Abstract
Genetic code expansion via stop codon suppression is a powerful technique for engineering proteins in mammalian cells with site-specifically encoded noncanonical amino acids (ncAAs). Current methods rely on very few available tRNA/aminoacyl-tRNA synthetase pairs orthogonal in mammalian cells, the pyrrolysyl tRNA/aminoacyl-tRNA synthetase pair from Methanosarcina mazei (Mma PylRS/PylT) being the most active and versatile to date. We found a pyrrolysyl tRNA/aminoacyl-tRNA synthetase pair from the human gut archaeon Methanomethylophilus alvus Mx1201 (Mx1201 PylRS/PylT) to be active and orthogonal in mammalian cells. We show that this PylRS enzyme can be engineered to expand its ncAA substrate spectrum. We find that due to the large evolutionary distance of the two pairs, Mx1201 PylRS/PylT is partially orthogonal to Mma PylRS/PylT. Through rational mutation of Mx1201 PylT, we abolish its noncognate interaction with Mma PylRS, creating two mutually orthogonal PylRS/PylT pairs. Combined in the same cell, we show that the two pairs can site-selectively introduce two different ncAAs in response to two distinct stop codons. Our work expands the repertoire of mutually orthogonal tools for genetic code expansion in mammalian cells and provides the basis for advanced in vivo protein engineering applications for cell biology and protein production.
Genetic code expansion allows for the addition of new chemical functionalities to proteins in living cells in the form of noncanonical amino acids (ncAAs). ncAAs are site-specifically installed through repurposing of a genetically encoded nonsense stop codon, most often amber (TAG). So-called amber suppression relies on introduction of a tRNACUA/aminoacyl-tRNA synthetase pair into the cell that is orthogonal to, i.e., does not cross-react with, all endogenous tRNAs and aminoacyl-tRNA synthetases.
Nature created a repertoire of alternatives to the standard genetic code over billions of years of evolution. It is the rare outliers to the universal code that have provided useful molecular tools for synthetic biology.1 The pyrrolysyl-tRNA (PylT)/pyrrolysyl-tRNA synthetase (PylRS) pair has become the most versatile tool for genetic code expansion in E. coli, yeast, mammalian cells, and metazoan organisms. Pyrrolysine (Pyl, N-ε-4-methyl-pyrroline-5-carboxylate-l-lysine) is a biosynthetic amino acid, genetically encoded in a small number of methanogenic bacteria and archaea. In these organisms, a dedicated PylRS/PylTCUA pair directs Pyl incorporation in response to amber stop codons.2 PylRS, the PylS gene product, accepts a range of structurally similar ncAAs in addition to its natural substrate. Further, PylRS has been a successful target for rational protein design and directed evolution, expanding the repertoire of accepted ncAA substrates.3 This includes ncAAs that carry chemical groups for bioorthogonal reactivity; photocaged amino acids or photoactivated cross-linkers for light-induced reactions; and amino acids with native post-translational modifications (PTMs).4 The PylRS/PylT pair supports highly efficient recoding in mammalian cells5,6 enabling application of genetic code expansion technology to address biological questions in the context of the living cell.
In principle, reassignment of more than one natural codon could further augment the ability to engineer proteins harboring multiple ncAAs in vivo. Since PylT is not hardwired to recode amber codons, other stop codons can be recoded in mammalian cells.7,8 Limiting to this application is the availability of mutually orthogonal tRNA/aminoacyl-tRNA synthetase pairs that are also orthogonal to the host cell. Thus, a key aim in the field is to find or rationally generate new mutually orthogonal pairs.9,10
The two widely used PylRS/PylT pairs belong to the archaea Methanosarcinales, M. mazei (Mma) and M. barkeri (Mba), predominantly found in semiaquatic environments. Recently, a number of new, evolutionary distant, Pyl-encoding archaea have been characterized from the human gut microbiome.11−14 Here, we explored the utility of the PylRS/PylT pair of Candidatus Methanomethylophilus alvus Mx1201 (Mx1201) in mammalian cells. There were three rationales for this: First, we speculated that proteins of gut-resident archaea might have evolved to optimally perform at human body temperature as opposed to the environmental species that need to be adaptive over a wide temperature range.15 Second, the Mx1201 PylS gene encodes a smaller protein (31 kDa), which may be easier to express in a heterologous system. PylRS expressed in mammalian cells shows predominantly nuclear localization which has been linked to inefficient function.16 Third, divergent evolution from the Methanosarcina PylRS/PylT pairs could manifest in mutual orthogonality. Mutually orthogonal PylRS/PylT pairs would enable incorporation of two distinct ncAAs using a dedicated tRNA each, within the same host system.
Results and Discussion
Mx1201 PylRS/PylT Pair Has Unique Properties
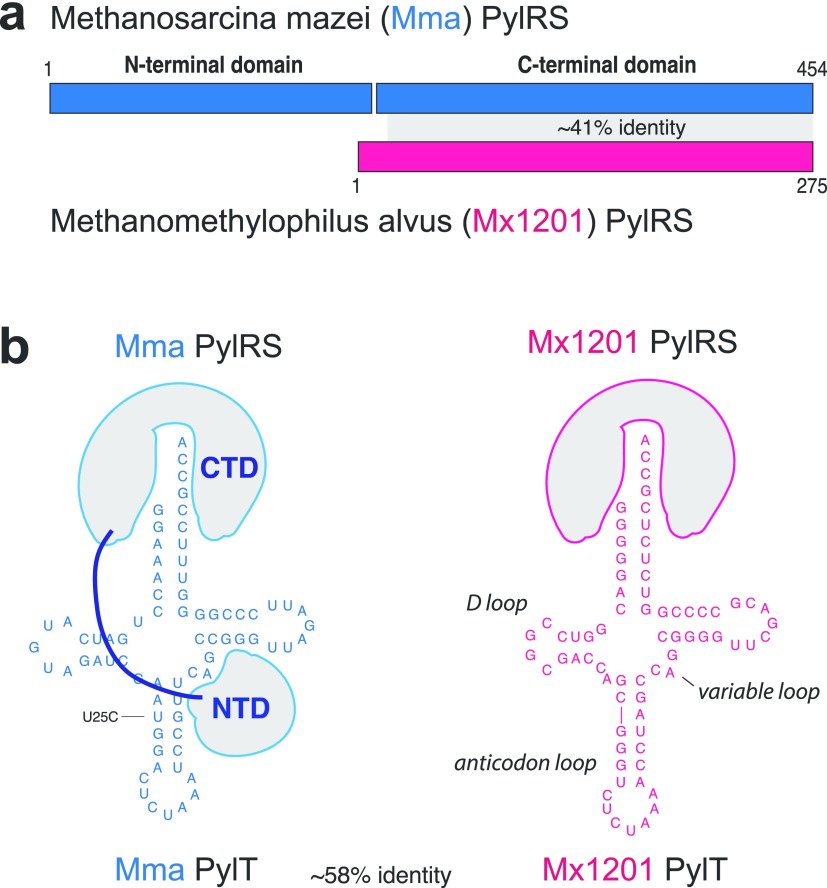
Mx1201 PylS encodes for a 275 amino acid protein, roughly half the size of Mma PylRS (Figure 1a). Mx1201 PylRS is homologous to the C-terminal domain (CTD) of Mma PylRS only, and there is no gene product in the Mx1201 genome with homology to the PylRS N-terminal domain (NTD) found in all previously characterized archaeal PylRS variants.11,17 The PylRS CTD harbors the catalytic site, binding both Pyl and the anticodon stem of PylT. The NTD has been shown to bind the variable loop region on the opposite side of PylT (Figure 1b) and has been considered essential for aminoacylation activity in vivo.18,19 Notably, bacterial PylS genes encode two separate subunits PylSn and PylSc that structurally correspond to the two domains described above for archaea, suggesting that the complementing roles of PylRS CTD and NTD are conserved across the two kingdoms. In contrast to this, Methanomethylophilus alvus Mx1201 and a few related species were the first genomes to be discovered lacking any detectable PylSn homologue,11 indicating that Mx1201 PylRS may have evolved to function entirely without NTD. Mx1201 PylRS and PylT show overall low sequence identity with the Mma PylRS/PylT pair (Figure 1a,b). Mx1201 PylT is one of the most distant homologues of known archaeal PylT (Supporting Information Figure 1), has a considerably divergent acceptor stem, and appears to have an even further shortened D-loop together with a “broken” anticodon stem when compared to Mma PylT (Figure 1b).
Figure 1.
Divergent features of Mx1201 and Mma PylRS/PylT pairs. (a) Methanosarcina mazei (abbreviated as Mma, shown in blue) PylRS and Methanomethylophilus alvus Mx1201 (abbreviated as Mx1201, shown in coral) PylRS domain structures and homology region. (b) Cloverleaf structure of Mma and Mx1201 PylTs. Cartoons of cognate PylRS show sites of recognition between N- and C-terminal domains (NTD, CTD) and tRNA.
Mx1201 PylRS/PylT Is Orthogonal to Endogenous tRNAs and RS in Mammalian Cells
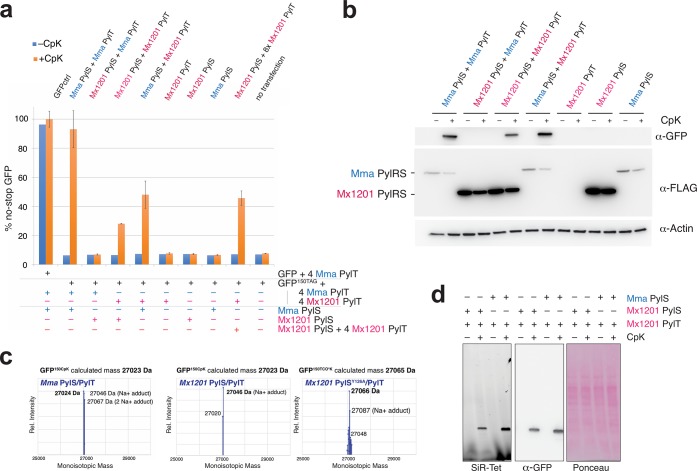
Previously, we have employed an efficient plasmid transfection system to direct ncAA incorporation into a GFP reporter protein in mammalian cells.6,20 Here, we cloned Mma and Mx1201 PylS and PylT genes into a similar plasmid design, expressing PylS from an EF1 promoter and 4 × PylT from human U6 or 7SK promoters in tandem repeats (Supporting Information Figure 2 and Table 1). For an initial combinatorial characterization, a plasmid expressing Mma or Mx1201 PylS was cotransfected with a second plasmid expressing four copies of either PylT variant together with the sfGFP150TAG reporter. The Mma PylT used in this study has a base substitution in the anticodon stem, U25C, previously found to increase amber suppression efficiency in E. coli and mammalian cells.6,21 Transient transfection was performed in human embryonic kidney (HEK293T) cells. Amber suppression was measured by GFP fluorescence in cell lysates and by Western blotting. We used cyclopropene-l-lysine (CpK, Supporting Information Figure 3) as ncAA, which is efficiently incorporated with wildtype Mma PylRS/PylT.6,22 First, we sought to test if the Mx1201 PylRS/PylT pair was functional in mammalian cells. Expression of Mx1201 PylRS/PylT pair indeed allowed selective incorporation of CpK into the GFP reporter, with 28% yield as compared to a no-stop GFP control (Figure 2a,b; Supporting Information Figure 4a). In comparison, the Mma PylRS/PylT pair reached 93% (Figure 2a).
Figure 2.
Mx1201 PylRS/PylT pair is active and orthogonal in mammalian cells; Mx1201 PylRS and Mma PylT are orthogonal, whereas Mma PylRS charges both Mma and Mx1201 PylT. (a) Fluorescence plate reader assay from HEK293T cell lysates transiently transfected with GFP150TAG reporter carrying a Mma or Mx1201 PylT cassette, in combination with Mma or Mx1201 PylRS, at a 9:1 ratio. GFP fluorescence is shown as a percentage of fluorescence measured with a GFP construct without TAG stop codon (GFPctrl) in the same experiment. For each combination, quadruplicate transfections were performed. For three of the four samples, the medium was supplemented with 0.2 mM CpK; all samples were harvested 24 h post-transfection. See Supporting Information Figure 4 for fluorescence microscopy pictures of transfected cells. (b) Western blot from HEK293T cell lysates transfected as above, showing the expression of GFP, FLAG-tagged synthetase variants, and a β-actin loading control. (c) Intact mass determination of purified GFP containing CpK at position 150 produced with Mma and Mx1201 PylRS/PylT, as well as TCO*K-containing GFP produced with Mx1201 PylRSY126A/PylT. All deconvoluted monoisotopic masses in the 25–30 kDa mass range are graphed, and the predicted monoisotopic mass is given for comparison. (d) In-gel far-red fluorescence image and Western blot against GFP from HEK293T cell lysates. Lysates have been labeled with silicon rhodamine tetrazine (SiR-Tet) fluorescent dye.
The PylRS/PylT pairs encoded by Mx1201 and Mma differ substantially in their primary sequences. We sought to understand if these PylRS and PylT would cross-react or were in fact nonfunctional accross species, thus mutually orthogonal. Interestingly, Mma PylRS aminoacylates Mx1201 PylT more efficiently (48% suppression) than Mx1201 PylRS, suggesting optimal enzymatic activity even for the noncognate PylT. This result also implies that key structural recognition features are conserved between the two distant PylT relatives. To the contrary, Mx1201 PylRS did not elicit any measurable amber suppression with Mma PylT.
Western blotting and immunostaining using an N-terminal FLAG-tag confirmed expression of Mx1201 PylRS (Figure 2b). Mx1201 PylRS protein levels appeared much higher than for Mma PylRS in our lysates (Figure 2b).
However, it should be noted that Mma PylRS has a distinctive nuclear localization (Supporting Information Figure 6a), and we only solubilized 50% of the Mma PylRS protein using RIPA buffer (Supporting Information Figure 4b). Mx1201 PylRS is soluble and mostly cytosolic (Supporting Information Figures 4b, 6). In summary, Mx1201 PylRS is stable and correctly localized in mammalian cells but does not generate aminoacylation activity equivalent to Mma PylRS. Given the known importance of the PylRS NTD for PylT binding,18,19 we speculate that CTD-only Mx1201 PylRS has a reduced affinity for PylT. In this case, raising cellular PylT concentration would be critical for enhancing activity of the Mx1201 PylRS/PylT pair. Indeed, supplying 4 × Mx1201 PylT on both plasmids raised the amber suppression efficiency to 46% of a no-stop GFP (Figure 2a).
To confirm orthogonality of Mx1201 PylRS/PylT in mammalian cells, we further needed to ensure that Mx1201 PylT is not charged by an endogenous aminoacyl-tRNA synthetase, and that Mx1201 PylRS does not charge other tRNAs with CpK. sfGFP150TAG expression was undetectable in the absence of CpK as judged by fluorescence measurement or Western blot (Figure 2a, b). Further, we determined the identity of CpK incorporated into the sfGFP150TAG construct by mass spectrometry (Figure 2c, Supporting Information Figure 4c). Vice versa, Mx1201 PylRS does not charge other mammalian tRNAs, as shown by selective SiR-tetrazine reaction with the amber-suppressed GFP and the absence of additional labeled endogenous proteins in whole-cell lysates (Figure 2d).
Together, these results show that the Mx1201 PylRS/PylT pair is functional and orthogonal in mammalian cells.
Mx1201 PylRS Can Be Engineered for Expanded Substrate Specificity
The key advantage of the PylRS/PylT system in mammalian cells over other orthogonal tRNA systems lies in the ability to incorporate structurally diverse ncAAs with useful functions for probing, controlling, and engineering proteins in living cells. This is both facilitated by the relative promiscuity of the wildtype PylRS enzyme and the ability to generate active site mutations with expanded substrate repertoire.
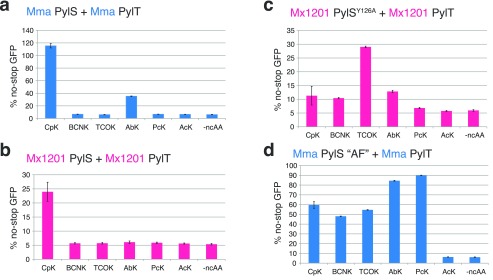
Exploring the substrate preferences of PylRS/PylT pairs from additional species may further expand the available repertoire of PylRS variants. Having established a new PylRS/PylT pair in mammalian cells, we sought to understand the substrate specificity of Mx1201 PylRS on a small set of ncAAs functionally interesting for mammalian cell biology (Supporting Information Figure 3). While Mma PylRS accepted CpK as well as AbK (Figure 3a), Mx1201 PylRS did not incorporate any of the tested ncAAs except CpK (Figure 3b).
Figure 3.
Substrate specificities of Mx1201 PylRS/PylT and Mx1201 PylRSY126A/PylT. Fluorescence plate reader assay from HEK293T cell lysates transiently transfected in a 4:1 ratio with a GFP150TAG reporter and (a) Mma PylRS/PylT, (b) Mx1201 PylRS/PylT, (c) Mx1201 PylRSY126A/PylT, or (d) Mma PylRS AF/PylT. GFP fluorescence is shown as a percentage of fluorescence measured with a GFP construct without a TAG stop codon in the same experiment. Cells were grown for 24 h in the absence (-ncAA) or presence of one of the following ncAAs: 0.2 mM CpK, 0.5 mM BCNK, 0.1 mM TCO*K, 0.5 mM AbK, 0.5 mM PcK, and 10 mM AcK. For full names and structures of amino acids, refer to Supporting Information Figure 3.
Sequence alignment and modeling suggests that, despite overall low sequence identity, the Pyl-binding pocket of Mx1201 PylRS is highly similar to other archaeal and bacterial PylRS homologues (Supporting Information Figure 5a). Few exceptions apply, such as Met129 and Val168 at the distal end contacting the pyrrole ring, where most other PylRS, including Mma PylRS, feature a highly conserved Leu and Cys residue, respectively (Supporting Information Figure 5b). Thus, there may be subtle variations in the substrate binding pocket underlying the more restricted repertoire of Mx1201 PylRS.
To expand the scope of the Mx1201 PylRS/PylT pair in mammalian cells, we tested the possibility of engineering the Mx1201 PylRS substrate binding pocket. We generated a variant, Mx1201 PylRSY126A, corresponding to a Mma PylRS Y306A mutant. The latter residue caps the Pyl-binding pocket in available PylRS structures and has previously been described to limit PylRS from incorporating ncAAs with longer and/or larger side chains than Pyl.23,24Mx1201 PylRSY126A has a reduced activity toward CpK, but gained activity (yield 29% of no-stop GFP) for axial trans-cyclooct-2-ene–lysine (TCO*K; Figure 3c). Comparing this result with the prior PylRS variant described for TCO*K (Figure 3d), PylRS-AF,23,25Mx1201 PylRSY126A has roughly half the activity but dramatically increased specificity for TCO*K over other ncAAs tested. The utility of the system is exemplified by selective bioorthogonal labeling of histone H3.2 using TCO*K and SiR-Tet fluorescent dye (Supporting Information Figure 6b).
Mma PylRS Requires Its NTD for Activity toward Mx1201 PylT
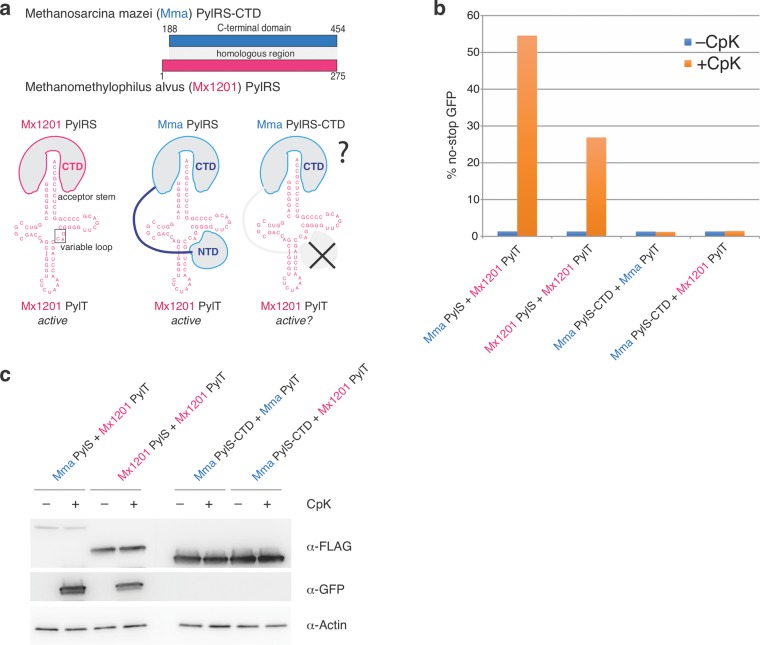
Following up on our finding that Mma PylRS accepts Mx1201 PylT (Figure 1a, b), we sought to understand how Mma PylRS recognizes the noncognate PylT. Specifically, we wondered if Mma PylRS N- and C-terminal domains differentially contributed to cognate Mma PylT or noncognate Mx1201 PylT binding. We created a new construct, Mma PylRS-CTD, by deleting the first 187 residues of Mma PylRS (Figure 4a). This fragment contains the entire region homologous to the Mx1201 PylRS enzyme (see Figure 1a).
Figure 4.
Mma PylRS interacts with noncognate Mx1201 PylT through its N-terminal domain. (a) Top scheme shows the C-terminal domain (CTD) construct used for Mma PylRS, containing the region corresponding to full-length Mx1201 PylRS. Bottom scheme indicates known identity elements (acceptor stem and variable loop) on putative Mx1201 PylT structure and putative binding regions for Mx1201 PylRS and Mma PylRS. (b) Fluorescence plate reader assay from HEK293T cell lysates transiently transfected as in Figure 1. GFP fluorescence is shown as a percentage of fluorescence measured with a GFP construct without a TAG stop codon in the same experiment. Cells were grown in the presence or absence of 0.2 mM CpK for 48 h. (c) Western blot showing the expression of FLAG-tagged synthetase variants, GFP and a β-actin loading control.
Mma PylRS-CTD was inactive with its cognate PylT (Figure 4b, c). The essentiality of the Mma PylRS NTD has never been explicitly tested in mammalian cells but mirrors observations made in E. coli and in vitro.18 Unlike the full-length enzyme, Mma PylRS-CTD also did not elicit measurable activity with Mx1201 PylT (Figure 4b, c). These results show that Mma PylRS activity toward Mx1201 PylT is fully dependent on its N-terminal domain. The results imply two fundamentally different binding modes employed by Mma and Mx1201 PylRS: Mx1201 PylRS has evolved efficient tRNA binding through its catalytic domain, making an N-terminal domain obsolete. This binding mode must be facilitated by specific features of the cognate Mx1201 PylT, since Mx1201 does not accept Mma PylT (Figure 2a). Mma PylRS, on the other hand, employs the canonical NTD interaction with the variable loop region of PylT, a known identity element of PylRS/PylT pairs.7,26 Supporting this hypothesis, the variable loop itself (41CAG43) and adjacent bases of Mx1201 PylT are conserved to their Mma PylT counterparts (Figure 4a).
Disrupting NTD Interaction of Mx1201 PylT
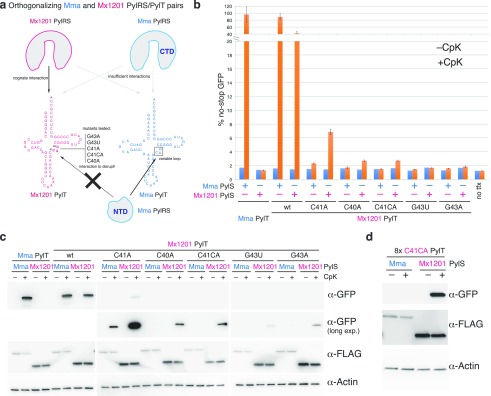
The above findings provide key prerequisites for the creation of mutually orthogonal PylRS/PylT pairs in mammalian cells: two evolutionarily distant PylRS enzymes (and engineered variants) with partially but not fully overlapping substrate specificities that use distinct surfaces for their recognition of the respective cognate PylT. We hypothesized that rationally designed mutations in the variable loop region of Mx1201 PylT would directly interfere with the noncognate recognition by Mma PylRS NTD (Figure 5a). Disrupting this interaction would make the Mx1201 and Mma tRNA/aminoacyl-tRNA synthetase pairs mutually orthogonal.
Figure 5.
Mutations in the Mx1201 PylT variable loop disrupt recognition by Mma PylRS, resulting in orthogonal PylRS/PylT pairs. (a) Scheme depicting predominant modes of recognition between PylRS and cognate PylTs: Mx1201 PylRS, in the absence of an NTD, likely retains interactions only with the acceptor stem. Mma PylRS CTD recognizes neither Mma PylT nor Mx1201 PylT in the absence of its NTD. Further indicated are mutations introduced in the variable loop of Mx1201 PylT that are predicted to abolish interaction with the Mma PylRS NTD. (b) Fluorescence plate reader assay of HEK293T cell lysates transiently transfected in 5:1:4 ratio with a GFP150TAG reporter and the indicated synthetase and tRNA. GFP fluorescence is shown as a percentage of fluorescence measured with a GFP construct without a TAG stop codon in the same experiment. For each combination, quadruplicate transfections were performed. For three of the four samples, the medium was supplemented with 0.2 mM CpK; all samples were harvested 48 h post transfection. Note the broken y axis. (c) Western blot showing the expression of FLAG-tagged synthetase variants, GFP, and a β-actin loading control. (d) Western blot of HEK293T cell lysates transiently transfected in 1:9 ratio of GFP150TAG reporter and either Mx1201 PylTC41CA/Mma PylRS or Mx1201 PylTC41CA/Mx1201 PylRS, showing the expression of FLAG-tagged synthetase variants, GFP, and a β-actin loading control. CpK was added at the time of transfection; samples were harvested 48 h post transfection.
A recent crystal structure of the PylRS-NTD–PylT complex reveals that tRNA recognition relies predominantly on steric constraints that would exclude any more spacious variable loop.19 We reasoned that alterations of the variable loop analogous to those shown to abrogate PylRS binding and aminoacylation7,26 would provide candidates for disrupting the interaction between Mx1201 PylT and Mma PylRS.
As a caveat to this simple approach, even isolated base changes may affect folding of the tRNA in unexpected ways, particularly since the short variable loop participates in a tightly packed tertiary core.27 We chose to test four single base substitutions, C40A, C41A, G43A, and G43U, and one insertion C41CA (Figure 5a).
For a more facile generation and screening of Mx1201 PylT mutants, we moved to a three-plasmid expression system where PylRS and sfGFP150TAG were expressed from individual plasmids without PylT, and PylT was supplied on a third plasmid (Figure 5b,a; Supporting Information Figure 2). As in previous experiments, Mx1201 PylT was more active with Mma PylRS (89% of no-stop GFP) than its cognate Mx1201 PylRS (42% of no-stop GFP).
None of the Mx1201 PylT mutants exhibited a similar efficiency, suggesting that mutations in the variable loop affected both cognate and noncognate interactions with Mx1201 PylRS and Mma PylRS, respectively. Nevertheless, C40A, C41A, and C41CA mutations preserved measurable activity with Mx1201 PylRS. Of these, C41A retained the highest activity with Mx1201 PylRS (7% of no-stop GFP) while disproportionally reducing Mma PylRS activity (Figure 5b,c). Since we aimed to create a fully orthogonal pair, we focused on C41CA, which showed lower activity with Mx1201 PylRS but lost all detectable activity with Mma PylRS even by sensitive Western blot (Figure 5c). Moving the Mx1201 PylTC41CA mutant back to our efficient two-plasmid transfection system (Supporting Information Figure 2), we were able to create robust amber suppression with Mx1201 PylRS and confirm orthogonality to Mma PylRS (Figure 5d, Supporting Information Figure 7). In conclusion, we have created a new tRNA/aminoacyl-tRNA synthetase pair, Mx1201 PylRS/Mx1201 PylTC41CA, that is orthogonal in mammalian cells, and also mutually orthogonal to the Mma PylRS/PylT pair. A recent first characterization of the Mx1201 PylRS/PylT pair in E. coli published during the preparation of this manuscript28 used a similar rational and directed evolution approach for creating orthogonal PylRS/PylT pairs, yielding orthogonal Mx1201 PylT variants that preserved high activity in E. coli. We thus additionally tested six of the most active variable loop variants in our mammalian system (Supporting Information Figure 8). Notably, the activity of two variants with the 4-bp variable loop was somewhat increased over our Mx1201 PylTC41CA variant, but those carrying a more extended 5- or 6-bp variable loop did not elicit any activity (Supporting Information Figure 8b).
Site-Specific Incorporation of Two Distinct ncAAs
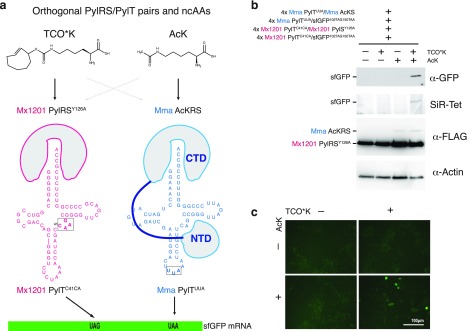
Next, we aimed to implement dual site-specific protein modification at independent sites and with distinct ncAAs in mammalian cells using the two orthogonal PylRS/PylT pairs. This first required to further modify one pair to recode another stop codon. We chose ochre (TAA) because it is only marginally more abundant than amber in mammalian cells. We thus created Mma PylTUUA, where the anticodon was changed from CUA to UUA. We used a fluorescent reporter, sfGFP102TAG150TAA, containing two stop codons, 102TAG and 150TAA.
To incorporate two distinct ncAAs, the two orthogonal PylRS must each incorporate one of the ncAAs with high selectivity (Figure 6a). Since we have shown that wild type Mx1201 PylRS and the engineered variant PylRSY126A both have narrow substrate specificities, they can be combined in many unique combinations with known Mma PylRS variants evolved for specific ncAAs, e.g., photocaged lysine (PcK), acetyl-lysine (AcK), diazirine-lysine (AbK).29−31
Figure 6.
Dual ncAA incorporation in mammalian cells using mutually orthogonal PylRS/PylT pairs. (a) Scheme for dual ncAA incorporation using mutually orthogonal PylRS/PylT pairs in mammalian cells. Two PylRS variants with narrow substrate specificity were chosen to incorporate two different ncAAs (TCO*K and AcK) in response to distinct stop codons. Chemical structures of ncAA are given. PylRS and PylT variants used in this experiment are indicated. (b) Western blot and SiR-Tet labeling of HEK293T cells transiently transfected in a 4:4:1:1 ratio with the following components: GFP102TAG150TAA reporter with 4 × Mx1201 PylTC41CA, GFP102TAG150TAA reporter with 4 × Mma PylTUUA, Mx1201 PylRSY126A with 4 × Mx1201 PylTC41CA, and Mma AcKRS with 4 × Mma PylTUUA. Lysates were analyzed for expression of FLAG-tagged synthetase variants, GFP, and a β-actin loading control. Notably, detectable AcKRS protein levels are dependent on the presence of AcK substrate. (c) Fluorescence images of transfected HEK293T cells used for panel b prior to cell lysis.
Here, we chose to incorporate AcK and TCO*K into the same protein, GFP, in response to UAA and UAG stop codons, respectively; we created a pair with Mma PylTUUA and AcKRS29 and combined it with the Mx1201 PylRSY126A/Mx1201 PylTC41CA pair in the same cell (Figure 6a). Indeed, we were able to observe production of GFP in cells transfected with the two PylRS/PylT pairs and the sfGFP102TAG150TAA reporter in the presence of both ncAAs, but not with either individually (Figure 6b, c). SiR-Tet labels sfGFP only in the presence of both ncAAs. This suggests orthogonal recognition of both the tRNAs and ncAAs by their respective synthetases and selective incorporation in response to the specified stop codons. We scaled up expression and purified the product (Supporting Information Figure 9a,b), we determined the yield to be roughly 1.5 μg per 25 mL/175m2 HEK293T culture. ESI-MS confirmed the incorporation of both ncAAs, resulting in sfGFP102TCO*K,150AcK (Supporting Information Figure 9d). Notably, ESI-MS also showed an additional mass species corresponding to sfGFP102AcK, 150AcK, likely a consequence of Mma PylTUUA mispairing to a low extent with the UAG stop codon as recently reported.32 A tetrazine pull-down further confirmed that a small fraction of purified sfGFP did not contain TCO*K (Supporting Information Figure 9c. Since we did not observe suppression of the sfGFP102TAG150TAA reporter with AcK alone before (Figure 6b), the increased amount of transfected DNA and longer expression time (60 h) used for the scaled-up sfGFP production may have exacerbated misincorporation of AcK. Thus, all components of the suppression system need to be finely tuned for maximal selectivity of the orthogonal pairs.
Conclusions
We have set out to identify new orthogonal tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells. Exploring the function of a distant homologue of the Mma PylRS/PylT pair from Methanomethylophilus alvus Mx1201, we found that the Mx1201 PylRS/PylT pair is active and orthogonal in mammalian cells. Mx1201 PylRS is also naturally orthogonal to Mma PylT. We identify several Mx1201 PylT mutants that retain their cognate interaction with Mx1201 PylRS but reduce or abrogate noncognate recognition by Mma PylRS. Combined in the same cell, we show that the two pairs can introduce two different ncAAs in response to two distinct stop codons. Our findings shed light into a new clade of PylRS enzymes with unexpected properties, functionally divergent from the previously studied archaeal and bacterial systems. Our work expands the repertoire of mutually orthogonal tools for genetic code expansion in mammalian cells and provides the basis for advanced in vivo protein engineering applications for cell biology and protein production.
Materials and Methods
DNA Constructs
The sfGFP 150L control reporter construct and the H32 K56TAG amber suppression construct have been described previously.6,20 A series of plasmids for amber suppression (pAS) was created based on the PB510B-1 (System Biosciences) piggybac plasmid (Table 1 and Supporting Information Figure 2). pUC-based plasmids were used to express tRNA from single copy genes (Table 2 and Supporting Information Figure 2). All DNA constructs were verified by Sanger sequencing.
Table 1. pAS Plasmids for EF1α Controlled Expression of PylS and Reporter Genes.
| plasmid number | 4 × tRNA cassette | EF1α promoter |
|---|---|---|
| 1 | – | FLAG_Mma PylRS |
| 2 | 7SK-Mma PylT | FLAG_Mma PylRS |
| 3 | 7SK-Mma PylT | FLAG_Mma PylRS/AF |
| 4 | 7SK-Mma PylT | FLAG_Mma PylRS CTD |
| 5 | 7SK-Mx1201 PylT C41CA | FLAG_Mma PylRS |
| 6 | 7SK-Mma PylTUUA | FLAG_AcKRS |
| 7 | – | FLAG_Mx1201 PylRS |
| 8 | U6-Mx1201 PylT | FLAG_Mx1201 PylRS |
| 9 | U6-Mx1201 PylT | FLAG_Mx1201 PylRS Y126A |
| 10 | 7SK-Mx1201 PylT | FLAG_Mx1201 PylRS |
| 11 | 7SK-Mx1201 PylT | FLAG_Mx1201 PylRS Y126A |
| 12 | 7SK-Mx1201 PylT C41CA | FLAG_Mx1201 PylRS |
| 13 | 7SK-Mx1201 PylT C41CA | FLAG_Mx1201 PylRS Y126A |
| 14 | – | sfGFP 150TAG |
| 15 | 7SK-Mma PylT | sfGFP 150TAG |
| 16 | 7SK-Mma PylTUUA | sfGFP 102TAG 150TAA |
| 17 | U6-Mx1201 PylT | sfGFP 150TAG |
| 18 | 7SK-Mx1201 PylT | sfGFP 150TAG |
| 19 | 7SK-Mx1201 PylT | HA histone H3.2 K56TAG |
| 20 | 7SK-Mx1201 PylT C41CA | sfGFP 150TAG |
| 21 | 7SK-Mx1201 PylT C41CA | sfGFP 102TAG 150TAA |
Table 2. PylT Expression Plasmids Are pUC-based.
| plasmid number | single copy tRNA gene |
|---|---|
| 22 | 7SK-Mma PylT |
| 23 | 7SK-Mx1201 PylT |
| 24 | 7SK-Mx1201 PylT C41CA |
| 25 | 7SK-Mx1201 PylT G43U |
| 26 | 7SK-Mx1201 PylT C40A |
| 27 | 7SK-Mx1201 PylT G43A |
| 28 | 7SK-Mx1201 PylT C41A |
Cell Culture and Transfection
HEK293T cells were maintained in adherent culture at 37 °C and 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM; AqDMEM high glucose, Sigma) supplemented with 10% (v/v) FBS. For transfection (1.5–2.0) × 105 HEK293T cells were seeded per well in 24-well plates 24 h prior to transfection. Cells were transiently transfected with TransIT-LT1 (Mirus) following the manufacturer’s instructions. ncAAs were added at the time of transfection as indicated and cells harvested after 24 or 48 h. Large-scale expressions were performed using a modified protocol increasing the total DNA amount 2-fold and replacing TransIT-LT1 with 1 mg mL–1 polyethylenimine (PEI). Cells were harvested after 60 h.
Amino Acids
Noncanonical amino acids (ncAAs) used in this study are summarized in Table 3. To incorporate ncAA into proteins, amino acid working solutions were prepared from 100 mM stock solutions and added to the cultured cells together with the transfection mixture.
Table 3. Noncoding Amino Acids Used in This Study.
| name | CAS number | final conc. | |
|---|---|---|---|
| AbK | Nε-[(2-(3-methyl- 3H-diazirin-3-yl)ethoxy)carbonyl]-l-lysine | 1253643-88-7 | 0.5 mM |
| AcK | Nε-acetyl-l-lysine | 692-04-6 | 10 mM |
| BCNK | Exo-BCN – l-Lysine | 1384100-45-1 | 0.5 mM |
| CpK | Nε-[[(2-methyl-2-cyclopropene-1-yl) methoxy] carbonyl-l-lysine | 1610703-09-7 | 0.2 mM |
| PcK | photocaged l-lysine | 0.5 mM | |
| TCO*K | axial trans-cyclooct-2-ene-l-lysine | 1384100-45-1 | 0.1 mM |
Mass Spectrometry
HEK293T cells were transfected and cultured in the presence of ncAA for 72 h. After cell lysis in RIPA buffer with added cOmplete protease inhibitor (Roche), the insoluble fraction was removed by centrifugation. GFP was captured on GFP-Trap_M magnetic beads (ChromoTek) and eluted with 1% (v/v) acetic acid.
ES-MS in Figure 2 was performed using a TriVersa NanoMate chip-based electrospray device (Advion, Ithaca, NY) coupled to the LTQ Velos Orbitrap Elite (Thermo Scientific, Bremen, Germany). The NanoMate delivered 2 μL of sample solution to the tip engaged with the back of the ESI chip and nanospray ionization was initiated applying 1.9 kV and 0.8 psi gas pressure. The mass spectrometer was operated in positive ion mode with activated protein mode settings. Data were analyzed using the Protein Deconvolution v3.0 software (Thermo Scientific) to calculate monoisotopic masses.
For ESI-MS of dual ncAA sfGFP (Supporting Information Figure 9d), purified sfGFP was desalted and concentrated using C18 ZipTips (Merck), eluted into 50% (v/v) acetonitrile/1% (v/v) formic acid, and directly infused into an Orbitrap Fusion Tribrid mass spectrometer equipped with an offline nanospray source using borosilicate capillaries (Thermo Scientific). The capillary voltage was 1.5 kV and the pressure in the ion-routing multipole was maintained at 0.11 Torr. Spectra were acquired in the Orbitrap mass analyzer operated in high mass mode at a resolution of 50.000 between 800 and 3000 m/z. Data were analyzed using Excalibur (Thermo Scientific) and UniDec (unidec.chem.ox.ac.uk) software packages.
Quantification of GFP Expression
Transfected HEK293T cells were grown in the presence of ncAA as indicated for 24 h or 48 h. Cells were lysed in RIPA buffer with 1 × cOmplete protease inhibitor (Roche); the insoluble fraction was removed by centrifugation. GFP bottom fluorescence of an aliquot was measured in a Tecan Infinity M200 pro platereader (excitation 485 nm, emission 518 nm). Fluorescence measurements were normalized to total protein content of each sample as determined by Pierce BCA assay kit (Fisher Scientific) on the same sample.
Live-Cell Imaging for GFP Expression
GFP expression was visualized by imaging in a ZOE Fluorescent Cell Imager (BioRad).
Western Blot and Pulldowns
Equal amounts of protein, as determined by Pierce BCA assay (Fisher Scientific), were separated on 4–20% Tris-glycine gels (BioRad) and transferred to nitrocellulose membranes.
Expression of the sfGFP reporter and aminoacyl-tRNA synthetases was confirmed by immunoblotting with antibodies against GFP (Santa Cruz, sc-9996), FLAG-HRP (Sigma, A8592), β-actin (cell signaling), and corresponding secondary HRP-conjugated antibodies when needed (BioRad). For GFP pulldown, GFP-Trap_M magnetic beads (ChromoTek) were used. For tetrazine pulldown, tetrazine-agarose was used (CLK-1199-2, Jena Bioscience).
Immunostaining and Fluorescence Microscopy
Transfected cells were cultured in the presence of ncAA for 24 h. Before fixation (4% (v/v) formaldehyde) and permeabilization (0.1% (v/v) triton) the ncAA was removed for 8 h. Samples were blocked in 2% (w/v) BSA in TBS-T and subsequently incubated in the presence of 0.5 μM SiR-tetrazine (Spirochrome). After washing with TBS-T, samples were incubated with primary antibodies mouse anti-GFP (B-2, Santa Cruz #9996) and rabbit anti-FLAG (D6W5B, Cell Signaling #14793) and subsequently incubated with secondary antibodies antimouse Alexa Fluor 488, antirabbit Alexa Fluor 555 (Life Technologies), and DAPI (Sigma-Aldrich). After washing, cells were imaged on a Nikon eclipse Ti2 inverted widefield microscope, using a 20 × 0.75 NA or a 40 × 1.15 NA objective.
Acknowledgments
We thank M. Landreh for ESI-MS measurements and analysis. Further mass spectrometry service was provided by Proteomics Biomedicum core facility, Karolinska Institutet, Stockholm. We thank A. Vegvari for data analysis and advice. We thank the J. Bartek lab for access to Tecan Infinity M200 Pro plate reader and Nikon eclipse Ti2 microscope.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.8b00571.
Supporting Figures 1–9, uncropped gel, and Western blot images (PDF)
Accession Codes
Plasmid sequences are deposited on Mendeley Data (DOI: 10.17632/bnm4x5vjrs.1).
Author Contributions
S.J.E. and B.M. conceived and planned experiments. B.M. and J.H. carried out experiments and analyzed data. L.L. performed immunofluorescence microscopy and analyzed data. B.M., J.H., and S.J.E. prepared figures and wrote the manuscript.
Research was funded by Karolinska Institutet SFO Molecular Biosciences, Vetenskapsrådet (2015–04815), H2020 ERC-2016-StG (715024 RAPID), Ming Wai Lau Center for Reparative Medicine, Ragnar Söderbergs Stiftelse.
The authors declare no competing financial interest.
Supplementary Material
References
- Baranov P. V.; Atkins J. F.; Yordanova M. M. (2015) Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat. Rev. Genet. 16, 517–529. 10.1038/nrg3963. [DOI] [PubMed] [Google Scholar]
- Polycarpo C.; Ambrogelly A.; Bérubé A.; Winbush S. M.; McCloskey J. A.; Crain P. F.; Wood J. L.; Söll D. (2004) An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. U. S. A. 101, 12450–12454. 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W.; Tharp J. M.; Liu W. R. (2014) Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta, Proteins Proteomics 1844, 1059–1070. 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. W. (2017) Expanding and reprogramming the genetic code. Nature 550, 53–60. 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- Elsässer S. J.; Ernst R. J.; Walker O. S.; Chin J. W. (2016) Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat. Methods 13, 158–164. 10.1038/nmeth.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmied W. H.; Elsässer S. J.; Uttamapinant C.; Chin J. W. (2014) Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 136, 15577–15583. 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogelly A.; Gundllapalli S.; Herring S.; Polycarpo C.; Frauer C.; Söll D. (2007) Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. U. S. A. 104, 3141–3146. 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H.; Chatterjee A.; Choi S.; Bajjuri K. M.; Sinha S. C.; Schultz P. G. (2013) Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chem., Int. Ed. 52, 14080–14083. 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Xiao H.; Schultz P. G. (2012) Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 109, 14841–14846. 10.1073/pnas.1212454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H.; Slusarczyk A. L.; Chin J. W. (2010) De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J. Am. Chem. Soc. 132, 2142–2144. 10.1021/ja9068722. [DOI] [PubMed] [Google Scholar]
- Borrel G.; Gaci N.; Peyret P.; O’Toole P. W.; Gribaldo S.; Brugère J.-F. (2014) Unique characteristics of the pyrrolysine system in the 7th order of methanogens: implications for the evolution of a genetic code expansion cassette. Archaea 2014, 374146. 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G.; Harris H. M. B.; Tottey W.; Mihajlovski A.; Parisot N.; Peyretaillade E.; Peyret P.; Gribaldo S.; O’Toole P. W.; Brugère J.-F. (2012) Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J. Bacteriol. 194, 6944–6945. 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G., Harris H. M. B., Parisot N., Gaci N., Tottey W., Mihajlovski A., Deane J., Gribaldo S., Bardot O., Peyretaillade E., Peyret P., O’Toole P. W., and Brugère J.-F. (2013) Genome Sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a Third Thermoplasmatales-Related Methanogenic Archaeon from Human Feces, Genome Announcements 1, 10.1128/genomeA.00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B.; Fardeau M.-L.; Ollivier B.; Raoult D.; Drancourt M. (2012) Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 1902–1907. 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- Gunnigle E.; McCay P.; Fuszard M.; Botting C. H.; Abram F.; O’Flaherty V. (2013) A functional approach to uncover the low-temperature adaptation strategies of the archaeon Methanosarcina barkeri. Appl. Environ. Microbiol. 79, 4210–4219. 10.1128/AEM.03787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikić I.; Estrada Girona G.; Kang J. H.; Paci G.; Mikhaleva S.; Koehler C.; Shymanska N. V.; Ventura Santos C.; Spitz D.; Lemke E. A. (2016) Debugging Eukaryotic Genetic Code Expansion for Site-Specific Click-PAINT Super-Resolution Microscopy. Angew. Chem., Int. Ed. 55, 16172–16176. 10.1002/anie.201608284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Crnković A., Umehara T., Ivanova N. N., Kyrpides N. C., and Söll D. (2017) RNA-Dependent Cysteine Biosynthesis in Bacteria and Archaea, mBio 8, 10.1128/mBio.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring S.; Ambrogelly A.; Gundllapalli S.; O’Donoghue P.; Polycarpo C. R.; Söll D. (2007) The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 581, 3197–3203. 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T.; Miller C.; Guo L.-T.; Ho J. M. L.; Bryson D. I.; Wang Y.-S.; Liu D. R.; Söll D. (2017) Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 13, 1261–1266. 10.1038/nchembio.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer S. J. (2018) Generation of stable amber suppression cell lines. Methods Mol. Biol. 1728, 237–245. 10.1007/978-1-4939-7574-7_15. [DOI] [PubMed] [Google Scholar]
- Chatterjee A.; Sun S. B.; Furman J. L.; Xiao H.; Schultz P. G. (2013) A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837. 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T. S.; Townsley F. M.; Bianco A.; Ernst R. J.; Sachdeva A.; Elsässer S. J.; Davis L.; Lang K.; Pisa R.; Greiss S.; Lilley K. S.; Chin J. W. (2014) Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 32, 465–472. 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T.; Ishii R.; Fukunaga R.; Kobayashi T.; Sakamoto K.; Yokoyama S. (2008) Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 15, 1187–1197. 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Kavran J. M.; Gundllapalli S.; O’Donoghue P.; Englert M.; Söll D.; Steitz T. A. (2007) Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U. S. A. 104, 11268–11273. 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikić I.; Plass T.; Schraidt O.; Szymański J.; Briggs J. A. G.; Schultz C.; Lemke E. A. (2014) Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem., Int. Ed. 53, 2245–2249. 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- Jiang R.; Krzycki J. A. (2012) PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J. Biol. Chem. 287, 32738–32746. 10.1074/jbc.M112.396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K.; O’Donoghue P.; Gundllapalli S.; Araiso Y.; Ishitani R.; Umehara T.; Söll D.; Nureki O. (2009) Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature 457, 1163–1167. 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis J. C. W.; Chin J. W. (2018) Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 10, 831–837. 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H.; Peak-Chew S. Y.; Chin J. W. (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234. 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Gautier A.; Nguyen D. P.; Lusic H.; An W.; Deiters A.; Chin J. W. (2010) Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc. 132, 4086–4088. 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- Hancock S. M.; Uprety R.; Deiters A.; Chin J. W. (2010) Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 132, 14819–14824. 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.; Addy P. S.; Mukherjee R.; Chatterjee A. (2017) Defining the current scope and limitations of dual noncanonical amino acid mutagenesis in mammalian cells. Chem. Sci. 8, 7211–7217. 10.1039/C7SC02560B. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.