Significance
Endosymbiotic theory suggests that mitochondria evolved from free-living prokaryotes which entered the host cell and were retained as endosymbionts. Here, we model this earliest stage of the endosymbiotic theory of mitochondrial evolution by engineering endosymbiosis between two genetically tractable model organisms, Escherichia coli and Saccharomyces cerevisiae. In this model system, we engineered E. coli strains to survive in the yeast cytosol and provide ATP to a respiration-deficient yeast mutant. In a reciprocal fashion, yeast provided thiamin to an endosymbiotic E. coli thiamin auxotroph. This readily manipulated chimeric system was stable for more than 40 doublings and should allow us to investigate various aspects of the endosymbiotic theory of mitochondrial evolution.
Keywords: mitochondria, endosymbiotic theory, ADP/ATP translocase, evolution
Abstract
It has been hypothesized that mitochondria evolved from a bacterial ancestor that initially became established in an archaeal host cell as an endosymbiont. Here we model this first stage of mitochondrial evolution by engineering endosymbiosis between Escherichia coli and Saccharomyces cerevisiae. An ADP/ATP translocase-expressing E. coli provided ATP to a respiration-deficient cox2 yeast mutant and enabled growth of a yeast–E. coli chimera on a nonfermentable carbon source. In a reciprocal fashion, yeast provided thiamin to an endosymbiotic E. coli thiamin auxotroph. Expression of several SNARE-like proteins in E. coli was also required, likely to block lysosomal degradation of intracellular bacteria. This chimeric system was stable for more than 40 doublings, and GFP-expressing E. coli endosymbionts could be observed in the yeast by fluorescence microscopy and X-ray tomography. This readily manipulated system should allow experimental delineation of host–endosymbiont adaptations that occurred during evolution of the current, highly reduced mitochondrial genome.
Endosymbiotic theory suggests that mitochondria were once free-living prokaryotes which entered the host cell and were retained as endosymbionts (1–4). The earliest recognized instance of endosymbiosis, which dramatically shaped the emergence of present-day eukaryotic cells, occurred more than 1.5 billion years ago (5). Previous studies hypothesized that an alphaproteobacterium became established in an archaeal host cell as an endosymbiont and triggered the evolution of the mitochondrion, an organelle specialized for efficient energy production, particularly ATP synthesis (3, 6). However, a recent study suggests that mitochondria evolved from a proteobacterial lineage that branched off before the divergence of all sampled alphaproteobacteria (7). The recent discovery of the Asgard superphylum archaea, which encode in their genomes homologs of cytoskeletal proteins and vesicular trafficking machinery, indicates that the last archaeo–eukaryotic common ancestor could have already featured precursors to the endomembrane system of modern eukaryotes (8).
Significant details have emerged about the nature of the premitochondrial endosymbiont. Studies involving the reconstruction of its genome from endosymbiont genes retained in eukaryotes and genes found in present-day alphaproteobacteria point to a bacterium related to intracellular endosymbionts from the Rickettsiales order (class of Alphaproteobacteria), some of which are human and animal pathogens (9–11). This premitochondrial endosymbiont likely possessed metabolic pathways that included glycolysis, the tricarboxylic acid cycle (TCA), the pentose phosphate pathway, and the fatty acid biosynthesis pathway. The reconstructed endosymbiont is also predicted to have an electron transport chain capable of functioning under low oxygen tension and an ADP/ATP translocase, the protein functionally homologous to the one used by many intracellular bacteria for ATP import from the host cytoplasm.
Herein, we attempted to experimentally recapitulate the early stages of mitochondrial evolution by generating Escherichia coli endosymbionts capable of providing ATP to host yeast cells deficient in ATP synthesis. Such an experimental system with easy-to-manipulate genomes may allow us to attempt evolution of a bacterial endosymbiont with a minimal genome in yeast that recapitulates key features of modern mitochondria.
Results
Experimental Approach.
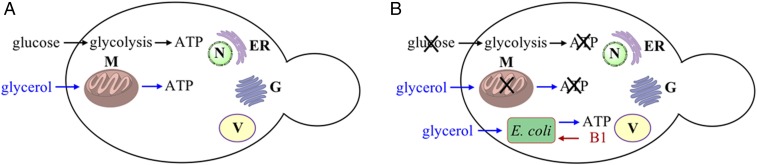
Our initial strategy involved engineering E. coli strains that are auxotrophic for an essential cofactor and that would depend on the host for this cofactor. We further engineered the bacteria to express a functional ADP/ATP translocase and GFP as a marker. As the host cells, we used Saccharomyces cerevisiae strains that had mitochondrial defects and were deficient in utilizing nonfermentable carbon sources (e.g., glycerol) for growth. Introduction of such engineered E. coli cells into the mutant S. cerevisiae cells followed by selection for yeast cells that can grow on a nonfermentable carbon source for multiple generations was expected to afford a stable yeast–E. coli chimera (Fig. 1).
Fig. 1.
Strategy to engineer S. cerevisiae–E. coli endosymbiont chimera. (A) Wild-type S. cerevisiae can grow on medium with glucose or glycerol due to ATP production by glycolysis in the cytoplasm and oxidative phosphorylation in mitochondria. (B) Yeast cells with a defect in oxidative phosphorylation cannot utilize glycerol for ATP synthesis and cannot grow in the absence of glucose. Introduction of E. coli-expressing ADP/ATP translocase and SNARE proteins into such mutant yeast can restore yeast growth with glycerol as the sole carbon source. Growth of intracellular E. coli is dependent on thiamin diphosphate (vitamin B1) provided by yeast. ER, endoplasmic reticulum; G, Golgi apparatus; M, mitochondria; N, nucleus; V, vacuole.
Engineering an E. coli Strain Expressing Functional ADP/ATP Translocase.
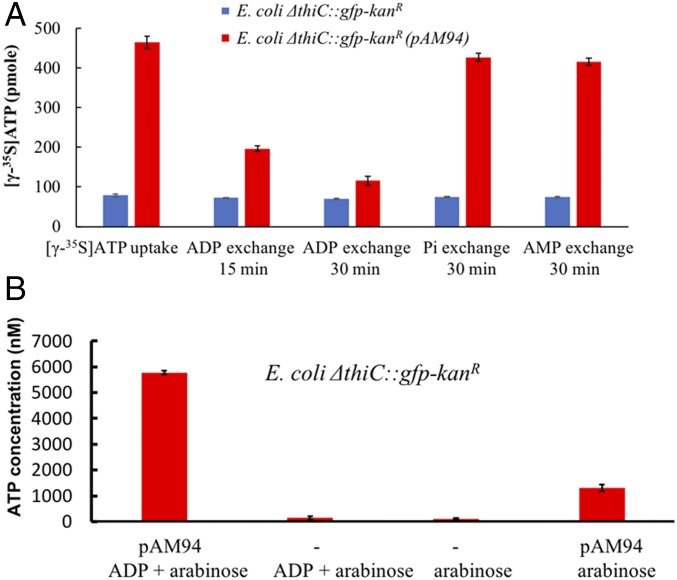
We first engineered an E. coli DH10B strain that had key features of an endosymbiont as outlined above. Because thiamin pyrophosphate is an essential bacterial cofactor, deletion of thiamin biosynthetic genes results in thiamin auxotrophy (12, 13). To create growth dependency of an experimental bacterial endosymbiont on the yeast host, the thiamin biosynthetic gene thiC was deleted and replaced with a gene cassette that coded for superfolder gfp and kanamycin resistance genes to afford the E. coli ΔthiC::gfp-kanR strain. We confirmed that the resulting E. coli strain required exogenous thiamin for growth (growth was enabled by the endogenous expression of TbpA, a thiamin transporter, in E. coli ΔthiC::gfp-kanR strain; see SI Appendix, Fig. S1), and that it expressed GFP (SI Appendix, Fig. S2 A and B). Next, we constructed an expression plasmid (pAM94) that encoded the ADP/ATP translocase gene from the intracellular bacterium Protochlamydia amoebophila strain UWE25, a symbiont of Acanthamoeba spp. (14), under control of a pBAD promoter. This construct was transformed into E. coli ΔthiC::gfp-kanR and the cellular activity of the ADP/ATP translocase was assayed. When translocase expression was induced by addition of 1 mM arabinose, and cells expressing the translocase were incubated with [γ-35S]ATP, we observed significant ATP uptake by the bacteria. In contrast, no ATP uptake was observed by the parent E. coli strain, which lacked the translocase-encoding plasmid (Fig. 2A and SI Appendix, Fig. S15A). To test release of ATP from the translocase-expressing E. coli cells, ADP, AMP, or potassium phosphate were separately added to the suspension of cells preloaded with [γ-35S]ATP. A significant, time-dependent drop in cellular radioactivity was observed on incubation with ADP, but not AMP or phosphate, indicating ADP-specific release of intracellular [γ-35S]ATP (Fig. 2A). Next, we analyzed the efflux of intracellular ATP on ADP stimulation during E. coli growth. Levels of extracellular ATP were quantified by a luciferase assay. We observed that cells expressing the ADP/ATP translocase released a significant amount of ATP on addition of ADP to the growth medium. For comparison, the media from E. coli lacking the plasmid pAM94 (expressing the ADP/ATP translocase) produced ∼50-fold lower luciferase signal with or without ADP addition (Fig. 2B and SI Appendix, Fig. S15B). Low levels of ATP were observed in the growth medium of cells expressing the ADP/ATP translocase even without addition of ADP, likely due to a small amount of cell lysis. These studies demonstrated that the E. coli ΔthiC::gfp-kanR cells expressing the translocase released ATP into the extracellular milieu in response to the presence of extracellular ADP.
Fig. 2.
Release of ATP by E. coli cells encoding ADP/ATP translocase. (A) Cellular [γ-35S]ATP uptake/release by E. coli cells expressing the UWE25 ADP/ATP translocase (pAM94 plasmid) in the presence of 1 mM arabinose. Cellular [γ-35S]ATP was released when E. coli cells expressing the ADP/ATP translocase were challenged with extracellular ADP (10 mM), but not with phosphate (Pi) or AMP (each at 10 mM). (B) Release of ATP into the growth medium by E. coli cells expressing the UWE25 ADP/ATP translocase (pAM94 plasmid) in presence of 20 µM ADP and 1 mM arabinose. The ATP concentration in the medium was determined by luciferase assay. Data bars show a mean of three technical replicates; error bars represent SE of the mean.
Introduction of Engineered E. coli Cells into Yeast.
Next, we optimized the protocol for introducing bacteria into yeast cells. Previously, bacterial cells were fused with yeast to clone whole bacterial genomes in yeast (15, 16). Since we wanted to retain intact live bacteria within yeast cells, we decided to use an alternative protocol that was previously developed for introduction of mitochondria into yeast by polyethylene glycol (PEG)-induced fusion (17). To test the latter protocol, we first isolated mitochondria from the respiration-competent YPH500 S. cerevisiae strain, and used a ρ0 S. cerevisiae mutant as the recipient for isolated mitochondria. ρ0 strains completely lack mitochondrial DNA (mtDNA) and are unable to utilize nonfermentable carbon sources for growth. As reported previously, PEG-induced fusion of isolated, respiratory-competent mitochondria with the ρ0 yeast spheroplasts led to the formation of yeast cybrids (cytoplasmic hybrids), which were able to grow on a nonfermentable carbon source (minimal medium + 3% glycerol/0.1% glucose, selection medium I) due to the presence of heterologous, respiring mitochondria in cells (SI Appendix, Fig. S3). Next, we attempted to introduce the engineered E. coli cells (the ΔthiC::gfp-kanR strain expressing the UWE25 ADP/ATP translocase under control of pBAD promoter from the pAM94 plasmid; see SI Appendix, Fig. S4) into the ρ0 mutant by following the same protocol. Similar to fusion with purified mitochondria, we expected to select yeast cells harboring E. coli endosymbionts that could utilize glycerol for the synthesis of ATP and provide it to the S. cerevisiae ρ0 host. However, we did not observe formation of any yeast colonies after the yeast/E. coli fusion mixtures were plated on a selection medium (rich medium + 3% glycerol, 0.1% glucose medium, 1 mM arabinose; selection medium II).
Mitochondria of ρ0 yeast cells lack both the electron transport chain and F1F0 ATP synthase (Atp6, Atp8, and Atp9 subunits are encoded in mitochondrial DNA). As a result, the ρ0 yeast cells energize their inner mitochondrial membrane inefficiently by electrogenic exchange of mitochondrial ADP for cytosolic ATP (catalyzed by the mitochondrial ADP/ATP translocase). To sustain ADP/ATP exchange and maintain an energized inner mitochondrial membrane, intramitochondrial ATP in the ρ0 cells undergoes futile hydrolysis to ADP (18). We reasoned that such an unproductive consumption of ATP by ρ0 mitochondria can critically deplete cellular ATP in chimera cells and block their growth. We therefore turned to a yeast mutant with a more limited mitochondrial defect as a host, S. cerevisiae cox2-60 (NB97), which possesses mtDNA but has an insertion in the COX2 gene. As a result, the cox2-60 strain lacks Cox2 protein, does not assemble a functional cytochrome c oxidase complex and, similar to ρ0 mutants, it does not grow in media with a nonfermentable carbon source (19). However, F1F0 ATP synthase expressed in mitochondria of the cox2-60 strain couples hydrolysis of intramitochondrial ATP to proton transport across the inner mitochondrial membrane resulting in more efficient generation of the inner membrane electrochemical potential. When S. cerevisiae cox2-60 spheroplasts were fused with the engineered E. coli cells containing the pAM94 plasmid, a few yeast colonies grew on the selection medium that had plated yeast/E. coli fusion mixtures, but not on plates with control mixtures that had omitted E. coli cells. However, most of the colonies were E. coli. To suppress E. coli (a thiamin auxotroph) growth, we also plated the fusion mixtures on a minimal selection medium, but did not observe formation of any colonies, neither yeast nor E. coli.
Introduction of Chlamydia SNARE Proteins into E. coli.
Intracellular pathogenic bacteria encode SNARE-like proteins that manipulate the host vesicular trafficking machinery and allow the pathogens to escape the lysosomal degradation pathway (20). Multiple SNARE-like proteins are expressed by such intracellular bacteria and are proposed to interfere with SNARE-mediated membrane fusion processes (21, 22). While the establishment of the alphaproteobacterial endosymbiont in the archaeal host could predate the emergence of eukaryotic vesicular trafficking (11, 23), successful engineering of endosymbiosis between present-day eukaryotic cells and bacteria might depend on the expression of SNARE-like proteins by experimental bacterial endosymbionts. Initially, we transformed the E. coli ΔthiC::gfp-kanR strain with pAM126 plasmid (SI Appendix, Fig. S4), which is derived from pAM94, but also contains the incA gene encoding a SNARE-like protein from the intracellular pathogen Chlamydia trachomatis behind the pBAD promoter (19). IncA is thought to inhibit endocytic SNARE-mediated fusion processes during the infectious cycle of C. trachomatis. Fusions performed with this E. coli strain and the S. cerevisiae cox2-60 mutant yielded again mostly E. coli colonies on plates with selection medium II. Next, we incorporated an additional SNARE-like gene, the incA homolog from Chlamydia caviae, which is proposed to have a broader inhibitory role than C. trachomatis incA (20, 24), into plasmid pAM126 to obtain pAM132 (SI Appendix, Fig. S4). Plasmid pAM132 was transformed into E. coli ΔthiC::gfp-kanR cells, which were then fused with the S. cerevisiae cox2-60 cells. This strategy resulted in significantly more yeast colonies upon plating, although we still observed formation of E. coli colonies as well. The yeast colonies were replated on selection medium III (rich medium + 3% glycerol + 1 mM arabinose) in the presence of carbenicillin (50 mg/L) and continued to grow for five rounds of replating. A low concentration of the antibiotic was added to eliminate the presence of extracellular bacteria if any, while the intracellular bacteria were expected to survive multiple rounds of replating on media containing antibiotic due to the lower intracellular levels of the carbenicillin in the yeast cytoplasm (25). We did not observe any extracellular E. coli cells within such secondary colonies microscopically (SI Appendix, Fig. S5A). Analysis of genomic DNA from these yeast cells by PCR indicated the presence of both the yeast and E. coli genomes; we were able to amplify the yeast MATa mating gene specific to S. cerevisiae cox2-60 (SI Appendix, Fig. S5B) and the gfp gene specific to the E. coli cells used in the fusion (SI Appendix, Fig. S5C).
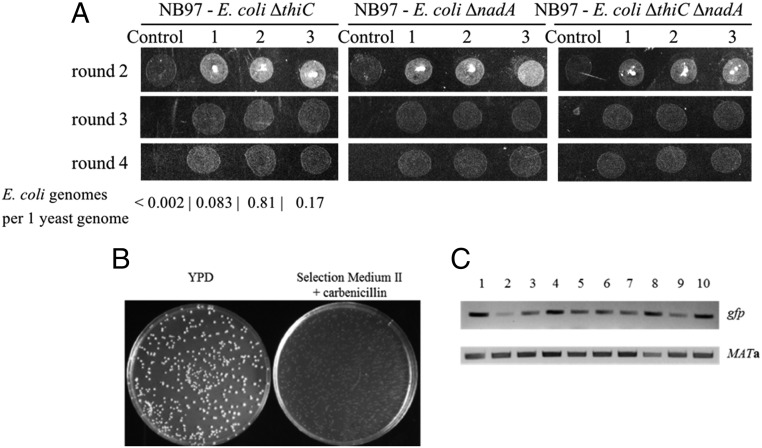
To generate more stable endosymbionts, we incorporated a third SNARE-like gene from C. trachomatis, CT_813, into plasmid pAM132 to afford pAM136 (SI Appendix, Fig. S4) and transformed it into E. coli ΔthiC::gfp-kanR cells. CT_813 is proposed to recruit ADP ribosylation factor GTPases and induce rearrangement of microtubules and Golgi around the Chlamydia inclusions (26). Fusions with S. cerevisiae cox2-60 cells again yielded yeast colonies growing on nonfermentable growth medium, but this time we did not observe E. coli colonies on the plates. We replated the colonies for four consecutive rounds (3 d per passage × 4 passages, >40 doublings) on selection medium III plates in the presence of carbenicillin (Fig. 3A). We again confirmed the presence of both yeast and E. coli genomes in the cells from the colonies by PCR amplification of the MATa gene for yeast and the gfp, 16S rDNA and glyA genes for E. coli (SI Appendix, Fig. S6) and by qPCR (Fig. 3A). Finally, culturing the yeast colonies from the fourth round of replating (SI Appendix, Fig. S7A) in a glucose-rich medium (rich medium + 2% glucose, a fermentative, nonselection medium) lacking carbenicillin produced mixed S. cerevisiae and E. coli cultures with individual E. coli cells being carbenicillin sensitive, kanamycin resistant, and gfp positive (SI Appendix, Fig. S7 B and C). All these data strongly suggest that the yeast colonies cultured on selection medium III continued to harbor intracellular E. coli cells during multiple rounds of replating. We also observed that intracellular E. coli cells continued to propagate in yeast when chimeras were grown on a less stringent selection medium II.
Fig. 3.
S. cerevisiae–E. coli chimeras have a partially rescued respiration-competent phenotype. (A) Growth of S. cerevisiae cox2-60–E. coli chimeras on medium containing glycerol as the sole carbon source, selection medium III. No growth was observed for parent cox2-60 yeast lacking intracellular E. coli (control). Three different chimera colonies growing during successive rounds of plating are shown for each S. cerevisiae–E. coli chimera. Number of E. coli genomes per one yeast genome was determined by qPCR for E. coli ΔthiC chimeras from the fourth round of growth. (B) A single cell suspension of S. cerevisiae cox2-60–E. coli nadA chimera culture formed a comparable number of colonies on nonselective (YPD) and selective medium (selection medium II) plates. (C) Total DNAs isolated from colonies grown on selection medium II in B contain E. coli-encoded gfp gene. Ten random colonies (labeled 1–10) were PCR amplified for presence of gfp and MATa genes.
To investigate the expression of SNAREs in the engineered E. coli cells, each of the SNAREs was C-terminally FLAG tagged. The cells were lysed, and SNARE expression was analyzed by Western blot. We observed the expression of C. trachomatis IncA and CT_813, but not C. caviae IncA (SI Appendix, Fig. S11 A–D). We also N-terminally FLAG-tagged C. caviae IncA, but still did not observe its expression. Our inability to detect FLAG-tagged C. caviae IncA could indicate that the protein is not expressed in E. coli. Alternatively, considering that its inclusion enhanced the stability of chimeras, it is possible that the N- and C-terminal-tagged IncA variants failed to express. To determine if the tagged C. trachomatis IncA and CT_813 are present in the E. coli membranes, we isolated the respective membrane fractions and detected by Western blotting the presence of both C. trachomatis IncA and CT_813 (SI Appendix, Fig. S11 E and F).
Next, we investigated the minimum number of SNAREs necessary to establish endosymbiosis within the yeast cells. To this end, we designed a construct, pAM162, which is similar to pAM136, but lacks C. caviae incA, and transformed it into E. coli ΔthiC::gfp-kanR and E. coli ΔnadA::gfp-kanR. Fusions with S. cerevisiae cox2-60 cells again yielded yeast colonies growing on nonfermentable growth medium. We replated these colonies for three consecutive rounds on selection medium III plates in the presence of carbenicillin (SI Appendix, Fig. S12A). We again confirmed the presence of both yeast and E. coli genomes in the cells from the colonies by PCR amplification of the MATa gene for yeast and the gfp gene for E. coli (SI Appendix, Fig. S12B). We further designed one other construct, pAM163, which is similar to pAM162 but lacks C. trachomatis incA, and transformed it into E. coli ΔthiC::gfp-kanR and E. coli ΔnadA::gfp-kanR. Fusions with S. cerevisiae cox2-60 cells yielded very few yeast colonies on nonfermentable growth medium and poor growth was observed on replating of these colonies on selection medium III, similar to fusions with pAM126 (containing only C. trachomatis incA SNARE). These studies indicated that a combination of ADP/ATP translocase together with the two C. trachomatis SNAREs is sufficient for establishing endosymbiosis. However, given that the three-SNARE system afforded the most stable endosymbionts, we further characterized this latter system, although the nature by which C. caviae IncA affects endosymbiont stability requires additional investigation.
Finally, to confirm that ADP/ATP translocase is required for establishing endosymbiosis, we eliminated the ADP/ATP translocase gene from pAM136 to afford pAM150 (SI Appendix, Fig. S4). Plasmid pAM150 was transformed into E. coli ΔthiC::gfp-kanR cells and fusions with S. cerevisiae cox2-60 were plated on selection medium II. This experiment yielded very few, slow growing yeast colonies. These colonies were replated on selection medium III, but no growth was observed for such yeast cells highlighting a key role of ADP/ATP translocase in establishing E. coli as an endosymbiont in S. cerevisiae cox2-60.
Significantly, and similar to the earlier yeast–E. coli chimera versions, we observed slow chimera growth (∼6 h per doubling at 25 °C) with the E. coli symbionts harboring plasmid pAM136. This observation could be due to several factors such as the rate of ATP production by the E. coli endosymbiont, inefficient partitioning of intracellular E. coli cells during yeast cell division, overgrowth of the E. coli endosymbiont causing inhibition or death of the host yeast, or depletion of metabolites from the yeast cytoplasm by the E. coli endosymbiont.
Imaging E. coli Endosymbionts Within Yeast Cells by Fluorescence Microscopy.
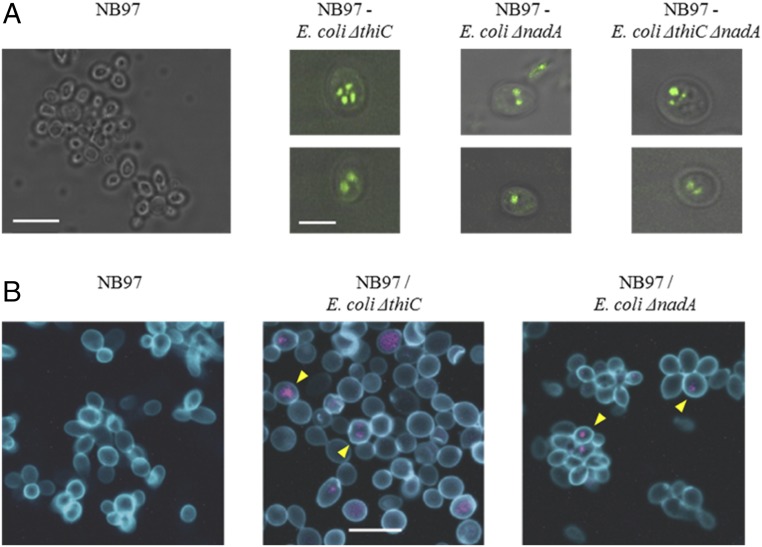
To obtain further evidence for the presence of the E. coli endosymbionts in yeast cells, we imaged the yeast cells from a third round of replating with total internal reflection fluorescence (TIRF) microscopy to detect the expression of the bacteria-encoded GFP protein within the yeast cells. We observed the presence of compartmentalized GFP-positive signals in the fused yeast cells, but not in the parental control NB97 yeast cells (Fig. 4A). Similarly, cox2-60 S. cerevisiae–E. coli chimeras and control S. cerevisiae were imaged by fluorescence confocal microscopy. The S. cerevisiae cell wall was labeled with FITC-labeled Con A (a lectin which binds to the yeast cell wall) (27), and the E. coli endosymbionts were labeled with a EUB338-Cy3 FISH probe, which can detect bacterial rRNA (28). We detected FISH probe signals only in the yeast–E. coli chimera cells, but not in control cells (Fig. 4B), further confirming the presence of E. coli endosymbionts in yeast cells.
Fig. 4.
Imaging intracellular endosymbiont E. coli by fluorescent microscopy. (A) TIRF microscopic images of chimeric cells (Right) and control yeast cells (Left). Two representative cells of indicated chimera type are shown. All panels are merged images of TIRF (green) and differential interference contrast (grayscale). (Scale bar in the NB97 panel, 10 µm; scale bar in the NB97–E. coli ΔthiC panel, 5 µm.) (B) Confocal fluorescence microscopy images of control and chimeric yeast–E. coli cells. Yeast cell wall was stained with Con A-FITC (blue) and bacterial rRNA with EUB338-Cy3 probe (purple). Yellow arrowheads indicate examples of EUB338-positive yeast cells. (Scale bar in the Middle, 10 µm.)
Mimicking Mitochondrial Genome Reduction with Additional E. coli Auxotrophies.
Having established the presence of E. coli endosymbionts in yeast cells, we next investigated if we could begin to replicate genome reduction events that occurred during mitochondrial evolution. As a first step, we investigated if a strain of E. coli auxotrophic for another key cofactor could generate endosymbionts within yeast. We deleted the NAD biosynthetic gene nadA by replacing it with the gfp-kanR gene cassette in E. coli DH10B (to afford E. coli ΔnadA::gfp-kanR) and confirmed that the resulting E. coli strain is a NAD auxotroph (growth in media with NAD+ enabled by E. coli pnuC transporter) (29). We then transformed this strain with the pAM136 plasmid (coding for the ADP/ATP translocase and three SNARE-like proteins), fused it with the S. cerevisiae cox2-60 spheroplasts, and plated the fusion mixtures on selection medium II. As before, most of the colonies that formed on the plates were yeast. We again confirmed the presence of E. coli and yeast genomes by PCR (SI Appendix, Fig. S8). These chimeras were also stable for four rounds of replating (>40 generations, Fig. 3A) and displayed a compartmentalized, E. coli-encoded GFP signal by TIRF microscopy (Fig. 4A) as well as bacterial RNA FISH signal by confocal fluorescence microscopy (Fig. 4B). We also assessed the fraction of S. cerevisiae cox2-60–E. coli ΔnadA::gfp-kanR chimeric cells present in the yeast culture grown on selection medium II. We plated a single cell suspension of one such culture on plates with nonselective (YPD + carbenicillin, allows growth of all yeast cells) and selective (selection medium II + carbenicillin, allows growth of chimeric cells only) media. We observed formation of a comparable number of colonies on both media (Fig. 3B), suggesting that most of yeast cells in the culture contained intracellular E. coli. This conclusion was further confirmed by detecting by PCR the E. coli gfp gene in 10 randomly selected yeast colonies from selection medium II (Fig. 3C).
Next, we generated a thiamin/NAD double auxotroph of E. coli DH10B, transformed it with pAM136, and used these cells in fusions with yeast as before. Formation of yeast colonies was again observed on selection plates (selection medium II) and the chimeras could be replated for four rounds on the selection medium III in the presence of carbenicillin (Fig. 3A). Additionally, we observed the E. coli-encoded GFP signal in these chimeric yeast cells by TIRF microscopy (Fig. 4A). Further, we confirmed the presence of both genomes by PCR amplification of the MATa gene for yeast and gfp and kanR genes for E. coli (SI Appendix, Fig. S9). Finally, we transformed a serine auxotroph of E. coli (E. coli ΔserA::kanR) with pAM136 plasmid and fused it with the S. cerevisiae cox2-60 spheroplasts to select chimeras on selection medium II, which were further replated on selection medium III for three rounds of growth. The presence of E. coli and yeast genomes was demonstrated by PCR during each round of replating (SI Appendix, Fig. S10). These preliminary experiments suggest that it might be relatively straightforward to eliminate a significant fraction of the E. coli genome by complementing essential bacterial metabolites and other cellular building blocks with those from the yeast cytosol.
Imaging Chimera Cells by Soft X-Ray Tomography.
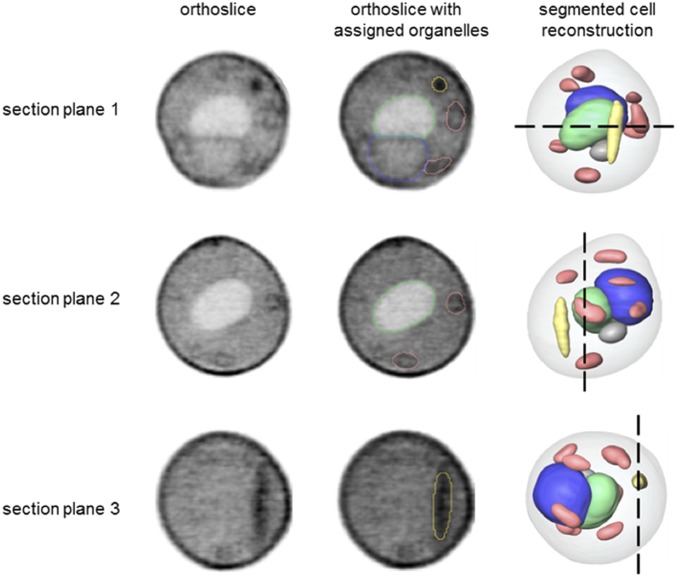
To characterize the S. cerevisiae–E. coli ΔnadA chimera at an ultrastructural level, we imaged the chimera cells by soft X-ray tomography. This technique is uniquely suited for high-resolution, quantitative imaging of intact cells in near native state and allows imaging and characterization of organelles and intracellular structures (30). In addition to major yeast organelles, the imaged chimera cell also contained an elongated subcellular structure that had a relatively high X-ray linear absorption coefficient value (LAC = 0.41 µm−1), suggesting high protein/lipid content (Fig. 5 and Movie S1). Both the object shape and LAC value were similar to those of freely growing E. coli cells (LAC = 0.49 µm−1) imaged by soft X-ray tomography (31). For comparison, we did not observe any such subcellular structures in a S. cerevisiae cox2-60 cell imaged by the same technique, although all major cell organelles were clearly identified (SI Appendix, Fig. S13 and Movie S2).
Fig. 5.
Ultrastructural features of S. cerevisiae–E. coli ΔnadA chimera cells. Segmented reconstruction of a S. cerevisiae–E. coli ΔnadA cell viewed from three different perspectives (section planes 1–3). Orthoslices (Left) at positions indicated by the black dashed lines in a reconstructed cell (Right) are shown for each perspective. The same orthoslices are overlaid in the Center column with outlines indicating segmented organelle assignment. The gray values were generated using LACs, with black corresponding to the highest LAC value. Organelle color key: green, vacuole; blue, nucleus; salmon, mitochondria; and yellow, high LAC value, bacteria-like structure that remained unassigned after segmentation.
Discussion
We have been able to generate E. coli endosymbionts that require cofactors/amino acid from the host S. cerevisiae cytosol and are capable of supplying ATP to the yeast. Another requirement for establishing the S. cerevisiae–E. coli chimera is the expression of SNARE-like proteins from intracellular pathogens, which are likely required to avoid lysosomal degradation. Although the detailed mechanism by which the SNARE-like proteins stabilize the endosymbiont remains to be elucidated, their presence is clearly required. Previous studies showed that C. trachomatis IncA and C. caviae IncA inhibit membrane fusion driven by endocytic SNAREs when included in v-SNARE (Syntaxin7/Syntaxin8/Vti1b) or t-SNARE-containing (VAMP8) liposomes (20). In addition, the SNARE VAMP8 has been shown to play a role in maturation of the late endosomes in S. cerevisiae (32). Therefore, it is possible that IncA may protect a membrane compartment against lysosomal degradation by interfering with yeast SNARE fusion function. CT_813 recruits ADP ribosylation factor GTPases and induces rearrangement of microtubules and Golgi around Chlamydia inclusions (26), processes which are proposed to play an important role in establishment of the replicative niche and may also play a role in establishing the yeast–E. coli endosymbiont. Further studies are required to define the precise roles of these SNARE-like proteins in establishing a stable yeast endosymbiont, as well as their potential relationship to the genetic elements that facilitated the establishment of stable endosymbionts within archaeal hosts.
The growth of E. coli within the yeast cytosol is likely to facilitate efforts to reduce the size of its genome. As a first step in this direction, we showed that the endosymbiont can be manipulated in terms of auxotrophy. It is likely that many other biosynthetic pathways (e.g., amino acids, cofactors, and other metabolites) can be eliminated from E. coli if endogenous or engineered bacterial transporters allow their products to be taken up from the yeast cytosol. Such biosynthetic pathways constitute a significant part of the E. coli genome; for example, the genes encoding the NAD biosynthetic pathway are ∼7 kb of chromosomal DNA, while the transporter is encoded by only a 0.7-kb gene. This engineered system should allow us to explore other aspects of the endosymbiotic theory of organelle evolution, including those factors that control the stability of symbionts with dramatically different replication rates. Indeed, we have recently isolated a bacterial mutant that is stable inside yeast cells for more than 120 d and are characterizing this mutant by whole genome sequencing and gene expression and metabolome analysis. Finally, it may be possible to complement loss of additional genes from yeast mitochondria with intracellular bacteria that supply additional functions beyond ATP production.
This synthetic symbiont system described here also affords an unexpected insight in mitochondrial evolution. Colonization of an archaeal host by an intracellular bacterium utilizing an ADP/ATP translocator for energy parasitism would likely be unfavorable for the host archaeal cells growing in its natural ecological niche (possibly rich in hydrogen) (33, 34). However, the orthogonal features of the parasitic bacterium energy metabolism (e.g., generation of ATP through TCA and oxygen-dependent oxidative phosphorylation) together with its ADP/ATP translocase could allow expansion of such colonized archaeal cells into new ecological niches native to the endosymbiont (e.g., containing oxygen and TCA substrate molecules), but prohibitive to nonparasitized host cells. In the new niche, the tables are turned: the archaeal host cell becomes the parasite (cannot produce its own ATP) and the endosymbiotic bacterium becomes the prey as the direction of the ATP flow is reversed (just like in our experimental system where yeast is the parasite). Such ecological expansion of archaea that harbored a parasitic endosymbiont expressing an ADP/ATP translocase could be the first step in evolutionary transition from premitochondrial endosymbiont to protomitochondrion.
Materials and Methods
A detailed experimental section describing E. coli engineering, S. cerevisiae–E. coli fusion protocols, imaging methods, Western blots, genomic DNA analysis, design and construction of recombinant plasmids, and gene deletion cassettes appears in SI Appendix. See SI Appendix, Figs. S1–S15 and Tables S1–S3 and Movies S1 and S2.
Supplementary Material
Acknowledgments
We thank Ashok Deniz, Jordan Kolarov, and Ladislav Kovac for helpful discussions and John Cortez for technical assistance with fluorescence confocal microscopy. The S. cerevisiae NB97 strain was a generous gift from Nathalie Bonnefoy. This work was supported by the California Institute for Biomedical Research, DOE/DE-SC0011787, NIH (P41GM103445), DOE (DE-AC02-5CH11231), and the Chan Zuckerberg Initiative, the Human Cell Atlas.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813143115/-/DCSupplemental.
References
- 1.Margulis L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant and Animal Cells on the Precambrian Earth. Yale Univ Press; New Haven, CT: 1970. [Google Scholar]
- 2.Kutschera U, Niklas KJ. The modern theory of biological evolution: An expanded synthesis. Naturwissenschaften. 2004;91:255–276. doi: 10.1007/s00114-004-0515-y. [DOI] [PubMed] [Google Scholar]
- 3.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archibald JM. Endosymbiosis and eukaryotic cell evolution. Curr Biol. 2015;25:R911–R921. doi: 10.1016/j.cub.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 5.Javaux EJ, Knoll AH, Walter MR. Morphological and ecological complexity in early eukaryotic ecosystems. Nature. 2001;412:66–69. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- 6.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. J Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature. 2018;557:101–105. doi: 10.1038/s41586-018-0059-5. [DOI] [PubMed] [Google Scholar]
- 8.Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Wu M. Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS One. 2014;9:e110685. doi: 10.1371/journal.pone.0110685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson SG, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 11.Ball SG, Bhattacharya D, Weber AP. EVOLUTION. Pathogen to powerhouse. Science. 2016;351:659–660. doi: 10.1126/science.aad8864. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki T, Nakata T, Nose Y. Genetic mapping with a thiamine-requiring auxotroph of Escherichia coli K-12 defective in thiamine phosphate pyrophosphorylase. J Bacteriol. 1968;95:1483–1485. doi: 10.1128/jb.95.4.1483-1485.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Begley TP, et al. Thiamin biosynthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz-Esser S, et al. ATP/ADP translocases: A common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 2004;186:683–691. doi: 10.1128/JB.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karas BJ, et al. Transferring whole genomes from bacteria to yeast spheroplasts using entire bacterial cells to reduce DNA shearing. Nat Protoc. 2014;9:743–750. doi: 10.1038/nprot.2014.045. [DOI] [PubMed] [Google Scholar]
- 16.Lartigue C, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 17.Sulo P, Griač P, Klobucníková V, Kovác L. A method for the efficient transfer of isolated mitochondria into yeast protoplasts. Curr Genet. 1989;15:1–6. doi: 10.1007/BF00445745. [DOI] [PubMed] [Google Scholar]
- 18.Traba J, Satrústegui J, del Arco A. Transport of adenine nucleotides in the mitochondria of Saccharomyces cerevisiae: Interactions between the ADP/ATP carriers and the ATP-Mg/Pi carrier. Mitochondrion. 2009;9:79–85. doi: 10.1016/j.mito.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Supekova L, Supek F, Greer JE, Schultz PG. A single mutation in the first transmembrane domain of yeast COX2 enables its allotopic expression. Proc Natl Acad Sci USA. 2010;107:5047–5052. doi: 10.1073/pnas.1000735107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paumet F, et al. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One. 2009;4:e7375. doi: 10.1371/journal.pone.0007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delevoye C, et al. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 2008;4:e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wesolowski J, Paumet F. SNARE motif: A common motif used by pathogens to manipulate membrane fusion. Virulence. 2010;1:319–324. doi: 10.4161/viru.1.4.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin WF, Tielens AGM, Mentel M, Garg SG, Gould SB. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol Mol Biol Rev. 2017;81:e00008–e00017. doi: 10.1128/MMBR.00008-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delevoye C, Nilges M, Dautry-Varsat A, Subtil A. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: Oligomerization of IncA mediates interaction between facing membranes. J Biol Chem. 2004;279:46896–46906. doi: 10.1074/jbc.M407227200. [DOI] [PubMed] [Google Scholar]
- 25.Prasad R, Goffeau A. Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu Rev Microbiol. 2012;66:39–63. doi: 10.1146/annurev-micro-092611-150111. [DOI] [PubMed] [Google Scholar]
- 26.Wesolowski J, et al. Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and Golgi complex positioning. MBio. 2017;8:e02280-16. doi: 10.1128/mBio.02280-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tkacz JS, Cybulska EB, Lampen JO. Specific staining of wall mannan in yeast cells with fluorescein-conjugated concanavalin A. J Bacteriol. 1971;105:1–5. doi: 10.1128/jb.105.1.1-5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaudinn C, et al. Imaging of endodontic biofilms by combined microscopy (FISH/cLSM–SEM) J Microsc. 2009;235:124–127. doi: 10.1111/j.1365-2818.2009.03201.x. [DOI] [PubMed] [Google Scholar]
- 29.Ollagnier-de Choudens S, Loiseau L, Sanakis Y, Barras F, Fontecave M. Quinolinate synthetase, an iron-sulfur enzyme in NAD biosynthesis. FEBS Lett. 2005;579:3737–3743. doi: 10.1016/j.febslet.2005.05.065. [DOI] [PubMed] [Google Scholar]
- 30.Parkinson DY, et al. Nanoimaging. Springer; New York: 2013. Nanoimaging cells using soft X-ray tomography; pp. 457–481. [DOI] [PubMed] [Google Scholar]
- 31.McDermott G, et al. Visualizing and quantifying cell phenotype using soft X-ray tomography. BioEssays. 2012;34:320–327. doi: 10.1002/bies.201100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryor PR, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5:590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 34.Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. Lokiarchaeon is hydrogen dependent. Nat Microbiol. 2016;1:16034. doi: 10.1038/nmicrobiol.2016.34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.