Abstract
Objectives
The aim of this study was to analyze the prevalence of the CTX-M, TEM, SHV, VIM, NDM, and OXA genes in carbapenemase-producing Escherichia coli and their transmissibility at a tertiary care hospital in south India.
Materials and Methods
Twenty-one carbapenem-resistant E. coli (carbapenem-resistant Enterobacteriaceae; CRE) were collected from the Sri Sathya Sai Institute of Higher Medical Sciences (Puttaparthi India). Resistance to antibiotics was analyzed by Vitek-2, and the identity of the isolates was confirmed by 16S rDNA sequencing. RAPD and enterobacterial repetitive intergenic consensus (ERIC)-PCR were performed for molecular typing. Metallo-β-lactamase production was confirmed by a double disc synergy test. The presence of the extended-spectrum β-lactamases CTX-M, TEM, and SHV and of the carbapenemases NDM, VIM, and OXA was determined by PCR. Carbapenemase variants were further confirmed by sequencing. The transmissibility of the genes was tested by conjugation.
Results
Twelve of the 21 (57%) carbapenem-resistant E. coli isolates were community acquired, indicating the spread of CRE in environmental samples. TEM and NDM-5 were found to be the major β-lactamases produced by the pathogens. OXA-181 was found in 5 of the isolates. All 21 isolates were found to harbor more than one of the tested β-lactamases, and all of the isolates were found to have the capacity to participate in conjugation; 15 of the transconjugants were found to have acquired the tested β-lactamases, substantiating their ability to be transferred to other strains of bacteria.
Conclusion
Monitoring of community-acquired carbapenem-resistant bacteria is very important as the association of resistance determinants with mobile genetic elements would present a serious clinical challenge.
Keywords: Carbapenem resistance, Escherichia coli, Transconjugation, New Delhi metallo-β-lactamase
Significance of the Study
The increasing rates of carbapenem-resistant Enterobacteriaceae in the community and in hospitals led us to determine the prevalence of NDM and OXA in twenty-one Escherichia coli isolates and their ability to be transferred to laboratory strains. We hope that this study will emphasize the importance of antibiotic stewardship programs in health care facilities and improvement of social hygiene in the community.
Introduction
Escherichia coli is the most common cause of infections in hospitals and in the community [1, 2]. It is also one of the main pathogens causing nosocomial infections, primarily in immunocompromised patients [3]. Carbapenems such as imipenem, meropenem, and ertapenem are the last drugs of choice for treating seriously ill patients who harbor organisms producing extended-spectrum β-lactamases (ESBL) such as CTX-M, TEM, and SHV [4]. More recently, carbapenem-resistant Enterobacteriaceae (CRE) are being increasingly reported [5]. The most prevalent mechanism of resistance among these is the production or acquisition of carbapenemases. Some carbapenemases that are encountered commonly include KPC, NDM, OXA, VIM, and IMP; these genes are generally located on mobile genetic elements which enable their transmission [6, 7, 8]. Globally, variants of OXA have been reported among Enterobacteriaceae. However, reports on the prevalence of OXA from India are sparse [9, 10, 11].
The rapid dissemination of antibiotic resistance genes globally is exemplified by the NDM carbapenemase. The spread of NDM-1-positive isolates in India and the global spread of NDM metallo-β-lactamase (MBL) was compiled and elucidated in a study conducted by Johnson and Woodford [12]. The prevalence of NDM-1 in Enterobacteriaceae among patients from Pakistan, India, and Bangladesh and among those in the UK who travelled to the Indian subcontinent has been reported. This study explained the epidemiology of NDM-1 and its association with the Indian subcontinent [13].
We aimed to investigate the prevalence of antibiotic resistance genes among CRE E. coli obtained from infected patients visiting a tertiary care hospital in Andhra Pradesh, India, and to characterize them with regard to their ability to transfer antibiotic resistance to laboratory strains. We believe that the importance of this study stems from the increased occurrence and spread of CRE in the Indian subcontinent [13].
Materials and Methods
Setting
This study included 20 patients with urinary tract infections visiting the Department of Urology and 1 patient with a fracture visiting the Department of Orthopedics of the Sri Sathya Sai Institute of Higher Medical Sciences (Puttaparthi, India). Urine samples and pus from these patients were cultured in the Department of Microbiology. Only one isolate from each patient was included in this study.
Bacterial Identification and Antibiotic Susceptibility Testing
During January 2013 to December 2014, a total of 233 carbapenem-resistant clinical isolates were obtained. Among these, 101 were found to be Enterobacteriaceae. The number of E. coli resistant to carbapenems was 21. Identification of the isolates was performed by Vitek-2 and confirmed by 16s rDNA sequencing. Antibiotic susceptibility testing (AST) and determination of minimum inhibitory concentrations were done using AST-N280 Vitek cards and interpreted as per Clinical Laboratory Standards Institute (CLSI) guidelines [14].
Phenotypic Detection of Carbapenemase Expression
Imipenem susceptibility testing was performed using the disc diffusion method as per CLSI guidelines [14]. A double disc synergy test using imipenem and imipenem-EDTA discs was performed as described earlier [15] by placing the 2 discs 30 mm apart on Müller-Hinton agar medium inoculated with a 0.5 McFarland suspension of the test organism. An increase in zone size of more than 5 mm for the imipenem-EDTA disc as compared to the imipenem disc was considered positive for the production of an MBL.
Phylogenetic Grouping
Random amplified polymorphic DNA (RAPD) [16, 17, 18] and enterobacterial repetitive intergenic consensus (ERIC)-PCR [19, 20, 21] were performed to understand the clonal relatedness of the study isolates. Genomic DNA was recovered using NucleoSpin® tissue kits (Macherey-Nagel). RAPD and ERIC-PCR were performed using the primers mentioned in Table 1. The PCR conditions for the phylogenetic analyses involving RAPD and ERIC-PCR are described in Table 2.
Table 1.
List of primers used for PCR amplification
| Primer | Sequence | Size, bp |
|---|---|---|
| OPA 2 | 5′-TGCCGAGCTG-3′ | |
| ERIC-F | 5′-ATGTAAGCTCCTGGGGATTCA-3′ | |
| ERIC-R | 5′-AAGTAAGTGACTGGGGTGAGC-3′ | |
| NDM-F | 5′-GGT TTG GCG ATC TGG TTT TC-3′ | 621 |
| NDM-R | 5′-CGG AAT GGC TCA TCA CGA TC-3′ | |
| OXA-F | 5′-GCG TGG TTA AGG ATG AAC AC-3′ | 389 |
| OXA-R | 5′-CAT CAA GTT CAA CCC AAC CG-3′ | |
| VIM-F | 5′-TTTGGTCGCATATCGCAACG-3′ | 500 |
| VIM-R | 5′-CCATTCAGCCAGATCGGCAT-3′ | |
| CTX-M F | 5′-SCSATGTGCAGYACCAGTAA-3′ | 543 |
| CTX-M R | 5′-CCGCRATATGRTTGGTGGTG-3′ | |
| TEM-F | 5′-TCGGGGAAATGTGCGCG-3′ | 971 |
| TEM-R | 5′-TGCTTAATCAGTGAGGCACC-3′ | |
| SHV-F | 5′-TTATCTCCCTGTTAGCCACC-3′ | 948 |
| SHV-R | 5′-GATTTGCTGATTTCGCTCGG-3′ |
Table 2.
PCR amplification conditions
| Primer | Initial denaturation | Denaturation 2 | Annealing | Extension 1 | Final extension | Cycles, n |
|---|---|---|---|---|---|---|
| RAPD | 95°C for 5 min | 95°C for 1 min | 36°C for 1 min | 72°C for 2 min | 72°C for 15 min | 40 |
| ERIC-PCR | 94°C for 7 min | 94°C for 30 s | 50°C for 1 min | 72°C for 3 min | 72°C for 15 min | 30 |
| NDM | 94°C for 10 min | 94°C for 30 s | 52°C for 40 s | 72°C for 50 s | 72°C for 5 min | 36 |
| OXA | 95°C for 5 min | 94°C for 45 s | 52°C for 1 min | 72°C for 1 min | 72°C for 10 min | 36 |
| CTX-M | 95°C for 5 min | 94°C for 45 s | 55°C for 1 min | 72°C for 1 min | 72°C for 5 min | 30 |
| TEM | 95°C for 5 min | 94°C for 45 s | 55°C for 1 min | 72°C for 1 min | 72°C for 5 min | 30 |
| SHV | 95°C for 5 min | 94°C for 30 s | 55°C for 2 min | 72°C for 1 min | 72°C for 10 min | 30 |
| VIM (touchdown PCR) | 95°C for 10 min | 95°C for 30 s | 69°C to 61°C (30 s) | 72°C for 30 s | 72°C for 10 min | 2 at each annealing temperature (18 cycles in total) |
PCR Amplification of β-Lactamase Genes
PCR amplification of the CTX-M, TEM, SHV, NDM, VIM, and OXA families of genes among the 21 carbapenem-resistant E. coli isolates was performed in a final volume of 50 μl containing PCR Master Mix – K0171 (25 μL; Thermo Scientific, USA), 10 pmol of each primer, and 50 ng of the template DNA. PCR amplifications were performed using primers (Table 1) and conditions as mentioned in Table 2. NDM- and OXA-positive amplicons were purified using a GeneJET PCR Purification Kit (Thermo Scientific). Sequencing was performed to identify the variants of the NDM and OXA amplicons only, while only PCR amplification was performed to confirm the presence of CTX-M, TEM, and SHV. Template DNA extracted from E. coli ATCC 25922, sensitive to carbapenem and cephalosporins, was used as a negative control.
Nucleotide Sequence Accession Numbers
The nucleotide sequence data reported in the current study has been assigned by the GenBank database under the accession numbers KY817193.1-KY817205.1 (NDM), MG865741-MG865744 (NDM), KY777229.1, KY777228.1, KY801334.1, KY817191.1, and KY817192.1 (OXA).
Conjugal Transfer
The DNA transfer capacity of the study isolates was tested by selecting dual resistant transconjugants after broth mating experiments with a recipient plasmid-free, azide-resistant, conjugation-compatible E. coli J53AziR strain [22]. Exponentially growing cells (0.8 at OD600) were mixed at a ratio of 1: 1 (donor:recipient) and transconjugants were selected on LB agar containing ampicillin (100 μg/mL) and sodium azide (100 μg/mL). The presence of bla genes in the transconjugants was confirmed by PCR amplification of the respective genes.
Results
Clinical Characteristics
Twenty of the isolates were obtained from patients with UTI, while 1 sample was obtained from surgical site pus of a 78-year-old patient who presented to the Department of Orthopedics with an intertrochanteric fracture. The age of the patients ranged from 2 to 78 years. Five of the patients were female while the remaining 16 were male. Urine from a catheter was collected from 4 of the patients, while it was collected from right kidney in 1 patient. In all of the other patients, mid-stream urine was collected for culture. Clinical abnormalities in these patients included intertrochanteric fracture, calculus, voiding dysfunction, hyperplasia of the prostate, and urethral stricture among other structural and functional abnormalities of the urinary tract and kidney (Table 3).
Table 3.
Clinical details of the study patients
| Isolate | Age, years | Gender | Specimen | Diagnosis |
|---|---|---|---|---|
| VIR 1 | 57 | F | fresh catheter urine | urinary tract infection |
| VIR 2 | 53 | M | midstream urine | calculus in the urethra |
| VIR 3 | 62 | M | midstream urine | calculus of the kidney |
| VIR 4 | 18 | M | midstream urine | voiding dysfunction |
| VIR 5 | 61 | M | midstream urine | diverticulum of the bladder |
| VIR 6 | 51 | M | suprapubic catheter urine | hyperplasia of the prostate, obstructive and reflux uropathy |
| VIR 7 | 4 | F | midstream urine | obstructive and reflux uropathy |
| VIR 8 | 66 | M | midstream urine | hyperplasia of the prostate |
| VIR 9 | 49 | M | right kidney urine | calculus of the kidney with calculus of the ureter |
| VIR 11 | 2 | M | midstream urine | urethral stricture |
| VIR 12 | 27 | M | midstream urine | redundant prepuce, phimosis, and bladder-neck obstruction |
| VIR 13 | 26 | F | midstream urine | multiple left secondary calculus |
| VIR 14 | 12 | M | fresh catheter urine | calculus of the ureter |
| VIR 16 | 55 | M | midstream urine | congenital posterior urethral valves |
| VIR 17 | 51 | M | suprapubic catheter urine | vesicoureteral-reflux-associated uropathy |
| VIR 18 | 52 | F | midstream urine | trauma with complete urethral stricture |
| VIR 19 | 66 | M | midstream urine | urethral stricture |
| VIR 20 | 4 | F | midstream urine | urethral stricture |
| VIR 21 | 65 | M | midstream urine | grade 1 prostatomegaly with significant postvoid residue |
| VIR 22 | 74 | M | fresh catheter urine | obstructive and reflux uropathy |
| VIR 25 | 78 | M | surgical site pus | intertrochanteric fracture |
Twelve of the patients were found to have community-acquired infections, while 9 patients had nosocomial infections. The demographic details of the patients include the following data: West Bengal (n = 6), Karnataka (n = 1), Orissa (n = 3), Telangana (n = 1), and Andhra Pradesh (n = 10). Eight of the patients from Andhra Pradesh belong to the Anantapur district.
Strain Characterization and Profiling of Antibiotic Resistance
All study isolates were confirmed as E. coli by Vitek-2 and 16S rDNA sequencing. Antibiotic sensitivity analyses revealed that the isolates were resistant to penicillins (ampicillin and ampicillin-clavulanic acid), cephalosporins (cefuroxime, ceftriaxone, cefoperazone-sulbactam, and cefepime), carbapenems (ertapenem, imipenem, and meropenem), and quinolones (ciprofloxacin and nalidixic acid). More than 52% (11 isolates against amikacin and 15 isolates against gentamycin) of the isolates were resistant to aminoglycosides. Seventeen (81%) were found to be resistant to co-trimoxazole, while only 4 isolates were resistant against nitrofurantoin. All of the isolates except Vir-4 (intermediate resistance) were found to be sensitive to tigecycline. All of the isolates were sensitive to colistin (online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000489885). The MBL-producing capacity of the isolates was confirmed by double disc synergy testing involving imipenem and imipenem-EDTA discs.
All of the isolates were found to carry NDM and/or OXA carbapenemase genes. NDM was found in 17 (81%) of the isolates, while OXA-181 was present in 5 (24%). Among the isolates positive for the NDM gene, NDM-5 was prevalent in 12 isolates, while NDM-7 was found in 3 isolates; NDM-4 and NDM-6 variants were present in 1 isolate each. One isolate, i.e., Vir14, was found to possess both NDM-4 and OXA-181. VIM was not identified in any of the 21 isolates. Of the 21 E. coli isolates, 13 (62%) were found to possess the CTX-M type of ESBL, while TEM β-lactamases were found in 20 (95%). Twelve of the 21 (57%) isolates were found to concomitantly encode CTX-M and TEM genes. None of the isolates were found to possess SHV ESBL's.
Transmissibility of the Resistance Determinants and Genetic Relatedness
After mating the study isolates with the recipient EJ53 E. coli, the resultant transconjugants resistant to sodium azide (100 μg/mL) and ampicillin (100 μg/mL) were screened for the presence of CTX-M, TEM, NDM, and OXA β-lactamases. PCR amplification of genomic DNA from the transconjugants revealed the presence of the NDM gene in 9 isolates, OXA in 1 isolate, CTX-M in 2 isolates, and TEM in 13 isolates. Analyses of the transconjugants of the 6 isolates (Vir-5, Vir-12, Vir-13, Vir-17, Vir-19, and Vir-20) did not reveal the presence of any of the four β-lactamases, indicating that they may not be on mobilizable elements or, because the selection of the transformants was performed in the presence of ampicillin, they may not have been transferred (Table 4). Further analyses are required to characterize the mobilizable elements of the study isolates.
Table 4.
Presence of bla genes in the E. coli isolates and their transconjugants
| Isolate | CTX-M | TEM | OXA | NDM | Transconjugants |
|||
|---|---|---|---|---|---|---|---|---|
| CTX-M | TEM | NDM | OXA | |||||
| vir 1 | negative | positive | negative | NDM 6 | positive | positive | ||
| vir 2 | positive | positive | negative | NDM 7 | positive | |||
| vir 3 | positive | positive | negative | NDM 5 | positive | |||
| vir 4 | negative | positive | negative | NDM 5 | positive | |||
| vir 5 | positive | positive | positive | negative | ||||
| vir 6 | negative | positive | negative | NDM 5 | positive | positive | ||
| vir 7 | negative | positive | negative | NDM 5 | positive | positive | ||
| vir 8 | positive | positive | positive | negative | positive | |||
| vir 9 | positive | positive | positive | negative | positive | |||
| vir 11 | positive | positive | negative | NDM 7 | positive | positive | positive | |
| vir 12 | positive | positive | negative | NDM 5 | ||||
| vir 13 | positive | positive | negative | NDM 5 | ||||
| vir 14 | positive | positive | positive | NDM 4 | positive | |||
| vir 16 | negative | positive | negative | NDM 7 | positive | |||
| vir 17 | negative | positive | negative | NDM 5 | ||||
| vir 18 | positive | positive | negative | NDM 5 | positive | positive | positive | |
| vir 19 | positive | negative | negative | NDM 5 | ||||
| vir 20 | negative | positive | negative | NDM 5 | ||||
| vir 21 | positive | positive | positive | negative | positive | |||
| vir 22 | positive | positive | negative | NDM 5 | positive | positive | ||
| vir 25 | negative | positive | negative | NDM 5 | positive | positive | ||
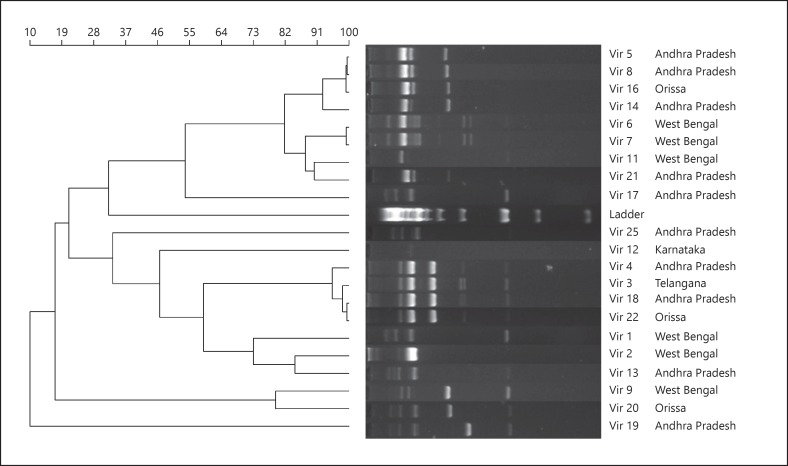
All of the 21 E. coli isolates were analyzed for their genetic relatedness using both ERIC-PCR and RAPD with 3 primers. Our analyses revealed that RAPD using the primer OPA-2 was able to provide a higher discrimination compared to the ERIC-PCR method. Although the ERIC-PCR method showed the presence of 11 clusters (data not provided), we found that the majority of the isolates (Vir-9, Vir-7, Vir-16, Vir-12, Vir-22, Vir-21, Vir-20, and Vir-18) were grouped together into a single cluster as ERIC-PCR could not produce good banding patterns for these isolates. On the other hand, RAPD using the OPA-2 primer was able to produce satisfactory banding patterns in the majority of the isolates (Fig. 1). A total of 6 clusters were found by RAPD analysis. In the first cluster, 4 isolates, i.e., Vir-5, Vir-8, Vir-16, and Vir-14, were found. Of these, Vir-14 branched out, indicating its divergence from the remaining 3 isolates. Incidentally, Vir-14 was the only isolate to possess CTX-M, TEM, NDM-4, and OXA-181 β-lactamases. The second cluster contained Vir-6 and Vir-7, both isolated from patients hailing from West Bengal. The third cluster had Vir-11 and Vir-21. The fourth cluster had Vir-4, Vir-3, Vir-18, and Vir-22 branching together. In the fifth cluster, Vir-2, Vir-13, and Vir-1 were found together. Vir-1 was found to further branch out from the remaining 2 isolates. The last cluster had Vir-9 and Vir-20. Vir-19 was found to be different from the remaining isolates based on its unique banding pattern. The RAPD banding pattern was not satisfactory among 3 isolates (Vir-17, Vir-25, and Vir-12) and so they could not be clustered with the other isolates.
Fig. 1.
RAPD profile of 21 carbapenem-resistant E. coli using the OPA-2 primer. The dendrogram was generated using UPGMA the method from the RAPD band patterns with the Opa-2 primer for the 21 CRE E. coli isolates.
Discussion
The emergence of multidrug-resistant E. coli and their rapid spread and ability to continuously evolve pose a serious threat to global health [5, 23, 24]. Carbapenemase-producing Enterobacteriaceae due to the presence of NDM and OXA β-lactamases have been reported in various medical centers globally as well as in India [25, 26, 27, 28, 29]. The spread of NDM-producing Enterobacteriaceae from the Indian subcontinent through visiting tourists has been documented in 2010 [13]. Ninety-five percent of the isolates at our center were uropathogenic E. coli (data not included) with high levels of antibiotic resistance. Similar trends were reported from 2 large hospitals located in Kuwait [30]. In this paper, 21 carbapenem-resistant E. coli were analyzed for the presence of ESBL (CTX-M, TEM, and SHV) and carbapenemase (NDM, OXA, and VIM) genes. The demographics of the patients indicate the widespread presence of CRE E. coli in community settings. Further, 57% of the study isolates were community acquired, indicating the spread of CRE in the general community, and they may not be solely hospital acquired. The AST pattern obtained from the VITEK 2 compact system of these isolates revealed that they are resistant to a majority of the antibiotics, except colistin, leaving clinicians with limited therapeutic options to treat the CRE infections. RAPD and ERIC-PCR revealed that the CRE E. coli involved in the current study belong to multiple distinct clades.
Eighty-one percent of the isolates were NDM positive, and nearly a quarter of the isolates were positive for OXA carbapenemases. Interestingly, 1 isolate (Vir-14) was positive for all of the 4 tested ESBL and carbapenemases. Since the first description of NDM, several variants of the gene have been reported [31]. More recently, 7 NDM (1–7) variants were described from an Indian health care center [29]. In the current study, the majority of the isolates 17 were found to possess the NDM gene. Of these, NDM-5 was found in 12 isolates, NDM-7 was detected in 3 isolates, and NDM-4 and NDM-6 variants were present in 1 isolate each. OXA-181 was first reported in 2011 [10]. All of the OXA genes in our population were found to be OXA-181. These findings confirm that NDM and OXA-181 are endemic to the Indian subcontinent and are highly prevalent in this region.
In the current study, all of the isolates were found to be compatible for conjugal transfer when mated with sodium-azide-resistant, plasmid-deficient E. coli J53 [22, 29]. ESBL and/or carbapenemases were detected in 15 (71%) of the transconjugants, indicating that the majority of these resistance determinants are transferable and capable of bestowing enhanced resistance features to the bacterial host. These findings suggest that NDM, TEM, and CTX-M genes are major causes of ESBL and carbapenem resistance in the Indian subcontinent and the predominant β-lactamase among our isolates is TEM (95%) [9].
These pathogens are a reservoir of antibiotic resistance genes and, together with their capacity to disseminate via horizontal gene transfer, they pose a grave threat to infection control strategies, with very few treatment options. Further, the spread of these resistance determinants in environmental samples should be a cause for alarm. A wide range of bacteria including E. coli, Klebsiella pneumoniae, Citrobacter freundii, Shigella boydii, Vibrio cholerae, and Aeromonas caviae harboring the blaNDM-1 gene were isolated from 2 drinking water and 12 seepage samples [32]. Consistent with previous reports, our analysis revealed the presence of non NDM-1 variants (NDM-4, NDM-5, NDM-6, and NDM-7) among patients with serious urological disorders [29]. If this increase in antibiotic resistance continues and spreads to bacteria which were previously sensitive and easily treatable, it may lead us to dire straits without any available treatment options for simple infections.
Conclusion
Molecular epidemiology of carbapenem-resistant E. coli harboring multiple resistance-encoding genes is rapidly evolving. In this study, CRE E. coli were predominantly isolates from community-acquired clinical samples and the pathogens were found to express resistance against a variety of antibiotics except colistin. CTX-M, TEM, NDM, and OXA were identified among the 21 study isolates. Isolates positive for OXA and/or NDM were found to harbor genes encoding other resistance determinants. NDM-5 and OXA-181 were found to be prevalent in the study isolates. The widespread coexistence of ESBL and carbapenemases on transferrable elements is a major clinical challenge for control of infections.
Disclosure Statement
None.
Funding Sources
This research project received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Supplementary Material
Supplementary data
Acknowledgement
We thank the Department of Microbiology of Sri Sathya Sai Institute of Higher Medical Sciences for providing the specimen and clinical information. We greatly acknowledge Dr. Kashinath Prasad (Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India) for providing the E. coli J53AzR strain. We acknowledge UGC-SAP (DRS), DST-FIST, and DBT-BIF, and the Government of India for infrastructural support to the Department of Biosciences of the Sri Sathya Sai Institute of Higher Medical Sciences.
References
- 1.Morrissey I, Hackel M, Badal R, et al. A review of ten years of the Study for Monitoring Antimicrobial Resistance Trends (SMART) from 2002 to 2011. Pharmaceuticals. 2013;6:1335. doi: 10.3390/ph6111335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez-Baño J, Alcalá JC, Cisneros JM, et al. Community infections caused by extended-spectrum β-lactamase-producing Escherichia coli. Arch Intern Med. 2008;168:1897–1902. doi: 10.1001/archinte.168.17.1897. [DOI] [PubMed] [Google Scholar]
- 3.Kang HY, Jeong YS, Oh JY, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005;55:639–644. doi: 10.1093/jac/dki076. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lascols C, Hackel M, Marshall SH, et al. Increasing prevalence and dissemination of NDM-1 metallo-β-lactamase in India: data from the SMART study (2009) J Antimicrob Chemother. 2011;66:1992–1997. doi: 10.1093/jac/dkr240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents. 2010;36:S8–S14. doi: 10.1016/S0924-8579(10)70004-2. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Wang Y, Walsh TR, et al. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother. 2017:p 61. doi: 10.1128/AAC.02233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolejska M, Villa L, Poirel L, et al. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J Antimicrob Chemother. 2013;68:34–39. doi: 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Ros A, Carricajo A, et al. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob Agents Chemother. 2011;55:447–448. doi: 10.1128/AAC.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castanheira M, Deshpande LM, Mathai D, et al. Early Dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother. 2011;55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh SK, Gupta M. BlaOXA-48 carrying clonal colistin resistant-carbapenem resistant Klebsiella pneumoniae in neonate intensive care unit, India. Microb Pathog. 2016;100:75–77. doi: 10.1016/j.micpath.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediat ed carbapenem resistance. J Med Microbiol. 2013;62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 13.Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement: CLSI document M100-S22. Wayne, Clinical and Laboratory Standards Institute, 2012.
- 15.Dongeun Yong KL, Yum JW, Shin HB, et al. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40:3798–3801. doi: 10.1128/JCM.40.10.3798-3801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumari N, Thakur SK. Randomly amplified polymorphic DNA: a brief review. Am J Anim Vet Sci. 2014;9:6–13. [Google Scholar]
- 17.Venieri D, Vantarakis A, Komninou G, et al. Differentiation of faecal Escherichia coli from human and animal sources by random amplified polymorphic DNA-PCR (RAPD-PCR) Water Sci Technol. 2004;50:193–198. [PubMed] [Google Scholar]
- 18.Kärkkäinen UM, Kauppinen J, Ikäheimo R, et al. Random amplified polymorphic DNA (RAPD) analysis of Escherichia coli strains: comparison of urinary and concomitant blood isolates of urosepsis patients. APMIS. 1996;104:437–443. doi: 10.1111/j.1699-0463.1996.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilson LA, Sharp PM. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: evolution and implications for ERIC-PCR. Mol Biol Evol. 2006;23:1156–1168. doi: 10.1093/molbev/msj125. [DOI] [PubMed] [Google Scholar]
- 20.Ramazanzadeh R, Zamani S, Zamani S. Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)-PCR technique in Sanandaj hospitals. Iran J Microbiol. 2013;5:126–131. [PMC free article] [PubMed] [Google Scholar]
- 21.Ardakani MA, Ranjbar R. Molecular typing of uropathogenic Ecoli strains by the ERIC-PCR method. Electron Physician. 2016;8:2291–2296. doi: 10.19082/2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain A, Ewers C, Nandanwar N, et al. Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum- β-lactamase-producing lineage. Antimicrob Agents Chemother. 2012;56:6358–6365. doi: 10.1128/AAC.01099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Zarouni M, Senok A, Al-Zarooni N, et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae: in vitro susceptibility to fosfomycin, nitrofurantoin and tigecycline. Med Princ Pract. 2012;21:543–547. doi: 10.1159/000339200. [DOI] [PubMed] [Google Scholar]
- 24.Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Carrër A, Poirel L, Yilmaz M, et al. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother. 2010;54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goren MG, Chmelnitsky I, Carmeli Y, et al. Plasmid-encoded OXA-48 carbapenemase in Escherichia coli from Israel. J Antimicrob Chemother. 2011;66:672–673. doi: 10.1093/jac/dkq467. [DOI] [PubMed] [Google Scholar]
- 27.Ktari S, Mnif B, Louati F, et al. Spread of Klebsiella pneumoniae isolates producing OXA-48 β-lactamase in a Tunisian university hospital. J Antimicrob Chemother. 2011;66:1644–1646. doi: 10.1093/jac/dkr181. [DOI] [PubMed] [Google Scholar]
- 28.Livermore DM, Walsh TR, Toleman M, et al. Balkan NDM-1: escape or transplant? Lancet Infect Dis. 2011;11:164. doi: 10.1016/S1473-3099(11)70048-2. [DOI] [PubMed] [Google Scholar]
- 29.Rahman M, Shukla SK, Prasad KN, et al. Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Al Sweih N, Jamal W, Rotimi VO. Spectrum and antibiotic resistance of uropathogens isolated from hospital and community patients with urinary tract infections in two large hospitals in Kuwait. Med Princ Pract. 2005;14:401–407. doi: 10.1159/000088113. [DOI] [PubMed] [Google Scholar]
- 31.Sassi A, Loucif L, Gupta SK, et al. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob Agents Chemother. 2014;58:5606–5608. doi: 10.1128/AAC.02818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh TR, Weeks J, Livermore DM, et al. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data