell fate. In plants, extreme specialization is displayed by the cells of the vascular tissues, which are vital for the distribution of water, nutrients, and signaling molecules. Xylem tracheary elements are characterized by lignified secondary cell-wall thickenings that protect against collapse and provide mechanical support for vertical growth. Positioned between xylem and the nutrient-transporting phloem are the cells of the procambium, which give rise to the lateral meristems during secondary growth (1). In Arabidopsis, root vascular tissue patterning is set up in the embryo by mutual antagonism of auxin and cytokinin signaling domains (2–5), but can adapt to environmental conditions later in development (6). After xylem precursor cells are displaced from the root meristem, an intricate gene-regulatory network connected to patterning mechanisms mediates differentiation into tracheary elements (7–10). Thus, primary root vascular patterning can be traced back to early specification events in the embryo. In contrast, during secondary growth, (pro)cambial cells adjacent to the existing tracheary elements acquire a xylem cell fate dependent on positional information (11).
Brassinosteroid (BR) hormone signaling (12, 13) is implicated in xylem differentiation and vascular patterning (14, 15). BRs are perceived by BRASSINOSTEROID INSENSITIVE 1 (BRI1) (16), which belongs to the large group of plant receptor-like kinases (RLK) with a leucine-rich repeat (LRR) extracellular domain, a transmembrane domain, and a cytosolic kinase domain related to animal Irak and Pelle kinases (17). Upon ligand binding, BRI1 heterodimerizes with members of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) LRR-RLK family such as BRI1-ASSOCIATED KINASE 1 (BAK1) (18, 19) and activates a signaling cascade that negatively regulates BRASSINOSTEROID INSENSITIVE 2 (BIN2) (20), a GSK3-like kinase that phosphorylates the BR-responsive transcription factors BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1 EMS SUPPRESSOR 1 (BES1)/BZR2 (21, 22). Inhibition of BIN2 activity allows BZR1 and BES1 to translocate to the nucleus, where they mediate BR-responsive transcriptional changes (23–25). A so far somewhat enigmatic relationship exists between BR and PHYTOSULFOKINE (PSK) signaling. PSKs are small secreted peptides that have been implicated in a variety of fundamental processes and are perceived by two close relatives of BRI1, PHYTOSULFOKINE RECEPTOR-1 and -2 (26–29). PSK activity depends on proteolytic processing of the precursor peptides, the sulfation of two tyrosine residues in the mature pentapeptide (YIYTQ) by TYROSYLPROTEIN SULFOTRANSFERASE (TPST) (30), and functional BR signaling (31, 32). At present, it is not clear how BR and PSK signaling interact, but the receptors for both growth factors share the requirement for a SERK coreceptor (33, 34).
Recently, we demonstrated that feedback information from the cell wall is integrated with BR signaling at the level of the receptor complex through RECEPTOR-LIKE PROTEIN (RLP) 44 (35). RLP44 is genetically required for the BR-mediated response to cell-wall modification and is sufficient to elevate BR signaling when overexpressed. RLP44 was shown to be in a complex with BRI1 and BAK1. Thus, we hypothesized that RLP44 modulates BR signaling strength in response to cues from the cell wall (35). However, it is not clear whether the RLP44-BR–signaling module plays additional roles in plant physiology (36). Here, we show that RLP44 is required for the maintenance of cell fate in the root vasculature by connecting components of the BR and PSK signaling pathways. RLP44 controls xylem differentiation in a BRI1-dependent manner by directly interacting with PSKR1 and promoting its interaction with BAK1. In addition, the rlp44 phenotype can be rescued by application of PSK peptide, and mutants affected in PSK signaling show an rlp44-like xylem phenotype, suggesting that RLP44 has a positive effect on PSK signaling, which, in turn, promotes procambial identity.
Results
RLP44 Is Expressed in the Developing Root Vasculature.
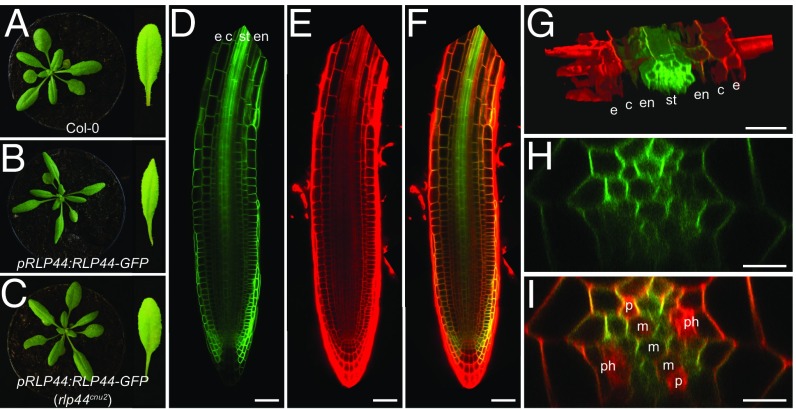
We previously demonstrated that RLP44 is present in a complex with BRI1 and BAK1 and is able to promote BR signaling upon cues from the cell wall or when overexpressed (35). To study the function of RLP44 in more depth, we generated transgenic plants expressing a translational GFP fusion of RLP44 under the control of the RLP44 promoter (pRLP44:RLP44-GFP). These plants displayed elongated, narrow leaf blades and elongated petioles, reminiscent of BRI1-overexpressing plants (Fig. 1 A and B) (37), as previously observed for RLP44 overexpression (35). We crossed a pRLP44:RLP44-GFP line with the RLP44 loss-of-function mutant rlp44cnu2 (35), resulting in plants with a wild-type–like appearance (Fig. 1C), demonstrating that the fusion protein is functional and confirming that the transgenic RLP44 expression (SI Appendix, Fig. S1A) is causative for the observed morphological effects. In the root apical meristem of pRLP44:RLP44-GFP and pRLP44:RLP44-GFP (rlp44cnu2), fluorescence was markedly enriched in the stele toward the more mature part of the root (Fig. 1 D–G and SI Appendix, Fig. S1 B–E) in accordance with previously published transcriptome data (37) and β-glucuronidase reporter activity under control of the RLP44 promoter (SI Appendix, Fig. S1 F and G). In the differentiating part of the root stele, RLP44-GFP fluorescence was present in all cell types, including the undifferentiated procambial cells (Fig. 1 H and I and SI Appendix, Fig. S1 C and D).
Fig. 1.
RLP44 is expressed in the root vascular tissue. (A) Col-0. (B) pRLP44:RLP44-GFP in wild-type background shows a growth phenotype reminiscent of enhanced BR signaling. (C) Mutation of endogenous RLP44 in pRLP44:RLP44-GFP (rlp44cnu2) reconstitutes wild-type–like phenotype. (D–F) pRLP44:RLP44-GFP expression (D) in root meristem counterstained with propidium iodide (E) and merged (F). c, cortex; e, epidermis; en, endodermis; st, stele. (Scale bars: 100 µm.) (G) Projection of a confocal stack through the differentiation zone before maturation of the casparian strip of a pRLP44:RLP44-GFP root showing fluorescence predominantly in the stele. Labeling as in D. (H and I) Optical section through the stele of a pRLP44:RLP44-GFP–expressing root in the differentiation zone (H), counterstained with propidium iodide (I), indicating differentiated phloem (ph) and protoxylem (p) and as-yet-undifferentiated metaxylem (m). (Scale bar: 10 µm.)
RLP44 Controls Xylem Cell Fate.
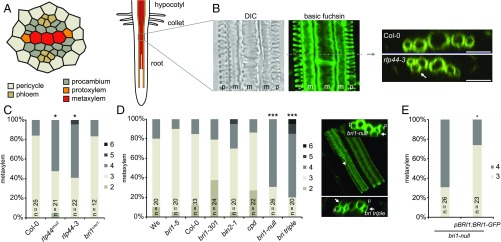
Because our reporter lines suggested expression of RLP44 in the stele, we assessed the role of RLP44 in vascular development. We visualized lignified secondary cell walls in rlp44 loss-of-function mutants through basic fuchsin staining. Strikingly, we observed supernumerary metaxylem-like cells, frequently outside the primary xylem axis in the position of the procambium (Fig. 2 A and B and SI Appendix, Fig. S2A), a phenotype we never observed in wild-type roots. Quantification of metaxylem cells in seedling roots of both rlp44cnu2 and the T-DNA insertion line rlp44-3 6 d after germination showed a significant increase (Fig. 2C), suggesting that RLP44 controls xylem cell fate. Expression of RLP44 under control of its own promoter complemented this phenotype (SI Appendix, Fig. S2B). Since we had previously identified RLP44 as an activator of BR signaling, we analyzed the root xylem of a number of BR-related mutants spanning a broad range of growth phenotypes. Hypomorphic bri1 mutants such as bri1cnu1 (38), bri1-301 (20), and bri1-5 (39), the more severe signaling mutant bin2-1 (20), as well as the BR-deficient biosynthetic mutants constitutive photomorphogenic dwarf (cpd) (40) and dwarf4-102 (41) did not show a pronounced increase in xylem cell number (Fig. 2D and SI Appendix, Fig. S2 C and D). In sharp contrast, bri1 null alleles such as a previously characterized T-DNA mutant (termed bri1-null) (42) and the bri1 brl1 brl3 triple mutant (called bri-triple from hereon) (43) displayed a marked increase in the number of differentiated xylem cells (Fig. 2D and SI Appendix, Fig. S3), whereas expression of BRI1 under the control of its own promoter in bri1-null restored wild-type–like xylem (Fig. 2E). Taken together, our results show that the xylem differentiation phenotype does not correlate with the severity of BR-deficiency–related growth phenotypes (SI Appendix, Fig. S2E). This is exemplified by the comparison between cpd and bri1-null, with cpd displaying wild-type–like xylem cell numbers, despite exhibiting a bri1-null–like growth phenotype. Thus, the control of xylem cell number requires the presence of both BRI1 and RLP44. To test whether increased levels of BRs in BRI1 loss-of-function mutants (39) contribute to the xylem phenotype, we depleted endogenous BRs in the wild-type and bri-triple plants with the BR biosynthesis inhibitor propiconazole (PPZ) (44), rendering wild-type plants indistinguishable from the mutant (SI Appendix, Fig. S4A). However, metaxylem cell number was not significantly affected in either genotype by PPZ treatment (SI Appendix, Fig. S4B), despite a slightly elevated number of metaxylem cells in wild type. However, PPZ treatment occasionally led to gaps in the protoxylem (SI Appendix, Fig. S4C), a phenotype also found in bri-triple (Fig. 2D and SI Appendix, Fig. S3) and in dwf4-102 (SI Appendix, Fig. S4D), but not in any other mutant (SI Appendix, Fig. S4 D–F), suggesting that BR signaling has a role in the maintenance of protoxylem and that the cpd mutant is not strictly equivalent to dwf4-102 for unknown reasons. Conversely, neither root-growth–promoting nor root-growth–inhibiting doses of brassinolide (BL) (SI Appendix, Fig. S4G) affected xylem cell numbers (SI Appendix, Fig. S4H). Activating BR signaling downstream of BRI1 by inhibiting BIN2 and other GSK3-like kinases through bikinin treatment (45) partially rescued the short-root phenotype of bri-triple (SI Appendix, Fig. S5A), but did not significantly alter the metaxylem cell number in either mutant or wild type (SI Appendix, Fig. S5B). This indicates that BRI1, rather than canonical downstream BR-signaling components, is critical for normal xylem cell fate. To assess whether BRI1 kinase activity is required for the control of cell fate, we analyzed bri1-1 (46, 47), which harbors a point mutation in the kinase domain (A909T) and is expected to prevent adenine nucleotide binding and thus to render the protein kinase dead (48). The bri1-1 mutant, which is morphologically indistinguishable from the transcriptional knockout bri1-null, showed supernumerary xylem cells despite the presence of BRI1 protein (SI Appendix, Fig. S5C), suggesting that BRI1 kinase activity is required for the control of xylem cell fate.
Fig. 2.
RLP44 and BRI1 are required for the control of xylem cell fate. (A) Overview of xylem differentiation in the Arabidopsis root and schematic representation of the stele. Gray square in root schematic indicates point of xylem observation. (B) Basic fuchsin staining of 6-d-old Arabidopsis root. DIC image shows secondary cell-wall thickenings of protoxylem and metaxylem (Left), and basic fuchsin labels lignified secondary cell walls (Middle). Confocal stacks allow xylem number quantification of the indicated genotypes in orthogonal view (Right). Note ectopic metaxylem in procambial position (arrow). (Left) A median plane image. (Middle) A maximum projection. (Scale bar: 50 µM.) (C and D) Frequency of roots with the indicated number of metaxylem cells in rlp44 and BR-related mutants. Right in D shows orthogonal view and maximum projection of bri-triple root. Note ectopic metaxylem (arrows) and disrupted protoxylem (arrowhead). Asterisks indicate statistically significant difference from Col-0 based on Dunn’s post hoc test with Benjamini–Hochberg correction after Kruskal–Wallis modified U test (*P < 0.05; ***P < 0.001). (E) Transgenic expression of BRI1 under control of its own regulatory 5′ sequence rescues the ectopic xylem phenotype of bri1-null.
BRI1 Is Required for Normal RLP44 Expression.
To analyze how BRI1 and RLP44 could be linked in the control of xylem cell fate, we investigated RLP44 expression in BR-related mutants. Interestingly, the expression of RLP44 was reduced in bri1-null, suggesting that reduced RLP44 levels could at least partially explain the xylem phenotype of this mutant (SI Appendix, Fig. S6 A and B). Consistent with this notion, uncoupling RLP44 transcription from BRI1 control through driving the expression of an RLP44 transgene by the 35S promoter could alleviate the bri1-null xylem phenotype (SI Appendix, Fig. S6C), but not its growth defects (SI Appendix, Fig. S7), suggesting that BRI1 and RLP44 indeed act in the same pathway regulating xylem cell fate. As RLP44 expression was only mildly affected in bri1 hypomorphs, cpd, dwf4-102, bin2-1 or by BR depletion (SI Appendix, Figs. S6 A and B and S8 A–C), BR signaling output-independent control of RLP44 expression by BRI1 may explain the presence and absence of vascular cell-fate defects in the various BR-related mutants. Conversely, BL or bikinin treatment, as well as BRI1-independent activation of BR-signaling outputs through hyperactive versions of the transcription factors BES1 and BZR1, did not alter RLP44 transcript levels in an appreciable manner (SI Appendix, Fig. S8A). These results corroborate previous genome-wide transcriptome analyses showing that expression of RLP44 is strongly reduced in the null mutant bri1-116, but in contrast to that of bona fide BR target genes, is not recovered in the bri1-116 bzr1-1D double mutant, which has constitutively activated BR-signaling outputs (24) SI Appendix, Fig. S8D). In line with this, RLP44 is not among the experimentally defined targets of BZR1 or BES1 (24, 25). Finally, the limited effects of BR-signaling–related cues on RLP44 transcript levels are consistent with publicly available transcriptome data (49), SI Appendix, Fig. S9). Taken together, our findings indicate that the phenotype of bri1 loss-of-function mutants is at least partially independent from BR-signaling outputs and suggest that RLP44 exerts its function downstream of BRI1 through other signaling components.
Vascular Cell-Fate Determination by RLP44 and BRI1 Is Independent of BR-Signaling–Mediated Control of Cell Proliferation.
We next asked whether the increase in xylem cell number observed in the rlp44 mutant could be caused by enhanced cell proliferation. In rlp44-3, vascular cell number was indistinguishable from wild type in the differentiation zone, suggesting normal meristematic activity (SI Appendix, Fig. S10A). The bri1cnu1 mutant, which did not display ectopic xylem cells, showed a significant increase in total vascular cell number (SI Appendix, Fig. S10A), consistent with the described role of BR signaling in controlling formative cell divisions (50). These results suggest that increased proliferation in the vasculature is not a prerequisite for an increase in metaxylem. In line with this, depletion of BRs by PPZ resulted in a pronounced increase of vascular cell number (SI Appendix, Fig. S10 B and C). When PPZ-treated roots were supplemented with 0.5 nM of BL, both root growth and vascular cell number were fully recovered (SI Appendix, Fig. S10 B and C). A higher dose of 5 nM BL suppressed root growth and led to a strongly decreased vascular cell number (SI Appendix, Fig. S10C). The rlp44cnu2 mutant displayed a wild-type–like response to the manipulation of BR levels in terms of cell number (SI Appendix, Fig. S10C), further supporting the independence of xylem cell fate from BR-signaling–mediated control of cell proliferation. Moreover, the expression domain of the xylem precursor marker pTMO5:NLS-3xGFP (51) was unaltered in rlp44cnu2 root meristems (SI Appendix, Fig. S11), suggesting that the acquisition of xylem cell fate in the mutant is a late event occurring outside of the meristem.
RLP44 Controls Xylem Cell Fate by Promoting PSK Signaling.
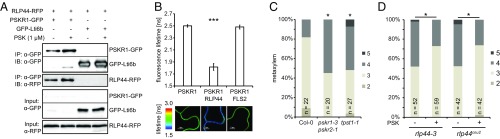
The results described so far suggested that the maintenance of procambial cell identity in the root requires the presence of both BRI1 and RLP44, with RLP44 acting downstream of BRI1. Thus, we speculated that, devoid of a kinase domain, RLP44 is likely required to interact with and influence the activity of another signaling component(s), which, in turn, control(s) xylem cell fate. Interestingly, in addition to BRI1 and its close homologs BRL1, BRL2, and BRL3, the LRR X clade of RLKs harbors the receptors for the peptide growth factor PSK, PSKR1, and -2 (17). As PSK signaling has also been implicated in promoting the transdifferentiation of Zinnia elegans mesophyll cells into tracheary elements (52, 53), depends on functional BR signaling (29), and BRI1 (14), PSKR1 (54), PSK4, and PSK5 (SI Appendix, Fig. S12A) are coexpressed with RLP44 in the vasculature, we tested the association of RLP44 with PSKR1. Coimmunoprecipitation experiments in Nicotiana benthamiana showed that PSKR1-GFP (34) was present in RLP44-RFP (35) immunoprecipitates (Fig. 3A). In addition, Foerster resonance energy transfer-fluorescence lifetime imaging microscopy (FRET-FLIM) analysis showed a pronounced reduction in fluorescence lifetime when PSKR1-GFP was coexpressed with RLP44-RFP, suggesting a direct interaction (Fig. 3B and SI Appendix, Fig. S12B), which was not affected by exogenous application of PSK peptide (SI Appendix, Fig. S12 C and D). Supporting a role of PSK signaling in the control of xylem cell fate, the pskr1-3 pskr2-1 double mutant (54) showed increased metaxylem cell numbers, reminiscent of rlp44 (Fig. 3C). A similar phenotype was observed in the tpst-1 mutant, which is impaired in the biosynthesis of PSK and other sulfated peptides (Fig. 3C) (30, 55). While exogenous PSK had no effect on wild-type xylem, it partially rescued metaxylem cell number in tpst-1 (SI Appendix, Fig. S12E) and reverted rlp44 xylem back to a wild-type pattern (Fig. 3D). Consistent with RLP44 acting through PSK signaling, the pskr1-3 pskr2-1 rlp44cnu2 triple mutant did not show an enhanced phenotype compared with pskr1-3 pskr2-1 (SI Appendix, Fig. S12F). In addition, RLP44 overexpression, which was able to rescue the bri1-null metaxylem phenotype (SI Appendix, Fig. S6C), did not rescue that of pskr1-3 pskr2-1 (SI Appendix, Fig. S12G). In accordance with this, rlp44cnu2 is quantitatively challenged in the root growth response to exogenous PSK (SI Appendix, Fig. S12H). Similar to RLP44 and in contrast to BR deficiency conditions, PSK-related mutants did not show gaps in the protoxylem (SI Appendix, Fig. S12I). Taken together, our results suggest that RLP44 acts through PSK receptors and is required to quantitatively control PSK-signaling strength.
Fig. 3.
RLP44 interacts with PSKR1 to promote PSK signaling and procambial identity. (A) Coimmunoprecipitation after transient expression in N. benthamiana leaves demonstrates the presence of RLP44-RFP in PSKR1-GFP immunoprecipitates. (B) FRET-FLIM analysis of the PSKR1-GFP/RLP44-RFP interaction in N. benthamiana leaves. Bars denote average of seven to eight measurements ±SD. Asterisks indicate statistically significant difference from PSKR1-GFP and PSKR1-GFP coexpressed with FLS2-RFP according to pairwise t test (***P < 0.001). (C) Quantification of metaxylem cell number in Col-0 and PSK-signaling–related mutants. (D) Application of PSK peptide rescues the ectopic xylem phenotype of rlp44 mutants. Asterisks in C and D indicate statistically significant difference from Col-0 based on Dunn’s post hoc test with Benjamini–Hochberg correction after Kruskal–Wallis modified U test (*P < 0.05).
RLP44 Promotes the Association of PSKR1/BRI1 and Their Coreceptor.
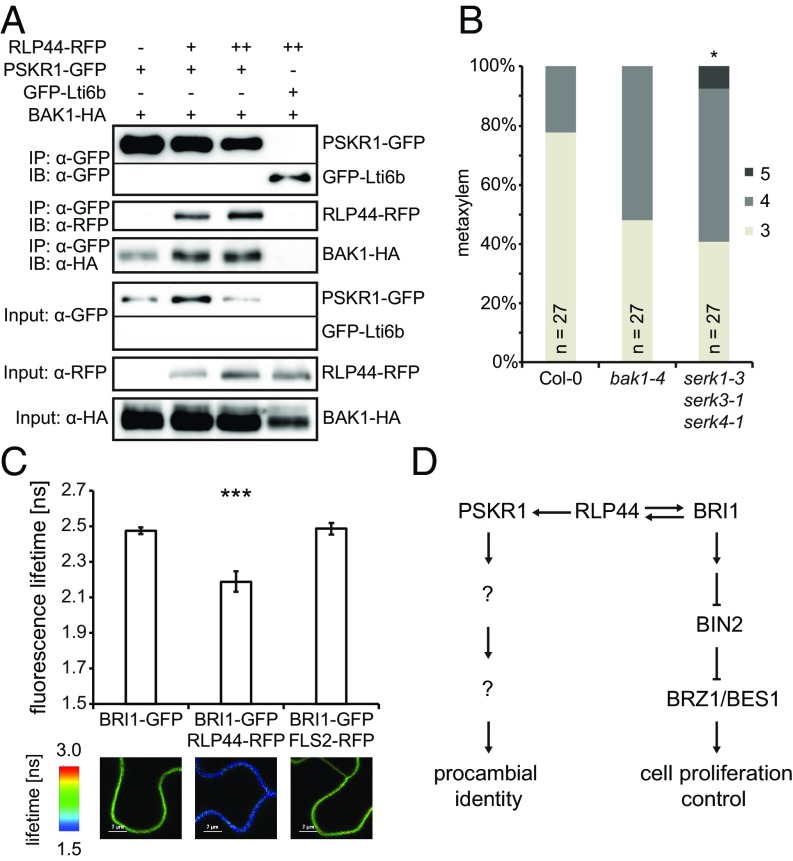
To elucidate how RLP44 might promote PSK signaling, we assessed whether its presence affects the association between PSKR1 and its coreceptor BAK1, both of which also directly interact with RLP44 (Fig. 3) (35). Indeed, more BAK1 was detected in immunoprecipitates of PSKR1-GFP when RLP44-RFP was coexpressed (Fig. 4A), suggesting that RLP44 might act as a scaffold in the complex. Supporting this notion, BAK1 levels in immunoprecipitates of PSKR1-GFP were reduced in the rlp44cnu2 mutant (SI Appendix, Fig. S13A). Consistent with an essential role of BAK1/SERK3 and other SERKs as coreceptors in PSK signaling (33, 34), the serk1-3 serk3-1 serk4-1 triple mutant (56) showed increased metaxylem cell numbers (Fig. 4B). Because we had previously demonstrated that RLP44 can activate BR signaling upon cues from the cell wall (35), we assessed whether BR-signaling activation by RLP44 might occur through a similar mechanism. RLP44 and BRI1 showed direct interaction in yeast-mating–based split ubiquitin assays (SI Appendix, Fig. S13B) and FRET-FLIM analysis after transient expression in N. benthamiana (Fig. 4C and SI Appendix, Fig. S13 C and D). Furthermore, endogenous BRI1 and BAK1 were detected in immunoprecipitates of RLP44-GFP expressed under the control of its own promoter in the rlp44cnu2 mutant background (SI Appendix, Fig. S13E). Similar to what was observed for PSKR1, the presence of RLP44 increased the association of BRI1 with its coreceptor BAK1 (SI Appendix, Fig. S13F) in a line that expresses BRI1-mCitrine and BAK1-HA under control of their own promoters in the bri1-null background. In summary, our data suggest that RLP44 acts as a scaffold to stabilize the PSKR1-BAK1 and BRI1-BAK1 complexes, respectively. While the interaction between RLP44 and BRI1 might not play a role in the context of vascular cell-fate determination, RLP44 is controlled by BRI1 at the transcriptional level and is required to promote PSK signaling in the vasculature, which, in turn, suppresses the progression from procambial to xylem identity (Fig. 4D).
Fig. 4.
RLP44 promotes the association of LRR-RLKs and their coreceptor. (A) Coimmunoprecipitation analysis after transient expression in N. benthamiana leaves demonstrates an increased amount of BAK1-HA in PSKR1-GFP immunoprecipitates in the presence of RLP44-RFP. RLP44 levels were adjusted through increasing the density of Agrobacteria (denoted by + or ++). (B) Quantification of metaxylem cell number in bak1-4 and triple-serk mutants. Asterisks in C and D indicate statistically significant difference from Col-0 based on Dunn’s post hoc test with Benjamini–Hochberg correction after Kruskal–Wallis modified U test (*P < 0.05). (C) FRETFLIM analysis of the RLP44-BRI1 interaction in N. benthamiana leaves. Bars denote average of six to seven measurements ±SD. Asterisks indicate statistically significant difference from BRI1-GFP and BRI1-GFP coexpressed with FLS2-RFP after pairwise t test (***P < 0.001). (D) Model of RLP44-mediated activation of PSK and BR signaling. RLP44 is capable of activating both signaling pathways, depending on the conditions, and is under transcriptional control by BRI1. Thus, BRI1 is required for RLP44-mediated control of procambial cell fate through PSK signaling.
Discussion
RLP44 Controls Vascular Cell Fate Through PSK Signaling.
The expanded family of plant RLK proteins and their ligands play central roles in intercellular communication, cell identity maintenance, and the regulation of cell expansion and proliferation (57). Currently, our view of these pathways is evolving to integrate the extensive cross-talk and interdependence of diverse signaling pathways (58). Here, we report that BR and PSK signaling are linked at the level of their plasma membrane receptors through RLP44 and that this signaling module is required to control xylem cell fate. Our genetic and biochemical data support a scenario where PSK-signaling strength is quantitatively controlled by RLP44, which itself is dependent on the presence of BRI1. While we cannot rule out posttranslational control of RLP44 by BRI1, for example, through phosphorylation of the cytoplasmic domain or through a role of BRI1 in the correct receptor complex assembly, this dependency is at least partially based on BRI1-mediated control of RLP44 expression. More work will be needed to understand how these BRI1-dependent, but apparently BR-signaling output-independent functions, such as the control of RLP44 expression, are achieved at the molecular level, but the branching of signaling transduction pathways immediately downstream or even at the level of plasma membrane receptor complexes is emerging as a common feature of RLK-dependent signaling (58). BRI1 and PSKR1/2 share the requirement for interaction with SERK coreceptors to form an active, heteromeric signaling complex (33, 34, 59). Consistent with this, the presence of RLP44 promoted the interaction between BAK1 and both BRI1 and PSKR1. Which RLK pathway is activated by RLP44 at a given time could depend on the conditions, in line with the initial identification of RLP44 as an essential factor for BR-signaling activation upon challenge of cell-wall integrity (35). These results and our model are in agreement with the emerging theme of dynamic, promiscuous, and flexible interactions of plasma membrane proteins to integrate signaling information and fine-tune cellular responses to external cues (60–62). Interestingly, the mechanism by which RLPs influence signaling seems to differ widely, ranging from direct participation in ligand binding (63, 64), to the control of signaling specificity through blocking access of RLK ligands (64), to the guarding of extracellular proteins targeted by pathogens (65). Here, we propose a scaffolding function of RLP44 for the interaction between PSKR1 and its coreceptor BAK1, expanding the mechanistic diversity of RLPs.
The Role of BR Signaling in Vascular Development.
It has been described that BR signaling plays an important role in the development of vascular tissue (14, 15). In addition, it has been reported that BR signaling is kept at low levels in procambial cells of leaf and hypocotyl to prevent their differentiation into xylem cells (66). Our results suggest that, in the primary xylem of the root, BR signaling plays only a minor role in controlling differentiation, in marked contrast to the strong patterning defects of BR signaling and biosynthetic mutants in the shoot (15). Conversely, at least in the root, the presence of BRI1 has a negative effect on xylem cell fate through RLP44- and PSK-signaling–mediated maintenance of procambial identity. Therefore, our results identify a role of BRI1 in root development that is independent of its role as a BR receptor.
PSK Signaling Likely Promotes Procambial Identity.
Alongside classical plant hormones, signaling peptides play major roles in plant development and stress responses (67, 68). The sulfated peptide PSK has been implicated diverse processes (26, 68). Here, we propose that PSK signaling controls xylem cell fate through promoting the maintenance of procambial identity. A number of observations support this hypothesis. First, PSK treatment rescued the ectopic xylem phenotype in rlp44 mutants. Second, PSK-related mutants showed increased xylem differentiation in procambial position, and PSK genes are coexpressed with RLP44 in procambial cells (37). Third, PSK expression is transiently increased before the acquisition of a procambial intermediate state by cells transdifferentiating into tracheary elements (52, 69), which could explain why PSK promotes tracheary element formation in Z. elegans only when applied early to the cell culture (52, 53). Finally, PSK signaling promotes callus growth and longevity, in line with a role in the maintenance of cell identity (29). However, it is unclear how PSK signaling affects cellular behavior, in part due to a lack of knowledge about potential downstream targets. To gain a deeper understanding of xylem differentiation, it will be important to unveil how the BRI1-RLP44-PSK–signaling module described here integrates with the fundamental patterning mechanisms and the gene regulatory networks controlling cell fate (2, 8).
Materials and Methods
The sources of mutants used in this study are described in SI Appendix, Table S1. Seeds were sterilized with 1.2% NaOCl in 70% ethanol and washed twice with absolute ethanol before being dried under the sterile hood. Plants were grown in 1/2 strength MS medium supplemented with 1% sucrose and 0.9% plant agar. If appropriate, 24 epi-BL, PPZ, or bikinin were added to the medium after sterilization at the indicated concentrations. After a 48- to 72-h incubation in the dark, plants were grown at 23 °C during a 16-h light period.
Details regarding the construction of plasmids and generation of transgenic plants, analysis of xylem and vascular cell number, immunoprecipitation, qRT-PCR, interaction assays, and microscopy are provided in SI Appendix, Materials and Methods. Mutants and transgenic lines used in this study are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Sigal Savaldi-Goldstein for the bri-triple mutant; Michael Hothorn for antisera against BRI1 and BAK1 and mutants; and Karin Schumacher for antiserum against RFP. Research in our laboratories was supported by German Research Foundation (DFG) Grants WO 1660/6-1 (to S.W.) and HA 2146/22-1 and CRC 1101-D02 (to K.H.). S.W. is supported by the DFG through Emmy Noether Programme Grant WO 1660/2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814434115/-/DCSupplemental.
References
- 1.Lucas WJ, et al. The plant vascular system: Evolution, development and functions. J Integr Plant Biol. 2013;55:294–388. doi: 10.1111/jipb.12041. [DOI] [PubMed] [Google Scholar]
- 2.Bishopp A, et al. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol. 2011;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 3.De Rybel B, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Wang X, Lee JY, Lee JY. Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell. 2013;25:187–201. doi: 10.1105/tpc.112.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohashi-Ito K, et al. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol. 2014;24:2053–2058. doi: 10.1016/j.cub.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 6.Ramachandran P, Wang G, Augstein F, de Vries J, Carlsbecker A. Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Development. 2018;145:dev159202. doi: 10.1242/dev.159202. [DOI] [PubMed] [Google Scholar]
- 7.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor-Teeples M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–575. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong R, Ye ZH. Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 2015;56:195–214. doi: 10.1093/pcp/pcu140. [DOI] [PubMed] [Google Scholar]
- 11.Baum SF, Dubrovsky JG, Rost TL. Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am J Bot. 2002;89:908–920. doi: 10.3732/ajb.89.6.908. [DOI] [PubMed] [Google Scholar]
- 12.Belkhadir Y, Jaillais Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015;206:522–540. doi: 10.1111/nph.13269. [DOI] [PubMed] [Google Scholar]
- 13.Singh AP, Savaldi-Goldstein S. Growth control: Brassinosteroid activity gets context. J Exp Bot. 2015;66:1123–1132. doi: 10.1093/jxb/erv026. [DOI] [PubMed] [Google Scholar]
- 14.Caño-Delgado A, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 15.Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA. 2009;106:13630–13635. doi: 10.1073/pnas.0906416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 17.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 19.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 21.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 22.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 23.Chaiwanon J, Wang ZY. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol. 2015;25:1031–1042. doi: 10.1016/j.cub.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 26.Sauter M. Phytosulfokine peptide signalling. J Exp Bot. 2015;66:5161–5169. doi: 10.1093/jxb/erv071. [DOI] [PubMed] [Google Scholar]
- 27.Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- 29.Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006;142:45–53. doi: 10.1104/pp.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:15067–15072. doi: 10.1073/pnas.0902801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartmann J, Stührwohldt N, Dahlke RI, Sauter M. Phytosulfokine control of growth occurs in the epidermis, is likely to be non-cell autonomous and is dependent on brassinosteroids. Plant J. 2013;73:579–590. doi: 10.1111/tpj.12056. [DOI] [PubMed] [Google Scholar]
- 32.Heyman J, et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science. 2013;342:860–863. doi: 10.1126/science.1240667. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature. 2015;525:265–268. doi: 10.1038/nature14858. [DOI] [PubMed] [Google Scholar]
- 34.Ladwig F, et al. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell. 2015;27:1718–1729. doi: 10.1105/tpc.15.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf S, et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci USA. 2014;111:15261–15266. doi: 10.1073/pnas.1322979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf S. Plant cell wall signalling and receptor-like kinases. Biochem J. 2017;474:471–492. doi: 10.1042/BCJ20160238. [DOI] [PubMed] [Google Scholar]
- 37.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 38.Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol. 2012;22:1732–1737. doi: 10.1016/j.cub.2012.07.036. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi T, et al. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 41.Nakamoto D, Ikeura A, Asami T, Yamamoto KT. Inhibition of brassinosteroid biosynthesis by either a dwarf4 mutation or a brassinosteroid biosynthesis inhibitor rescues defects in tropic responses of hypocotyls in the Arabidopsis mutant nonphototropic hypocotyl 4. Plant Physiol. 2006;141:456–464. doi: 10.1104/pp.105.076273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA. 2011;108:8503–8507. doi: 10.1073/pnas.1103556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vragović K, et al. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci USA. 2015;112:923–928. doi: 10.1073/pnas.1417947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartwig T, et al. Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One. 2012;7:e36625. doi: 10.1371/journal.pone.0036625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Rybel B, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bojar D, et al. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014;78:31–43. doi: 10.1111/tpj.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hruz T, et al. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang YH, Breda A, Hardtke CS. Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development. 2017;144:272–280. doi: 10.1242/dev.145623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 52.Motose H, et al. Involvement of phytosulfokine in the attenuation of stress response during the transdifferentiation of zinnia mesophyll cells into tracheary elements. Plant Physiol. 2009;150:437–447. doi: 10.1104/pp.109.135954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsubayashi Y, Takagi L, Omura N, Morita A, Sakagami Y. The endogenous sulfated pentapeptide phytosulfokine-alpha stimulates tracheary element differentiation of isolated mesophyll cells of zinnia. Plant Physiol. 1999;120:1043–1048. doi: 10.1104/pp.120.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kutschmar A, et al. PSK-α promotes root growth in Arabidopsis. New Phytol. 2009;181:820–831. doi: 10.1111/j.1469-8137.2008.02710.x. [DOI] [PubMed] [Google Scholar]
- 55.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 56.Hohmann U, et al. Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc Natl Acad Sci USA. 2018;115:3488–3493. doi: 10.1073/pnas.1714972115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Smet I, Voss U, Jürgens G, Beeckman T. Receptor-like kinases shape the plant. Nat Cell Biol. 2009;11:1166–1173. doi: 10.1038/ncb1009-1166. [DOI] [PubMed] [Google Scholar]
- 58.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 59.Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 2013;341:889–892. doi: 10.1126/science.1242468. [DOI] [PubMed] [Google Scholar]
- 60.Smakowska-Luzan E, et al. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature. 2018;553:342–346. doi: 10.1038/nature25184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stegmann M, et al. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science. 2017;355:287–289. doi: 10.1126/science.aal2541. [DOI] [PubMed] [Google Scholar]
- 62.Gui J, et al. OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev Cell. 2016;38:201–213. doi: 10.1016/j.devcel.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Albert I, et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants. 2015;1:15140. doi: 10.1038/nplants.2015.140. [DOI] [PubMed] [Google Scholar]
- 64.Lin G, et al. A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 2017;31:927–938. doi: 10.1101/gad.297580.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rooney HC, et al. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- 66.Kondo Y, et al. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat Commun. 2014;5:3504. doi: 10.1038/ncomms4504. [DOI] [PubMed] [Google Scholar]
- 67.Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell. 2015;27:2095–2118. doi: 10.1105/tpc.15.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsubayashi Y. Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol. 2014;65:385–413. doi: 10.1146/annurev-arplant-050312-120122. [DOI] [PubMed] [Google Scholar]
- 69.Kondo Y, Fujita T, Sugiyama M, Fukuda H. A novel system for xylem cell differentiation in Arabidopsis thaliana. Mol Plant. 2015;8:612–621. doi: 10.1016/j.molp.2014.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.