Significance
An integrative approach is presented for studying cell biology in vivo, assessing protein dynamics and cell behavior, and offering in situ analyses of cytokinesis, daughter-cell polarity, and stereotyped tissue morphogenesis. Tagging endogenous Discs Large 1 (Dlg1) in cartilage using intrabody technology permits in situ 3D time-lapse imaging and reveals Dlg1 enrichment at the midbody during cytokinesis. Functional significance is tested by disrupting Dlg1 multimerization and its midbody localization by using an ablating intrabody, DLGE3. Building on prior work on Dlg1 in epithelia, our work reveals that Dlg1 propagates cell polarity in proliferative mesenchymal tissues and suggests that multiple mechanisms act in concert at distinct phases of the cell cycle to transmit and maintain cell polarity.
Keywords: intrabody, FingR, cell polarity, DLGE3, bone morphogenesis
Abstract
Vertebrate embryogenesis and organogenesis are driven by cell biological processes, ranging from mitosis and migration to changes in cell size and polarity, but their control and causal relationships are not fully defined. Here, we use the developing limb skeleton to better define the relationships between mitosis and cell polarity. We combine protein-tagging and -perturbation reagents with advanced in vivo imaging to assess the role of Discs large 1 (Dlg1), a membrane-associated scaffolding protein, in mediating the spatiotemporal relationship between cytokinesis and cell polarity. Our results reveal that Dlg1 is enriched at the midbody during cytokinesis and that its multimerization is essential for the normal polarity of daughter cells. Defects in this process alter tissue dimensions without impacting other cellular processes. Our results extend the conventional view that division orientation is established at metaphase and anaphase and suggest that multiple mechanisms act at distinct phases of the cell cycle to transmit cell polarity. The approach employed can be used in other systems, as it offers a robust means to follow and to eliminate protein function and extends the Phasor approach for studying in vivo protein interactions by frequency-domain fluorescence lifetime imaging microscopy of Förster resonance energy transfer (FLIM-FRET) to organotypic explant culture.
During development, multiple cellular processes work collectively to build tissues of characteristic sizes and architectures (1). The fundamental cellular processes of mitosis and cell polarity must be coordinated so that cells can amplify their numbers while maintaining their characteristic morphologies (2, 3). In many proliferative tissues, this coordination is achieved during metaphase and anaphase by stereotypically orienting the cell-division plane and generating daughter cells with specific spatial relationships and morphologies (2). The common finding that mitotic cells often orient their division planes along their own elongation axes has been formulated as Hertwig’s rule (4), but the mechanistic underpinnings that explain the cell’s inner workings in executing this rule remain ill-defined. In the zebrafish embryo and mammalian skin, cell shape and the cell-division plane are not always linked, but daughter cells maintain their mothers’ characteristic morphologies (5, 6). These exceptions to Hertwig’s rule (7) point to gaps in our understanding of how cell polarity, orientation, and shape are propagated.
The growth plate cartilage of long bones represents an excellent system to meet this challenge because of its proliferative capacity and simple tissue architecture. Along the proximal–distal axis (PDA) of the tissue, morphologically distinct chondrocytes display characteristic mitotic rates and directional growth (8, 9), suggesting that cell polarity and mitosis are linked (Fig. 1A). Progenitor chondrocytes located within the resting zone are round and divide relatively slowly. After entering the proliferative phase, they acquire an elliptical shape with their elongation axis oriented orthogonal to the PDA. These cells progressively mature into prehypertrophic (PHZ) and hypertrophic (HZ) chondrocytes which enlarge and become postmitotic in the conversion of the growth plate cartilage into calcified bone (8, 9).
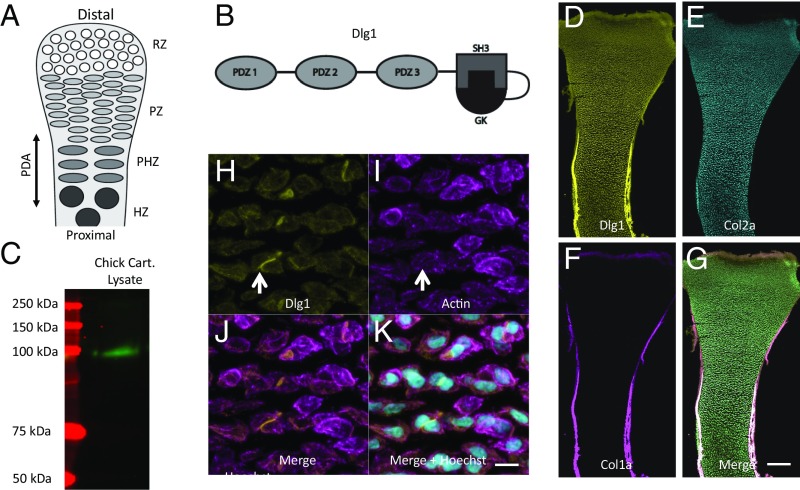
Fig. 1.
Dlg1 is expressed in the growth plate cartilage. (A) The growth plate cartilage contains four populations of chondrocytes that progressively differentiate along the tissue PDA: the resting (RZ), PZ, PHZ, and HZ zones. (B) Dlg1 is comprised of three PDZ domains and one SH3-GK domain. (C) Western blot probing of chondrocyte lysate with pan-MAGUK antibody showed a band at 140 kDa, which corresponds to Dlg1. n = 5 wild-type chicken humeri. (D–G) Hybridization chain reaction with anti-Dlg1 DNA was performed on frozen sections of chicken humerus and revealed that Dlg1 mRNA (D) was expressed in both cartilage (marked by Col2a1) (E) and PC/PO (marked by Col1a) (F). A merged image is shown in G. n = 7 wild-type chick humeri. (Scale bar: G, 200 μm, applies to D–G.) (H–K) To determine the expression pattern of Dlg1 protein, immunohistochemistry was performed on frozen sections of chicken humerus. Pan-MAGUK antibody, phalloidin, and Hoechst were used to label Dlg1 (H), actin (I), and nuclei (K). As illustrated in the resting zone, Dlg1 was localized on the cell membrane in most cells and was enriched in the middle between two cells. White arrows point to cell–cell contacts with concentrated Dlg1 but without an actin-enriched contractile ring. A merged image is shown in J. n = 5 wild-type chick humeri. (Scale bar: K, 15 μm, applies to H–K.)
The ability of the cells in the proliferative zone (PZ) to expand in the proper direction is critical for cartilage morphogenesis and homeostasis (10–14). In agreement with Hertwig’s rule, PZ cells divide across their elongation axes, generating daughter cells laterally displaced from the PDA (10, 11). This rule has been disrupted in chicken cartilage by expressing a constitutively activated calcium/calmodulin-dependent kinase (11) and in mouse cartilage by depleting glycosylphosphatidylinositol-anchored cell-surface proteins (15). In both cases, PZ cells become rounder, without discernible elongation axes, but appear to divide laterally. Hence, cell division and cell polarity must both be propagated to daughter cells during normal skeletogenesis; the molecular bases of this propagation have not been defined.
The properties and expression pattern of Discs large 1 (Dlg1) make it a candidate mediator for the coordination of mitosis and cell polarity in growth plate cartilage. Dlg1 is the only member of the membrane-associated guanylyl kinase-like (MAGUK) family of scaffolding proteins (16–19) to be expressed in bone. Each of the three key members of the MAGUK family (Dlg1/SAP-97, Dlg2/PSD-93, and Dlg4/PSD-95) contains three PDZ domains that mediate interactions with signaling proteins and cytoskeletal elements (20). In addition, they contain Src-homology 3 (SH3) and GTPase-deficient guanylate kinase (GK) domains (Fig. 1B) (20). The SH3 and GK domains adopt a clamshell-like tertiary structure that maintains a nonscaffolding monomeric state when in a closed configuration (21); when in an open state, these domains become available to multimerize via intermolecular SH3–GK associations (22). Thus, Dlg1 can assemble functional signaling specializations on the plasma membrane for facilitating high-fidelity and amplified signal transduction (23, 24). Previous work in cell culture and in neuroepithelia suggested that Dlg1 regulates oriented cell division by placing mitotic spindles along a cell’s elongation axis (25, 26). Interestingly, Dlg1-null mice display shorter and wider cartilage elements (a sign of misregulated polarized cell behaviors) than wild-type mice (27, 28). Determining how Dlg1 regulates cartilage elongation at both the molecular and cellular levels promises to fill gaps in our knowledge of the relationship between mitosis and cell polarity.
Here, we combine protein labeling and perturbation reagents with in vivo imaging to analyze the normal behavior of Dlg1 and to assess the consequences of perturbing its function in cell behavior. Our imaging approach (29) follows chondrocyte cell behaviors within explanted chicken cartilage in 4D (3D over time). To image native Dlg1, we tagged it with an affinity label: PSD95.FingR (Fibronectin intrabody generated by mRNA display), a genetically encoded recombinant antibody-like protein originally developed for neuroscience applications (30) through mRNA display technology (31, 32). Because it is promiscuous with Dlg1/SAP-97 (demonstrated in a heterologous expression system; ref. 30), we have renamed this probe “Dlg.FingR” for our purposes and used it to visualize the spatiotemporal dynamics of Dlg1 protein in developing chicken limbs. To perturb Dlg1 function in chicken embryos, which are not accessible via conventional genetic approaches, we constructed a synthetic E3 ubiquitin ligase for viral delivery. Recently, an ablating FingR capable of eliminating Gephyrin in neurons (GFE3) was created by fusing the RING domain of X-linked inhibitor of apoptosis protein (XIAP) to Gephyrin FingR (33). Experiments using GFE3 to degrade the inhibitory postsynaptic protein Gephyrin elucidated the importance of GABA receptor scaffolding at inhibitory synapses. We applied a similar strategy to construct an ablating intrabody, named “DLGE3,” to study Dlg1 function.
Our targeted perturbation of Dlg1 in chicken chondrocytes, in conjunction with genetic studies in Dlg1-null mice, reveals Dlg1 enrichment at the midbody during cytokinesis and shows that this transient membrane specialization controls neither oriented cell division nor daughter-cell separation; instead, this protein scaffold is critical for the polarity of daughter cells and provides the link needed to coordinate mitosis and cell polarity.
Results
Dlg1 Is the only Detectable MAGUK Protein Expressed in Cartilage.
Probing chicken chondrocyte lysate with a pan-MAGUK antibody in a Western blot assay revealed only one positive band, corresponding to the appropriate molecular weight of Dlg1 (Fig. 1C). The presence of Dlg1 in distinct cartilage zones and the surrounding perichondrium/periosteum (PC/PO) was confirmed through RNA in situ hybridization (Fig. 1 D–G and SI Appendix, Fig. S1). Pan-MAGUK immunofluorescent staining showed that Dlg1 could be found either dispersed around the plasma membrane or concentrated at cell–cell contacts (Fig. 1 H–K). Finally, mass spectrometry confirmed that Dlg1 is the sole member of the MAGUK family of proteins within cartilaginous tissue (SI Appendix, Table S1).
Dlg.FingR-GFP Reveals Dlg1 Is Transiently Enriched in the Midbody During Early Cytokinesis.
The concentration of Dlg1 at cell–cell contacts was most evident in near-neighboring pairs of cells that were not yet separated by extracellular matrix and thus were likely to be mitotic pairs. Actin-enriched contractile rings were absent in a subset of these contacts (arrows in Fig. 1 H and I), raising two possibilities: (i) that these cells had entered cytokinesis but had not yet undergone abscission through the contraction of the actin ring or (ii) that these cells had completed cytokinesis but remained associated through an adhesive interface. Rigorous testing of these two possibilities requires determining the spatiotemporal dynamics of Dlg1 at single-cell resolution.
For imaging the subcellular distribution of Dlg1, Dlg.FingR-GFP was introduced using replication-competent avian retrovirus (RCAS) (Fig. 2A), and chicken metacarpal tissues were explanted for our analyses. The transduced cells showed normal apoptosis (SI Appendix, Fig. S3) and differentiation (SI Appendix, Fig. S4); with respect to cell proliferation, we observed no change in the number of cells incorporating BrdU, indicating that there was no change in the number of cells entering S phase and preparing to divide (SI Appendix, Fig. S2). Taken together, these findings suggest that Dlg.FingR-GFP expression does not impact normal growth.
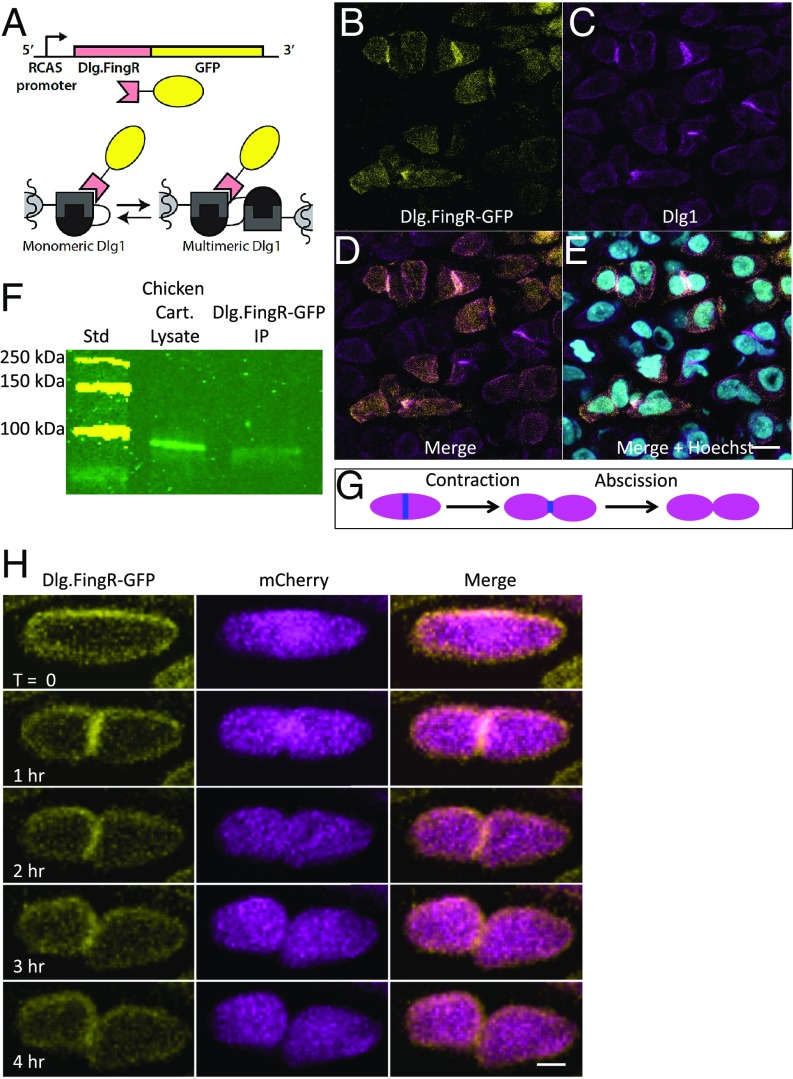
Fig. 2.
Dlg.FingR-GFP reveals the dynamic distribution of Dlg1 in PZ chondrocytes. (A) Schematic diagram showing recognition of Dlg1 with the FingR. The genetically encoded probe Dlg.FingR fused with GFP was cloned into the RCAS vector. This probe recognizes the SH3-GK domains of Dlg1 and therefore can localize to both the monomeric and the multimeric forms of these scaffolding proteins. (B–E) In chicken humerus sparsely transduced with RCAS-Dlg.FingR-GFP, the GFP signal (B) was colocalized with Dlg1 immunoreactivity (revealed by pan-MAGUK staining) (C) on the cell membrane and at cell–cell contacts. Some cells showed a cytoplasmic Dlg.FingR-GFP signal caused by Dlg.FingR-GFP that has not bound to Dlg1, which could not be detected by pan-MAGUK antibody. Because the tissues were partially infected, Dlg.FingR-GFP signal was not presented in all cells. n = 5 transduced chick humeri. (Scale bar: E, 15 μm, applies to B–E.) D and E show merged images, as labeled. (F) To determine the physical association between Dlg1 and Dlg.FingR, coimmunoprecipitation was performed on chicken humeri globally transduced with RCAS-Dlg.FingR-GFP. Chondrocytes were lysed at E8, and a portion of them was exposed to the bead-linked anti-GFP antibody. The resulting precipitates were run on a SDS/PAGE gel (lane 2) along with the lysates (lane 1), blotted, and probed with pan-MAGUK antibody. In lane 2, a specific band was observed at the Dlg1 position (140 KD). n = 3 transduced chick humeri. (G) Schematic diagram of cytokinesis. A midbody ring (blue) is formed between two future daughter cells. Subsequently, daughter cells pull away from each other, and the cytoplasmic connections become narrower (contraction) until they are cleaved (abscission). (H) Time-lapse images of a representative cytokinetic cell cotransduced with Dlg.FingR-GFP and mCherry. Dlg1 had a diffused localization with enrichment on the cell membrane. During early cytokinesis, when two daughter cells were still closely connected with each other, Dlg1 was mainly concentrated in the midbody. Afterward, Dlg1 in the midbody was down-regulated, and upon abscission Dlg1 was again enriched on the membrane. n = 3 transduced chick metacarpals. (Scale bar: 3 μm.)
Immunohistochemistry demonstrated the codistribution of the FingR-dependent GFP signal with Dlg1 (Fig. 2 B–E), and coimmunoprecipitation analysis validated the physical interaction of Dlg.FingR-GFP with Dlg1 (Fig. 2F), confirming that Dlg.FingR-GFP reveals the native distribution of Dlg1 within chondrocytes.
Live-image analysis of chicken metacarpal explants (29) double-labeled with Dlg.FingR-GFP and cytosolic mCherry revealed Dlg1 distributed around the cell perimeter at most cell-cycle stages (Movie S1). In addition, Dlg1 was enriched in the midbodies of mitotic cells (Fig. 2 G and H), beginning with the onset of cytokinesis (34, 35) and subsequently dissipating during contraction and abscission phases. Thus, Dlg.FingR-GFP offered clear evidence that the Dlg1 concentration at cell–cell contacts came from its transient midbody association during cytokinesis.
Functional Knockout of Dlg1 with Ablating DLGE3.
Misregulation of cell polarity likely produces the shorter and wider cartilage elements displayed by Dlg1-null mutant mice (27, 28); however, the underlying mechanisms can only be inferred in such purely genetic approaches. Cultured chicken metacarpals provide the access needed to analyze Dlg1 localization and cell behavior in living cartilage (29) but require a technique to eliminate Dlg1 function to assess Dlg1’s impact on cell behaviors. DLGE3, a Dlg1-ablating FingR designed in a fashion similar to that used in our previous work (Fig. 3A) (33), answers the need for a rapid knockdown of endogenous proteins (33), avoiding the challenges of applying RNAi or CRISPR approaches (36, 37) in older chicken embryos.
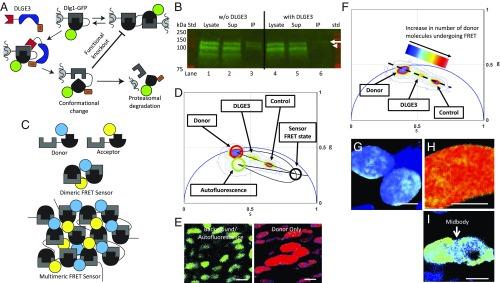
Fig. 3.
DLGE3 eliminates Dlg1 multimerization. (A) Schematic diagram showing the proposed action for DLGE3. One proposal is that polyubiquitination leads to proteasomal targeting and hydrolysis of ectopic Dlg1-GFP fusion (diagramed), as well as endogenous Dlg1 (not diagramed), resulting in a reduction of total Dlg1. Another proposal is that ubiquitination of SH3-GK renders the homo-multimerization domains nonfunctional. Red, blue, orange, green, and gray/black shapes depict Dlg.FingR, the RING-domain of XIAP, ubiquitin, GFP, and the SH3-GK domain, respectively. (B) To test these two possibilities, GFP-based coimmunoprecipitation was performed in HEK293 cells. In cells transfected with only myc-Dlg1 and Dlg1-GFP (lanes 1–3), multimers formed, and all three species were pulled down. In contrast, in the presence of DLGE3 (lanes 4–6), there was a reduction in the amount of endogenous and myc-Dlg1 (both −100 kDa in this gel) as well as the GFP fusion (∼127 kDa, arrow), and the other available Dlg1 species (plus a truncated ∼90 kDa species) failed to precipitate with the GFP fusion (arrowhead), indicating a reduction of Dlg1 multimers. n = 3 replicates for each condition. (C) Schematic diagram describing our dual-chain FRET sensor in which SH3-GK domains (gray/black) were fused to CFP (blue) and YFP (yellow) to create donors and acceptors, respectively. These were introduced into chicken metacarpal explants for FLIM-FRET analysis. In the case of Dlg1 multimerization, donor and acceptor will be close enough so that FRET will occur. (D) Phasor distribution of lifetimes for the FLIM-FRET experiments on chick metacarpal explants with background/autofluorescence (green circle), donor only (red circle), and FRET state of the sensor (black circle on the FRET trajectory). Control cells expressing sensors only (including donor and acceptor) had a shorter lifetime, whereas the cells with both sensors and DLGE3 had significantly longer lifetimes. (E and G–I) This Phasor distribution includes background/autofluorescence in untransduced chondrocytes (green cells corresponded to phasor-plotted lifetimes within the green circle in D) and chondrocytes expressing donor only (red cells corresponded to lifetimes falling within the red circle in D) (E); cells with donor only (G); cells with sensors only (H); and cells with both sensors and DLGE3 (I). (Scale bars: 5 μm.) (F) Phasor distribution of lifetimes was heat-mapped and color-coded based on sensor populations undergoing FRET. This map depicts regions within the cells having lower (blue) and higher (red) numbers of active sensors. Consistent with the lifetime distribution (D), cells expressing DLGE3 had fewer active sensors and accordingly fewer SH3-GK multimers than control cells, demonstrating that DLGE3 prevents Dlg1 multimerization in chondrocytes.
Conceptually, it seemed that DLGE3 could block the wild-type functions of Dlg1 by two means. First, DLGE3 could polyubiquitinate Dlg1, directing its proteasomal degradation. Second, DLGE3-mediated ubiquitination of Dlg1 could deform the SH3-GK domain or create steric hindrance of Dlg1’s multimerization. A biochemical analysis of SH3-GK multimerization-mediated coimmunoprecipitation of Dlg1 offers a direct means to test these two possibilities. HEK293 cells were transfected with both myc-tagged Dlg1 and Dlg1-GFP. These cultures were then cotransfected with DLGE3; cells without DLGE3 served as our control (Fig. 3B). Anti-GFP immunoprecipitation of Dlg1-GFP offered a direct assay for the association of Dlg1 and Dlg1-GFP: If they are associated, they will coprecipitate; in the absence of multimerization, only Dlg1-GFP will immunoprecipitate. The analysis showed clear coprecipitation in the absence of DLGE3; coprecipitation was greatly inhibited by DLGE3 (arrow, Fig. 3B). In addition, the absolute Dlg1 abundance was decreased by DLGE3 (arrowhead, Fig. 3B), suggesting that both potential mechanisms are at play, resulting in significant decreases in both Dlg1 multimerization and Dlg1 protein expression.
We validated the biochemical findings with frequency-domain fluorescence lifetime imaging microscopy of Förster resonance energy transfer (FLIM-FRET) (38) imaging using a dual-chain FRET sensor (Fig. 3C and SI Appendix, Fig. S5) and analysis via the Phasor approach (39). Multimerization brings a subpopulation of the CFP donors sufficiently close to transfer their excited states to adjacent YFP acceptors, which decreases both the intensity and the lifetime of CFP fluorescence (40) and results in a demodulation shifting the pixel values toward the infinitely short lifetime, s = 0 and g = 1. Changes in FRET are most accurately measured using fluorescence lifetime, which is best represented within the Phasor plot (ref. 41; see Methods for details) (Fig. 3 D and E). The power of the Phasor approach versus intensity-based FRET analysis was recently highlighted by Sadybekov et al. (42). The phasor approach to frequency-domain FRET relies on excitation of a reference fluorescent molecule with a modulated, pulsed light source followed by measuring the demodulation of FRET fluorescence with respect to the reference frequency. Demodulation is summed for each pixel in the image and is assigned a sine and cosine value. The Phasor plot (Fig. 3 D and F) presents the CFP fluorescence lifetime of each pixel in the image in a 2D, semicircular universal plot. The pixel’s position in the Phasor plot is characteristic of the molecular species (39), and the position is altered by the species’ interactions with other fluorescent moieties. The amount of FRET is easily measured by the shape and extent of these positional changes. Pixels containing pure single exponential fluorescence decay reside on the edge of the semicircle. Pixels recording multiexponential decays reside within the semicircle and are a linear combination of each pure single exponential decay. Molecules not undergoing FRET occupy a region directly between the Phasor positions of CFP (donor only) and the cell’s autofluorescence (background). The Phasor positions for molecules undergoing FRET are distributed linearly toward the sensor’s “FRET state” which resides on the calculated FRET trajectory curve (Fig. 3 D and E) (41).
Phasor analysis of single chondrocytes in explanted chicken metacarpals showed that our FRET sensor had an average FRET efficiency of ∼70% and that 50–60% of the CFP population was undergoing FRET (Fig. 3 F–H). In cells coexpressing DLGE3, only 15–30% of the CFP sensors were undergoing FRET, a 40–75% reduction (Fig. 3 F and I). Given that FRET reflects the close apposition of YFP and CFP, the observed increase in CFP lifetime confirmed that Dlg1 multimerization is inhibited by DLGE3. Phasor analysis of lower-magnification images containing multiple chondrocytes confirmed the average FRET efficiency of ∼70% with 30–40% of the CFP population undergoing FRET (SI Appendix, Fig. S5 B–F). This represents a consistent FRET knockdown due to the presence of DLGE3. The variation in the distributions appears to result from the variability in dual sensor expression levels and a greater influence of background pixels. Our FLIM-FRET analyses consistently revealed CFP shifts to a longer lifetime upon the addition of DLGE3, consistent with the biochemical data, and suggest that DLGE3 inhibits Dlg1 multimerization within chondrocytes.
DLGE3 Prevents Midbody Targeting of Dlg1 in Chondrocytes.
We virally delivered DLGE3 and GFP using a bicistronic RCAS vector [DLGE3 and GFP coding sequences separated by an internal ribosomal entry site (IRES)] to determine whether DLGE3 reduces the total amount of Dlg1 in cartilage. Immunofluorescent staining of the tissues transduced with titrated virus showed that midbody-associated Dlg1 was absent in GFP-positive (e.g., DLGE3-expressing) chicken chondrocytes; otherwise, the total Dlg1 expression levels and distributions appeared similar to those in adjacent GFP-negative cells (Fig. 4 C–E).
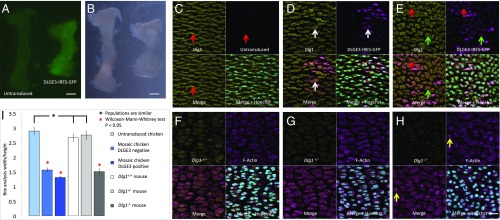
Fig. 4.
Dlg1 is required for normal chondrocyte cell morphology and cartilage dimensions. (A and B) Chicken humerus transduced with high-titer (1 × 109) RCAS-DLGE3-IRES-GFP virus was shorter and wider than the contralateral untransduced humerus, as demonstrated by both fluorescent (A) and differential interference contrast (B) images. (Scale bars: 300 μm.) (C–E) Cell morphology and Dlg1 expression in untransduced chicken humerus (C) and in humerus with low-titer (1 × 108) (D) and high-titer (E) RCAS-DLGE3-IRES-GFP virus. (C) Untransduced PZ cells were elliptically shaped with their elongation axes orthogonal along the PDA. Optical sections contain only partial midbodies, which are not pronounced. (D) In the sparsely transduced tissues, both transduced and adjacent untransduced cells were rounder and misoriented (GFP, purple). (E) A higher-efficiency transduction showed a more complete disorganization of tissue architecture even far away from the transduced source. Assaying protein level by the intensity of pan-MAGUK antibody staining did not reveal obvious reduction of total Dlg1 (demonstrated in SI Appendix, Fig. S9). In contrast, while Dlg1 was enriched in the midbodies of untransduced cells (red arrows in C and E), it was largely absent in the midbodies of DLGE3-IRES-GFP–expressing cells, with some exceptions likely due to heterogeneity of DLGE3 expression and expression timing (white arrows in D and green arrows in E), suggesting a correlation between midbody-bound Dlg1 and cellular phenotype. n = 5 untransduced and 5 transduced chick humeri. (F–H) Cell morphology and Dlg1 expression in the humerus of Dlg1+/+ (F), Dlg1+/− (G), and Dlg1−/− (H) mice. In both Dlg1+/+ (F) and Dlg1+/− (G) mice, PZ cells were elliptical and stacked into columns. In contrast, PZ cells were rounder and misoriented in Dlg1−/− mice (H). Pan-MAGUK antibody staining confirmed Dlg1 was partially and completely knocked out in Dlg1+/− mice (G) and Dlg1−/− mice (H), respectively. Two connected future daughter cells were aligned orthogonal to the PDA (yellow arrows in H). n = 3 Dlg1+/+, 3 Dlg1+/−, and 3 Dlg1−/− mouse humeri. (Scale bar: H, 20 μm, applies to C–H.) (I) Quantification of cell morphology using w/h analysis confirmed that the cell morphology was rounder in the transduced chicken cartilage and Dlg1−/− mouse cartilage than in their wild-type counterparts. n = 30 cells in each condition.
Others have shown that truncations of Dlg1 lacking SH3-GK domains, expressed in heterologous cell systems, not only failed to multimerize but also failed to localize to the midbodies of mitosing cells (34, 35). Considering that DLGE3 blocks Dlg1 multimerization in chondrocytes of chicken explants, with the most significant effect seen at the midbodies (white arrow, Fig. 3I), it is likely that Dlg1 multimerization mediates its midbody targeting.
Dlg1 Controls Chondrocyte Cell Polarity and Cartilage Dimension.
Chicken cartilage globally transduced with high-titer (1 × 109) virus became shorter and wider (Fig. 4 A and B and SI Appendix, Fig. S6), recapitulating the Dlg1−/− mouse phenotype (27, 28) on the macroscopic level. Microscopic inspection revealed that PZ cells were significantly rounder and more disorganized in both transduced chicken and Dlg1−/− mouse cartilage (Fig. 4 C–H). We quantitatively scored these changes in cell morphology by performing width-to-height (w/h) ratio analysis of cells in both tissues (see Fig. 6D). We measured the w/h ratio for cells by drawing minimal bounding rectangles around each chondrocyte. The normal oblong cells in the PZ, with their long axes oriented orthogonally to the PDA, had a w/h ratio >1, whereas, more rounded or misoriented cells had a w/h ratio closer to 1. These ratios provided quantitative evidence that the PZ cells in Dlg1−/− mouse (w/h ratio = 1.5 ± 0.10) and in transduced chicken (w/h ratio = 1.4 ± 0.05) were rounder than wild-type mouse and chick cells (w/h ratio = 2.7 ± 0.22 and 2.8 ± 0.20, respectively) (Fig. 4I), indicative of defective chondrocyte cell polarity (11–15, 43).
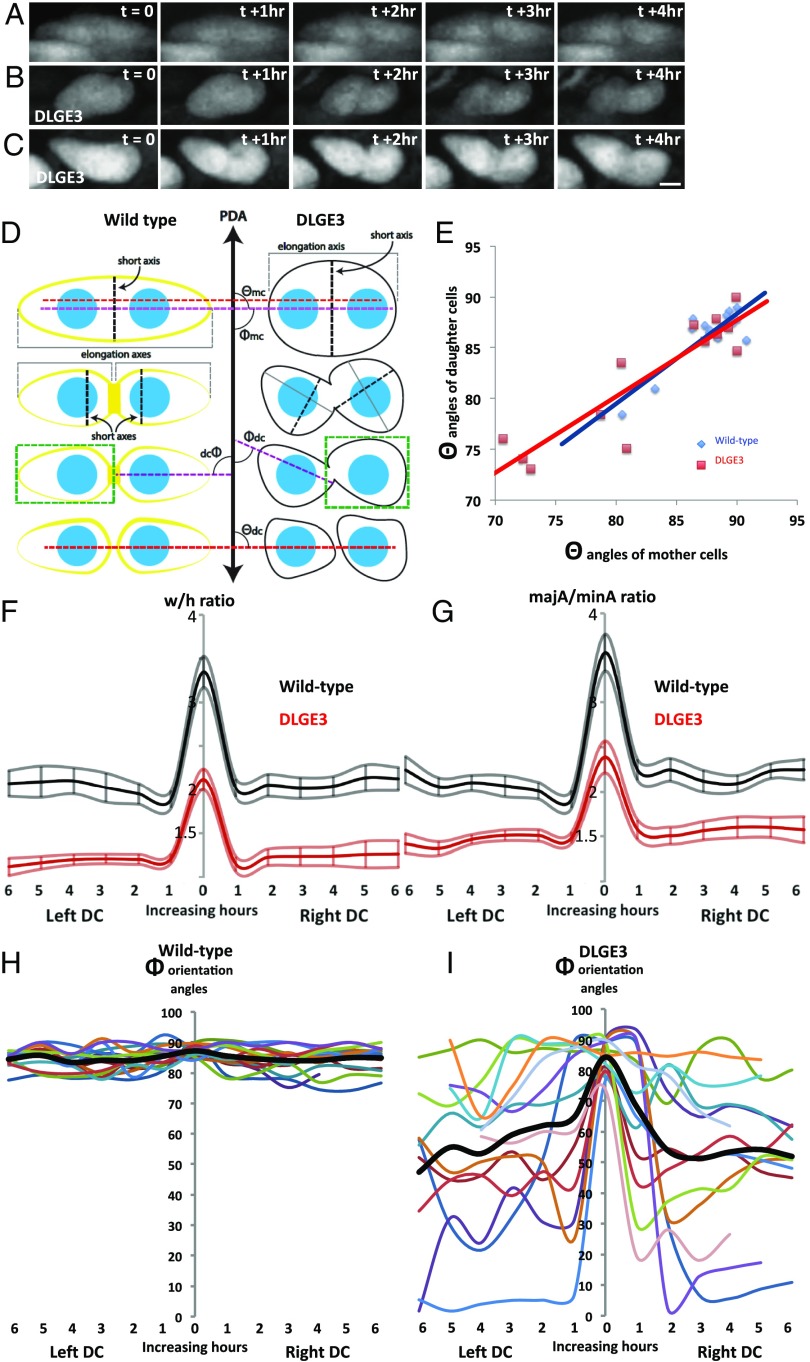
Fig. 6.
Dlg1 maintains daughter-cell orientation after cytokinesis. (A–C) Representative time-lapse images of mitosing cells in GFP-expressing chicken metacarpals (n = 3) (A) and cells expressing DLGE3-IRES-GFP (n = 3) (B and C). (Scale bar: C, 3 μm, applies to A–C.) (D) A schematic diagram showing w/h analysis (green box), majA/minA ratio analysis composed from the length of the elongation axis (solid gray line) and the short axis (dashed black line), division plane analysis (angle θ between the red dashed line and the PDA), and cell orientation analysis (angle Φ between the purple dashed line and the PDA). dc, daughter cell; mc, mother cell. W/h analysis is a compound measurement of cell elongation axis and elongation angle relative to the PDA; the angular component is removed from the majA/minA analysis. (E) Correlation analysis of θ angles shows that at time t = 0, the alignment of two daughter cells was parallel to the major (elongation) axes of mother cells in both GFP (R2 = 0.78) and DLGE3 (R2 = 0.82) tissues, suggesting that Dlg1 is not required for orienting the division planes. (F) W/h analysis showing both DLGE3-expressing mother cells (at t = 0) and daughter cells (at t = 1–6 h) (dark-red lines represent average ratios; light-red lines represent SE) were rounder than their GFP counterparts (black lines represent average ratios; gray lines represent SE). Note that the daughter cells’ morphology was rounder than mother cells’ regardless of Dlg1. (G) MajA/minA ratio analysis describes cell shape agnostic to the PDA and expresses a major component of w/h analysis. (H) Orientation analysis of Φ angles showing that after division GFP-positive daughter cells were aligned orthogonal to the PDA over time. (I) In contrast, the orientation of DLGE3-positive daughter cells had obvious deviations from this orthogonal angle (colored lines represent angles of individual cells; thick black lines represent the average angles). In F–I, n = 15 GFP cells and 13 DLGE3+GFP cells.
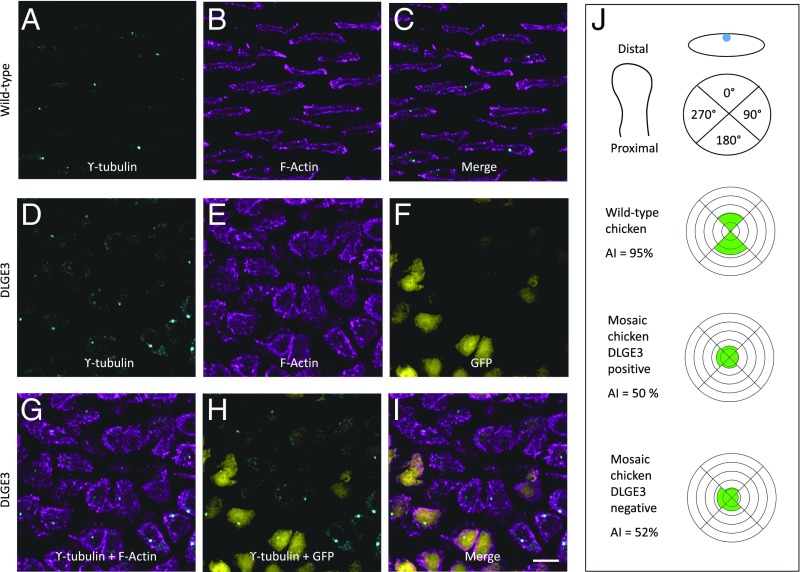
An independent assay of chondrocyte cell polarity is provided by ϒ-tubulin immunocytochemistry, highlighting the positions of the microtubule-organizing centers (MTOCs) (14). Similar to previous findings (14), MTOCs were localized in the middle of the PZ cell elongation axis in both untransduced chicken cells (Fig. 5 A–C) and wild-type mouse cells (SI Appendix, Fig. S7 A–C). In contrast, the MTOCs were mislocalized in chicken cells expressing DLGE3 (Fig. 5 D–I) and in Dlg1−/− mouse cells (SI Appendix, Fig. S6 D–F). Quantification of cell morphology and MTOC orientation in the sparsely transduced chicken cartilage revealed abnormal morphologies (Fig. 4I) and polarities (Fig. 5J) of both transduced and adjacent untransduced cells, similar to that seen in mouse ureter development (44).
Fig. 5.
Dlg1 is specifically required for normal chondrocyte cell polarity. (A–I) DLGE3 caused mislocalization of MTOCs in chicken. As revealed by γ-tubulin staining (A and D) and phalloidin staining (B and E), MTOCs in untransduced tissues were located in the middle of the longitudinal sides of PZ cells (A–C); however, this stereotypical subcellular localization of MTOCs was disrupted in DLGE3-IRES-GFP–positive tissues (D–I). n = 4 untransduced and 7 transduced chicken humeri. (Scale bar: I, 10 μm, applies to A–I.) (J) Quantitative analysis of MTOC orientation. Each PZ cell was divided into four quadrants, and the orientation of individual MTOCs was determined based on the quadrant in which it was located. Quadrants were determined based on the PDA of the tissue with 0° and 180° in the circle corresponding to the distal and proximal surfaces of the cell, respectively. The percentage of MTOCs in each quadrant is plotted in a rosette diagram; the spacing of concentric rings represents a 20% increase moving outward from the center of the plot. In untransduced cartilage, MTOCs were located on the proximal and distal sides of the PZ cells; in contrast, in mosaic cartilage, MTOC localization shifted to lateral sides of the cells. P = 0.0093 for untransduced cells (n = 53) vs. DLGE3-positive cells (n = 54) in the mosaic tissues and P = 0.0187 for untransduced cells vs. DLGE3-negative cells (n = 54) in the mosaic tissues. The asymmetrical index (AI), the percentage of the cells with proximal and distal localization of MTOCs, showed an obvious difference between untransduced and mosaic tissues.
There were no significant differences between transduced and untransduced tissues in other major cellular processes. The BrdU incorporation index (SI Appendix, Fig. S2 G–J), the levels of apoptosis (SI Appendix, Fig. S3 G–I), and the timing of differentiation (SI Appendix, Fig. S4 G–K) all appeared normal. The expression of Dlg1 in the PC/PO raised the formal possibility that the loss of Dlg1 in surrounding tissues might indirectly impact chondrocyte cell polarity (Fig. 1 D–G). We used highly diluted virus, so that only PC/PO cells, but not chondrocytes, were transduced, and found that PZ cells displayed normal morphology (SI Appendix, Fig. S8), ruling out an indirect effect from expression in PC/PO cells.
The apparent molecular and cellular effects do not correlate with decreased expression of Dlg1, as the Dlg1 levels were not noticeably reduced in chicken chondrocytes expressing DLGE3 (Fig. 4 C–E and SI Appendix, Fig. S9). Similarly, Dlg1+/− mouse tissues exhibited levels of Dlg1 decreased by one-half (44), but no cellular disruption was observed (Fig. 4 G and I). These findings, together with the fact that DLGE3 specifically eliminates midbody localization of Dlg1 in chicken cartilage, suggest that the midbody-localized Dlg1 subpopulation is most critical for chondrocyte cell polarity.
Dlg1 Controls Axial Orientation of Daughter Cells After Cytokinesis.
Examination of fixed and stained chicken tissues expressing DLGE3 and Dlg1−/− mouse tissues revealed many normal-appearing mitosing cells, suggesting a mechanism by which midbody-bound Dlg1 couples cytokinesis and chondrocyte daughter-cell polarity. These mitosing cells showed normal elongation aligned orthogonal to the PDA; furthermore, they had normal-appearing contractile rings at cell–cell contacts, and no cells were multinucleated (Fig. 4 D, E, and H and white arrows in SI Appendix, Fig. S10), suggesting that normal cell division orientation, contraction, and abscission are taking place in these tissues. Intriguingly, some recently separated daughter cells (inferred by their relative positions) appeared round and disorganized (blue arrows, SI Appendix, Fig. S10), indicating that these cells had not inherited the stereotypic morphology and polarity of their mother cells.
We performed live-imaging analyses of recently mitosed cells in chicken metacarpals transduced with RCAS-DLGE3-IRES-GFP to assess the role of Dlg1 (Fig. 6 A–C and Movies S2 and S3); control metacarpals were transduced with GFP alone. To estimate the orientation of cell division, we measured the correlation between the orientation angle of the mother cell to the PDA and the angle generated from the line drawn between two future daughter cells’ center of mass relative to the PDA (Fig. 6D). All mitosing control cells (GFP expressing) and most DLGE3-IRES-GFP–positive cells divided orthogonal to the PDA and produced daughter cells in similar orientations (Fig. 6E). Even DLGE3+GFP-expressing cells misaligned to the PDA had division planes aligned with each cell’s major axis (Fig. 6E), as expected from Hertwig’s rule. We confirmed this finding through imaging analysis of mitotic spindle formation in both wild-type and DLGE3-transduced tissues; the results showed that during cell division spindle microtubules aligned parallel to the major axes of mother cells (SI Appendix, Fig. S11). We therefore concluded that, although Dlg1 multimerization is necessary for PZ cell polarity, disrupting the multimerized state of Dlg1 is not sufficient to perturb oriented cell divisions.
To further explore the morphological changes in daughter cells, we followed recently mitosed cells for 6 h after cytokinesis and applied w/h analysis to quantify changes in cell morphology. The morphologies of both DLGE3-positive mother cells and daughter cells were rounder and skewed in comparison with their GFP-positive counterparts (Fig. 6F). To better capture the roundness and skewness of these cells without respect to the PDA (unlike the w/h analysis), we measured the ratio between cell major and minor axes (majA/minA), additionally measuring the angle of their major axes to the PDA (Fig. 6D). The majA/minA analysis showed that both GFP- and DLGE3-expressing daughter cells remained elliptically shaped after cell division (Fig. 6G); however, the orientations of DLG3-positive daughter cells were more inconsistent than those of GFP-expressing daughter cells (Fig. 6 H and I). Given that both DLGE3- and GFP-expressing mother cells were oriented orthogonally to the PDA (compare the black lines at time 0 in Fig. 6 H and I), this finding strongly suggests that Dlg1 plays a major role in the maintenance of daughter-cell orientation.
Thus, collectively, based on the midbody localization of Dlg1 during cytokinesis, the function of DLGE3 in inhibiting multimerization of Dlg1, and the specific loss of normal orientation after cytokinesis by DLGE3-expressing daughter cells, we propose that multimerization of Dlg1 controls daughter-cell polarity.
Discussion
In this study of the interaction between chondrocyte cell division and cell polarity, we exploit the advantages of two complementary animal model systems: chicken and mouse. Chicken offers an oviparous system in which viral transduction permits convenient and rapid in vivo tests of FRET sensors as well as tagging and ablating reagents. Transduction with titrated virus offers mosaics in which cells expressing and not expressing the reagents can be compared directly, simplifying tests of cell–cell interaction. Mouse provides a defined genetic system with which to validate the knockdown phenotypes observed in chicken. The consistent results obtained from the two systems highlight a conserved role of Dlg1 in cartilage morphogenesis in higher vertebrates.
Our work offers several insights into Dlg1’s function in controlling cell division and cell polarity (Fig. 7 A and B). First, it extends previous studies in cell culture describing Dlg1 enrichment in the midbody ring during cytokinesis and its dispersal around the cell perimeter during other phases of the cell cycle (34). While this might have suggested that midbody-localized Dlg1 has a role in controlling abscission (35), our dynamic imaging reveals that the timing of the enrichment is incorrect for this role. The newly developed DLGE3 permits us to perturb Dlg1 and demonstrates that stereotypical subcellular localization of Dlg1 is multimerization dependent. Thus, Dlg1 at the midbody is required for the maintenance of cell polarity but not for abscission of daughter cells. It is also possible that plasma membrane-localized Dlg1 may play a polarity-propagating role in premitotic cells; this possibility should be further investigated in future studies but does not weaken our conclusion regarding to the roles of midbody-bound Dlg1 during mitosis.
Fig. 7.
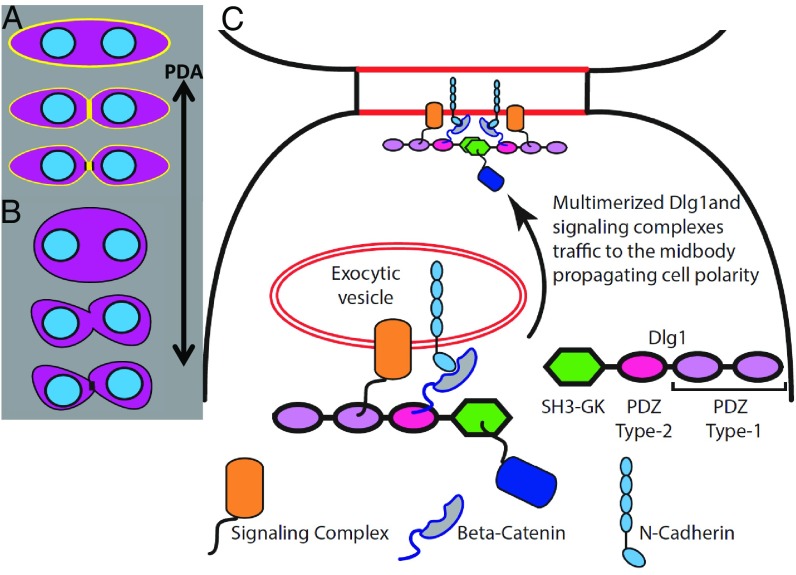
Model of Dlg1 regulation of chondrocyte cell polarity. (A) Wild-type PZ cells undergo oriented cell division and produce two daughter cells with their elongation axes orthogonal to the PDA. Dlg1 (yellow) is transiently enriched in the midbody during cytokinesis and reconcentrates onto the membrane after cytokinesis. (B) When Dlg1’s midbody targeting is prevented, mother cells are rounder and sometimes are misoriented but still place their division planes in normal orientation. After cytokinesis, the two daughter cells lose their polarity to a greater extent than the mother cell. (C) Schematic diagram of a hypothetical model showing the molecular mechanism by which Dlg1 may maintain cell polarity information. This process is likely accomplished through midbody targeting of cell polarity proteins such as the Frizzled/Disheveled/Vangl complex (orange), and cell adhesion complex (i.e., β-catenin/N-cadherin or β-catenin/protocadherin-γC3 demonstrated in our mass spectrometry data) (gray with blue outline). These proteins bind to PDZ domains (magenta and lilac) or to SH3-binding proteins (dark blue) with polyproline motifs to further interact with the SH3-GK domain (green) of Dlg1. Subsequently, these signaling and cell-adhesion molecules are likely trafficked to midbodies via multimerized Dlg1.
Second, the loss of axial information propagates to daughter cells, and the cumulative effects disrupt normal tissue architecture and alter normal tissue dimensions. Considering that Dlg1 is transiently enriched in the midbody during early cytokinesis, we propose a hypothetic model to explain how Dlg1 recruits cell-polarity proteins and signaling complexes into the midbody ring (Fig. 7C) for transmitting cell polarity (45–47). In this model, it is likely that Dlg1 fills the same trafficking role in chondrocytes that Dlg3 and Dlg4 play in neurons (21–24). Candidate proteins are members of the planar cell polarity (PCP) signaling pathway (16, 25), β-catenin, which binds to Dlg1 in cartilage (SI Appendix, Fig. S12), and others that have been demonstrated to associate with Dlg1 in various tissues (12, 17, 47, 48). As observed in other systems (7) organized by PCP signaling, the effects of perturbing Dlg1 could spread into neighboring cells. Similarly, we observed misorientation of untransduced cells adjacent to DLGE3-positive cells. This nonautonomy suggests communication between cells, perhaps via PCP signaling, protocadherin (SI Appendix, Table S1), diffusible ligand(s), or collagen 2a deposition. Future studies are needed to test these hypotheses for the propagation of disorder and to shed light on the roles of ligand–receptor signaling and physical interactions with matrix and adjacent cells in developing cartilage.
Third, our study of vertebrate limb skeletons suggests that polarity and mitosis might be separately controlled, as mitotic cells expressing DLGE3 exhibit normal division planes but produce daughter cells with abnormal axial orientation. We propose that oriented cell division alone is not sufficient to generate daughter cells with normal morphology; rather, a Dlg1-mediated protein complex located in the midbody during cytokinesis ensures that daughter cells maintain the correct polarity information within the collagen matrix. Our work expands cell-polarity mechanisms beyond the establishment of division orientation at metaphase and anaphase (2) and suggests that multiple mechanisms act at distinct phases of the cell cycle to transmit cell polarity.
This study enriches our knowledge of the cellular mechanisms underlying limb skeletal elongation. The abnormal cartilage morphology of the Dlg1-mutant mouse in the previous end-point analyses could have been generated by a variety of defects at any developmental stage. Our Dlg1 knockout in chicken embryos affords mosaic analysis within otherwise normal tissue development and is temporally isolated to bone elongation and not differentiation. This permits us to correctly identify the contributions of multiple cellular factors and highlight the importance of cell polarity.
FLIM-FRET of live tissue served as an important adjunct to our molecular biology approaches. The Phasor approach to frequency-domain analysis of FRET permitted us to study molecular interactions in situ (38). Unlike the standard method of estimating FRET through measurements of fluorescence intensity (49), the Phasor approach yields quantitative data on donor–acceptor interactions that are insensitive to fluorophore concentration, making it more suitable for systems such as our transduced chicken embryos with heterogeneous protein-expression levels. The robustness of this approach in cartilage suggests that it can be adapted to determine protein dynamics in a multitude of cell and developmental biology experiments.
mRNA display (50) and the FingR approach to labeling can be applied to any protein that has near-native conformation in vitro and has a reasonably hydrophobic surface when presented as an antigen for mRNA display (30–32). Well-characterized components of subcellular structures have been the clearest choice for FingR targets, to ease characterization of the FingRs. The only example of a nonstructurally targeted FingR (51) was selected against the neuronal Ca2+/calmodulin-dependent kinase (CaMKIIα). Due to the abundance of CaMKIIα within neurons and an activity-dependent rearrangement of CaMKIIα, we were able to successfully characterize this FingR.
Ablating FingRs is a nascent technology, and it is important to experimentally test whether total ablation (33) or functional ablation, as in our case, is achieved with any particular FingR. As regards the FingR/ubiquitin ligase combination, it is clear that ubiquitination of proteins does not universally lead to the ubiquitinated protein’s degradation (52–55); therefore, confirmation of ablation will need to be performed on a target-by-target and FingR-by-FingR basis.
Materials and Methods
DLGE3 Construction.
The DNA sequence for the E3 ligase RING-domain of mouse XIAP (base pairs 1818–1998 encoding the final 60 amino acids of the wild-type protein) with a 5′ sequence that codes for a 28-residue flexible Gly-Ser-Ala linker was ordered as a G-block synthesis from IDT. This fragment was cloned downstream of the PSD-95.FingR in the pCAG-PSD-95.FingR-eGFP-CCR5TC (Addgene plasmid no. 46295) vector in place of the GFP and zinc-finger. The new chimeric construct was PCR amplified and subcloned into pLVX IRES GFP. The bicistronic construct was PCR amplified, digested, and ligated into the RCAS viral vector.
Viral Production and Infection.
RCAS(A)-mCherry, RCAS(A)-Dlg.FingR-GFP, RCAS(A)-DLGE3-IRES-GFP, and RCAS-β-tubulin-mRFP were transfected into DF1 cells (ATCC) in a 6-cm culture dish using a standard transfection protocol. The transfected cells were split sequentially into 10-cm and 15-cm dishes. When the cells were confluent in 15-cm dishes, the cell-culture medium was harvested once per day for 3 d and was concentrated at 49,000 × g for 1.5 h. After the spin, the supernatant was discarded, and the pellet was dissolved in minimal volume and further injected into specific pathogen-free chicken (Charles River) right forelimbs at E3 [Hamburger–Hamilton (HH) stage 19–20]. The experiments were approved by the University of Southern California Institutional Review Board. IACUC does not regulate the use of chicken embryos.
Immunofluorescence.
Tissues were fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4 °C. The tissue was then cryosectioned to 15-μm thickness and was immobilized on treated microscope slides and stored at −20 °C. Upon removal from frozen storage, the sections were fixed again with 4% PFA in PBS for 5 min at room temperature and were washed three times in PBS for 5 min each. Our samples were permeabilized with a blocking buffer [1× PBS with 10% (vol/vol) normal goat serum, 1% BSA, 0.1% vol/vol Triton X-100, 0.025% sodium azide] for 1 h at room temperature with agitation and then were stained with primary antibody [1:100 dilution for mouse anti–pan-MAGUK (73-029, Antibodies Inc.); all GFP signal is native fluorescence] in blocking buffer at room temperature overnight with agitation. Three washes of 5-min duration each were performed at room temperature with PBS. Secondary staining was carried out for 1 h at room temperature upon the addition of goat anti-mouse IgG1–Alexa-Fluor 594 (1:500 dilution; A-21125; Thermo Fisher Scientific) and phalloidin–Alexa-Fluor 647 (1:100 dilution; A-22287; Thermo Fisher Scientific) to blocking buffer. After 1 h the tissues were washed once with PBS plus a 1:100 dilution phalloidin–Alexa-Fluor 647 for 5 min and then with PBS only for 5 min, followed by a 5-min washing with a 1:10,000 dilution of Hoechst 33342 (62249; Thermo Fisher Scientific) in PBS. To detect chondrocyte apoptosis, cleaved casepase-3 antibody (1:200 dilution; no. 9661; Cell Signaling) was used. The coverslips were mounted with Fluoromount-G (17984-25; Electron Microscopy Sciences, Inc.) and were imaged on an LSM 780 inverted laser-scanning confocal microscope (Carl Zeiss).
To measure mitotic rates, tissues were labeled with the nucleotide analog BrdU for 8 h. Sectioned tissues were digested with trypsin and depurinated with hydrochloric acid. BrdU signal was detected using anti-BrdU antibody (1:200 dilution; Developmental Studies Hybridoma Bank) and goat anti-mouse IgG1–Alexa-Fluor 594 (1:500 dilution; A-21125; Thermo Fisher Scientific).
Hybridization Chain Reaction to Detect mRNA.
Chicken humeri at E7.5 were dissected, fixed in 4% paraformaldehyde, and frozen in O.C.T. (Tissue-Tek) for making sections (20-μm thickness). Tissue sections were hybridized to anti-chick Dlg1, Col2a, and Ihh DNA probes labeled with digoxigenin. Tissue sections of the postimaging chicken metacarpal (expressing GFP) were stained with phalloidin–Alexa-Fluor 546 (1:100; Molecular Probes).
Western Blot.
All denatured protein samples were loaded into precast SDS/PAGE gels (4569033 Any kDa; Bio-Rad) and were run alongside dual-color Precision Plus (1610374; Bio-Rad) protein standards. The Bio-Rad mini-PROTEAN tetra cell system (1658029FC) was used for electrophoresis and transblotting per the manufacturer’s protocol. Nitrocellulose containing the transferred protein was first washed/blocked three times (5 min each washing) in Blotto [20 mm Tris, 250 mm NaCl (pH 8.0), 5% nonfat milk, 1% Tween-20, 0.025% sodium azide]. Primary antibodies were added to an aliquot of Blotto (with the same pan-MAGUK antibody and dilution as described previously) or mouse anti–β-catenin antibody (1:500 dilution; ab6301; Abcam) and were added to the blots for overnight staining with agitation at room temperature. The membranes were washed in Tris buffer with 1% Tween-20 three times (5 min each washing) and then were placed in secondary antibodies [goat anti-mouse IgG–Alexa-Fluor 750 (1:10,000 dilution; A-21037; Thermo Fisher Scientific)] for imaging on the LI-COR Odyssey imaging system or donkey anti-mouse IgG–HRP (1:10,000 dilution; ab97023; Abcam) and conjugated for chemiluminescent imaging for 1 h at room temperature. The stained nitrocellulose was washed three times in standard Tris buffer (5 min each washing) and either was washed in deionized water once for 5 min and then dried for LI-COR near-IR fluorescent imaging or was placed into the Western Lightning Ultra ECL reagents (NEL111001EA, PerkinElmer) to be imaged on Kodak X-ray film.
Immunoprecipitation.
Humeri were dissected from the limbs of E8.5 chickens (five limbs in each condition), chopped into small pieces, and placed in 600 μL of ice-cold homogenization buffer (HB) [20 mM Tris, 250 mM NaCl (pH 8.0), 1% (vol/vol) Tween-20, and SIGMAFAST protease inhibitor mixture (S8830; Sigma Aldrich)]. The tissues were Dounce homogenized on ice every 15 min for 1 h and then were centrifuged at 5,000 relative centrifugal force and 4 °C for 10 min. The supernatant was collected and precleared against Protein A/G beads (20421; Thermo Fisher Scientific) and diluted 1:4 in HB. For GFP immunoprecipitation anti-GFP agarose (20-μL bead volume; D153-8; MBL) was equilibrated and washed three times in HB using Spin-X spin filters (CLS8160; Sigma Aldrich). Then 500 μL of the diluted supernatant was added to the beads and mixed end-over-end at 4 °C overnight. For Dlg1 immunoprecipitation lysates were treated with 10 μg/mL mouse anti-pan-MAGUK primary antibody (73-029; Antibodies Inc.) and were mixed end-over-end at 4 °C overnight. Anti–pan-MAGUK was precipitated onto Protein A/G beads mixed end-over-end at 4 °C for 1 h. For all immunoprecipitations, the flowthroughs and five washes were collected from the Spin-X columns, and Laemmli loading dye was added to a 1× final concentration. The beads were collected in 2× Laemmli loading dye, and all samples were boiled for 5 min.
HEK293 cells (CRL-1573; ATCC) were cultured in DMEM (30-2002; ATCC) with 5 μg/mL gentamicin (G1272; Sigma Aldrich) and 1× GlutaMAX (35050061; Gibco) at 37 °C under 5% CO2. All plasmid constructs were introduced to the HEK293 cells using Effectene transfection reagent (301425; Qiagen)in a 60-mm dish format. The plasmids were allowed to express for 24 h. Then the transfected cell cultures were expanded into three 60-mm dishes and were cultured for an additional 48 h. The cells were washed three times in ice-cold PBS and then were sequentially scraped from the bottom of the dishes in ice-cold HB (600 μL per condition). The immunoprecipitation proceeded as described previously.
Mass Spectroscopy Sample Preparation.
E7.5 humeri were dissected and lysed in 10 mM Tris⋅HCl lysis buffer (150 mM NaCl, 10% glycerol, 2% deoxycholate, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, pH 8.0) supplemented with SIGMAFAST protease inhibitor mixture (S8830; Sigma Aldrich). The samples were separated by 1D SDS/PAGE using a 4–12% gradient NuPAGE gel (Invitrogen) with NuPAGE MES running buffer (Invitrogen) for 1 h at 200 V. The gel was then stained in colloidal Coomassie stain (Invitrogen) and destained with water and ammonium bicarbonate. The desired band (∼80–125 kDa) was excised, finely diced, and transferred to an Eppendorf tube. The gel pieces were reduced, alkylated, and digested by trypsin overnight at 37 °C. Digested peptides were extracted from gel, lyophilized, and desalted. A sample was resuspended in 0.2% formic acid and subjected to LC-MS/MS analysis on an EASY-nLC 1000 nanoflow LC system (Thermo Fisher Scientific) coupled to a Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific) equipped with a Nanospray Flex ion source.
Briefly, for the EASY-nLC 1000 system, solvent A consisted of 97.8% H2O, 2% acetonitrile (ACN), and 0.2% formic acid, and solvent B consisted of 19.8% H2O, 80% ACN, and 0.2% formic acid. A 50-µm self-pack PicoFrit column (New Objective, Inc.) with a coated tip was packed in-house with 1.9 μm ReproSil-Pur C18-AQ resin (120-Å pore size; Dr. Maisch) to 20 cm. Samples were directly loaded onto the column, and the column was heated to 65 °C. The peptides were separated with a 60-min gradient at a flow rate of 220 nL/min. The Orbitrap Fusion was operated in a data-dependent Top speed method with a cycle time of 3 s.
Mass Spectroscopy Analysis.
Thermo raw files were searched using MaxQuant, version 1.6.1.0 (1, 2). Spectra were searched against the chicken entries in UniProt (29,729 sequences) and a contaminant database. Default settings were used, and iBAQ was enabled. Protein and peptide score thresholds were set to achieve an estimated 1% false-discovery rate as estimated by the target-decoy approach.
Metacarpal Culture and 4D Imaging of Cartilage Elongation.
The virus-infected metacarpals were dissected at E7.5 (HH 32–33) for organ culture. Molten agarose was poured into the FluoroDish (World Precision Instruments), and the custom-designed mold was immediately inserted into it. When the agarose was solidified, the mold was pulled out, leaving grooves in the agarose for holding the metacarpals. The metacarpals were submerged in DMEM/F12 growth medium containing 0.2% BSA, 50 mM ascorbate acid (Sigma-Aldrich), 10 mM β-glycerophosphate, and 1% glutamine-penicillin-streptomycin (Invitrogen) in a humidified chamber at 37 °C on the stage of the LSM 780 inverted laser-scanning microscope (Carl Zeiss).
Non-Descanned Multiphoton FLIM.
Fluorescence lifetime images were acquired with a Zeiss LSM-780 inverted microscope coupled to a Ti:Sapphire laser system (Coherent Chameleon Ultra II, 80-fs pulses with repetition rate of 80 MHz) and an ISS A320 FastFLIM data acquisition card (1). A 40× 1.1 NA water-immersion objective Korr C-Apochromat microscope (Zeiss) optimized for two-photon imaging was used. For image acquisition an image size of 256 × 256 pixels and a scan speed of 12.6 μs per pixel were used. A short-pass dichroic filter (760 nm) was used to separate the fluorescence signal from the laser light. For the acquisition of FLIM images, fluorescence light was separated into donor and acceptor fluorescence by a 509-nm long-pass CFP/YFP filter and then was detected by two hybrid photomultiplier tube detectors (Hamamatsu R10467U-40), one having a CFP 470/22 and the other a YFP 542/27 band-pass filter for the detection of donor and acceptor signal, respectively. FLIM data were acquired by VistaVision software from ISS, Inc., and were processed by SimFCS software developed at the Laboratory of Fluorescence Dynamics (LFD), University of California, Irvine. Calibration of the FLIM system was performed by measuring the known lifetime of coumarin 6 in 99% ethanol solution, characterized by a single exponential decay time of 2.55 ns. Typically, the acquisition time was about 30 s.
The Phasor Approach to FLIM Data Analysis.
Every pixel of the FLIM image was transformed in one pixel in the phasor plot as previously described and reported in detail (2, 3). The coordinates g and s in the phasor plot were calculated from the fluorescence-intensity decay of each pixel of the image by using Fourier transformations. The analysis of the phasor distribution was performed by cluster identification. We calculated the FRET efficiency trajectory according to the classical definition of FRET efficiency:
| [1] |
The phasor of the donor in the absence of the acceptor was obtained from an independent preparation in which the acceptor is absent. The phasor corresponding to the quenched donor is calculated according the quenching Eq. 1. The positions of all possible phasors that are quenched with different efficiencies describe a curved trajectory in the phasor plot. The experimental position of the phasor of a given pixel along the trajectory determines the amount of quenching and therefore the FRET efficiency. The contributions of the background and of the donor without acceptor are evaluated using the rule of linear combination (4, 5) with the background phasor and the unquenched donor determined independently. All phasor transformations and the data analysis of FLIM data were performed using SimFCS software developed at the LFD (https://www.lfd.uci.edu/).
Supplementary Material
Acknowledgments
We thank Prof. Francesca Mariani for her critical reading of the manuscript, Prof. Cheng-Ming Chuong for sharing equipment, Ang Li for statistical analyses and sharing reagents, Francesco Cutrale for helpful discussions and editing, Prof. Rich Roberts for the mRNA display without which FingRs would not exist, and Annie Moradian and Michael Sweredoski of the Proteome Exploration Laboratory at the California Institute of Technology for services, troubleshooting, and experimental advice regarding mass spectroscopy. This study was supported by the University of Southern California Translational Imaging Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713959115/-/DCSupplemental.
References
- 1.Sasai Y. Next-generation regenerative medicine: Organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Morin X, Bellaïche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Panbianco C, Gotta M. Coordinating cell polarity with cell division in space and time. Trends Cell Biol. 2011;21:672–680. doi: 10.1016/j.tcb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Hertwig O. Ueber den Werth der ersten Furchungszellen fur die Organbildung des Embryo. Experimentelle Studien am Froschund Tritonei. Arch Mikrosk Anat. 1893;42:662–807. German. [Google Scholar]
- 5.Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature. 2006;439:220–224. doi: 10.1038/nature04375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devenport D, Oristian D, Heller E, Fuchs E. Mitotic internalization of planar cell polarity proteins preserves tissue polarity. Nat Cell Biol. 2011;13:893–902. doi: 10.1038/ncb2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong Y, Mo C, Fraser SE. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430:689–693. doi: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- 8.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre V, Bhattaram P. Vertebrate skeletogenesis. Curr Top Dev Biol. 2010;90:291–317. doi: 10.1016/S0070-2153(10)90008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodds G. Row formation and other types of arrangement of cartilage cells in endochondral ossification. Anat Rec. 1930;46:385–399. [Google Scholar]
- 11.Li Y, Dudley AT. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;136:1083–1092. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao B, et al. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell. 2011;20:163–176. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aszodi A, Hunziker EB, Brakebusch C, Fässler R. Beta1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 2003;17:2465–2479. doi: 10.1101/gad.277003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Pabic P, Ng C, Schilling TF. Fat-Dachsous signaling coordinates cartilage differentiation and polarity during craniofacial development. PLoS Genet. 2014;10:e1004726. doi: 10.1371/journal.pgen.1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahrens MJ, Li Y, Jiang H, Dudley AT. Convergent extension movements in growth plate chondrocytes require gpi-anchored cell surface proteins. Development. 2009;136:3463–3474. doi: 10.1242/dev.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanaka T, Ohno S. Role of Lgl/Dlg/Scribble in the regulation of epithelial junction, polarity and growth. Front Biosci. 2008;13:6693–6707. doi: 10.2741/3182. [DOI] [PubMed] [Google Scholar]
- 17.Walch L. Emerging role of the scaffolding protein Dlg1 in vesicle trafficking. Traffic. 2013;14:964–973. doi: 10.1111/tra.12089. [DOI] [PubMed] [Google Scholar]
- 18.Ahn SY, Kim Y, Kim ST, Swat W, Miner JH. Scaffolding proteins DLG1 and CASK cooperate to maintain the nephron progenitor population during kidney development. J Am Soc Nephrol. 2013;24:1127–1138. doi: 10.1681/ASN.2012111074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka-Kogo A, et al. Requirement of DLG1 for cardiovascular development and tissue elongation during cochlear, enteric, and skeletal development: Possible role in convergent extension. PLoS One. 2015;10:e0123965. doi: 10.1371/journal.pone.0123965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 21.McGee AW, Bredt DS. Identification of an intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. J Biol Chem. 1999;274:17431–17436. doi: 10.1074/jbc.274.25.17431. [DOI] [PubMed] [Google Scholar]
- 22.McGee AW, et al. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol Cell. 2001;8:1291–1301. doi: 10.1016/s1097-2765(01)00411-7. [DOI] [PubMed] [Google Scholar]
- 23.Ponting C, Phillips C, Davies KE, Blake D. PDZ domains: Targeting signalling molecules to sub-membranous sites. BioEssays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 24.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 25.Manneville JB, Jehanno M, Etienne-Manneville S. Dlg1 binds GKAP to control dynein association with microtubules, centrosome positioning, and cell polarity. J Cell Biol. 2010;191:585–598. doi: 10.1083/jcb.201002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saadaoui M, et al. Dlg1 controls planar spindle orientation in the neuroepithelium through direct interaction with LGN. J Cell Biol. 2014;206:707–717. doi: 10.1083/jcb.201405060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivera C, et al. Requirement for Dlgh-1 in planar cell polarity and skeletogenesis during vertebrate development. PLoS One. 2013;8:e54410. doi: 10.1371/journal.pone.0054410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruana G, Bernstein A. Craniofacial dysmorphogenesis including cleft palate in mice with an insertional mutation in the discs large gene. Mol Cell Biol. 2001;21:1475–1483. doi: 10.1128/MCB.21.5.1475-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, et al. Dynamic imaging of the growth plate cartilage reveals multiple contributors to skeletal morphogenesis. Nat Commun. 2015;6:6798. doi: 10.1038/ncomms7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross GG, et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 33.Gross GG, et al. An E3-ligase-based method for ablating inhibitory synapses. Nat Methods. 2016;13:673–678. doi: 10.1038/nmeth.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unno K, Hanada T, Chishti AH. Functional involvement of human discs large tumor suppressor in cytokinesis. Exp Cell Res. 2008;314:3118–3129. doi: 10.1016/j.yexcr.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massimi P, Gardiol D, Roberts S, Banks L. Redistribution of the discs large tumor suppressor protein during mitosis. Exp Cell Res. 2003;290:265–274. doi: 10.1016/s0014-4827(03)00317-3. [DOI] [PubMed] [Google Scholar]
- 36.Das RM, et al. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev Biol. 2006;294:554–563. doi: 10.1016/j.ydbio.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Véron N, Qu Z, Kipen PA, Hirst CE, Marcelle C. CRISPR mediated somatic cell genome engineering in the chicken. Dev Biol. 2015;407:68–74. doi: 10.1016/j.ydbio.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Jameson D, Gratton E, Hall R. The measurement and analysis of heterogeneous emissions by multifrequency phase and modulation fluorometry. Appl Spectrosc Rev. 1984;20:55–106. [Google Scholar]
- 39.Digman MA, Caiolfa VR, Zamai M, Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luedtke R, Owen CS, Vanderkooi JM, Karush F. Proximity relationships within the Fc segment of rabbit immunoglobulin G analyzed by resonance energy transfer. Biochemistry. 1981;20:2927–2936. doi: 10.1021/bi00513a033. [DOI] [PubMed] [Google Scholar]
- 41.Hinde E, Digman M, Welch C, Hahn K, Gratton E. Biosensor FRET detection by the phasor approach to fluorescence lifetime imaging microscopy (FLIM) Microsc Res Tech. 2012;75:271–281. doi: 10.1002/jemt.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadybekov A, Tian C, Arnesano C, Katritch V, Herring BE. An autism spectrum disorder-related de novo mutation hotspot discovered in the GEF1 domain of Trio. Nat Commun. 2017;8:601. doi: 10.1038/s41467-017-00472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuss P, et al. Regulation of cell polarity in the cartilage growth plate and perichondrium of metacarpal elements by HOXD13 and WNT5A. Dev Biol. 2014;385:83–93. doi: 10.1016/j.ydbio.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 44.Kim ST, Ahn SY, Swat W, Miner JH. DLG1 influences distal ureter maturation via a non-epithelial cell autonomous mechanism involving reduced retinoic acid signaling, Ret expression, and apoptosis. Dev Biol. 2014;390:160–169. doi: 10.1016/j.ydbio.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahoney ZX, et al. Discs-large homolog 1 regulates smooth muscle orientation in the mouse ureter. Proc Natl Acad Sci USA. 2006;103:19872–19877. doi: 10.1073/pnas.0609326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geldmacher-Voss B, Reugels AM, Pauls S, Campos-Ortega JA. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development. 2003;130:3767–3780. doi: 10.1242/dev.00603. [DOI] [PubMed] [Google Scholar]
- 47.Roberts S, Delury C, Marsh E. The PDZ protein discs-large (DLG): The ‘Jekyll and Hyde’ of the epithelial polarity proteins. FEBS J. 2012;279:3549–3558. doi: 10.1111/j.1742-4658.2012.08729.x. [DOI] [PubMed] [Google Scholar]
- 48.Hering H, Sheng M. Direct interaction of Frizzled-1, -2, -4, and -7 with PDZ domains of PSD-95. FEBS Lett. 2002;521:185–189. doi: 10.1016/s0014-5793(02)02831-4. [DOI] [PubMed] [Google Scholar]
- 49.Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi TT, Roberts RW. In vitro selection of protein and peptide libraries using mRNA display. Methods Mol Biol. 2009;535:293–314. doi: 10.1007/978-1-59745-557-2_17. [DOI] [PubMed] [Google Scholar]
- 51.Mora RJ, Roberts RW, Arnold DB. Recombinant probes reveal dynamic localization of CaMKIIα within somata of cortical neurons. J Neurosci. 2013;33:14579–14590. doi: 10.1523/JNEUROSCI.2108-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziv I, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics. 2011;10:M111.009753. doi: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 54.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 55.Yuan WC, et al. K33-linked polyubiquitination of Coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell. 2014;54:586–600. doi: 10.1016/j.molcel.2014.03.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.