Abstract
Objective
To characterize all 4 mutations described for CD59 congenital deficiency.
Methods
The 4 mutations, p.Cys64Tyr, p.Asp24Val, p.Asp24Valfs*, and p.Ala16Alafs*, were described in 13 individuals with CD59 malfunction. All 13 presented with recurrent Guillain-Barré syndrome or chronic inflammatory demyelinating polyneuropathy, recurrent strokes, and chronic hemolysis. Here, we track the molecular consequences of the 4 mutations and their effects on CD59 expression, localization, glycosylation, degradation, secretion, and function. Mutants were cloned and inserted into plasmids to analyze their expression, localization, and functionality.
Results
Immunolabeling of myc-tagged wild-type (WT) and mutant CD59 proteins revealed cell surface expression of p.Cys64Tyr and p.Asp24Val detected with the myc antibody, but no labeling by anti-CD59 antibodies. In contrast, frameshift mutants p.Asp24Valfs* and p.Ala16Alafs* were detected only intracellularly and did not reach the cell surface. Western blot analysis showed normal glycosylation but mutant-specific secretion patterns. All mutants significantly increased MAC-dependent cell lysis compared with WT. In contrast to CD59 knockout mice previously used to characterize phenotypic effects of CD59 perturbation, all 4 hCD59 mutations generate CD59 proteins that are expressed and may function intracellularly (4) or on the cell membrane (2). None of the 4 CD59 mutants are detected by known anti-CD59 antibodies, including the 2 variants present on the cell membrane. None of the 4 inhibits membrane attack complex (MAC) formation.
Conclusions
All 4 mutants generate nonfunctional CD59, 2 are expressed as cell surface proteins that may function in non–MAC-related interactions and 2 are expressed only intracellularly. Distinct secretion of soluble CD59 may have also a role in disease pathogenesis.
Complement activation triggers membrane attack complex (MAC) assembly to form pores in cell membrane lipid bilayers of susceptible bacteria.1 However, unregulated MAC formation may cause host tissue damage.2 The glycosyl phosphatidylinositol (GPI)-anchored cell surface membrane glycoprotein CD59 inhibits the final step of MAC formation to protect host cells from MAC-mediated injury.3
Several mutations in the CD59-coding sequence are known in human patients (figure 1A). We and others have previously reported of 13 patients4–11 aged 1–4.5 years who suffered from chronic hemolysis and recurrent episodes of Guillain-Barré syndrome (GBS)-like disease from early infancy, suggesting chronic inflammatory demyelinating polyradiculoneuropathy (CIDP), and recurrent strokes (table e-1, links.lww.com/NXG/A87). The mutations included p.Cys64Tyr, p.Asp24Val, p.Asp24Valfs*, and p.Ala16Alafs*, all leading to CD59 loss of function. In all mutations, no surface protein was detected by anti-CD59 antibody staining. We were interested, in the current study, to verify whether indeed no proteins were produced, whether any proteins that were produced reached the membrane, and whether proteins or truncated proteins exist intracellularly or secreted outside the cells. These potential differences may have functional implications and clinical manifestations.
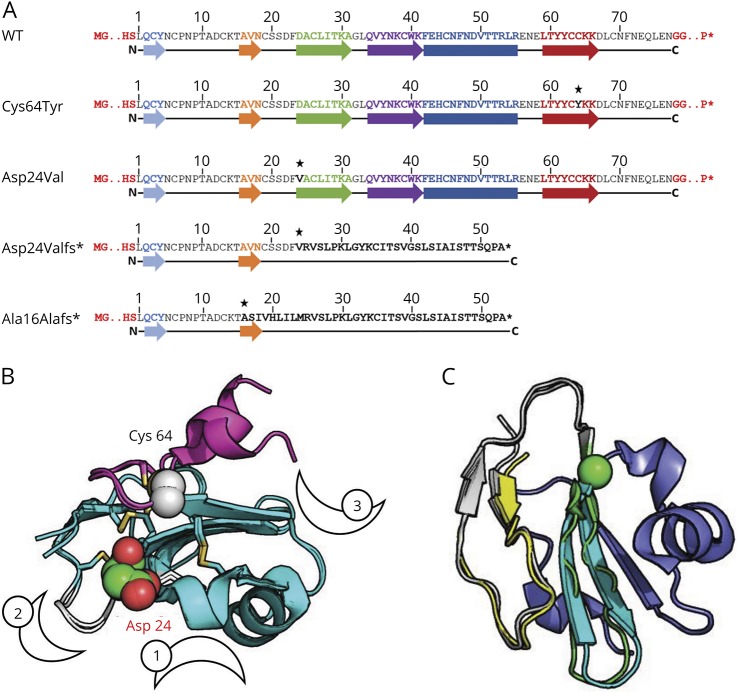
Figure 1. Sequence and structure of CD59 WT and mutants.
(A) The sequences of WT CD59, point mutations Cys64Tyr and Asp24Val, and frameshift mutations Asp24Valfs* and Ala16Alafs*: The mature membrane surface CD59 primary sequence consists of 77 residues after removal of a 25-residue N-terminal signal sequence and a C-terminal GPI-anchoring signal (not shown). Colored arrows and boxes represent β-strands and α-helices, respectively, as observed in WT CD59 (see B). Point mutations are marked with an asterisk. The sequence of the 2 frameshift mutants reveals significantly shortened proteins that lack most of the CD59 sequence, in particular the GPI-anchoring signal. (B) The structure of CD59 highlights the vicinity of the point mutations to known sites of activity and interaction. The mutated Cys64 and Asp24 positions are highlighted as spheres and labeled. The 3 characterized interfaces of CD59 are marked by numbered crescents: (1) The classic site characterized originally described by Bodian et al.21 with the central Trp40 residue shown in sticks, (2) a loop spanning residues 20–24 that modulates CD59 activity20 (colored in white), and (3) the edge β-strand that interacts with ILY18 (to the left of the crescent). Disulfide bridges that stabilize CD59 are shown in sticks. Two solved structures (2j8b and 2uwr) are shown, highlighting the relative flexibility of the C-terminal helix (colored in magenta). (C) The frameshift mutants contain only a small part of the original CD59 sequence: The structure of CD59 is shown, with the region with a sequence in common with the Ala16Alafs* and Asp24Valfs* mutants colored in yellow and yellow-white, respectively. The I-TASSER model for the Asp24Valfs* sequence aligns 2 additional β-strands (colored in green, ending with the sphere at CD59 position Trp40), before it diverges, and the remaining part (in blue) is unique to CD59. GPI = glycosyl phosphatidylinositol; WT = wild type.
Methods
Generation of myc-tagged wild-type (WT) and mutant hCD59 expression plasmids
To verify the membrane and intracellular localization of the mutated protein, we isolated, myc-tagged, and expressed WT CD59 and each mutant in mammalian cell lines. Total RNA was extracted from human white blood cell samples using TRI Reagent (Sigma, St Louis, MO). cDNAs generated by SuperScript II Reverse Transcriptase (Life Technologies, Carlsbad, CA) from RNAs isolated from both a Cys64Tyr homozygous patient and a healthy individual were used as templates for PCR reactions (details appear in a supplementary section of Methods links.lww.com/NXG/A88).
Antibodies
To test staining of WT and mutant CD59 by anti-CD59 antibodies, human CD59 antibodies were used in immunofluorescence labeling assays of overexpressing COS7 cells and detected by flow cytometry and fluorescent microscopy. Details appear in supplementary section.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting and immunoprecipitation
SDS-PAGE, Western blot, and immunoprecipitation were used for CD59 detection (details appear in the supplementary section).
Bortezomib (proteasome inhibitor) treatment
To test the effect of ubiquitination on expression, we inhibited ubiquitination by bortezomib. Forty-eight hours after transfection, culture medium was supplemented with 10 nM bortezomib (Velcade, Cell Signaling) or vehicle-only control. A mouse antibody against the myc-tag peptide was used for Western blot analysis.
In vitro cell lysis assay assessing MAC attack in WT and mutants
To evaluate deposition of MAC and cell lysis, we used in vitro cell lysis assays. Transfections of Chinese hamster ovary (CHO) cells were performed using Lipofectamine 2000 reagent. Details appear in the supplementary section. Cells were harvested 24 hours after transfection, replated on 24-well plates (3 × 105 cells in 1 mL per well), and left until the plates were confluent. Cells were then washed twice with serum-free medium and incubated in 0.25 mL complete medium (7.5% FCS heat inactivated) containing a 1/750 dilution of calcein AM (Molecular Probes, Eugene, OR, 1 mg/mL stock in dimethyl sulfoxide) for 1 hour at 37°C. After 1 wash with phosphate buffered saline (PBS), duplicate wells were incubated with 1/5 dilution of normal human serum and 40 μL of rabbit anti-CHO immunoglobulin G (IgG). After incubation for 1 hour at 37°C, all fluid was removed from the cells and transferred to 96-well plates for calcein measurement.
The remaining cells were lysed with 0.25 mL PBS containing 0.1% Triton X-100 during a 15-minute incubation period at room temperature, and the lysate was removed to 96-well plates for calcein release measurement. Calcein fluorescence of supernatants was read in a Cytation 3 Cell Imaging Multi-Mode Reader (BioTek, Winoosky, VT) with excitation and emission filters set at 485 and 530 nm, respectively. Percent lysis for each well was calculated as calcein release/total calcein loading. Mean values and SDs were determined from duplicate samples. Analysis of variance (ANOVA) and Student t test were used for statistical analysis.
Structural biology
Pymol (Schrödinger LLC, New York, NY) was used for inspection of structures and figure generation. Models of the frameshift peptide sequences were generated using the I-TASSER server.12–14
Standard protocol approvals, registrations, and patient consents
Human DNA and tissue samples were obtained in protocols approved by our institutional review board. All patients or their families provided informed consent.
Data availability
Study data for the primary analyses presented in this report are available upon reasonable request from the corresponding and senior author.
Results
Predicted effect of CD59 mutations on its 3D protein structure
Figure 1 shows the sequences (figure 1A) and structures (figure 1, B and C) of CD59 with residues mutated in the 2 described point mutations (figure 1B). The similarities between the 2 frameshift mutants are highlighted (figure 1C). The mature membrane surface–expressed CD59 sequence consists of 77 residues after removal of a 25-residue N-terminal signal sequence and a C-terminal GPI-anchoring signal.15
Inspection of solved structures of CD59,16 pdb IDs 2j8b, 2uwr, 2ux2,17 4bik,18 and 2ofs19 shows that in the p.Cys64Tyr mutant, a local disulfide bridge near the carboxy terminus of the mature protein is interrupted (Cys64-Cys69) (figure 1B). This would probably merely loosen the loop that is followed by the C-terminal helix, which adopts different orientations depending on the solved structure (e.g., 2j8b vs 2uwr), indicating that it is not strongly stabilized in the WT structure. As for p.Asp24Val, Asp24 was previously reported to play an important role in CD59 stability and activity: p.Asp24Ala strongly destabilizes CD59,20 whereas p.Asp24Arg permits protein folding and expression, but abolishes CD59 activity.21 Indeed, Asp24 is part of an exposed loop (Ser20-Asp24), located between the second and third β-strands (figure 1B), that is involved in modulating CD59 activity. Single mutations to alanine in that loop increased, whereas multiple alanine substitutions decreased its activity.20 The 2 other mutants, p.Ala16Alafs and p.Asp24Valfs, contain frameshifts that lead to premature termination of the protein chain (figure 1A). The frameshift mutants result in two 53 residue proteins with very different structure compared with WT CD59, but their sequences are similar (figure 1A), including a common C-terminus that lacks the GPI anchor attachment signal and thus cannot attach to the membrane. Consequently, neither is expected to successfully proceed via the Golgi apparatus to the cell surface. The resulting proteins most probably do not fold into a stable structure, in particular the Ala16Alafs* mutation (figure 1C), as estimated by the low quality of the models predicted by the I-TASSER server12–14 (see Methods).
Western blot analysis of CD59 mutants
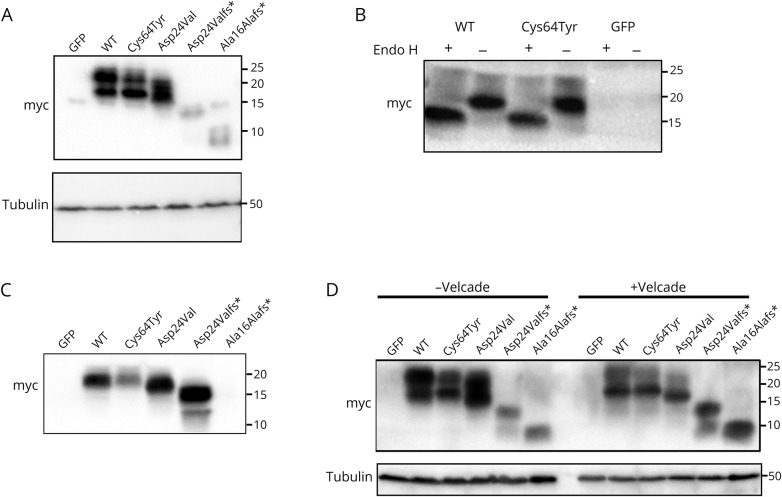
To characterize the expression of CD59 mutants, we performed Western blot analysis. We myc-tagged WT and all 4 CD59 mutants, p.Cys64Tyr, p.Asp24Val, p.Asp24Valfs*, and p.Ala16Alafs*, as well as GFP as a control, and expressed each in HEK293T cells. Lysates were separated in SDS-PAGE gels and immunoblotted with the anti-myc antibody. Western blot analysis of whole-cell lysate probed with the anti-myc antibody revealed similar patterns of the myc antibody for WT CD59 myc protein and the missense mutant p.Cys64Tyr myc protein. In both, 2 bands were seen, 1 between 20 and 25 kDa and the other between 15 and 20 kDa (figure 2A). The missense mutant p.Asp24Val myc protein appeared as bands between 15 and 20 kDa. For frameshift mutations p.Asp24Valfs* and p.Ala16Alafs*, very weak bands between 10 and 15 kDa were identified (figure 2A). To test whether the 2 bands seen in WT and p.Cys64Tyr proteins result from N-glycosylation in the Golgi apparatus, cell lysates were treated with Endo H. The data showed that the 20-kDa band represents glycosylated protein, and the Cys64Tyr mutant is glycosylated to the same extent as the WT protein (figure 2B).
Figure 2. Western blot profile.
See Methods for details. Anti-myc was used in all assays for detection. (A) Western blot expression pattern of WT and mutants Cys64Tyr, Asp24Val, Asp24Valfs*, and Ala16Alafs*, as well as GFP. Anti-myc was used for detection (upper panel) and anti-tubulin as a loading control (lower panel). (B) Western blot expression pattern of WT and Cys64Tyr mutant and GFP after using the Endo H enzyme. (C) Western blot expression pattern of conditioned medium. (D) Proteosomal degradation profile. Transfected cells treated with bortezomib (Velcade) and lysate sample were separated on 15% polyacrylamide gels. PVDF membranes were used for protein transfer. Anti-tubulin was used as a loading control (lower panel). PVDF = polyvinylidene difluoride; WT = wild type.
p.Cys64Tyr reduces the ability of CD59 to be secreted from the cell
Different processes, including cell damage and activation, induce the release of membrane-anchored proteins from the cell surface (shedding) or secretion from within the cell. CD59 can be released into the circulation or the interstitial fluid in a soluble form (sCD59) and can be detected in various body fluids including urine, milk, serum, and plasma.22–25 Various studies have designated sCD59 as biomarker for disease activity. Elevated circulating sCD59 concentrations have been found in acute myocardial infarction, and higher serum titers of glycated sCD59 have been described in diabetes mellitus.26 To examine whether CD59 is secreted, we checked the conditioned media of HEK293T cells transfected with all the above myc constructs for the presence of CD59. Conditioned media from WT, p.Cys64Tyr, p.Asp24Val, p.Asp24Valfs*, and p.Ala16Alafs* CD59, as well as GFP-transfected cells, were collected and subjected to immunoprecipitation using anti-myc antibodies. Western blot analysis of the precipitated proteins showed a single band between 15 and 20 kDa (equivalent to the glycosylated protein) for WT and p.Cys64Tyr and a single band of slightly lower MW for p.Asp24Val; p.Asp24Valfs* supernatant showed a single major band at ∼15 kDa, whereas no secreted protein was detected for p.Ala16Alafs*. Of note, much lower levels of secreted protein were detected for p.Cys64Tyr CD59 myc compared with WTCD59-myc (figure 2C). Both frameshift mutants have a signal peptide but lack the GPI anchor attachment signal and are therefore expected to be secreted from the cell. Of interest, whereas p.Asp24Valfs* was indeed secreted, no secretion was seen for p.Ala16Alafs* (figure 2C).
Proteosomal degradation profile
We further examined whether the CD59 mutants p.Cys64Tyr, p.Asp24Val, p.Asp24Valfs*, and p.Ala16Alafs* are abnormally degraded via the ubiquitin-proteasome pathway. For this, we treated the lysate of HEK293T cells transfected with the above myc-tagged mutants with bortezomib, a proteasome inhibitor. Cell lysates were separated by SDS-PAGE and immunoblotted with an anti-myc antibody (figure 2D). This experiment revealed that the p.Cys64Tyr myc and p.Asp24Val myc mutants, similar to the WT protein, do not accumulate in cells on inhibition of the proteasome. In contrast, the frameshift mutants, p.Asp24Valfs* myc and p.Ala16Alafs* myc, do accumulate, indicating that they are subject to proteasome degradation (figure 2D).
Intracellular and membrane localization of CD59 mutants
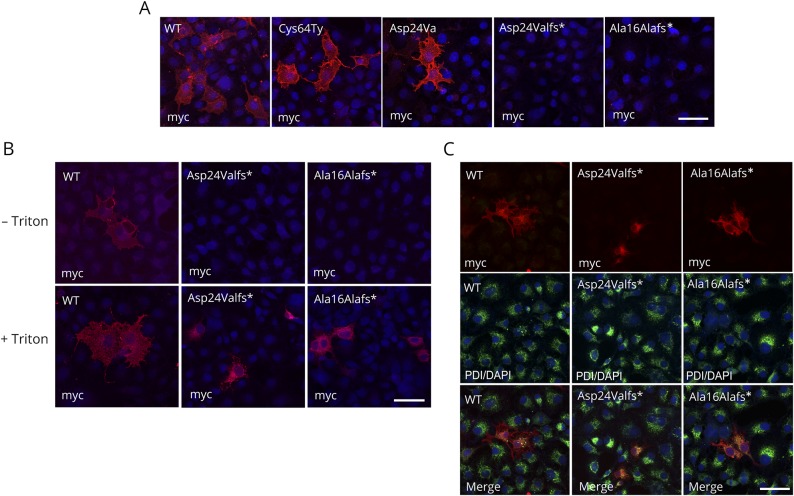
To examine CD59 localization, we expressed in COS7 cells myc-tagged WT CD59 and the 4 mutants of interest, as well as GFP as a negative control, and detected surface-expressed proteins by staining with the specific anti-myc antibody. In cells expressing WT, p.Cys64Tyr, and p.Asp24Val constructs, cell surface localization was detected by the myc antibody, whereras no surface staining was seen in cells expressing p.Asp24Valfs* and p.Ala16Alafs* constructs (figure 3A). Because we could not detect any surface staining in the 2-frameshift mutants, we repeated the immunolabeling procedures on fixed cells, with or without membrane permeabilization by Triton-X100. When immunolabeling procedures were performed without Triton, the myc antibody clearly detected WT CD59 on the cell surface, but not the frameshift mutants p.Asp24Valfs* and p.Ala16Alafs* (figure 3B upper panel). After Triton permeabilization, however, the myc antibody stained the 2 frameshift mutant CD59 proteins intracellularly, presumably in the endoplasmic reticulum (ER) (figure 3B lower panel). To verify the ER staining of the frameshift mutations, we double labeled the transfected COS7 cells with anti-myc and an anti-PDI antibody, an ER marker. Immunolabeling revealed clear colocalization demonstrating that both frameshift mutants localized to the ER (figure 3C). To summarize, although both missense mutants p.Cys64Tyr and p.Asp24Val are expressed on the cell surface, the frameshift mutants p.Asp24Valfs* and p.Ala16Alafs* fail to reach the cell surface and remain in the ER.
Figure 3. Surface and intracellular localization of hCD59 mutants.
(A) Characterization of WT and mutant Cys64Tyr, Asp24Val, Asp24Valfs*, and Ala16Alafs* constructs by myc antibody (red) and DAPI (blue). Staining with myc tag and secondary antibodies was added after fixation without Triton. (B) Characterization of WT and Asp24Valfs* and Ala16Alafs* CD59 mutants by anti-myc antibodies with or without Triton permeabilization. Differential recognition of WT and the Asp24Valfs* and Ala16Alafs* CD59 mutants by anti-myc. In both procedures, anti-myc detected the WT constructs with or without Triton, but in the mutant construct, ER staining was seen only with Triton. (C) Characterization of WT and Asp24Valfs* and Ala16Alafs* CD59 mutants by anti-myc and PDI antibodies. Upper panel: myc antibody (red); middle panel: PDI antibody (green) and DAPI (blue); and lower panel: merge. Cells were treated with methanol for permeabilization. Scale bars = 50 μm. PDI = protein disulfide isomerase; WT = wild type.
These results are in agreement with Western blot expression results (figure 2, C and D), indicating that the missense mutants are expressed and stable at the cell surface, whereas the frameshift mutants are retained in the ER, from which they are secreted from the cell (in the case of p.Asp24Valfs*) or sent to proteosomal degradation (both p.Asp24Valfs* and p.Ala16Alafs*).
CD59 mutants are not detected by anti-CD59 antibodies
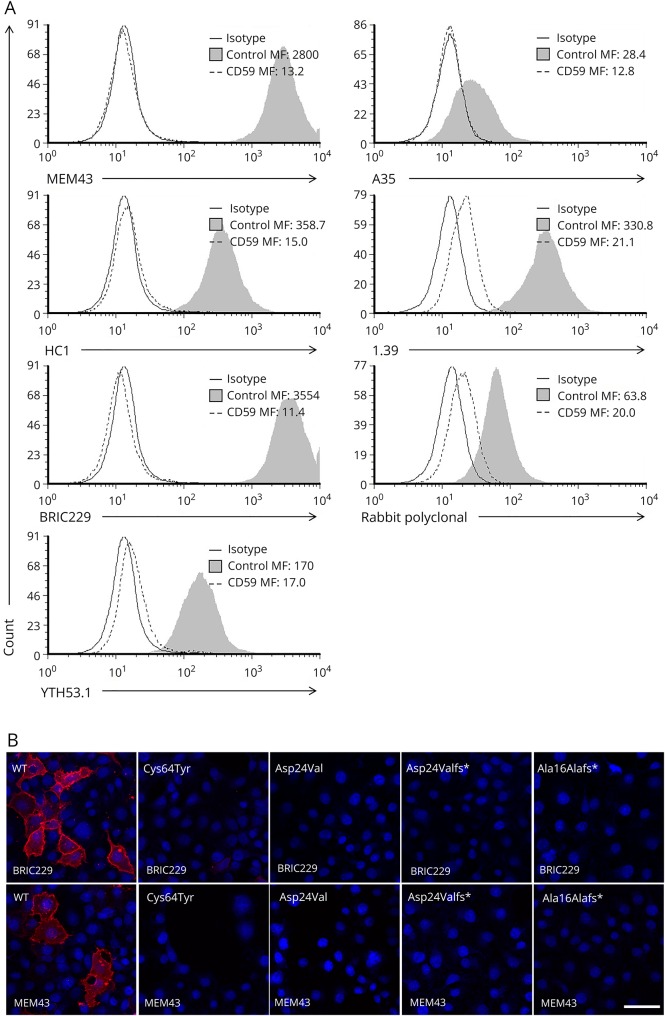
To investigate whether any of the available anti-CD59 antibodies detect the surface-expressed mutants, patient-derived Cys64Tyr-expressing and healthy control lymphoblasts were exposed to 5 different hCD59 monoclonal antibodies representing mapped epitopes MEM43, HC1, BRIC229, YTH53.1, and A35,21 1 monoclonal antibody with an unknown epitope, 1.39, and a rabbit polyclonal antiserum raised against human CD59. By flow cytometry, detection of CD59 by each of these 7 antibodies occurred only in the control lymphoblasts and not in lymphoblasts expressing CD59 mutants (figure 4A). We further verified these results using cells transfected with WT mutant, both missense mutants p.Cys64Tyr and p.Asp24Val and the 2 frameshift mutants p.Asp24Valfs* and p.Ala16Alafs* by immunofluorescence. Transfected cells were immunolabeled with each of the above human CD59 antibodies and, in parallel, with the anti-myc antibody. Immunolabeling with the monoclonal antibodies, MEM43, BRIC229 (figure 4B), HC1, 1.39 (figure e-2) and rabbit polyclonal antibodies (data not shown) revealed clear cell surface detection of the WT but not the mutant constructs (figure 4B). For technical reasons, A35 and YTH53.1 were not included in the staining. Results of the immunofluorescence labeling, including the myc antibody, are summarized in table e-2 (links.lww.com/NXG/A87). Taken together, known anti-CD59 antibodies did not detect surface expression of CD59 protein in either of the 2 missense mutations. This may suggest that the missense mutations result in substantial structural changes that abolish the protein's interaction with various antibodies and may similarly abolish protein-protein interactions necessary for its different functions.
Figure 4. Mutant detection by anti-CD59 antibodies using flow cytometry and fluorescent microscopy.
(A) A panel of anti-CD59 antibodies in Cys64Tyr and healthy control lymphoblasts: MEM43, HC1, BRIC229, YTH53.1, A35, 1.39, and rabbit polyclonal antibodies, assayed by flow cytometry. (B) Staining of WT and mutant constructs Cys64Tyr, Asp24Val, Asp24Valfs*, and Ala16Alafs* by BRIC229 or MEM43f (red), DAPI (blue). Procedures were performed by live staining without fixation and Triton treatment and assayed by fluorescent microscopy. Scale bars = 50 μm.
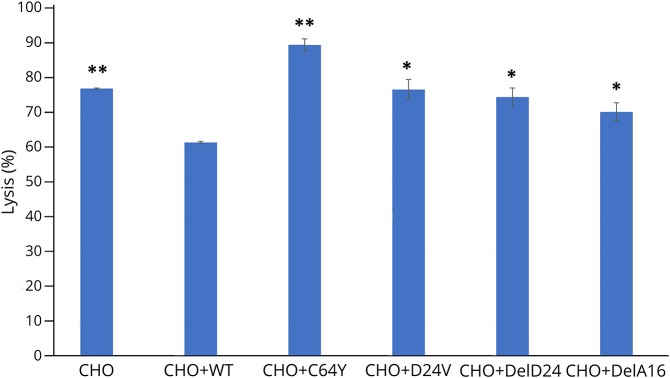
CD59 p.Cys64Tyr and p.Asp24Val mutants are nonfunctional in protecting from MAC attack
All 7 patients described with the p.Cys64Tyr mutation, and all 3 patients described with the Asp24Val mutation exhibited paroxysmal nocturnal hemoglobinuria (PNH)-like RBC hemolysis and a hypercoagulability state6,9 due to increased MAC formation. We expressed WT and mutant hCD59 constructs in the CHO cells and examined MAC-dependent lysis of the transfected cells. As shown in figure 5, WT CD59 transfection protected CHO cells from lysis (p < 0.05, ANOVA and Student t test); however, all mutants, including p.Cys64Tyr and p.Asp24Val, showed significantly increased MAC-dependent cell lysis (p < 0.05, ANOVA and Student t test for frameshift mutants and p.Asp24Val) compared with the level of MAC-dependent cell lysis in the presence of WT hCD59. For the p.Cys64Tyr mutant, this measured lysis (p < 0.002, ANOVA and Student t test) even surpassed the lysis of CHO cells without any additional expression, indicating perhaps an agonist effect on MAC-dependent lysis that needs further investigation.
Figure 5. Lysis functional assay in hCD59 mutants.
WT and mutant constructs Cys64Tyr, Asp24Val, Asp24Valfs*, and Ala16Alafs* were transfected to CHO cells. Forty-eight hours after transfection, cells were marked by calcein AM, and calcein fluorescence of supernatants was read using the Cytation 3 Cell Imaging Multi-Mode Reader with the excitation filter set at 485 nm and emission filter at 530 nm. Percent lysis for each well was calculated as calcein release/total calcein loading (*p < 0.05, **p < 0.002, ANOVA and Student t test). WT = wild type.
Taken together, we conclude that although both p.Cys64Tyr and p.Asp24Val mutant CD59 are surface expressed as demonstrated by myc staining, they are not detected by any of a panel of anti-CD59 antibodies using flow cytometry or immunofluorescence labeling and do not protect against MAC killing. The data indicate that these mutant proteins are misfolded and, similar to the frameshift mutations, show no CD59 function in relation to protection against MAC.
Discussion
Secondary CD59 deficiency is a common finding in patients with PNH. This condition is characterized by clonal expansion of hematopoietic stem cells that have acquired a somatic mutation in the PIGA gene (phosphatidylinositol N-acetylglucosaminyltransferase subunit A). PIGA encodes a GPI biosynthesis protein, phosphatidylinositol N-acetylglucosaminyltransferase subunit A.27,28 The CD59 protein inhibits the final and most important step of MAC formation. Erythrocytes that are deficient in GPI-anchored membrane proteins, including CD59, undergo complement-mediated hemolysis.29,30
The relationship of CD59 deficiency to the erythrocyte phenotypes of PNH has been established.31–34 Indeed, in a murine model, targeted deletion of the CD59 gene resulted in spontaneous intravascular hemolysis and hemoglobinuria, findings characteristic of PNH.35
PNH is a rare disease, affecting approximately 1 per million in Caucasian populations. Primary CD59 deficiency caused by germline mutations is extremely rare, described so far in 13 individuals globally, and characterized by hemolytic anemia (13/13), recurrent strokes (7/13), and peripheral demyelination (12/13) imitating recurrent GBS or CIDP.6,29,30
The 4 CD59 mutations described so far have similar clinical manifestations, despite large differences between the mutant proteins. The frameshift mutants p.Asp24Valfs* and p.Ala16Alafs* are predicted to generate truncated proteins with a distinct C-terminal sequence (common to both mutants) and as we show here undergo either ubiquitination or secretion and do not reach the membrane surface. In contrast, the mutants p.Cys64Tyr and p.Asp24Val do reach the cell surface but are nonfunctional in protecting the host from MAC attack; nevertheless, these proteins have the potential capability to perform non–MAC-related functions.
The MAC inhibitory function of CD59 is related to its capacity to bind C8 (within C5b-8) and block recruitment of C9, essential for MAC formation, accounting for the observed effect on lytic sensitivity.36,37 The region in CD59 responsible for interaction with these complement proteins has been defined by analysis of engineered CD59 mutants, and includes positions Trp40, Arg53, and Glu56, which colocalize to the glycosylated, membrane-distal face of its extracellular domain, and a distal position Asp24.21 Of note, the latter is the position mutated in the p.Asp24Val point mutation. In another study, this residue was shown to be part of a loop spanning residues 20–24 that effectively modulates CD59 activity; engineered mutations in this loop both reduced and increased CD59 activity, highlighting its role in fine-tuning CD59 function.20
A recently solved structure of a complex between the pathogen pore-forming intermedilysin (ILY) and CD59 (PDB ID 4bki)18 revealed another distinct binding site on CD59 that promotes polymerization of ILY on the cell surface and catalyzes pore formation.18 Cys64 is located in this third interface (figure 1B), suggesting that, in addition to a destabilizing effect leading to loss of recognition by antibodies and loss of MAC regulatory function, the Cys64Tyr mutation could also specifically affect other interactions. The fact that hCD59 Cys64Tyr overexpression caused increased MAC-dependent lysis beyond the control, in marked contrast to the lytic inhibitory effect of WT, may indicate an involvement of this interface in the regulation of MAC formation. The interface may also mediate functional interactions with complement or other players, such as activation of T cells38,39 and Islet cell secretion of insulin.40
The mutations reported in this study, together with previous studies on CD59 activity, highlight the versatility of CD59 activity and regulation. CD59 likely acts via at least 3 distinct interfaces (figure 1B): (1) The binding site was originally defined21 between the β-sheet and α-helices 1&2; (2) loop 20–24, which fine tunes activity20; and (3) the β-strand interface binding to the pathogen intermedilysin.18 The latter 2 are directly affected by the CD59 point mutations described in this study. In humans, and in contrast to CD59 knockout experiments in mice, the present study shows a much more complex picture of mutated CD59 expression and mode of action.
Acknowledgment
This research was supported by the Legacy Heritage BioMedical Program of the Israel Science Foundation (grant no. 1070/15 to D.M.). The authors wish to thank Shifra Fraifeld, Hadassah-Hebrew University Medical Center, for her editorial assistance.
Glossary
- CIDP
chronic inflammatory demyelinating polyradiculoneuropathy
- ER
endoplasmic reticulum
- GBS
Guillain-Barré syndrome
- MAC
membrane attack complex
- PNH
paroxysmal nocturnal hemoglobinuria
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- WT
wild type
Author contributions
D. Mevorach, E. Peles, and Y. Eshed-Eisenbach were responsible for study concept and design. N. Karbian, Y. Eshed-Eisenbach, A. Tabib, O. Schueler-Furman, and D. Mevorach were responsible for data acquisition and analysis. H. Hoizman participated in data acquisition. N. Karbian, Y. Eshed-Eisenbach, O. Schueler-Furman, B.P. Morgan, and D. Mevorach were responsible for drafting the manuscript and figures.
Study funding
This research was supported by the Legacy Heritage Biomedical Program of the Israel Science Foundation (grant no. 1070/15 to D.M.). No author has a financial relationship with any company in relation with this study.
Disclosure
N. Karbian, Y. Eshed-Eisenbach, A. Tabib, and H. Hoizman report no disclosures. P. Morgan has served on the scientific advisory boards of Roche, Alexion, and Achillion; has been a consultant for GSK (payments to university), Roche, Alexion, and Achillion; and has received research support from GSK, the Medical Research Council, and Wellcome Trust. O. Schueler-Furman reports no disclosures. E. Peles has served on the editorial boards of the Journal of Cell Biology, ASN NEURO, Molecular and Cellular Neuroscience, Neuronal and Glial Biology, Faculty of 1,000, and F1000 Research and has received research support from the NIH and the Israel Science Foundation. D. Mevorach has received consultation fees from Enlivex; has received research support from the Israel Science Foundation; and holds stock/stock options in Enlivex. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Podack ER. Molecular composition of the tubular structure of the membrane attack complex of complement. J Biol Chem 1984;259:8641–8647. [PubMed] [Google Scholar]
- 2.Morgan BP. Regulation of the complement membrane attack pathway. Crit Rev Immunol 1999;19:173–198. [PubMed] [Google Scholar]
- 3.Meri S, Morgan BP, Davies A, et al. . Human protectin (CD59), an 18,000-20,000 MW complement lysis restricting factor, inhibits C5b-8 catalysed insertion of C9 into lipid bilayers. Immunology 1990;71:1–9. [PMC free article] [PubMed] [Google Scholar]
- 4.Nevo Y, Ben-Zeev B, Tabib A, et al. . CD59 deficiency is associated with chronic hemolysis and childhood relapsing immune-mediated polyneuropathy. Blood 2013;121:129–135. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Zeev B, Tabib A, Nissenkorn A, et al. . Devastating recurrent brain ischemic infarctions and retinal disease in pediatric patients with CD59 deficiency. Eur J Paediatr Neurol 2015;19:688–693. [DOI] [PubMed] [Google Scholar]
- 6.Mevorach D, Reiner I, Grau A, et al. . Therapy with eculizumab for patients with CD59 p.Cys89Tyr mutation. Ann Neurol 2016;80:708–717. [DOI] [PubMed] [Google Scholar]
- 7.Ardicli D, Taskiran EZ, Kosukcu C, et al. . Neonatal-onset recurrent Guillain-Barre syndrome-like disease: clues for inherited CD59 deficiency. Neuropediatrics 2017;48:477–481. [DOI] [PubMed] [Google Scholar]
- 8.Hochsmann B, Dohna-Schwake C, Kyrieleis HA, Pannicke U, Schrezenmeier H. Targeted therapy with eculizumab for inherited CD59 deficiency. N Engl J Med 2014;370:90–92. [DOI] [PubMed] [Google Scholar]
- 9.Haliloglu G, Maluenda J, Sayinbatur B, et al. . Early-onset chronic axonal neuropathy, strokes, and hemolysis: inherited CD59 deficiency. Neurology 2015;84:1220–1224. [DOI] [PubMed] [Google Scholar]
- 10.Yamashina M, Ueda E, Kinoshita T, et al. . Inherited complete deficiency of 20-kilodalton homologous restriction factor (CD59) as a cause of paroxysmal nocturnal hemoglobinuria. N Engl J Med 1990;323:1184–1189. [DOI] [PubMed] [Google Scholar]
- 11.Motoyama N, Okada N, Yamashina M, Okada H. Paroxysmal nocturnal hemoglobinuria due to hereditary nucleotide deletion in the HRF20 (CD59) gene. Eur J Immunol 1992;22:2669–2673. [DOI] [PubMed] [Google Scholar]
- 12.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods 2015;12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudd PM, Morgan BP, Wormald MR, et al. . The glycosylation of the complement regulatory protein, human erythrocyte CD59. J Biol Chem 1997;272:7229–7244. [DOI] [PubMed] [Google Scholar]
- 16.Berman HM, Westbrook J, Feng Z, et al. . The protein data bank. Nucleic Acids Res 2000;28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leath KJ, Johnson S, Roversi P, et al. . High-resolution structures of bacterially expressed soluble human CD59. Acta Crystallogr Sect F Struct Biol Cryst Commun 2007;63:648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson S, Brooks NJ, Smith RA, Lea SM, Bubeck D. Structural basis for recognition of the pore-forming toxin intermedilysin by human complement receptor CD59. Cell Rep 2013;3:1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Fedarovich A, Tomlinson S, Davies C. Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex. Acta Crystallogr D Biol Crystallogr 2007;63:714–721. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Smith CA, Song H, Morgan BP, Abagyan R, Tomlinson S. Insights into the human CD59 complement binding interface toward engineering new therapeutics. J Biol Chem 2005;280:34073–34079. [DOI] [PubMed] [Google Scholar]
- 21.Bodian DL, Davis SJ, Morgan BP, Rushmere NK. Mutational analysis of the active site and antibody epitopes of the complement-inhibitory glycoprotein, CD59. J Exp Med 1997;185:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budding K, van de Graaf EA, Kardol-Hoefnagel T, et al. . Soluble CD59 is a novel biomarker for the prediction of obstructive chronic lung allograft dysfunction after lung transplantation. Sci Rep 2016;6:26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakulinen J, Meri S. Shedding and enrichment of the glycolipid-anchored complement lysis inhibitor protectin (CD59) into milk fat globules. Immunology 1995;85:495–501. [PMC free article] [PubMed] [Google Scholar]
- 24.Vakeva A, Lehto T, Takala A, Meri S. Detection of a soluble form of the complement membrane attack complex inhibitor CD59 in plasma after acute myocardial infarction. Scand J Immunol 2000;52:411–414. [DOI] [PubMed] [Google Scholar]
- 25.Meri S, Lehto T, Sutton CW, Tyynela J, Baumann M. Structural composition and functional characterization of soluble CD59: heterogeneity of the oligosaccharide and glycophosphoinositol (GPI) anchor revealed by laser-desorption mass spectrometric analysis. Biochem J 1996;316(pt 3):923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh P, Sahoo R, Vaidya A, et al. . A specific and sensitive assay for blood levels of glycated CD59: a novel biomarker for diabetes. Am J Hematol 2013;88:670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyata T, Takeda J, Iida Y, et al. . The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science 1993;259:1318–1320. [DOI] [PubMed] [Google Scholar]
- 28.Takeda J, Miyata T, Kawagoe K, et al. . Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell 1993;73:703–711. [DOI] [PubMed] [Google Scholar]
- 29.Mevorach D. Paroxysmal nocturnal hemoglobinuria (PNH) and primary p.Cys89Tyr mutation in CD59: differences and similarities. Mol Immunol 2015;67:51–55. [DOI] [PubMed] [Google Scholar]
- 30.Tabib A, Karbian N, Mevorach D. Demyelination, strokes, and eculizumab: lessons from the congenital CD59 gene mutations. Mol Immunol 2017;89:69–72. [DOI] [PubMed] [Google Scholar]
- 31.Holguin MH, Fredrick LR, Bernshaw NJ, Wilcox LA, Parker CJ. Isolation and characterization of a membrane protein from normal human erythrocytes that inhibits reactive lysis of the erythrocytes of paroxysmal nocturnal hemoglobinuria. J Clin Invest 1989;84:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada N, Harada R, Fujita T, Okada H. A novel membrane glycoprotein capable of inhibiting membrane attack by homologous complement. Int Immunol 1989;1:205–208. [DOI] [PubMed] [Google Scholar]
- 33.Sugita Y, Nakano Y, Tomita M. Isolation from human erythrocytes of a new membrane protein which inhibits the formation of complement transmembrane channels. J Biochem 1988;104:633–637. [DOI] [PubMed] [Google Scholar]
- 34.Holguin MH, Wilcox LA, Bernshaw NJ, Rosse WF, Parker CJ. Relationship between the membrane inhibitor of reactive lysis and the erythrocyte phenotypes of paroxysmal nocturnal hemoglobinuria. J Clin Invest 1989;84:1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holt DS, Botto M, Bygrave AE, Hanna SM, Walport MJ, Morgan BP. Targeted deletion of the CD59 gene causes spontaneous intravascular hemolysis and hemoglobinuria. Blood 2001;98:442–449. [DOI] [PubMed] [Google Scholar]
- 36.Rollins SA, Sims PJ. The complement-inhibitory activity of CD59 resides in its capacity to block incorporation of C9 into membrane C5b-9. J Immunol 1990;144:3478–3483. [PubMed] [Google Scholar]
- 37.Ninomiya H, Sims PJ. The human complement regulatory protein CD59 binds to the alpha-chain of C8 and to the “b”domain of C9. J Biol Chem 1992;267:13675–13680. [PubMed] [Google Scholar]
- 38.Deckert M, Ticchioni M, Mari B, Mary D, Bernard A. The glycosylphosphatidylinositol-anchored CD59 protein stimulates both T cell receptor zeta/ZAP-70-dependent and -independent signaling pathways in T cells. Eur J Immunol 1995;25:1815–1822. [DOI] [PubMed] [Google Scholar]
- 39.Longhi MP, Harris CL, Morgan BP, Gallimore A. Holding T cells in check--a new role for complement regulators? Trends Immunol 2006;27:102–108. [DOI] [PubMed] [Google Scholar]
- 40.Krus U, King BC, Nagaraj V, et al. . The complement inhibitor CD59 regulates insulin secretion by modulating exocytotic events. Cell Metab 2014;19:883–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Study data for the primary analyses presented in this report are available upon reasonable request from the corresponding and senior author.