Abstract
Objective
To evaluate cerebral degenerative changes in ALS and their correlates with survival using 3D texture analysis.
Methods
A total of 157 participants were included in this analysis from four neuroimaging studies. Voxel‐wise texture analysis on T1‐weighted brain magnetic resonance images (MRIs) was conducted between patients and controls. Patients were divided into long‐ and short‐survivors using the median survival of the cohort. Neuroanatomical differences between the two survival groups were also investigated.
Results
Whole‐brain analysis revealed significant changes in image texture (FDR P < 0.05) bilaterally in the motor cortex, corticospinal tract (CST), insula, basal ganglia, hippocampus, and frontal regions including subcortical white matter. The texture of the CST correlated (P < 0.05) with finger‐ and foot‐tapping rate, measures of upper motor neuron function. Patients with a survival below the media of 19.5 months demonstrated texture change (FDR P < 0.05) in the motor cortex, CST, basal ganglia, and the hippocampus, a distribution which corresponds to stage 4 of the distribution TDP‐43 pathology in ALS. Patients with longer survival exhibited texture changes restricted to motor regions, including the motor cortex and the CST.
Interpretation
Widespread gray and white matter pathology is evident in ALS, as revealed by texture analysis of conventional T1‐weighted MRI. Length of survival in patients with ALS is associated with the spatial extent of cerebral degeneration.
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that is characterized by progressive motor decline. It is accompanied by cognitive impairment in up to 50% of patients.1 Clinical diagnosis requires concurrent evidence of upper and lower motor neuron (UMN and LMN) signs on physical examination. Patients have a median survival of 26 months from diagnosis, with considerable variability in longevity amongst individuals.2 Marked clinical heterogeneity poses significant challenges to development of therapies in this disease with no curative treatment.
The identification of prognostic factors is vital for the evaluation of novel therapeutics, as they could assist in the selection of more homogeneous patient subtypes for clinical trials.3 Older age, shorter time to diagnosis, bulbar onset, as well as the presence of comorbid cognitive deficits, particularly executive dysfunction, are regarded as negative prognostic factors in ALS.3, 4, 5
Magnetic resonance imaging (MRI) is considered the leading tool for biomarker discovery in ALS.6 Imaging studies have corroborated the pathological elements of neurodegeneration in ALS in vivo. Namely, degeneration of the motor (motor cortex and corticospinal tract (CST))7, 8, 9 and extra‐motor (frontotemporal)7, 10 regions. Few studies have emerged that examine the role of neuroimaging in the prognostication of ALS.11, 12, 13 These studies have associated degeneration of motor regions as predictive of survival; however, the implications of extra‐motor degeneration have not yet been explored.
Despite the contributions of neuroimaging studies toward the understanding of ALS, the clinical purpose of MRI is limited to ruling out ALS‐mimics. The pathological processes in the gray and white matter – neuronal loss,14, 15 protein inclusions,16 gliosis,14, 17 and demyelination17, 18 – cause unique alterations in the MRI signal intensity; however, these changes are inaccurately measured by current analytical methods.19, 20
Texture analysis of MR images has been utilized to detect and characterize cerebral changes on routine MRI scans.21 It is a quantitative approach that can detect subtle local signal intensity and pattern variations in an image. In focal cortical dysplasia, texture analysis techniques have demonstrated superior classification accuracy when compared to visual inspection.22 Studies in multiple sclerosis have identified the pathological correlates of altered texture on T2‐weighted (T2W) MRI.23 In an initial study of 19 patients, a 3‐dimensional (3D) texture analysis approach localized changes in the CST and the motor cortex from T1W images in ALS.24
The objectives of this work are twofold: (1) to evaluate the spatial distribution of cerebral degeneration in a large cohort of patients with ALS as reflected by changes in texture of T1W images, and (2) to investigate the neuroanatomical correlates of survival with texture. Based on the previous literature, it was hypothesized that (1) texture analysis will detect motor and extra‐motor pathology, and (2) it will demonstrate differences in the spatial distribution of cerebral degeneration between long‐ and short‐surviving patients. The successful application of texture analysis in ALS could aid in detecting pathological changes on conventional MRI and provide prognostic indicators with clinical relevance.
Methods
Participants
Participants were recruited from the University of Alberta between 2003 and 2018 as part of four different prospective MRI studies (herein referred to as Study 1, 2, 3, and 4). Patients with ALS were recruited for these studies from a multidisciplinary ALS clinic if they had no history of other neurological or psychiatric disorders. Healthy controls with no neurological or psychiatric disorders were also recruited in each of the four studies. All participants provided informed written consent prior to their involvement in their respective studies, which were approved by the local research ethics review board.
Patients had UMN and LMN signs on clinical examination and met criteria for possible, probable lab‐supported, probable, or definite ALS according to the El Escorial criteria.25 Patients with frontotemporal dementia (FTD), or those with a family history of ALS were also included. All patients underwent neurological assessments according to their specific study protocol. For the present work, UMN examination data were included for analysis. Patients from Study 2, 3, and 4 had performed finger‐ and foot‐tapping tests as a measure of their UMN function. Finger‐ and foot‐tapping rates were calculated by averaging the total number of taps in 10 seconds over two trials. Scores from the left and right side were averaged to give a single score for each patient's finger‐ and foot‐tapping rate. Patient's functional disability was measured using the ALS functional rating scale‐revised (ALSFRS‐R). The scale ranges from 0 to 48, with lower scores reflecting greater disability. Two patients were assessed using the ALSFRS (range 0–40), an older version that does not assess respiratory symptoms. Symptom duration was recorded as the date from their reported onset of symptoms to the date of their MRI scan. Disease progression rate was calculated by (48 – patient ALSFRS‐R score)/symptom duration for every patient. For the two patients with ALSFRS scores, disease progression rate was calculated as (40 – patient ALSFRS score)/symptom duration.
MRI Protocols
Participants in Study 1 and 2 were scanned in a Siemens 1.5 T scanner, and participants in Study 3 and 4 were scanned on a Siemens 3 T and a Varian 4.7 T scanner, respectively. T1W images for all participants were acquired with a 3D magnetization‐prepared rapid gradient‐echo sequence. The images in Study 1, 3, and 4 were acquired axially and images for Study 2 were acquired coronally (Table 1).
Table 1.
Participants’ characteristics and T1W scan protocols
| Study | Patients (M/F) | Mean age (years) | Controls (M/F) | Mean age (years) | TR (ms) | TE (ms) | Voxel size (mm 3) | B0 (T) |
|---|---|---|---|---|---|---|---|---|
| 1 | 27 (19/8) | 61.2 | 13 (5/8) | 53.6 | 1600 | 3.8 | 1 × 1 × 1.5 | 1.5 |
| 2 | 19 (10/9) | 57.7 | 23 (11/12) | 55.6 | 1600 | 3.8 | 1 × 1.5 × 1 | 1.5 |
| 3 | 20 (11/9) | 57.6 | 16 (7/9) | 57.0 | 2300 | 3.4 | 1 × 1 × 1 | 3.0 |
| 4 | 17 (10/7) | 57.1 | 22 (8/14) | 56.6 | 508 | 4.5 | 1 × 1 × 1 | 4.7 |
MRI, magnetic resonance imaging; M, male; F, female; TR, repetition time; TE, echo time; ms, milliseconds; mm, millimeters; B0, magnetic field strength.
Image processing and texture analysis
Image processing and subsequent voxel‐wise analyses were conducted in Statistical Parametric Mapping 12 version 6685 (SPM12; http://www.fil.ion.ucl.ac.uk/spm/software/spm12/) and Computational Anatomy Toolbox 12.1 (CAT12; http://dbm.neuro.uni-jena.de/cat12/). T1W images were first assessed for quality assurance, which included a visual check for subject motion and scanner artifacts. They were then aligned along the anterior and posterior commissures to ensure anatomical alignment across participants for 3D texture analysis and image registration procedures. Images were corrected for non‐uniformity intensity bias and segmented into gray and white matter in their native space. The gray and white matter segments were combined to create a custom whole‐brain mask for each participant. T1W images were normalized using a high‐dimensional approach26 to the Montreal Neurological Institute (MNI) template provided by CAT12 – the forward deformation fields were saved for later transformations into the standard space. Normalized gray matter segments were smoothed with a 6‐mm full‐width at half maximum (FWHM) Gaussian kernel for statistical analyses.
Texture analysis was performed using the gray level co‐occurrence matrix (GLCM) method, a second‐order statistical approach for extracting texture features.21, 27 Methodological details regarding the 3D adaptation of GLCM to generate 3D texture maps from T1W images are provided elsewhere.28 Briefly, the GLCM was defined for each voxel and its adjacent voxels (referred to as the reference voxel and its neighborhood) in all three orthogonal planes for a T1W image. A GLCM is an N × N matrix, where N is the total number of gray levels in an image. To reduce computation time, T1W images were scaled down to eight gray levels. Each cell in the GLCM (i,j) specifies the number of times gray level i co‐occurs with gray level j over a distance d and in a particular direction θ within the neighborhood. In this study, a distance of one and four directions – 0°, 45°, 90°, and 135° – were considered for the construction of the GLCMs. The GLCMs for all four directions were summed and normalized to represent the probability of co‐occurrence between gray levels in the neighborhood. This was carried out in the axial, coronal, and sagittal planes to generate three GLCMs per voxel. A texture feature could then be calculated from the GLCMs and averaged over the three planes to compute a single 3D texture value for each voxel. Figure 1 illustrates a schematic representation of the creation of the GLCM.
Figure 1.
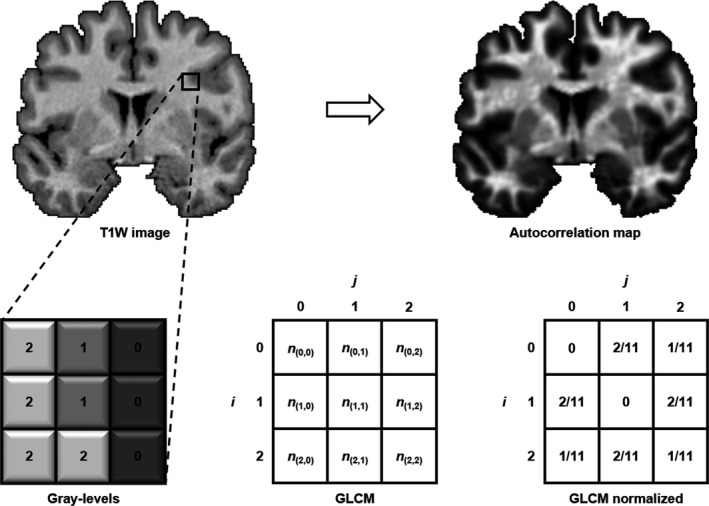
Schematic representation of the creation of a gray level co‐occurrence matrix (GLCM) in a neighborhood of pixels in an image. Each cell in the GLCM matrix represents the number of times a certain pixel combination (i,j) co‐occurs in a particular direction. In this example, the GLCM is evaluated for θ = 0° (left‐right direction). Values are then normalized in the GLCM by dividing them by the sum of all co‐occurrences in the GLCM. Texture autocorrelation is then calculated from the normalized GLCM in every direction in each orthogonal plane (i.e. in three dimensions) to produce an autocorrelation map for each T1W image. Areas of increased autocorrelation (hyperintensity on the maps) represent regions where there is an increased probability of co‐occurrence of gray levels.
In this work, whole‐brain autocorrelation maps were computed from the GLCMs of T1W images. The texture feature autocorrelation has been shown to be sensitive to cerebral degeneration in ALS and in Alzheimer's disease using 3D texture analysis.24, 28 It can be best understood as a measure of the linear dependency in the gray level, or as being sensitive to repetitive patterns in pairs of gray levels in a local neighborhood; the higher the autocorrelation value, the higher the likelihood that a pair of gray levels will co‐occur. Autocorrelation is calculated as follows:
where p(i,j) is the probability of co‐occurrence between gray levels i and j in a neighborhood. The texture maps were normalized to the MNI template by applying the forward deformation fields obtained earlier and smoothed with a 6‐mm FWHM Gaussian kernel for statistical analyses.
Statistical analysis
All statistical tests were conducted in MedCalc Statistical Software version 18.2.1 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org). One‐way analysis of variance (ANOVA) was used to ensure participant demographic characteristics did not differ between the MRI studies. Demographic differences between patients and controls were assessed with independent samples t‐tests and χ 2 tests. Statistical significance was defined at P < 0.05. Whole‐brain voxel‐wise analyses were performed in SPM12 for the normalized and smoothed texture maps. Statistical significance was defined as P < 0.05 after correcting for multiple comparisons with the false discovery rate (FDR) method with a cluster size of 10 voxels. A second‐level full factorial model was designed to examine whole‐brain voxel‐wise differences in autocorrelation between patients and controls while controlling for age, sex, and MRI protocol. Autocorrelation values from significant clusters in the motor regions were extracted and correlated with finger‐ and foot‐tapping scores. Partial correlations were used to control for MRI protocol and statistical significance was defined at P < 0.05. Voxel‐based morphometric (VBM) analysis of gray matter density was also conducted for comparison with texture analysis. A threshold mask of 0.2 was applied to the gray matter segments to exclude noise and other tissue classes from the analysis. The full factorial model additionally controlled for total intracranial volume to control for differences in head sizes.29
Patients were dichotomized at their median survival into long‐ and short‐surviving patients. The two groups’ texture maps were directly compared using a second‐level full factorial model while controlling for age and MRI protocol. Additional models compared autocorrelation in long‐surviving patients with controls and short‐surviving patients with controls. Patients’ five‐year survival analysis using Cox proportional‐hazards regression models was conducted in MedCalc. Univariate and stepwise multivariate models were carried out with age, sex, site of onset, ALSFRS‐R score, symptom duration, and disease progression rate as covariates. Survival was defined as the time from the T1W scan to death. Patients were censored if they were alive at the time of the analysis or lost to follow‐up.
Results
Demographics
A total of 157 participants (patients = 83, controls = 74) with T1W images were included in this study. Patients’ mean age, symptom duration, ALSFRS‐R score, and progression rate did not differ between the 4 MRI studies. Patients’ mean age (58.7 ± 10.8 years) did not differ significantly from controls’ mean age (55.8 ± 11.3 years). There were 50 male and 33 female patients, and 31 male and 43 controls (χ 2 P < 0.05). 76.8% of the patients had limb onset and 23.2% of the patients had bulbar onset. One patient did not have information regarding their site of onset. Three patients had concomitant FTD on clinical examination. The mean ALSFRS‐R score was 38.7 ± 5.6. Two patients completed the ALSFRS and had scores of 29 and 25. The mean and median symptom duration were 22.9 ± 15.4 months and 17.3 months (range 2–86 months), respectively. The mean and median disease progression rates were 0.67 ± 1.08 and 0.41 (range 0.07–9.09), respectively.
Texture differences between patients and controls
Significant regional differences in autocorrelation were observed in the whole‐brain voxel‐wise analysis (FDR P < 0.05). Autocorrelation was decreased in patients in bilateral motor cortex, insula, prefrontal cortex, bilateral hippocampus, thalamus, caudate, cingulate gyrus, and subcortical frontal regions (Fig. 2A, Figure S1). Autocorrelation was increased in the CST in patients bilaterally (Fig. 2B, Figure S1). No other regions of increased autocorrelation were observed. An F‐test was conducted to ensure that these voxel‐wise group differences were not biased by an interaction effect caused by the different MRI studies. No interaction effect was found between the main effect of group and the main effect of studies on autocorrelation (FDR P < 0.05). Additionally, supplementary whole‐brain voxel‐wise analyses were repeated separately for studies that acquired images at different resolutions to ensure the heterogeneity in voxel sizes did not impact the results. Study 1 and 2 were analyzed separately and Study 3 and 4 were pooled together because their images were acquired at 1 × 1 × 1 mm3 (Table 1). The results are reported in Figure S2.
Figure 2.
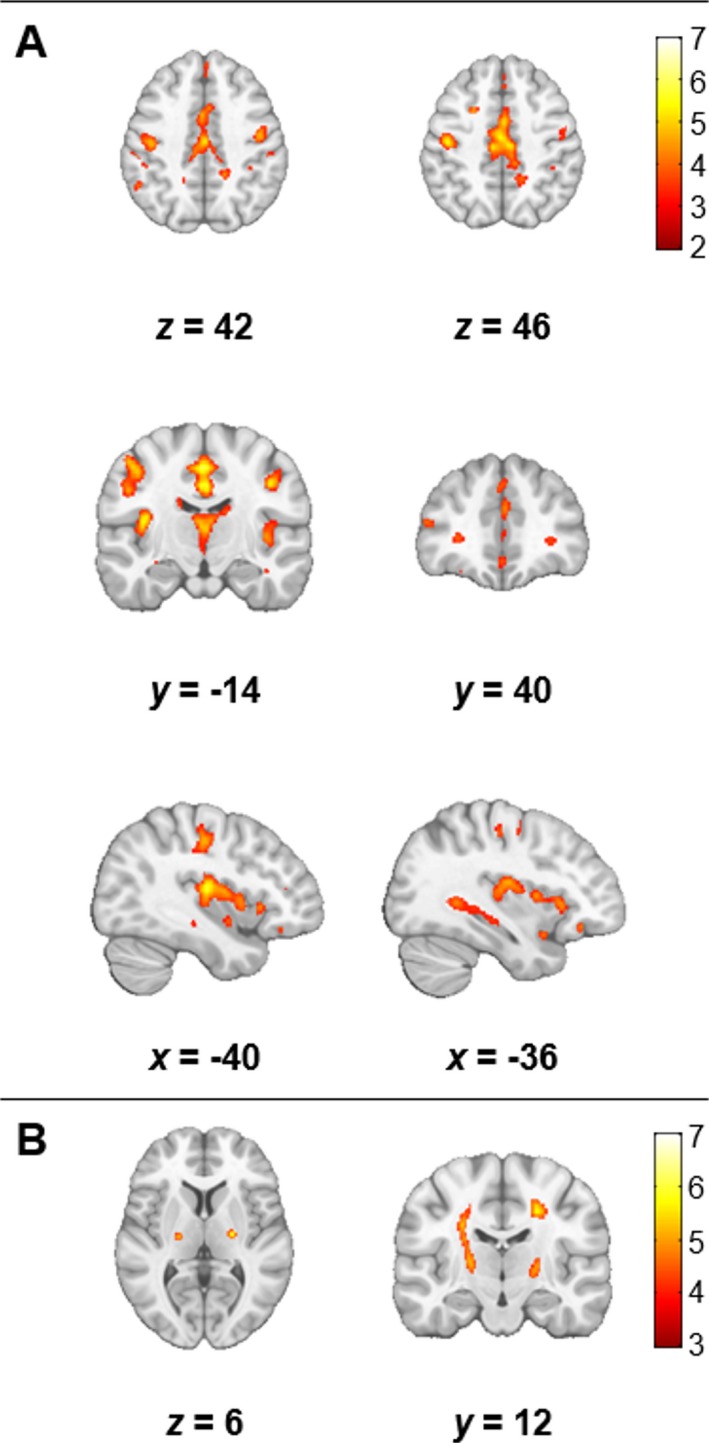
Significant differences in autocorrelation between patients and controls (FDR P < 0.05, cluster size > 10). Results are overlaid on the MNI template in neurological convention. Areas of the motor cortex, insula, thalamus, caudate, subcortical white matter, and hippocampus bilaterally had decreased autocorrelation in patients (A). However, autocorrelation was increased along the CST in patients (B). The color bars show the range of T values.
Correlation between UMN function and texture
Autocorrelation values were extracted from patients in Study 2, 3, and 4 from the significant clusters of between‐group differences in the motor cortex and the CST region. Finger‐tapping scores were available for 55 patients and foot‐tapping scores were available for 53 patients. Significant correlations (P < 0.05) were found between autocorrelation values from the CST and finger‐ (r = −0.28) and foot‐tapping scores (r = −0.31) (Fig. 3). No significant correlations were found between autocorrelation values from the motor cortex and either tapping scores.
Figure 3.
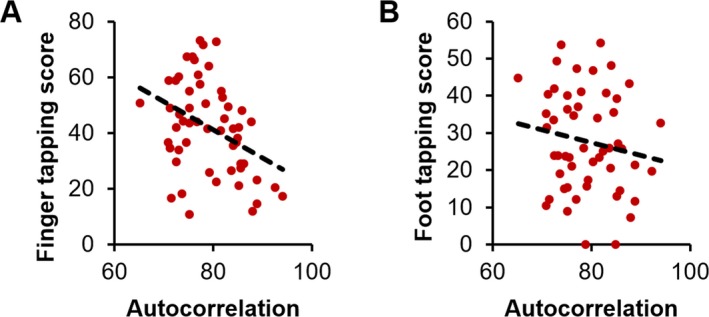
Clinical correlations in patients: autocorrelation in the CST varied with and finger‐ (A) and foot‐tapping rates (B).
Gray matter density differences between patients and controls
Significant reductions in gray matter density were detected in patients in the whole‐brain VBM analysis. Gray matter density was decreased bilaterally in the motor cortex, gyrus rectus, and frontal gyri (Fig. 4). The thalamus, cingulate gyrus, and the insula also demonstrated reductions in gray matter density (Fig. 4).
Figure 4.
Voxel based morphometry revealed bilaterally decreased gray matter density in the motor cortex, insula, and thalamus in patients (FDR P < 0.05, cluster size > 10). Results are overlaid on the MNI template in neurological convention. The color bars show the range of T values.
Regional gray matter density reductions did not completely overlap changes in autocorrelation. Notably, gray matter density was not reduced in the hippocampi of patients, even at lower statistical thresholds (uncorrected P < 0.001). Changes in the lateral motor cortex were evident in gray matter density and autocorrelation values; however, only autocorrelation demonstrated reductions in the medial motor cortex region. Furthermore, gray matter density was reduced largely in the anterior subsection of the insula, whereas autocorrelation was reduced more extensively in its anterior and posterior subsections.
Survival analysis
One patient had missing survival information and was excluded from this analysis (n = 82). The mean time from MRI scan to outcome did not differ significantly between MRI studies. At the time this analysis was performed, 64 patients had died, and 18 patients were censored (17 alive and one lost to follow‐up). The median survival was 19.5 months, which was used to dichotomize patients into long‐ and short‐survivors (Table 2). Long‐survivors had a mean survival of 40.3 ± 24.6 months, whereas short‐survivors had a mean survival of 11.9 ± 5.1 months. Short‐survivors were significantly older (62.0 ± 10.3 years) than long‐survivors (55.2 ± 10.3 years) at the time of the MRI scan. The two groups did not differ on any other demographic detail.
Table 2.
Patients’ survival and clinical characteristics
| All patients | Long‐survivors | Short‐survivors | P value | |
|---|---|---|---|---|
| Outcome | ||||
| Deceased | 78.0% (64) | 70.7% (29) | 85.4% (35) | |
| Censored | 22.0% (18) | 29.3% (12) | 14.6% (6) | |
| Survival (months) | 26.1 ± 22.5 | 40.3 ± 24.6 | 11.9 ± 5.1 | <0.01 a |
| Age (years) | 58.6 ± 10.8 | 55.2 ± 10.3 | 62.0 ± 10.3 | <0.01 a |
| Sex (M/F) | 50/32 | 27/14 | 23/18 | 0.37b |
| Site of onset (limb/bulbar) | 62/19 | 32/9 | 30/10 | 0.75b |
| ALSFRS‐R | 38.7 ± 5.6 | 39.6 ± 6.2 | 37.8 ± 4.9 | 0.17a |
| Symptom duration (months) | 23.1 ± 15.4 | 25.1 ± 18.8 | 21.0 ± 10.9 | 0.24a |
| Disease progression rate | 0.67 ± 1.1 | 0.55 ± 0.68 | 0.78 ± 1.39 | 0.34a |
Patients were dichotomized at the median survival of 19.5 months. Survival data were not available for one patient and was therefore excluded from the analysis. Where applicable, data are presented as mean ± standard deviation. The P value is presented for group tests between long‐ and short‐survivors. The values in bold represent significant differences at P < 0.05.
M, male; F, female; ALSFRS‐R, ALS functional rate scale‐revised.
Independent samples t‐test.
Chi‐squared test.
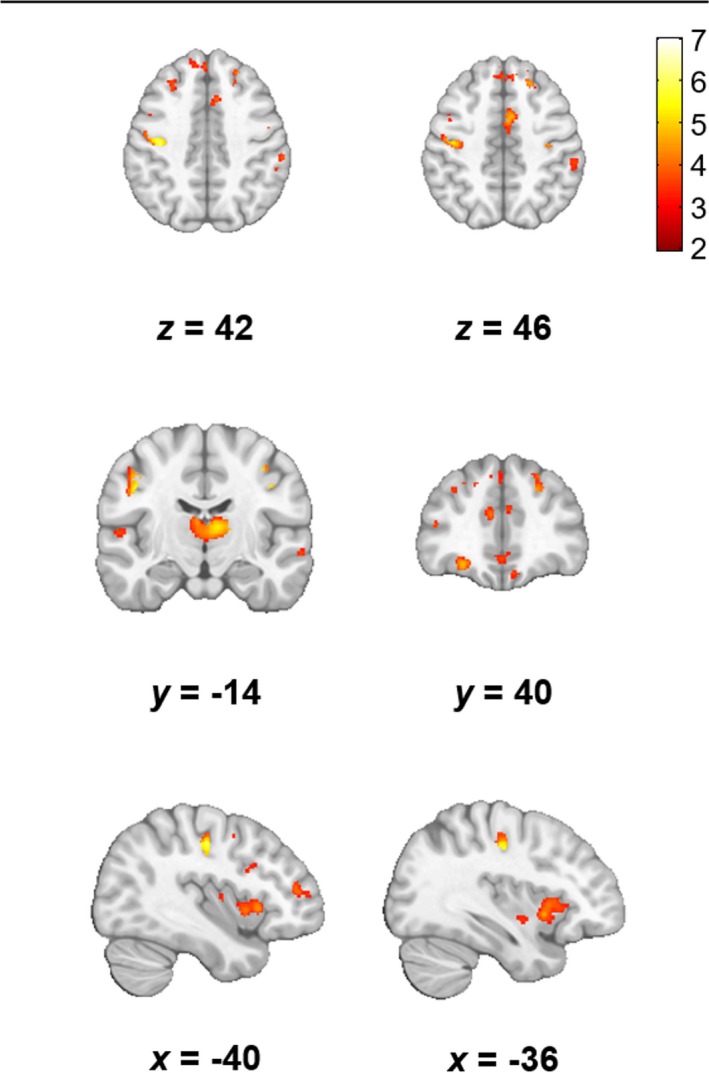
Relative to controls, the two groups of patients demonstrated significant differences in autocorrelation (FDR P < 0.05). Long‐survivors displayed bilateral decreases in the motor cortex and increases in the region of the CST in comparison with controls (Fig. 5A). Small significant clusters of decreased autocorrelation values were observed in the frontal regions. No differences were noted in the hippocampi, insula, or basal ganglia.
Figure 5.
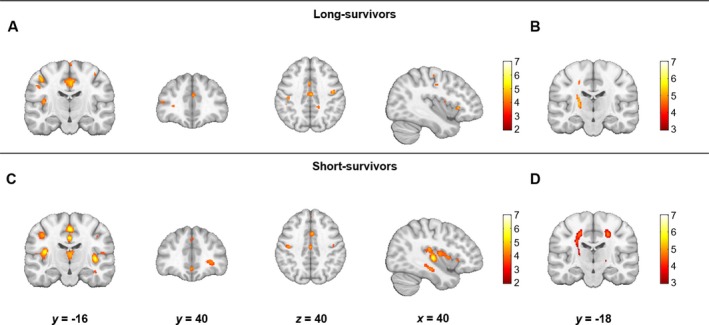
Differences in the texture feature autocorrelation between long‐survivors and controls (A, B) and short‐survivors and controls (C, D). Panels A and C show the areas of decreased autocorrelation and panels B and D show areas of increased autocorrelation (FDR P < 0.05, cluster size > 10). Results are overlaid on the MNI template in neurological convention. The color bars show the range of T values. The comparison between short‐survivors and controls for significant increases in autocorrelation (D) did not survive FDR correction and is reported at an uncorrected P < 0.001 with a cluster threshold of 10 voxels.
Short‐survivors had significantly decreased autocorrelation values in bilateral motor cortex and frontal regions. In contrast to the long‐survivor group, reductions were also found in the insula right hippocampus, and thalamus (Fig. 5B). Comparable to the long‐survivor group, autocorrelation was significantly increased in the CST in short‐survivors, although this was reported at a lower statistical threshold (uncorrected P < 0.001). A direct comparison between long‐ and short‐survivors did not reveal significant regional differences in autocorrelation.
Patients’ age, sex, site of onset, ALSFRS‐R score, symptom duration, and progression rate were evaluated as predictors of five‐year survival in univariate and multivariate models. Univariate analysis revealed age (HR = 1.04, 95% confidence interval (CI) = 1.01–1.06, P = 0.01) and symptom duration (HR = 0.98, 95% CI = 0.97–1.00, P = 0.03) as significant predictors of survival. The multivariate model also selected older age (HR = 1.05, 95% CI = 1.02–1.08, P < 0.01) and shorter symptom duration (HR = 0.98, 95% CI = 0.96–0.99, P < 0.01) as significant predictors when all the variables were entered in the model and selected with the stepwise method.
Discussion
In this work, an unbiased whole‐brain texture analysis approach was used to study cerebral degeneration in ALS and its impact on survival. Confirming the first hypothesis, both gray and white matter structures of motor and extra‐motor regions demonstrated texture differences between patients and controls. Affected gray matter regions closely mirrored the neuroanatomical pattern of pathological TDP‐43 inclusions in ALS16 and the involvement of the CST resembled the hallmark pathology of the disease.17 Secondly, shorter survival was associated with changes in the extra‐motor regions, particularly the basal ganglia and hippocampus. This suggests that survival is potentially mediated by the spatial burden of pathology in ALS. These findings are further discussed below.
Texture analysis and pathology of ALS
Autocorrelation was found to be altered in patients in the motor (motor cortex and CST) and extra‐motor regions (frontal, temporal, and subcortical structures). This is in accordance with the view that ALS is a multisystem neurodegenerative disease. Loss of pyramidal neurons in layer V of the motor cortex14, 15 and ubiquitin‐reactive cytoplasmic inclusions in the frontal and temporal regions are consistent pathological features of the disease.16, 18 In addition, ALS is regarded as a disorder on the larger ALS‐FTD spectrum with the identification of the pathological TDP‐43 inclusions as the unifying disease protein.30 Subsequent in vivo MRI studies have found similar pathological involvement of gray and white matter structures in the frontal and temporal regions.7, 9
Autocorrelation was decreased in the motor cortex, frontal regions, basal ganglia structures, and the hippocampus. This spatial pattern is in alignment with the progressive TDP‐43 pathology in ALS.16 Interestingly, VBM analysis did not detect change in the hippocampus at an identical statistical threshold, consistent with previous VBM studies.7, 10 Ubiquitin‐positive inclusions have been documented on histology in the granular cell layer of the hippocampus in ALS.31 This feature is not only seen in cases of ALS with clinical dementia, but also in patients without clinical evidence of dementia.32, 33 Furthermore, TDP‐43 inclusions in the hippocampus are characterized as the final pathological stage of the disease.16 It could be hypothesized that texture analysis is detecting imaging correlates of TDP‐43 inclusions in the gray matter that precede atrophic changes that are eventually detectable by VBM analysis. Indeed, in a recent longitudinal study, patients had demonstrable hippocampal atrophy at two‐year follow‐up.34 This view is further supported from studies in Alzheimer's disease that suggest that texture analysis is a marker for pre‐atrophy amyloid beta plaque accumulation in the hippocampus because of its sensitivity to the subtle pathological variation in the MRI signal caused by the inclusions.35
The CST demonstrated pronounced increases in autocorrelation in patients along with correlations with UMN function. Pathological demyelination and gliosis of the CST in ALS have been well documented in autopsy14, 17, 18 and in in vivo MRI studies.8 It is postulated that T1W hyperintensity, otherwise known as T1‐shortening, along the CST may be due to the pathological changes including the presence of lipid‐laden macrophages and accumulation of intra‐axonal neurofilaments.36 The involvement of the corpus callosum is also a frequent finding in diffusion tensor imaging (DTI) studies and is proposed to be characteristic of Wallerian‐type degeneration37; however, systematic neuropathological evaluations of the corpus callosum are lacking to support that view. Few autopsy reports have provided initial support for gliosis, but not a demyelinating pathology in the corpus callosum.38, 39 Phosphorylated TDP‐43 inclusions, which are considered a hallmark pathological feature of ALS,16 are also reportedly absent in the corpus callosum.40 The corpus callosum did not exhibit any degenerative changes with texture analysis in this study. The contributions of the pathological mechanisms toward the increase in autocorrelation in the CST, but not in the corpus callosum cannot be concluded from this work. It can be speculated that autocorrelation is detecting a specific pathology of the CST, which is not present extensively in the corpus callosum; however, MRI‐histological studies are needed to investigate the relationship between textures and the underlying pathological processes. One such study in multiple sclerosis demonstrated that heterogeneity in MRI texture was predictive predominantly of demyelinating pathology, followed by axonal injury, and inflammation.41
Neuroanatomical correlates of survival in ALS
Survival analysis showed that shorter survival is associated with greater gray matter degeneration. Compared to patients with longer survival, those with shorter survival additionally had degenerative changes in the insula, subcortical frontal white matter, thalamus, and hippocampus. Dysfunction of the insula has been implicated as a contributing factor to verbal‐fluency deficits,42 which typify the overall executive dysfunction in ALS.43 Systematic investigations of the sub‐cortical structures have also revealed the involvement of the thalamus and the hippocampus, including its relation to memory‐deficits, in ALS.44, 45, 46 Thus, the extra‐motor changes in short‐survivors potentially reflect the extra‐motor phenotype of the disease. Indeed, there is a growing body of evidence that suggests that the presence of comorbid FTD, or executive dysfunction in the absence of other cognitive deficits, are predictors of poor survival in ALS.4, 5
Recently, Brettschneider et al. proposed a neuropathological staging system in ALS based on the sequential spatial burden of intraneuronal phosphorylated TDP‐43 inclusions.16 Lowest burden of TDP‐43 pathology was characterized by inclusions in the motor cortex (stage 1), followed by inclusions in the prefrontal neocortex, precerebellar nuclei, and red nuclei (stage 2).16 Later stages included TDP‐43 inclusions in the striatum (stage 3) and the hippocampi (stage 4).16 It is noteworthy that based on this staging scheme, short‐survivors demonstrated regions from all four stages of the disease, including the hippocampus, whereas long‐survivors did not demonstrate late‐stage structures. Patients in stage 4 also have been shown to have memory deficits associated with hippocampus and perforant pathway degeneration.47, 48 These results strongly suggest that survival is associated with the spatial burden of the cerebral degeneration in ALS. Initial support for this comes from a large autopsy study that demonstrated that ALS cases with temporal lobe involvement, specifically in the hippocampus, had significantly reduced survival and invariant UMN involvement compared to cases with only UMN involvement.18 Pending replication and further validation of these findings, future clinical trials evaluating novel therapeutics in ALS should consider prospectively subgrouping patients into short and long disease course patients based on the involvement of extra‐motor structures using texture analysis.
Age and symptom duration were associated with shorter survival in this study, which is in accordance with the current literature.3 There are only three studies in the literature that have investigated in vivo cerebral degeneration as a predictor of survival in ALS.11, 12, 13 The first study testing this utilized magnetic resonance spectroscopy of the motor cortex and found N‐acetylaspartate/choline ratio, a marker for neuronal loss, to be a significant predictor of survival (HR = 0.24, 95% CI = 0.08–0.72, P = 0.01).11 The second investigated white matter involvement with DTI and identified fractional anisotropy of the CST as a significant predictor of survival (HR = 0.94, 95% CI = 0.89–1.00, P = 0.06)12; however, this study had a small number of patients (n = 24) and more than 50% of them were censored at the time of analysis.12 Finally, the third study evaluated MRI measures of cortical and white matter motor structures along with clinical indices including age and ALSFRS‐R to predict 18‐month survival.13 The study demonstrated that the combination of MRI measures and clinical indices had better predictive accuracy (79.15%) than using clinical indices alone (66.67%); however, the results were not replicated in an independent, albeit small, validation sample.13 In this work, no overt differences in the degree of motor region involvement between the two survival groups were found and short‐survivors displayed more widespread degeneration when compared to controls. Previous studies have not evaluated the influence of extra‐motor degeneration on survival in ALS. Incorporating extra‐motor regions and multimodal data likely would improve the accuracy of survival models.
Summary and study limitations
Results here suggest that the spatial extent of cerebral involvement is an important prognostic factor, with involvement of extra‐motor regions leading to a worse outcome. This conclusion was achieved with the largest study to date in ALS that investigated in vivo neuroanatomical correlates of survival. The findings reported in this work indicate that 3D texture analysis of T1W images is efficacious in detecting pathological cerebral changes in ALS. This is an important step toward the development of cerebral markers of the disease. Texture features could play a role in detecting varying neurobiological processes that can lead to the identification of patient subtypes in vivo; a biomarker with this property could play a critical role in patient selection in clinical trials.
There are limitations to this study. First, this was an analysis utilizing a large heterogenous dataset. Although the potential effect of MRI protocols on autocorrelation was statistically controlled for and no demographic differences in the patient and control populations from the different studies were found, future studies should be performed on homogenously acquired data. Conversely, however, the heterogeneity of the data in light of the results also supports the clinical relevance and the robustness of texture analysis. Second, as the clinical datasets were not uniform between studies, correlations with clinical measures including cognitive assessments could not be performed. Lastly, though the regional group differences in autocorrelation between patients and controls and patients of long and short survivorship are in accordance with the current understanding of the pathology in ALS, little is understood about the histopathological basis of these findings. Future studies should further validate texture analysis in ALS with clinical and cognitive assessments and histological studies.
Author Contributions
A. I. contributed to the conception and design of the study, analysis of data, and drafting of the manuscript and figures; S.K., D.M., P.S., C.L., and W.J. contributed to the acquisition of data; D.E., contributed to analysis of data; and Y.Y. and S.K. contributed to the conception and design of the study.
Conflicts of Interest
Nothing to report.
Supporting information
Figure S1. Autocorrelation values extracted from the regions of significant differences in the pooled analysis from patients and controls in studies at different resolutions.Figure S2: Significant differences in autocorrelation between patients and controls (P < 0.001, cluster size > 10) in separate studies at different resolutions.
Acknowledgments
The authors would like to thank the participants and their families. This study was supported by funding from ALS Society of Canada, the ALS Association of America, the Canadian Institutes of Health Research, Brain Canada, the MSI Foundation, and the Shelly Mrkonjic ALS Research Fund. Data were made available in part from the Canadian ALS Neuroimaging Consortium (CALSNIC), for which data management and quality control were facilitated by the Canadian Neuromuscular Disease Registry (CNDR).
Funding Information
This study was supported by funding from ALS Society of Canada, the ALS Association of America, the Canadian Institutes of Health Research, Brain Canada, the MSI Foundation, and the Shelly Mrkonjic ALS Research Fund.
Funding Statement
This work was funded by ALS Society of Canada grant ; ALS Association of America grant ; Canadian Institutes of Health Research grant ; Brain Canada grant ; MSI Foundation grant ; Shelly Mrkonjic ALS Research Fund grant .
References
- 1. Ringholz G, Appel S, Bradshaw M, et al. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology 2005;65:586–590. [DOI] [PubMed] [Google Scholar]
- 2. Pupillo E, Messina P, Logroscino G, Beghi E. Long‐term survival in amyotrophic lateral sclerosis: a population‐based study. Ann Neurol 2014;75:287–297. [DOI] [PubMed] [Google Scholar]
- 3. Chiò A, Logroscino G, Hardiman O, et al. Prognostic factors in ALS: a critical review. Amyotrophic Lat Scler 2009;10:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olney R, Murphy J, Forshew D, et al. The effects of executive and behavioral dysfunction on the course of ALS. Neurology 2005;65:1774–1777. [DOI] [PubMed] [Google Scholar]
- 5. Elamin M, Phukan J, Bede P, et al. Executive dysfunction is a negative prognostic indicator in patients with ALS without dementia. Neurology 2011;76:1263–1269. [DOI] [PubMed] [Google Scholar]
- 6. Turner MR, Grosskreutz J, Kassubek J, et al. Towards a neuroimaging biomarker for amyotrophic lateral sclerosis. Lancet Neurol 2011;10:400–403. [DOI] [PubMed] [Google Scholar]
- 7. Chang JL, Lomen‐Hoerth C, Murphy J, et al. A voxel‐based morphometry study of patterns of brain atrophy in ALS and ALS/FTLD. Neurology 2005;65:75–80. [DOI] [PubMed] [Google Scholar]
- 8. Pyra T, Hui B, Hanstock C, et al. Combined structural and neurochemical evaluation of the corticospinal tract in amyotrophic lateral sclerosis. Amyotrophic Lat Scler 2010;11:157–165. [DOI] [PubMed] [Google Scholar]
- 9. van der Graaff Maaike M, Sage CA, Caan MW, et al. Upper and extra‐motoneuron involvement in early motoneuron disease: a diffusion tensor imaging study. Brain 2011;134:1211–1228. [DOI] [PubMed] [Google Scholar]
- 10. Agosta F, Pagani E, Rocca M, et al. Voxel‐based morphometry study of brain volumetry and diffusivity in amyotrophic lateral sclerosis patients with mild disability. Hum Brain Mapp 2007;28:1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kalra S, Vitale A, Cashman NR, et al. Cerebral degeneration predicts survival in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2006;77:1253–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agosta F, Pagani E, Petrolini M, et al. MRI predictors of long‐term evolution in amyotrophic lateral sclerosis. Eur J Neurosci 2010;32:1490–1496. [DOI] [PubMed] [Google Scholar]
- 13. Schuster C, Hardiman O, Bede P. Survival prediction in amyotrophic lateral sclerosis based on MRI measures and clinical characteristics. BMC Neurol 2017;17:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawamata T, Akiyama H, Yamada T, McGeer PL. Immunologic reactions in amyotrophic lateral sclerosis brain and spinal cord tissue. Am J Pathol 1992;140:691–707. [PMC free article] [PubMed] [Google Scholar]
- 15. Nihei K, McKee AC, Kowall NW. Patterns of neuronal degeneration in the motor cortex of amyotrophic lateral sclerosis patients. Acta Neuropathol 1993;86:55–64. [DOI] [PubMed] [Google Scholar]
- 16. Brettschneider J, Del Tredici K, Toledo JB, et al. Stages of pTDP‐43 pathology in amyotrophic lateral sclerosis. Ann Neurol 2013;74:20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawyer T, Netsky MG. Amyotrophic lateral sclerosis: a clinicoanatomic study of fifty‐three cases. AMA Arch Neurol Psychiatry 1953;69:171–192. [DOI] [PubMed] [Google Scholar]
- 18. Piao Y, Wakabayashi K, Kakita A, et al. Neuropathology with clinical correlations of sporadic amyotrophic lateral sclerosis: 102 autopsy cases examined between 1962 and 2000. Brain Pathol 2003;13:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hecht M, Fellner F, Fellner C, et al. MRI‐FLAIR images of the head show corticospinal tract alterations in ALS patients more frequently than T2‐, T1‐and proton‐density‐weighted images. J Neurol Sci 2001;186:37–44. [DOI] [PubMed] [Google Scholar]
- 20. Hecht M, Fellner F, Fellner C, et al. Hyperintense and hypointense MRI signals of the precentral gyrus and corticospinal tract in ALS: a follow‐up examination including FLAIR images. J Neurol Sci 2002;199:59–65. [DOI] [PubMed] [Google Scholar]
- 21. Kassner A, Thornhill RE. Texture analysis: a review of neurologic MR imaging applications. AJNR Am J Neuroradiol 2010;31:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bernasconi A, Antel SB, Collins DL, et al. Texture analysis and morphological processing of magnetic resonance imaging assist detection of focal cortical dysplasia in extra‐temporal partial epilepsy. Ann Neurol 2001;49:770–775. [PubMed] [Google Scholar]
- 23. Zhang Y, Moore G, Laule C, et al. Pathological correlates of magnetic resonance imaging texture heterogeneity in multiple sclerosis. Ann Neurol 2013;74:91–99. [DOI] [PubMed] [Google Scholar]
- 24. Maani R, Yang Y, Emery D, Kalra S. Cerebral degeneration in amyotrophic lateral sclerosis revealed by 3‐dimensional texture analysis. Front Neurosci 2016;10:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brooks BR, Miller RG, Swash M, Munsat TL. El escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotrophic Lat Scler 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 26. Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage 2007;38:95–113. [DOI] [PubMed] [Google Scholar]
- 27. Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;6:610–621. [Google Scholar]
- 28. Maani R, Yang YH, Kalra S. Voxel‐based texture analysis of the brain. PLoS ONE 2015;10:e0117759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malone IB, Leung KK, Clegg S, et al. Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. NeuroImage 2015;104:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP‐43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006;314:130–133. [DOI] [PubMed] [Google Scholar]
- 31. Okamoto K, Hirai S, Yamazaki T, et al. New ubiquitin‐positive intraneuronal inclusions in the extra‐motor cortices in patients with amyotrophic lateral sclerosis. Neurosci Lett 1991;129:233–236. [DOI] [PubMed] [Google Scholar]
- 32. Mackenzie IR, Feldman H. The relationship between extramotor ubiquitin‐immunoreactive neuronal inclusions and dementia in motor neuron disease. Acta Neuropathol 2003;105:98–102. [DOI] [PubMed] [Google Scholar]
- 33. Stewart H, Rutherford NJ, Briemberg H, et al. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol 2012;123:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menke R, Proudfoot M, Talbot K, Turner M. The two‐year progression of structural and functional cerebral MRI in amyotrophic lateral sclerosis. NeuroImage Clin 2018;17:953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sørensen L, Igel C, Liv Hansen N, et al. Early detection of alzheimer's disease using MRI hippocampal texture. Hum Brain Mapp 2016;37:1148–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Waragai M. MRI and clinical features in amyotrophic lateral sclerosis. Neuroradiology 1997;39:847–851. [DOI] [PubMed] [Google Scholar]
- 37. Filippini N, Douaud G, Mackay CE, et al. Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology 2010;75:1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murray C, Viehman A, Lippa CF. The corpus callosum in pick's disease, alzheimer's disease, and amyotrophic lateral sclerosis: gliosis implies possible clinical consequence. Am J Alzheimer's Dis Other Demen 2006;21:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugiyama M, Takao M, Hatsuta H, et al. Increased number of astrocytes and macrophages/microglial cells in the corpus callosum in amyotrophic lateral sclerosis. Neuropathology 2013;33:591–599. [DOI] [PubMed] [Google Scholar]
- 40. Fatima M, Tan R, Halliday GM, Kril JJ. Spread of pathology in amyotrophic lateral sclerosis: assessment of phosphorylated TDP‐43 along axonal pathways. Acta Neuropathol Commun 2015;3:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J, Tong L, Wang L, Li N. Texture analysis of multiple sclerosis: a comparative study. Magn Reson Imaging 2008;26:1160–1166. [DOI] [PubMed] [Google Scholar]
- 42. Abrahams S, Goldstein L, Kew J, et al. Frontal lobe dysfunction in amyotrophic lateral sclerosis: a PET study. Brain 1996;119:2105–2120. [DOI] [PubMed] [Google Scholar]
- 43. Abrahams S, Leigh P, Harvey A, et al. Verbal fluency and executive dysfunction in amyotrophic lateral sclerosis (ALS). Neuropsychologia 2000;38:734–747. [DOI] [PubMed] [Google Scholar]
- 44. Bede P, Elamin M, Byrne S, et al. Basal ganglia involvement in amyotrophic lateral sclerosis. Neurology 2013;81:2107–2115. [DOI] [PubMed] [Google Scholar]
- 45. Abdulla S, Machts J, Kaufmann J, et al. Hippocampal degeneration in patients with amyotrophic lateral sclerosis. Neurobiol Aging 2014;35:2639–2645. [DOI] [PubMed] [Google Scholar]
- 46. Christidi F, Karavasilis E, Zalonis I, et al. Memory‐related white matter tract integrity in amyotrophic lateral sclerosis: an advanced neuroimaging and neuropsychological study. Neurobiol Aging 2017;49:69–78. [DOI] [PubMed] [Google Scholar]
- 47. Kassubek J, Müller H, Del Tredici K, et al. Diffusion tensor imaging analysis of sequential spreading of disease in amyotrophic lateral sclerosis confirms patterns of TDP‐43 pathology. Brain 2014;137:1733–1740. [DOI] [PubMed] [Google Scholar]
- 48. Lulé D, Böhm S, Müller H, et al. Cognitive phenotypes of sequential staging in amyotrophic lateral sclerosis. Cortex 2018;101:163–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Autocorrelation values extracted from the regions of significant differences in the pooled analysis from patients and controls in studies at different resolutions.Figure S2: Significant differences in autocorrelation between patients and controls (P < 0.001, cluster size > 10) in separate studies at different resolutions.


