Significance
The glucocorticoid receptor (GR) is an important signaling protein and a prominent drug target. Cortisone, one of the drugs directed against the GR, is among the most prescribed drugs worldwide. Hormone binding leads to activation of the receptor, which is then transported into the nucleus, affecting gene transcription. Despite its importance, little is known about the structural prerequisites of hormone binding and the associated conformational changes. We used a mechanical single-molecule method that allowed us to observe in detail the steps of folding and hormone binding of GR. We identify a structural element that opens and closes upon hormone binding. Our results form the basis for understanding GR activation and its regulation by chaperone proteins.
Keywords: glucocorticoid receptor, ligand binding, cortisol signaling, protein misfolding, optical tweezers
Abstract
The glucocorticoid receptor (GR) is a prominent nuclear receptor linked to a variety of diseases and an important drug target. Binding of hormone to its ligand binding domain (GR-LBD) is the key activation step to induce signaling. This process is tightly regulated by the molecular chaperones Hsp70 and Hsp90 in vivo. Despite its importance, little is known about GR-LBD folding, the ligand binding pathway, or the requirement for chaperone regulation. In this study, we have used single-molecule force spectroscopy by optical tweezers to unravel the dynamics of the complete pathway of folding and hormone binding of GR-LBD. We identified a “lid” structure whose opening and closing is tightly coupled to hormone binding. This lid is located at the N terminus without direct contacts to the hormone. Under mechanical load, apo-GR-LBD folds stably and readily without the need of chaperones with a folding free energy of . The folding pathway is largely independent of the presence of hormone. Hormone binds only in the last step and lid closure adds an additional of free energy, drastically increasing the affinity. However, mechanical double-jump experiments reveal that, at zero force, GR-LBD folding is severely hampered by misfolding, slowing it to less than 1·s−1. From the force dependence of the folding rates, we conclude that the misfolding occurs late in the folding pathway. These features are important cornerstones for understanding GR activation and its tight regulation by chaperones.
Steroid hormone receptors are soluble, ligand-controlled transcription factors which shuttle between the cytosol and the nucleus (1). They are composed of an N-terminal domain, a DNA-binding domain, and the ligand binding domain (LBD) (2). Upon hormone binding, they interact with specific response elements on the DNA and activate gene transcription. The glucocorticoid receptor (GR) is a member of this family linked to a variety of diseases such as diabetes (3), rheumatoid arthritis (4), allergic rhinitis (5), asthma (6), leukemia (7), and depression (8). Using a stabilized variant, F602S, a dexamethasone (DEX)-bound structure of the GR-LBD in complex with the coactivator TIF-2 could be solved (9). Meanwhile, further structures with different ligands and coregulatory peptides have emerged (10).
An interesting aspect of GR biology is its strong regulation by the molecular chaperones Hsp70 and Hsp90 (11). This chaperone dependence suggests that GR, and especially the LBD, the main interaction site for the chaperone machinery (12), may exhibit pronounced folding defects. This view is supported by in vitro analyses showing that in the hormone-free state the GR-LBD is unstable and aggregation-prone, even if stabilizing mutations are introduced (13). Since the hormone is deeply buried inside the protein structure, the apo-GR-LBD is thought to be only weakly folded and highly dynamic (14).
Hydrogen/deuterium exchange mass spectrometry was used to gain insight into the dynamics of hormone binding (15), but the details of the conformational dynamics triggered by these interactions have remained elusive. In particular, the pathway of hormone binding into the binding pocket of GR has remained enigmatic. Whether the protein folds around the hormone or undergoes major structural rearrangements from a folded open to a hormone-bound closed structure is hence still unresolved. In this study, we have used single-molecule optical tweezers experiments to study the folding and hormone binding pathway of the GR-LBD.
Results
GR-LBD Folds and Unfolds Reversibly in Single-Molecule Optical Trap Experiments.
In recent ensemble measurements, the GR-LBD could not be reversibly unfolded and showed a strong tendency to aggregate (13). Therefore, we set out to investigate the folding/unfolding of GR-LBD under single-molecule conditions. We designed a construct carrying the stabilizing point mutation F602S as well as two cysteine residues at the termini to attach dsDNA linkers for the single-molecule mechanical measurements. To avoid cross-reactions, a surface-exposed internal cysteine (C638) was mutated to an aspartic acid residue (complete sequence in SI Appendix). The mutations F602S and C638D have been used previously to improve expression levels of the GR-LBD (9, 16, 17). Via the terminal cysteines, this protein (in the following called ) was covalently bound to DNA linkers and then attached to 1-µm-sized beads, allowing handling by optical tweezers (Fig. 1A and SI Appendix). A second variant () with six stabilizing mutations (sequence in SI Appendix), used in a previous study (13), was created and analyzed to check for consistency.
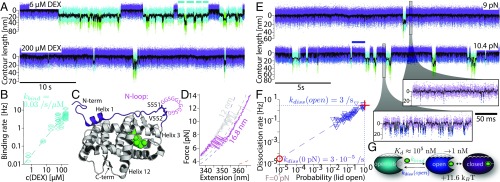
Fig. 1.
GRSD molecules fold and unfold reversibly in single-molecule experiments. (A) Scheme of optical-trap assay. DEX bound GR-LBD F602S [Protein Data Bank (PDB) ID code 1M2Z] linked to dsDNA linkers which are tethered to 1-µm silica beads. (B) Stretch (blue) and relax (orange) cycle in the presence of 200 µM DEX using a constant pulling velocity of 500 nm/s. Dashed colored lines show WLC fits for different contour length (details in SI Appendix). (C) Contour length vs. time trace of the transition between open (blue) and closed (purple) state while keeping the traps at a constant distance applying a force bias of around 10 pN (passive-mode). (D) Force dependence of closing (blue) and opening (purple) rates. Lines are extrapolations using a model described in SI Appendix.
First, we performed experiments in which we continuously stretched and relaxed the molecule in the optical trap in the presence of the hormone DEX. Mechanical stretching of (blue trace in Fig. 1B) leads to rapid folding/unfolding fluctuations at around 10 pN, in which parts of the structure fold/unfold in equilibrium (blue circle). At around 15 pN, the protein transitions to the fully unfolded state. Fits of the traces to the worm-like chain (WLC) model of polymer elasticity (18) (dashed lines in Fig. 1B; see also SI Appendix) provide an estimate for the number of amino acids unfolding in each transition. The 12 ± 1-nm contour length increase we find for the low-force equilibrium transition corresponds to an unfolding of about aa residues (19). The 86 ± 3-nm contour length gain we find for the fully unfolded protein corresponds well with the aa of the GR-LBD.
Reducing the force in the subsequent relax trace (orange trace in Fig. 1B) we observe folding transitions with the molecule contracting back to the length of the natively folded structure. The subsequent unfolding trace of the refolded structure is indistinguishable from the first unfolding trace, indicating the molecule has refolded to the native state. A sequence of consecutive stretch and relax cycles is shown in SI Appendix, Fig. S1.
To study the kinetic and energetic properties of the early equilibrium transition occurring at 10 pN (blue circle in Fig. 1B), we used passive-mode experiments. In this mode, the trap distance was kept constant. Hence, forces will fluctuate around a preset value whenever the protein contracts or expands (details in SI Appendix). To this end, we subjected to forces of around 10 pN and recorded the folding/unfolding dynamics (Fig. 1C). This assay allows a detailed kinetic and energetic characterization of the observed equilibrium transition.
To obtain folding/unfolding kinetics from these traces, we applied a two-state assignment of the raw data using a hidden-Markov-model (HMM) analysis (20) (details in SI Appendix). We define the purple, fully folded state as “closed” and the dark blue, partially unfolded state as “open” (Fig. 1C). Systematic variation of the applied force yields the force dependence of the opening and closing rate (Fig. 1D). We extrapolated these rates using a model accounting for the elasticity of the unfolded polypeptide (21–23) (described in SI Appendix). This extrapolation yielded a closing rate of ·s−1 and an opening rate of ·s−1 at 0 pN. The ratio of these rates also provides a free energy of () stored in this structural part. Apparently, this part binds strongly and can detach and rebind rapidly to the remaining folded part of GR without triggering further unfolding. In the following, we will call this structural part the “lid.” The variant behaves highly comparably to the , with rates differing by less than a factor of 3 (SI Appendix, Table S1).
The N-Terminal Helix Is a Gate for Hormone Binding and Release.
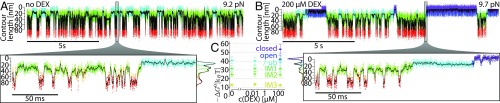
When observing the fluctuations of the lid at 10 pN over a longer timescale, we consistently detected an interruption of these fluctuations by another state (cyan in Fig. 2A, upper trace). The contour length is similar to the open state (see histograms in SI Appendix, Fig. S2A) but the lifetime is distinctly longer. From this state, further partial unfolding processes (green states) proceed. Higher DEX concentration resulted in shorter duration of the interrupting states (Fig. 2A, lower trace), indicating they reflect hormone-unbound states. To prove this, we varied hormone concentration and measured the total dwell time spent in the cyan states between two purple states (marked by bars in Fig. 2A, upper trace). A plot of the inverse of the cumulated dwell time vs. DEX concentration (Fig. 2B) shows a linear dependence. From this dependence, we conclude the cyan state is the apo state which binds hormone with a rate of kbind = 0.033 ± 0.003⋅s−1⋅μM−1. We hence call the cyan state open-unbound (open-ub). Lid closing occurs only upon hormone binding (transition to the purple state).
Fig. 2.
Hormone binding and unbinding kinetics are governed by opening and closing of an N-terminal lid. (A) Comparison of contour length vs. time traces at low (6 µM) and high (200 µM) DEX concentrations at constant trap distance (passive-mode). Lid fluctuations between closed (purple) and open (blue) state are interrupted by DEX unbound open-ub (cyan) and partially unfolded (green) phases. Cyan bars in the upper trace mark an example of total dwell time spent in the open-ub state between two closed states. (B) Concentration dependence of the DEX binding rate to the open-ub state. The dashed line is a linear fit. (C) Structure of GR-LBD F602S (PDB ID code 1M2Z). N-terminal 33-aa residues are colored in purple. DEX is colored in green. S551 and V552 are highlighted by stick representation. The loop insertion in the construct is symbolized by GGS in pink. (D) Comparison of force-extension curves obtained from (blue) and (gray) at 200 µM DEX using a constant pulling velocity of 50 nm/s. Dashed colored lines show WLC fits (compare Fig. 1B). (E) Comparison of contour length vs. time traces under two different force biases at 200 µM DEX. The purple bar marks an example for a DEX bound dwell time. (Lower Right) Zooms into the traces. (F) Dependence of dissociation rate on the population probability of the lid open state during the DEX bound phase. Dashed line is a linear fit . Red cross and circle show intersection at and . (G) Pathway for hormone binding.
Since the lid constitutes an important structural element for hormone binding we determined the location of the lid in the protein structure. To this end, we used a construct similar to the , but with a flexible insert of 11 residues (GGSGGSGGSGG) between the N-terminal helix 1 and helix 3 (Fig. 2C; sequence in SI Appendix). If the lid comprises the 33 N-terminal residues (purple in Fig. 2C), its opening should result in an increase of the contour length gain by about 4 nm. We call this construct . A zoom into the lid fluctuation region of a force vs. extension trace of pulled at 50 nm/s is shown in Fig. 2D. The fluctuations show a clear increase in contour length gain ( vs. ) (cf. gray trace of ). Passive-mode experiments yielded force-dependent opening and closing kinetics of , plotted in SI Appendix, Fig. S2. Consistent with the increase in contour length, midpoint forces are shifted to lower values (8 vs. 10 pN). Conservation of energy requires that an increase in contour length gain by inserting additional amino acids must lead to reduced forces. Moreover, the energy for lid closing as obtained by the zero force rates decreased slightly ( vs. ). showed DEX rebinding kinetics similar to with = 0.05 ± 0.01⋅s−1⋅μM−1, which confirms the proper functioning of the construct.
To find out whether the hormone dissociates from the lid-open or lid-closed state, we monitored the dissociation kinetics at different forces shifting the relative population of the open vs. closed state. Sample traces at two different forces are shown in Fig. 2E. An increase in force by 1.4 pN already results in a significant increase in the population of the open (dark blue) state (see zooms into the traces in Fig. 2E). Concomitantly, also more open-ub states (cyan) can be observed. We analyzed the dwell time of the open/closed state ensemble (an example is marked by a bar in Fig. 2E, lower trace) and how it is affected by a changing population of the open state. A log-log plot of the inverse dwell time vs. population probability of the open state exhibits direct proportionality (Fig. 2F), indicating that dissociation occurs exclusively from the open state under these force conditions. From Fig. 2F we can directly extract the dissociation rate from the open state to ·s−1. (red cross in Fig. 2F). The open-state population probability in the absence of force is only as calculated by the ratio of opening and closing rate at zero force (Fig. 1D). Extrapolation of the dissociation rate to zero force yields an effective off rate of ·s−1 (red circle in Fig. 2G), assuming that dissociation can only occur from the open state. From these measurements, we can also derive an affinity to the open state as well as a zero-force affinity. Unlabeled DEX binds to open (blue) state with µM and, at zero force, closing of the lid increases the effective affinity to nM.
For better comparison with literature values (13, 17, 24), which have been determined for fluorescein-labeled DEX (F-DEX), we repeated this analysis using F-DEX, arriving at a zero-force off rate of ·s−1 (SI Appendix, Fig. S3A). We did not observe an influence of the fluorescence label on the opening and closing kinetics (SI Appendix, Fig. S3B). In analogy to Fig. 2B we find a binding rate of F-DEX = 0.21 ± 0.04⋅s−1⋅μM−1 (SI Appendix, Fig. S3C). So, both binding and dissociation of F-DEX appear to be around a factor of 10 faster, yielding a similar affinity of nM. The rates of the variant are the same within a factor of 4 (SI Appendix, Table S1). Fig. 2G summarizes the emerging pathway of hormone binding and how it is coupled to lid closing.
The GR-LBD Folding Pathway Comprises Several Intermediates.
In a next set of experiments, we performed passive-mode measurements at higher applied forces to drive the to the fully unfolded state and study its refolding pathway and energy landscape. Contour length vs. time traces without addition of hormone in solution are shown in Fig. 3A. The contour length vs. time data were analyzed using HMM analysis (25) (details in SI Appendix). As we observe five distinct levels of contour length (histogram next to zoom in Fig. 3A), a model requires a minimum of five states. A model with five on-pathway states described our data. Remaining deviations from single exponentiality of the lifetime distributions in each state (example in SI Appendix, Fig. S4A) may hint toward the existence of further unresolvable states. The five states exchange on a millisecond timescale (see zoom in Fig. 3A) and we label them as follows: open-unbound (open-ub, cyan), Intermediate 1 (IM1, dark green), Intermediate 2 (IM2, light green), Intermediate 3 (IM3, yellow), and unfolded (unf, red). Apparently, even against significant mechanical force, GR can fold rapidly (less than a second) and multiple times from the fully unfolded to the open-ub state.
Fig. 3.
GR folds through many on-pathway intermediates before it binds hormone and reaches the native state. (A) Contour length vs. time traces in passive-mode without DEX added in solution. The trace is colored according to HMM state assignment (details in text and SI Appendix). The given force bias refers to the force acting on the closed state. A zoom into the data is shown below. Next to the zoom is a histogram of the smoothed contour length data with the contributions of the subpopulations highlighted in color. (B) Measurement similar to that in A, but with 200 µM DEX in solution. (C) Comparison of free energy differences of the states to the unfolded state at different hormone concentrations. The error bars combine statistical as well as the estimated systematic error of the force calibration.
Adding 200 µM of DEX leaves the pattern and kinetics unchanged in comparison with the hormone-free case but shows, in addition, the purple and dark blue states, representing the hormone-bound closed and open states as discussed (Fig. 3B).
An analysis of force-dependent population probabilities using the Boltzmann distribution (details in SI Appendix) yields quantitative information about the relative difference in free energy among the observed states at zero force (Fig. 3C and SI Appendix, Table S2). We find that the folding free energies of all intermediate states up to the open-ub state (cyan) do not depend on hormone concentration. Consequently, the unfolded and intermediate states (IM1, IM2, and IM3) cannot bind hormone. Hormone binding can only occur in the open-ub state and induces lid closing. Further supporting this conclusion, the binding probability for hormone strictly depends on the time spent in the open-ub state but not on the earlier folding intermediates (SI Appendix, Fig. S5C).
Surprisingly, we find that even apo- is a thermodynamically stable protein as already the open-ub state exhibits a folding free energy of . Binding of hormone further increases the stability ( at 200 µM DEX). The succession of stable folding intermediates we observe along the folding pathway of GR at the applied forces of 9–10 pN lets us conclude that the folding energy landscape is rough under these conditions. From the force-dependent kinetics of the transitions between the intermediates (example data in SI Appendix, Fig. S4B) we were able to compute this energy landscape under different mechanical forces (SI Appendix, Fig. S5A). Indeed, the energy landscape at 9 and 10 pN shows large barriers between the intermediates. However, when force is reduced to 0 pN, the barriers are reduced and folding should proceed rapidly to the native state. From this energy landscape, we compute an overall folding time on the order of a millisecond. A summary of the folding pathway as measured under mechanical forces (9–10 pN) is given in SI Appendix, Fig. S5B. Estimates for the number of amino acids contained in the folded portion of the intermediate states are given in SI Appendix, Table S3. For simplicity, we have depicted the structure growing around one nucleus from intermediate to intermediate. However, with the exception of the lid, we cannot tell their location within the protein structure.
Refolding in the Absence of Force Is Slowed Drastically by Misfolded Structures.
To measure folding rates at low loads, we employed a double-jump single-molecule assay, in which we continuously switch between high and low mechanical forces (26, 27). An example is shown in Fig. 4A, where we applied low force (6 pN) for 200 ms alternating with 30 ms at high force (12 pN). During the low-force phases, we allowed the protein to refold while it unfolds during the high-force phases. Low- and high-force phases were analyzed separately. A zoom into the concatenated high-force phases is shown in Fig. 4A, Upper Right. For clarity, the 200-ms gaps of the low-force phase are removed but marked with a blue dashed line. Note that, despite the similarity of these traces to those obtained in passive-mode experiments (Figs. 2 and 3), refolding occurs only during the low-force phases (at the blue dashed line). Hence, the signal observed during the high-force phase reports on the extent to which refolding had occurred during the preceding low-force phase. Comparable to the passive-mode experiments, we used an HMM model to classify states present in the high-force phase (details in SI Appendix). The refolding kinetics at low force can now be obtained from the merged high-force traces. Once the molecule is in an unfolded state (marked in red), we can calculate the refolding kinetics from the number of jumps to low force necessary until refolding to the native state occurs (details in SI Appendix). Monitoring the low-force phases (Fig. 4A, Lower Right) enabled us to follow directly the length contraction during refolding, albeit without an explicit state assignment due to the lower resolution at the low forces applied.
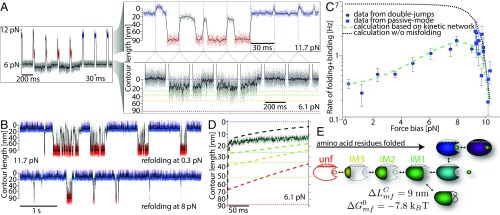
Fig. 4.
Folding is drastically slowed at low forces through the occurrence of misfolded intermediates. (A) Double-jump assay: (Left) Force vs. time trace while alternating between two trap distances. (Upper Right) Contour length vs. time trace of the high-force phases merged together. The vertical blue dotted lines mark the positions where a low-force phase was cut out. The trace is colored according to an HMM state assignment (details in SI Appendix). The gray colored data could not be classified unambiguously. (Lower Right) Zoom into the low-force region. For comparison, the colored dashed lines mark the contour length levels of the folding intermediates from Fig. 3. (B) Merged high-force phases of contour length vs. time traces obtained in double-jump experiments. The refolding force bias during the corresponding low-force phases was set to 0.3 pN (upper trace) and 8 pN (lower trace), respectively. DEX concentration was 200 µM. (C) Force dependence of the overall folding + binding rates from double-jump (blue circles) experiments and passive-mode (blue squares) measurements (as presented in Fig. 3B). The error bars reflect SEM. Black dotted line shows the expected force-dependent folding + binding rates calculated from the force-dependent kinetics as obtained from the equilibrium high-force folding measurements shown in Fig. 3 (example data in SI Appendix, Fig. S4B). The green dashed line shows a similar calculation including the possibility of misfolding branching from the IM1 state as depicted in E (details in SI Appendix). (D) Average contour length evolution for refolding against a force bias of 6.1 pN. The dashed colored lines show simulations of different models each assuming misfolding branching off a different on-pathway intermediate (colored accordingly; see text for details). Dotted colored lines mark contour length levels of the respective folding intermediates. (E) Model for folding and binding including an off-pathway misfolded structure.
Fig. 4B shows a direct comparison of the folding of at 0.3 pN (Upper) and 8 pN (Lower) of force. Surprisingly, refolding at zero force is significantly slower than refolding against 8 pN of force. Fig. 4C displays a quantitative analysis of refolding kinetics vs. applied force. We evaluated the overall rates for folding starting from the unfolded state including the binding of hormone () (details in SI Appendix). We chose to evaluate because hormone binding is a hallmark of the correctly folded native state. At forces higher than 8 pN, we found the steep decrease in generally predicted by simple models of force-dependent protein folding (28) (blue squares). Since force puts an energy penalty on the folding process it slows folding down, as also visualized by Fig. 3 A and B.
The decrease in toward lower forces is less intuitive. A rollover of refolding kinetics at low forces or denaturant conditions can be explained by an increased occurrence of off-pathway misfolded intermediates (29). In this case, increasing force will depopulate the misfolded state more than it slows down productive folding and leads to the given force dependence. Can we identify the misfolded state in our traces directly? The states populated after the refolding attempts do not exhibit well-defined contour length. Moreover, their lifetime distributions are not single-exponential (SI Appendix, Fig. S7B). We conclude that not a single well-defined but rather an ensemble of misfolded states is populated at low forces. We assessed the average properties of the misfolded states by averaging the time traces obtained during the low-force phases in which the protein starts its refolding attempt from the fully unfolded state (Fig. 4D). We find that the protein contracts rapidly (<10 ms) but even after 200 ms it is stuck in misfolded states, on average exhibiting a contour length 14 nm longer than the native length. The black dashed line in Fig. 4D shows the expected time-dependent contour length evolution based on the force-dependent kinetics of the on-pathway intermediates (SI Appendix, Fig. S4B). Apparently, after 200 ms, folding and DEX binding should have proceeded significantly further.
For simplicity, we modeled the force dependence of using a single misfolded state originating from one of the on-pathway intermediates. To this end, we numerically solved the kinetic equations of the folding network (details in SI Appendix) using the force-dependent folding/unfolding rates obtained from passive-mode traces (as shown in SI Appendix, Fig. S4B). In addition, we introduced a state describing an off-pathway misfolded structure with an additional length contraction () and free energy () (details in SI Appendix) branching off from one of the on-pathway states unf, IM3, IM2, or IM1. For each scenario, we could find a solution fitting the data in Fig. 4C, albeit with different parameters for and (SI Appendix, Fig. S8). The data in Fig. 4D put an additional constraint in choosing the best model for misfolding. In a second step, we therefore used the same numerical calculations to obtain the contour length vs. time evolution of the folding network with the respective intermediates (dashed colored lines in Fig. 4D). Only the model assuming that misfolding branches off IM1 describes our data. The length of the misfolded part is and the associated free energy of misfolding is (). Fig. 4E shows the full folding pathway including the misfolding species.
Discussion
Folding and Stability of Apo-GR-LBD.
Expression of soluble and functional GR-LBD has been difficult owing to its strong tendency to aggregate (30, 31). Refolding of denatured GR-LBD in vitro has so far not been achieved despite stabilizing mutations and the presence of hormone (13). Moreover, apo-GR-LBD appeared to be particularly unstable in ensemble measurements exhibiting unfolding and aggregation already below room temperature (13, 24). This observation supported the idea that GR-LBD needs assistance by chaperones to mature into its high-affinity hormone-binding state (32–34). In contrast, our single-molecule study shows that refolding and hormone binding of single, isolated molecules occurs readily without the need of additional assistance by chaperones. Refolding is possible even in the absence of hormone. Apo- exhibits a folding free energy of (), making it a remarkably stable protein. Folding of apo- to the open-ub state proceeds through at least three on-pathway intermediates, each of which adds of stability. All these intermediates form fast with zero-force rates between and (cf. SI Appendix, Fig. S4B). It is important to note that with an increasing number of states, a clear separation becomes challenging and we cannot exclude that more intermediates are populated, leading to an even rougher energy landscape than the one depicted in SI Appendix, Fig. S5A. We speculate that the intermediate states along the folding pathway may provide the receptor with the conformational plasticity needed to allow interaction with a multitude of different ligands (10, 35, 36), cofactors (17, 31, 32, 37), and chaperones.
Why then is apo- so aggregation-prone in bulk? So far there is no structure of apo-GR-LBD, but it is thought to be dynamic (14). The native state (lowest-energy state) of apo- is the open-ub state where all but the N-terminal 33-aa residues (lid) are folded. We propose that this unfolded part plays an important role for further aggregation in bulk and may need protection by chaperones. Consistent with this view, an important role has been attributed to the N-terminal region of GR-LBD for its interaction with Hsp70 (24).
Hormone Binding and the N-Terminal Lid.
The crystal structure of holo-GR-LBD shows the hormone (DEX) completely buried inside the protein with no obvious entry/exit paths (9). Our results reveal that the ligand does not bind to any of the early folding intermediates but only after folding into the open-ub state. Hence, we can exclude a folding scenario where hormone binds early and the receptor folds around this nucleus.
The mechanical single-molecule assay used in this work provides a direct readout for hormone binding together with primary structure information about the conformational changes involved. For hormone binding to the apo-GR-LBD, the C-terminal helix 12 has been widely discussed as an important structural factor since it is allosterically coupled to hormone binding (10, 15, 17, 32). Instead, we could identify the N-terminal helix 1 as a key structural lid-like element important for hormone binding. Hormone binding drives the structure toward the closed lid conformation by (), leading to the high-affinity structure with the hormone fully embedded. Interestingly, Dong et al. (38) found the LXXLL motif in residues 532–536 located in helix 1 important for hormone binding in vivo. Our results now offer a structural explanation for this finding. The strong coupling of hormone binding and closing of the N-terminal lid is surprising since the lid has no direct contacts with the hormone (9). Thus, we suggest that not only lid closing but also hormone binding is accompanied by a significant allosteric rearrangement. The on rates we find ( = 0.033⋅s−1⋅μM−1) for binding to the lid open state (open-ub) are slow for small-molecule binding (39, 40), strongly suggesting that major structural rearrangements are necessary for binding the hormone deep inside the protein structure.
Is the strong coupling between hormone binding and closing of the N-terminal lid also relevant under zero-force conditions? In the absence of force, the lid is predominantly closed with an open probability of only . If hormone dissociation can only proceed after opening of the lid, also the dissociation rate would be reduced by this factor. We find that the extrapolated off rate by assuming lid opening preceding dissociation [for F-DEX: ·s−1] is in good agreement with bulk measurements () (13, 24). This agreement is evidence that there is no other pathway for hormone dissociation in the absence of force that is significantly faster. Thus, even at zero force, lid opening has to precede dissociation. As a consequence, lid closing increases hormone affinity from to , which is at the lower end but still consistent with the values reported in the literature (6–250 nM) (13, 17, 24, 37).
Misfolding.
The fast folding we find for at high forces between 9 and 10 pN under equilibrium conditions (Fig. 3) suggests it should be a very fast folding protein also at low forces. According to the reconstructed energy landscape at zero force (SI Appendix, Fig. S5A) the rate of folding from the completely unfolded to the binding-competent open-ub state at zero force would be (SI Appendix, Fig. S6). However, in our double-jump experiments, we find that misfolded intermediates populated at low force reduce the overall folding rate to less than 1·s−1. A similar effect has been reported for Hsp90 (41), NSC-1 (42), and CaM (20). In contrast to the simpler proteins NSC-1 and CaM, where the misfolded state could be directly detected in equilibrium traces, the nature of the misfolded state is more elusive in the case of GR. While a drastic effect on folding is evident, we could not identify a single well-defined misfolded state in our traces. Instead, the multiexponential lifetimes of the intermediates forming at low forces (SI Appendix, Fig. S7B) indicate that the misfolded species rather constitute a broad ensemble of states. With their lengths often indistinguishable from those of the on-pathway intermediates, an unambiguous assignment of the regions involved is impossible. In the case of GR-LBD, it is only a small portion of the polypeptide chain with 9-nm contour length that misfolds late in the folding pathway. Those misfolds are dynamic and may initiate further aggregation, as is observed in bulk experiments due to the exposure of hydrophobic elements (13). Apparently, nature has not selected against this misfolded state. This may not have been necessary because chaperones prevent folding into this state or recognize and bind to this state.
In summary, our results reveal that the apo-GR-LBD is a stably folded protein in which mainly one specific element, the lid, needs to be positioned in response to hormone binding. However, once the apo form undergoes further unfolding, refolding becomes difficult due to the possibility to diverge from the correct folding pathway into misfolded species in which wrong intramolecular interactions are formed. Molecular chaperones thus have to maintain the hormone binding state of GR while at the same time preventing further unfolding and misfolding.
Materials and Methods
All protein constructs were prepared using standard recombinant techniques as described in SI Appendix. The experiments were carried out using custom-built dual-beam optical tweezers as in ref. 27 and as described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Katarzyna Tych and Marco Grison for technical help and discussions. This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 1035 Grants A5 (to M.R.) and A3 (to J.B.). D.R. was supported by a fellowship from the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807618115/-/DCSupplemental.
References
- 1.Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR. Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat Rev Mol Cell Biol. 2017;18:159–174. doi: 10.1038/nrm.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giguère V, Hollenberg SM, Rosenfeld MG, Evans RM. Functional domains of the human glucocorticoid receptor. Cell. 1986;46:645–652. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson PB, et al. Hepatic glucocorticoid receptor antagonism is sufficient to reduce elevated hepatic glucose output and improve glucose control in animal models of type 2 diabetes. J Pharmacol Exp Ther. 2005;314:191–200. doi: 10.1124/jpet.104.081257. [DOI] [PubMed] [Google Scholar]
- 4.Laan RF, Jansen TL, van Riel PL. Glucocorticosteroids in the management of rheumatoid arthritis. Rheumatology (Oxford) 1999;38:6–12. doi: 10.1093/rheumatology/38.1.6. [DOI] [PubMed] [Google Scholar]
- 5.Barnes PJ. Therapeutic strategies for allergic diseases. Nature. 1999;402(Suppl 6760):B31–B38. doi: 10.1038/35037026. [DOI] [PubMed] [Google Scholar]
- 6.Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renner K, Ausserlechner MJ, Kofler R. A conceptual view on glucocorticoid-induced apoptosis, cell cycle arrest and glucocorticoid resistance in lymphoblastic leukemia. Curr Mol Med. 2003;3:707–717. doi: 10.2174/1566524033479357. [DOI] [PubMed] [Google Scholar]
- 8.Chourbaji S, Gass P. Glucocorticoid receptor transgenic mice as models for depression. Brain Res Brain Res Rev. 2008;57:554–560. doi: 10.1016/j.brainresrev.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Bledsoe RK, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 10.Schoch GA, et al. Molecular switch in the glucocorticoid receptor: Active and passive antagonist conformations. J Mol Biol. 2010;395:568–577. doi: 10.1016/j.jmb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Vandevyver S, Dejager L, Libert C. On the trail of the glucocorticoid receptor: Into the nucleus and back. Traffic. 2012;13:364–374. doi: 10.1111/j.1600-0854.2011.01288.x. [DOI] [PubMed] [Google Scholar]
- 12.Howard KJ, Holley SJ, Yamamoto KR, Distelhorst CW. Mapping the HSP90 binding region of the glucocorticoid receptor. J Biol Chem. 1990;265:11928–11935. [PubMed] [Google Scholar]
- 13.Lorenz OR, et al. Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol Cell. 2014;53:941–953. doi: 10.1016/j.molcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: Implications for function. Annu Rev Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- 15.Frego L, Davidson W. Conformational changes of the glucocorticoid receptor ligand binding domain induced by ligand and cofactor binding, and the location of cofactor binding sites determined by hydrogen/deuterium exchange mass spectrometry. Protein Sci. 2006;15:722–730. doi: 10.1110/ps.051781406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauppi B, et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem. 2003;278:22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 17.Pfaff SJ, Fletterick RJ. Hormone binding and co-regulator binding to the glucocorticoid receptor are allosterically coupled. J Biol Chem. 2010;285:15256–15267. doi: 10.1074/jbc.M110.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bustamante C, Marko J, Siggia E, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 19.Dietz H, Rief M. Protein structure by mechanical triangulation. Proc Natl Acad Sci USA. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 21.Schlierf M, Berkemeier F, Rief M. Direct observation of active protein folding using lock-in force spectroscopy. Biophys J. 2007;93:3989–3998. doi: 10.1529/biophysj.107.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebhardt JCM, Bornschlögl T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci USA. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rognoni L, Stigler J, Pelz B, Ylänne J, Rief M. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc Natl Acad Sci USA. 2012;109:19679–19684. doi: 10.1073/pnas.1211274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stigler J, Rief M. Hidden Markov analysis of trajectories in single-molecule experiments and the effects of missed events. ChemPhysChem. 2012;13:1079–1086. doi: 10.1002/cphc.201100814. [DOI] [PubMed] [Google Scholar]
- 26.Schwaiger I, Schleicher M, Noegel AA, Rief M. The folding pathway of a fast-folding immunoglobulin domain revealed by single-molecule mechanical experiments. EMBO Rep. 2005;6:46–51. doi: 10.1038/sj.embor.7400317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grison M, Merkel U, Kostan J, Djinović-Carugo K, Rief M. α-actinin/titin interaction: A dynamic and mechanically stable cluster of bonds in the muscle Z-disk. Proc Natl Acad Sci USA. 2017;114:1015–1020. doi: 10.1073/pnas.1612681114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell G. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 29.Baldwin RL. On-pathway versus off-pathway folding intermediates. Fold Des. 1996;1:R1–R8. doi: 10.1016/S1359-0278(96)00003-X. [DOI] [PubMed] [Google Scholar]
- 30.Seitz T, et al. Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening. J Mol Biol. 2010;403:562–577. doi: 10.1016/j.jmb.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 31.Kroe RR, et al. Agonist versus antagonist induce distinct thermodynamic modes of co-factor binding to the glucocorticoid receptor. Biophys Chem. 2007;128:156–164. doi: 10.1016/j.bpc.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Bledsoe RK, Stewart EL, Pearce KH. Structure and function of the glucocorticoid receptor ligand binding domain. Vitam Horm. 2004;68:49–91. doi: 10.1016/S0083-6729(04)68002-2. [DOI] [PubMed] [Google Scholar]
- 33.Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 34.Cheung J, Smith DF. Molecular chaperone interactions with steroid receptors: An update. Mol Endocrinol. 2000;14:939–946. doi: 10.1210/mend.14.7.0489. [DOI] [PubMed] [Google Scholar]
- 35.Weikum ER, Okafor CD, D’Agostino EH, Colucci JK, Ortlund EA. Structural analysis of the glucocorticoid receptor ligand-binding domain in complex with triamcinolone acetonide and a fragment of the atypical coregulator, small heterodimer partner. Mol Pharmacol. 2017;92:12–21. doi: 10.1124/mol.117.108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suino-Powell K, et al. Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol. Mol Cell Biol. 2008;28:1915–1923. doi: 10.1128/MCB.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricketson D, Hostick U, Fang L, Yamamoto KR, Darimont BD. A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticoid receptor on Hsp90. J Mol Biol. 2007;368:729–741. doi: 10.1016/j.jmb.2007.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong DD, Jewell CM, Bienstock RJ, Cidlowski JA. Functional analysis of the LXXLL motifs of the human glucocorticoid receptor: Association with altered ligand affinity. J Steroid Biochem Mol Biol. 2006;101:106–117. doi: 10.1016/j.jsbmb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Pelz B, Žoldák G, Zeller F, Zacharias M, Rief M. Subnanometre enzyme mechanics probed by single-molecule force spectroscopy. Nat Commun. 2016;7:10848. doi: 10.1038/ncomms10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cappello G, et al. Myosin V stepping mechanism. Proc Natl Acad Sci USA. 2007;104:15328–15333. doi: 10.1073/pnas.0706653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jahn M, Buchner J, Hugel T, Rief M. Folding and assembly of the large molecular machine Hsp90 studied in single-molecule experiments. Proc Natl Acad Sci USA. 2016;113:1232–1237. doi: 10.1073/pnas.1518827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heidarsson PO, et al. Direct single-molecule observation of calcium-dependent misfolding in human neuronal calcium sensor-1. Proc Natl Acad Sci USA. 2014;111:13069–13074. doi: 10.1073/pnas.1401065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.