Abstract
Plastoceridae Crowson, 1972, Drilidae Blanchard, 1845 and Omalisidae Lacordaire, 1857 (Elateroidea) are families of the Coleoptera with obscure phylogenetic relationships and modified morphology showing neotenic traits such as soft bodies, reduced wing cases and larviform females. We shotgun sequenced genomes of Plastocerus, Drilus and Omalisus and incorporated them into data matrices of 66 and 4202 single-copy nuclear genes representing Elateroidea. Phylogenetic analyses indicate their terminal positions within the broadly defined well-sclerotized and fully metamorphosed Elateridae and thus Omalisidae should now be considered as Omalisinae stat. nov. in Elateridae Leach, 1815. The results support multiple independent origins of incomplete metamorphosis in Elateridae and indicate the parallel evolution of morphological and ecological traits. Unlike other neotenic elateroids derived from the supposedly pre-adapted aposematically coloured and unpalatable soft-bodied elateroids, such as fireflies (Lampyridae) and net-winged beetles (Lycidae), omalisids and drilids evolved from well-sclerotized click beetles. These findings suggest sudden morphological shifts through incomplete metamorphosis, with important implications for macroevolution, including reduced speciation rate and high extinction risk in unstable habitats. Precise phylogenetic placement is necessary for studies of the molecular mechanisms of ontogenetic shifts leading to profoundly changed morphology.
Introduction
The elateroid beetles (Coleoptera: Elateroidea) are a morphologically heterogeneous group and include two different body plans: the well sclerotized ‘true elateroids’ (Elateridae, Eucnemidae, Throscidae), many of them able to jump for predator evasion using the long prosternal process fitting in the mesosternal cavity, and the soft-bodied ‘cantharoids’, some of them with larviform or incompletely metamorphosed females, all with short prosternum and unable to jump (Cantharidae, Lycidae, Lampyridae, Phengodidae, etc.). Taxonomists have accepted the paradigm of two reciprocally monophyletic groups corresponding to these types, the Cantharoidea and Elateroidea, until the late 20th century when the cantharoid families were recognized as a sub-lineage within the more broadly defined Elateroidea1,2. Nevertheless, the concept of the ‘cantharoid’ clade was not questioned even after they were subsumed in the elateroids, on the assumption that soft-bodiedness originated only once3–5. When molecular data became available, multiple origins of soft-bodied elateroid families were proposed6. Nowadays, both the soft-bodied and the hard-bodied groups are known to be composed of distantly related families7–11.
The confusion about the relationships of soft-bodied groups also produced uncertainty for the placement of three small elateroid families, Omalisidae Lacordaire, 1857, Drilidae Blanchard, 1845 and Plastoceridae Crowson, 1972, which traditionally have been placed in the cantharoid clade3,4. In the latest morphological study, Omalisidae, a family of only about twenty known species were recovered as closely related to Lampyridae, Phengodidae and Telegeusidae (the fireflies and glow-worms)5, whereas molecular studies placed them in various positions either as a sister group to the hard-bodied Elateridae or as a terminal lineage within Elateridae, but in all cases very distant from Lampyridae and Telegeusidae6,8,12. Drilidae, the ‘false fireflies’, are a small group of predatory soft-bodied beetles. They have incompletely metamorphosed females with only head and appendages expressing the adult traits. Molecular data placed them in a derived position within Elateridae, specifically in the subfamily Agrypninae, as the tribe Drilini7,13. A recent analysis of basal coleopteran relationships that sampled both Drilidae and Omalisidae recovered them together with cardiophorine click beetles, but the conclusions were problematic for splitting the core groups of Elateridae8. The third ‘family’ under consideration here, Plastoceridae, represented by Plastocerus Schaum, 1852, have been firmly placed within the soft-bodied cantharoids ever since their formal recognition1. However, molecular data tentatively placed them within Elateridae near Oxynopterus Hope, 1842, leading to a proposed ranking as a subfamily of Elateridae14, in conflict with the earlier morphology-based analyses4,5. All molecular studies have been using only Sanger data, i.e., a low number of mitochondrial and rRNA markers. The molecular findings are counterintuitive and have been met with skepticism. Therefore, most taxonomists still assign Drilidae the status of a family and do not accept the results of the molecular studies and proposed changes in the formal classification5,8,15–18.
Among soft-bodied groups, several lineages exhibit larviform or incompletely metamorphosed females, which also occur in a few other beetles, e.g., Thylodrias Motschulsky, 1839, Micromalthus, Leconte, 1878 and Ozopemon Hagedorn, 1910, but they are most prevalent in Elateroidea6,19–24. The distinct appearance generally led to their taxonomic recognition at family rank1,5,15. Only females show the most extreme cases of larviform or partially metamorphosed morphology, although some traits may appear also in males25,26. The point at which metamorphosis is prematurely terminated across elateroid lineages is variable. The neotenic females of net-winged beetles (Lycidae), glow-worm beetles (Rhagophthalmidae) and some genera of fireflies (Lampyridae) are completely larviform, except for the fully developed reproductive organs. Various fireflies have only an adult-like pronotum and head, in drilids only the head and appendages are adult-like, omalisids exhibit a larviform abdomen, modified thorax and short elytra, and modifications in plastocerids are limited to the free abdominal sternites and lack the click mechanism14,27–34. The neotenics differ not only in their morphology, but also in ecological traits, which affect macroevolution: the lineages with brachelytrous, wingless or completely larviform females occupy small ranges due to the low ability to disperse, they usually move only slowly, and are often unpalatable and aposematically coloured. Due to limited dispersal capacity, they are mostly limited to environmentally stable habitats and regions where populations persist with low risk of extinction. The lineages exhibiting neotenic females usually represent only a fraction of the species diversity compared to their fully winged sister groups. Most neotenics are uncommon and, therefore, often only males are known while the females are rarely seen or may not be known at all, but inferred to be neotenic based on certain modifications in the males1,7,19,20,34. Taxa with incompletely metamorphosed females have generally been found to be related with soft-bodied lineages. Their characteristic traits, such as low dispersal capacity, restricted ranges and chemical protection as an alternative anti-predatory strategy, have been considered as pre-adaptations which increase the profitability of the shift to incomplete metamorphosis and higher investment in offspring19,20.
The aim of this study is to produce genomic data for three enigmatic neotenic and morphologically aberrant lineages, Omalisidae, Drilidae and Plastoceridae and use them to investigate their relationships to other elateroid families. As the previous studies provided ambiguous phylogenetic signal, whole genome data are the ultimate source of information which could shed light on their phylogenetic relationships. These elateroids are unique by their divergent morphology and relictual distribution. Their robust placement is crucial for future studies on the molecular mechanisms of incomplete metamorphosis leading to weakly sclerotized bodies and winglessness.
Results
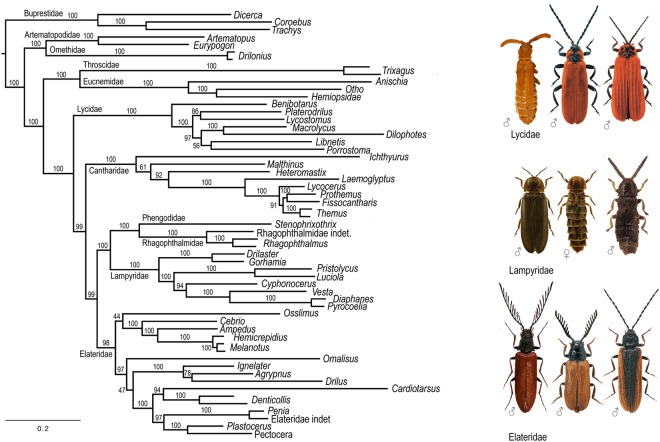
The Fig. 1 shows the ML tree topology inferred from the 66-gene analysis at nucleotide level. The three focal taxa were found within Elateridae: Plastocerus associated with Pectocera (Oxynopterinae), Drilus associated with Agrypnus (Agrypninae) and Omalisus as a sister to the latter clades combined. The AU tests rejected alternative topologies (Table 1).
Figure 1.
The maximum likelihood tree recovered from the 66-single copy protein coding genes at nucleotide level. Photographs of general appearance © authors.
Table 1.
Approximately unbiased test of alternative relationships of Drilus, Omalisus, and Plastocerus recovered by the analysis of the 66-taxa dataset.
| Topology | logL | deltaL | p-AU |
|---|---|---|---|
| ML topology as in Fig. 1 | −1035875 | 0.000 | 0.7247 |
| Constraint: | |||
| Omalisus + Plastocerus + Drilus | |||
| sister to Elateridae | −1038925 | 3046.7 | 0.0001 |
| Drilus sister to Cantharidae | −1037003 | 1124.9 | 0.0000 |
| Drilus sister to Elateridae | −1036926 | 1048.2 | 0.0000 |
| Drilus sister to Lampyridae | −1037133 | 1254.5 | 0.0000 |
| Drilus sister to Lycidae | −1037070 | 1192.1 | 0.0000 |
| Omalisus sister to Cantharidae | −1035963 | 84.5 | 0.0114 |
| Omalisus sister to Elateridae | −1035942 | 63.8 | 0.0045 |
| Omalisus sister to Lampyridae | −1036076 | 197.6 | 0.0000 |
| Omalisus sister to Lycidae | −1036033 | 154.6 | 0.0004 |
| Plastocerus sister to Cantharidae. | −1038735 | 2857.3 | 0.0000 |
| Plastocerus sister to Elateridae | −1037933 | 2054.8 | 0.0006 |
| Plastocerus sister to Lampyridae | −1038902 | 3023.6 | 0.0000 |
| Plastocerus sister to Lycidae | −1038975 | 3096.6 | 0.0000 |
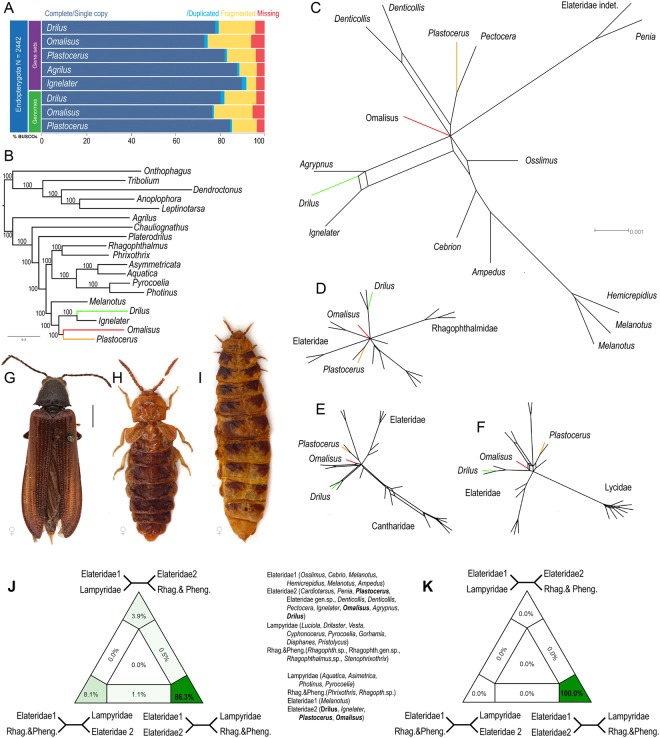
Newly generated shotgun genomic sequencing data provided high coverage of genomes at a sequencing depth of 69–312×. Data were used to create an ortholog set of 4202 genes from publicly available transcriptome and genome data of Coleoptera. The genome size was estimated to ~536 million base pairs (Mbp) for D. mauritanicus, 367 Mbp for P. angulosus and 270 Mbp for O. fontisbellaquei (Fig. S1). The genome completeness is summarized in Fig. 2A and the completeness of datasets in Figs S2–S7. The dataset based on 4202 orthologs recovers an almost identical topology at amino acid and nucleotide levels or filtering approach (Figs S8–S14). Taking into account the lower taxon sampling, Omalisus was recovered as sister to Plastocerus, unlike in the 66-gene dataset which placed it as sister to a wider clade that also includes Drilus, but with poor support. The genomic dataset includes only two elaterids, Melanotus and Ignelater, but they represent the two major clades of the family (Fig. 2). Because of the early branching of Melanotus, the three neotenic lineages are safely placed within the Elateridae (Fig. 2), with 100% support at all nodes.
Figure 2.
(A) Summarized benchmarks in the BUSCO assessment among assembled draft genomes and annotated gene sets. These estimations used 2442 expected Endopterygota genes as query; (B) Maximum likelihood tree obtained from the analysis of the 4202 ortholog dataset at nucleotide level; (C) Network obtained from the separate maximum likelihood analyses of Elateridae and all 66 single copy genes gene trees, (D) ditto, Elateridae and Rhagophthalmidae, (E) ditto, Elateridae and Cantharidae; (F) ditto, Elateridae and Lycidae; (G–I) General appearance of females: (G) Plastocerus angulosus, (H) Omalisus fontisbellaquei, (I) Drilus flavescens. Note the similar degree of morphological modifications in the males of some Lycidae and Lampyridae in Fig. 1. (J–K) 2D simplex graph. The support values in cells show support for each of the three topologies illustrated. (J) Topologies recovered using 66-taxa dataset as nucleotide level; (K) Topologies recovered using genomic dataset at nucleotide level. Photographs of general appearance of females © authors.
Further exploration of the 66-gene dataset based on individual gene trees from individual loci and supernetwork analysis, using various subsets of taxa in addition to the Elateridae, always grouped the three focal groups within Elateridae, in positions very compatible with the supermatrix approach on the full taxon set (Fig. 2C–F). The Four cluster Likelihood Mapping (FcLM) analysis identified predominant support for the monophyly of all Elateridae including the three focal taxa and Rhagophthalmidae/Phengodidae + Lampyridae as a sister to them (Fig. 2J,K). Further ML topologies recovered from 66-taxa dataset, the coalescent trees recovered from both datasets and the tests of the alternative relationships are shown in Figs S15–S23.
Discussion
The current study is based on the most extensive transcriptomic dataset of ‘cantharoid’ families to date. At the base of the tree, the results recover three ‘cantharoid’ lineages, i.e. Cantharidae, Lycidae and the bioluminescent clade, composed of the Lampyridae, Rhagophthalmidae, and Phengodidae. The monophyly of the latter is in contrast to some studies that separated these three families and suggested that bioluminescence in adult Lampyridae has a separate evolutionary origin from the other two families with exclusively larval bioluminescence6,7. It seems now unlikely and the current topology is in agreement with eight-gene analysis8 and with morphology4. The widely defined Elateridae, including the three small ‘cantharoid’ lineages Drilidae, Omalisidae and Plastoceridae is the sister of the bioluminescent clade (Figs 1 and 2). The families traditionally assigned to the morphology-based Cantharoidea1 acquired soft-bodiedness independently, or alternatively reverted to hard-bodied forms in the Elateridae (Fig. 1). Whatever the ancestral state, neotenic lineages may be derived both from soft-bodied and hard-bodied ancestors.
The morphology-based classifications have emphasized morphological divergence of Omalisidae, Drilidae, and Plastoceridae, and thus assigned them family rank4,5,8,15–18 although the possible relationships of Omalisus and Elateridae was mentioned already in the 19th century35. These taxa share some characters with the ‘cantharoid’ soft-bodied elateroids (Fig. 2H,I) and even if not all traits are present14, morphological phylogenetic analyses have never found the proximity of these taxa to the well-sclerotized click beetles4,5.
In contrast with morphology, molecular analyses indicate that phenotypically similar elateroids are not necessarily closely related. Initial hints of cantharoid non-monophyly6,12 were corroborated later7,8,10. Kundrata & Bocak13 were the first to hypothesize that the unrelated forms exhibiting incompletely metamorphosed females might have originated also within click beetles and proposed that Drilidae are a terminal branch in the elaterid subfamily Agrypninae, consistent with the tribal level (Drilini). These results have been treated as conjectural or have been ignored5,8,15–18. Recently, also based on the rRNA and mtDNA markers, the Plastoceridae was down-ranked to a subfamily in Elateridae and inferred as a sister lineage of Oxynopterus (Oxynopterinae)14, but also with only moderate support due to a limited amount of information. All earlier studies have been based on Sanger-era markers and some lacked adequate taxon sampling. Therefore, a robust placement was not obtained6–9,13,36,37. Similarly, the analysis of eight nuclear markers did not recover click beetles as a monophylum and no conclusive hypothesis could be proposed8. The rRNA-based analyses of Elateroidea produced ambiguous alignments when numerous length-variable sequences were combined in a single dataset which contained a broad set of Coleoptera, mis-aligning the comparatively short loops in the rRNA genes of elaterids6. Similarly, low genetic divergence is characteristic for the protein coding nuclear genes of Elateridae, which might have contributed to the failure to recover click beetle monophyly in the recent analysis of Zhang et al.10. Therefore, more conclusive evidence was sought in the genomic datasets. We investigate two contradicting phylogenetic hypotheses: (1) drilids, plastocerids and omalisids evolved within the elaterid clade, hence, they are in fact modified click beetles or (2) these lineages are deeply nested in Elateroidea and should be designated as families, as is still widely held.
The current study, based on tens of thousands nucleotide positions in the densely sampled 66-gene Elateroidea dataset and millions of positions in the genomic dataset, now confirms the placement of former Omalisidae, Drilidae and Plastoceridae in Elateridae with high support (Figs 1 and 2) and under a wide array of analytical approaches (Figs S8–S20) which were performed to test their impact on the resulting topology38. The topologies set Drilus and Plastocerus near Agrypnus and Pectocera, respectively, corroborating the earlier studies13,14, and Omalisus is robustly recovered as a deeply rooted branch in Elateridae, although in variable positions (Figs 1 and 2). Deep splits11 and rapid radiations39,40 often represent extremely difficult phylogenetic problems even when large datasets are analyzed (here, the unstable position of Omalisus or a conflicting signal for Melanotus; Figs S11–S21). We found that although the genomic and 66-gene datasets place Omalisus in Elateridae, the later cannot robustly identify its sister clade within this family (Figs 2, S15–S17, S20). Therefore, we studied in detail alternative positions of focal taxa and we found that the alternative hypotheses that force well-sclerotized elaterids to be monophyletic, or Drilus, Omalisus, and Plastocerus to be a sister to either Lampyridae, Cantharidae, Lycidae or Elateridae are all rejected by AU tests (Table 1). Similarly, the monophyly of all Elateridae including three focal families got predominant support (Fig. 2J,K). To sum up, it becomes difficult to dispute the possibility that the lineages with modified morphology (Plastocerus) and even with incompletely metamorphosed females (Drilus, Omalisus) are nested within the Elateridae. Thus, these taxa do not deserve the family rank despite their morphological uniqueness. Based on the current analysis, we propose to lower Omalisidae from the family rank to a subfamily Omalisinae Lacordaire, 1857 in Elateridae and we confirm the status of Drilini and Plastocerinae13,14.
The shift to incompletely metamorphosed females (Fig. 2H,I) is known to affect the macroevolution of a lineage19. The well-sclerotized elaterids contain the substantial part of the extant Elateroidea diversity, i.e. ~10,000 species37, and the morphologically modified elaterids are represented only by Plastocerinae (2 sp), Omalisinae (22 spp) and Drilini (>100 spp). Their evolutionary trajectory is determined by low species numbers, limited geographic ranges, and small population sizes14,34,41,42. Most omalisids and Plastocerus angulosus occur in the Mediterranean only and their ranges are usually restricted to costal refugia where broad leaf forests persisted during the last glacial maximum, indicating the constraints to their subsequent dispersal14,34. Drilini are widespread in the Afrotropical region, the Mediterranean and along the South Asian coast to Thailand, but similarly to omalisids they are known to have limited species ranges and are generally rare in ecosystems41. Hence, omalisids, drilids and plastocerids represent examples of lineages which diverted from a successful body-plan of click beetles whose morphology did not substantially change since the Late Triassic43 and which has been reproduced in thousands of extant species. Yet, they survive for a long time in stable ecosystems similarly to other neotenic elateroids19,32,33.
The secure placement of these three lineages within the true click beetles has important implications for the origin of neoteny and macroevolution of modified lineages. First, the derived positions within Elateridae demonstrates that astonishing differences in morphology can be achieved through what appears to be shifts in ontogenetic programs. The general appearance and individual morphological traits changed over short evolutionary time scales, and these differences lead to convergent morphologies observed in several unrelated groups throughout the Elateroidea. Second, these morphological shifts also have profound macroevolutionary consequences. The traditional placement suggested relationships of Elateridae, Eucnemidae and Throscidae2, and Elateridae became the dominant part of Elateroidea diversity with >40% of species, world-wide distribution and high dominance in many beetle communities since the Jurassic43. Strong body sclerotization and an effective escape mechanism apparently proved to be an evolutionarily successful design, to make Elateridae an example of a morphologically conservative lineage whose great species diversity was not accompanied by morphological diversity43,44. Yet, among these conservative groups some lineages arose which were very different morphologically and resemble distant neotenic relatives in Lycidae, Lampyridae and Rhagophthalmidae. It has generally been assumed that soft-bodiedness in Elateroidea might be the first symptom of unfinished metamorphosis19,30 raising the possibility for the more fully neotenic lineages raised within them. Soft-bodied elateroids move slowly, are commonly unpalatable45,46 and aposematically coloured47. These factors might increase the trade-off gains if dispersal capacity is further lowered in favor of higher fecundity due to incomplete metamorphosis20. Neotenics nested in click beetles now falsify the general validity of this hypothesis about ecological pre-adaptations required for the origin of arrested or prematurely terminated metamorphosis20. If there are life-history pre-adaptations for incomplete metamorphosis present in Elateroidea, they differ in individual groups and might not have only the ecological character. Based on the origins of neoteny in click beetles, the alternative hypothesis can be formulated: a rapid modification of the regulatory system of insect metamorphosis at molecular level48,49 might be a trigger for a subsequent improvement by the natural selection, such as large-bodied females, higher fecundity and evolution of alternative defensive strategies. To sum up, we can say that convergent evolution has produced multiple lineages that ‘replay life’s tape’50,51 exploring a life strategy that is favored by stable climatic and ecological conditions, and which can apparently start from different ancestral states. As this shift is triggered by prematurely arrested metamorphosis, the repeated origin may indicate the low stability of the molecular regulatory system in Elateroidea. The identification of the closest relatives of neotenics may therefore be the first step for a comparative biology of the mechanisms of metamorphosis.
Methods
Material, laboratory procedures, and draft genomes
Total genomic DNA was extracted from single adult specimens of Omalisus fontisbellaquei Geoffroy, 1785 (from Czechia), Drilus mauritanicus Lucas, 1842 (Spain) and Plastocerus angulosus Schaum, 1852 (Turkey) using the DNeasy kit (Qiagene Inc., Hilden, Germany). The voucher specimens have been deposited in the collection of Department of Zoology, Palacky University, Olomouc. Genomic DNA of all three specimens was shotgun sequenced on the Illumina X Ten platform (Illumina Inc., San Diego, CA) for 2 × 150 bp paired-end reads by Novogene Co., Ltd. (Beijing, China) and each individual was sequenced for 30–107 × 109 base pairs. Raw paired-end reads were filtered using fastp 0.13.252 under the following parameters -q 5 -u 50 -l 50 -n 15 and other settings as default. The filtering steps included the removal of read pairs if either one read contains adapter contamination; if the proportion of low quality bases is over 50%; or if either one read contains more than 15 N bases. The quality of reads was visualized with FastQC (http://www.bioinformatics. babraham.ac.uk/projects/fastqc). Sequence data were deposited in GenBank SRA (Accession Numbers AB123456-AB123456).
We performed k-mer counts on the filtered data in Jellyfish 2.2.753 using 31-mer sizes. Moreover, based on the distribution of k-mer occurrences, we estimated the genome size using GenomeScope54 and assembled draft genomes of O. fontisbellaquei, D. mauritanicus and P. angulosus using MEGAHIT 1.1.355,56, with all parameters set to default and k-mer sizes of 31, 59, 87, 115 and 143. Additionally, the genome of O. fontisbellaquei was processed using the Shovill pipeline (https://github.com/tseemann/shovill) and assembled with SPAdes 3.12.057 using k-mer size 31, 51, 71, 91 and 111 under default parameters. We produced statistics of draft genomes with the assembly-stats algorithm (https://github.com/rjchallis/assembly-stats) and the results of both methods used for O. fontisbellaquei were similar and are shown in Fig. S24. The SPAdes contigs were used for further analyses of Omalisus. Obtained contig sequences were used to train Augustus58 for species specific gene models with BUSCO 359, -long option, Endopterygota set of conserved genes (n = 2442) and -sp tribolium2012 as the closest relative. Predicted species specific gene models were then used for ab initio gene predictions in Augustus, and predicted protein coding sequences were used for subsequent analyses in Orthograph 0.6.160. The genome and coding gene set completeness was evaluated based on the predicted protein sets with BUSCO using expected 2442 Endopterygota single-copy orthologs as targets. BUSCO quantitatively assesses completeness using evolutionary conserved expectations of gene content. We compared completeness of predicted protein sets with Agrilus planipennis and Ignelater luminosus.
Data matrices
Two datasets were assembled for the phylogenetic analysis:
(1) Single-copy genes – 66-taxa dataset (Table S1). This dataset is based on PCR amplified sequences for 95 genes across Coleoptera10, of which 66 gene for 54 taxa were retained after the removal of 29 supposedly multi-copy genes, as described earlier11. The putative homologs for O. fontisbellaquei, D. mauritanicus and P. angulosus were added to the earlier published data. Exons were concatenated to produce a supermatrix of 53,253 aligned positions.
(2) Genome orthologs. Transcriptomes of Melanotus cribricollis61; Asymmetricata circumdata, Aquatica ficta, Pyrocoelia pectoralis, Rhagophthalmus sp62; Chauliognathus flavipes and Phrixothrix hirtus63 were downloaded from the NCBI SRA archive and assembled as described by Kusy et al.11. Additionally, we downloaded the gene set of I. luminosus from fireflybase.org64 and the transcriptome of Photinus pyralis65 from NCBI Transcriptome Shotgun Assembly database (Table S2). The ortholog set was obtained by searching the OrthoDB 9.1 database66 for one-to-one orthologs among Coleoptera in available genome sequences of A. planipennis67, Anoplophora glabripennis68, Dendroctonus ponderosae69, Leptinotarsa decemlineata67, Onthophagus taurus67, and Tribolium castaneum70,71 (Tables S3, S4). OrthoDB 9.1 specified 4225 protein coding single copy genes for the above species and the Coleoptera reference node. We used Orthograph 0.6.160 to search the above transcriptomes and predicted protein coding gene sets for the corresponding sequences. Default settings were used. We summarized Orthograph data reporting 4202 orthologs, removed terminal stop codons and masked internal stop codons at the translational level and nucleotide levels using the perl script summarize_orthograph_results.pl60. The amino acid sequences were aligned using MAFFT 7.394 with the L-INS-i algorithm72. Resulting alignments from each ortholog group were checked for the presence of outliers using the script https://github.com/mptrsen/scripts/blob/master/outlier_check.pl and following the methods reported by Misof et al.73 and Peters et al.74. We used Pal2Nal75 to generate multiple sequence alignments of nucleotides corresponding to amino acids. Aliscore 2.076,77 with the maximal number of pairwise comparisons, -e option and default settings were used to identify random or ambiguous similarity within alignments which were masked using Alicut 2.3 (https://github.com/mptrsen/scripts/blob/master/ALICUT_V2.3.pl) and Alinuc.pl73 to apply Aliscore results to the nucleotide data. The dataset of 4202 orthologs on amino acid and nucleotide levels were concatenated into supermatrices 1 and 2 (1–amino acids; 2–all nucleotides) with FASconCAT-G78. Additional filtered datasets were used for tree construction: (a) nucleotides, 1st + 2nd codon positions only; (b) amino acid raw data (no filtering e.g. outliers removal, Aliscore); (c) nucleotide raw data; (d) a subset of Supermatrix 1, orthologs available for all taxa; (e) a subset of Supermatrix 2 containing orthologs for all taxa. AliStat 1.7 (https://github.com/thomaskf/AliStat) was used to generate distributions of missing data per site in the supermatrices.
Phylogenetic analyses
IQ-TREE 1.6.679 and RaxML80 were used to calculate maximum likelihood (ML) trees, with partitions identified by the Model Finder tool of IQ-TREE and using the Bayesian Information Criterion81,82. The partitions, models and parameters are available upon request. The ultrafast bootstrap option was used with 3000 bootstrap iterations83. The IQ-TREE analyses were run with the -spp parameter allowing each partition to have its own evolutionary rate.
To investigate alternative and/or confounding signal in the 66-taxa dataset and genomic dataset, we used FcLM analysis73,84 implemented in IQ-TREE to study a possibility of the occurrence of incongruent signal in phylogenomic datasets that might not be revealed by a phylogenetic multi-species tree. Additionally, gene tree incongruence in 66 genes dataset was tested by visualizations of the dominant bipartitions among individual loci based on the individual IQ-TREE ML gene topologies by constructing supernetworks using the SuperQ method implemented in Spectre selecting the ‘balanced’ edge-weight with ‘JOptimizer’ optimization function, and applying no filter85,86. This methodology decomposes all gene trees into quartets to build supernetworks where edge lengths correspond to quartet frequencies. We tested the alternative tree topologies within Elateridae and three focal taxa Plastocerus, Drilus, and Omalisus and further, we tested a potential ambiguity of relationships of all these taxa and three putative relatives, i.e., Cantharidae, Lycidae, and Rhagophthalmidae. Resulting supernetworks were visualized in SplitsTree 4.14.687. Further, we used ASTRAL 5.6.188 and genomic dataset to construct coalescent species trees from individual IQ-TREE ML gene topologies at amino acid and nucleotide level.
The focal taxa, Plastocerus, Drilus and Omalisus have been placed in relationships with soft-bodied ‘cantharoid’ families1–5. We tested these alternative hypotheses using the densely sampled 66-taxa dataset and compared likelihoods of hypothesized relationships of focal taxa with Cantharidae, Lampyridae and Lycidae and alternatively the topology (Omalisus,(Plastocerus, Drilus)),(Elateridae) which accepts their relationships with Elateridae, but excludes them as sister lineages of Elateridae. The likelihood of these topologies was compared with the best ML topology using AU test89 implemented in IQTREE79 and using -au option and 1,000,000 replicates.
The click beetle Melanotus was earlier recovered as a lineage distantly related to other well-sclerotized Elateridae8,10. We encountered a similarly contentious position of Melanotus. The topology recovered from the dataset at amino acid level suggested that Melanotus does not belong to click beetles8,10. Therefore, we estimated the number of genes supporting the alternative relationships. We tested positions of Melanotus as (1) a sister to Lampyridae and (2) a sister to other Elateridae by evaluating which single gene partition of amino acid data favor alternative topologies by calculating log-likelihood scores for each gene partition using IQ-TREE option -wpl. As an input we provided both topologies. To interpret the results of the partition log-likelihood and to evaluate the contribution of each gene partition, we calculated differences of each pL score of topologies90,91.
Electronic supplementary material
Acknowledgements
The authors are obliged to V. Kuban (Brno) for information on P. angulosus, R. Kundrata and M. Baena for the specimen of D. mauritanicus. The study was funded by the IGA and GACR projects (PrF-2018, 18-14942S to L.B., M.Ma., M.Mo., and D.K.), Leverhulme Trust (F/00696/P to A.P.V. and L.B.) and the NHM Biodiversity Initiative.
Author Contributions
D.K. analyzed genomic data and carried out, sequence alignments and phylogenetic analyses, D.K., M.M. and M.B. participated in other data analyses, D.K. and M.M. carried out the statistical analyses; all coauthors helped draft the manuscript; L.B. provided specimens, L.B. and A.P.V. conceived, designed and coordinated the study and drafted the manuscript. All authors gave final approval for publication.
Data Accessibility
The DNA sequences reported in this article can be accessed in GenBank under accessions AB123456-789.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35328-0.
References
- 1.Crowson RA. A review of the classification of Cantharoidea (Coleoptera), with definition of two new families Cneoglossidae and Omethidae. Rev. Univ. Madrid. 1972;21:35–77. [Google Scholar]
- 2.Lawrence JF. Rhinorhipidae, a new beetle family from Australia, with comments on the phylogeny of the Elateriformia. Invertebr. Taxon. 1988;2:1–53. doi: 10.1071/IT9880001. [DOI] [Google Scholar]
- 3.Beutel RG. Phylogenetic analysis of Elateriformia (Coleoptera: Polyphaga) based on larval characters. J. Zool. Syst. Evol. Res. 1995;33:145–171. doi: 10.1111/j.1439-0469.1995.tb00222.x. [DOI] [Google Scholar]
- 4.Branham MA, Wenzel JW. The evolution of photic behaviour and the evolution of sexual communication in fireflies (Coleoptera: Lampyridae) Cladistics. 2003;84:565–586. doi: 10.1111/j.1096-0031.2003.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence JF, et al. Phylogeny of the Coleoptera based on morphological characters of adults and larvae. Ann. Zool. 2011;61:1–217. doi: 10.3161/000345411X576725. [DOI] [Google Scholar]
- 6.Bocakova M, Bocak L, Hunt T, Teravainen M, Vogler AP. Molecular phylogenetics of Elateriformia (Coleoptera): evolution of bioluminescence and neoteny. Cladistics. 2007;23:477–496. doi: 10.1111/j.1096-0031.2007.00164.x. [DOI] [Google Scholar]
- 7.Kundrata R, Bocakova M, Bocak L. The comprehensive phylogeny of the superfamily Elateroidea (Coleoptera: Elateriformia) Mol. Phyl. Evol. 2014;76:162–171. doi: 10.1016/j.ympev.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 8.McKenna DD, et al. The beetle tree of life reveals that Coleoptera survived end-Permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 2015;40:835–880. doi: 10.1111/syen.12132. [DOI] [Google Scholar]
- 9.Timmermans MJTN, et al. Family-Level Sampling of Mitochondrial Genomes in Coleoptera: Compositional Heterogeneity and Phylogenetics. Genome Biol. Evol. 2016;8:161–175. doi: 10.1093/gbe/evv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang SQ, et al. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Comm. 2018;9:205. doi: 10.1038/s41467-017-02644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusy D, et al. Genome sequencing of Rhinorhipus Lawrence exposes an early branch of the Coleoptera. Front. Zool. 2018;15:21. doi: 10.1186/s12983-018-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt T, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 13.Kundrata R, Bocak L. The phylogeny and limits of Elateridae (Insecta, Coleoptera): is there a common tendency of click beetles to soft-bodiedness and neoteny? Zool. Scr. 2011;40:364–378. doi: 10.1111/j.1463-6409.2011.00476.x. [DOI] [Google Scholar]
- 14.Bocak L, Motyka M, Bocek M, Bocakova M. Incomplete sclerotization and phylogeny: The phylogenetic classification of Plastocerus (Coleoptera: Elateroidea) PLoS One. 2018;13:e0194026. doi: 10.1371/journal.pone.0194026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beutel, R. G. & Leschen, R. A. B. Coleoptera, Beetles; Volume 1: Morphology and Systematics (Archostemata, Adephaga, Myxophaga, Polyphaga partim) 2nd edition. In: Kristensen N. P. & Beutel R. G., eds. Handbook of Zoology, Arthropoda: Insecta. Berlin/New York: Walter de Gruyter GmbH & Co. (2016).
- 16.Amaral DT, et al. Transcriptional comparison of the photogenic and non-photogenic tissues of Phrixothrix hirtus (Coleoptera: Phengodidae) and non-luminescent Chauliognathus flavipes (Coleoptera: Cantharidae) give insights on the origin of lanterns in railroad worms. Gene Rep. 2017;7:78–86. doi: 10.1016/j.genrep.2017.02.004. [DOI] [Google Scholar]
- 17.Toussaint E, et al. The peril of dating beetles. Syst. Entomol. 2017;42:1–10. doi: 10.1111/syen.12198. [DOI] [Google Scholar]
- 18.Martin GJ, Branham M, Whiting MF, Bybee SM. Total evidence phylogeny and the evolution of adult bioluminescence in fireflies (Coleoptera: Lampyridae) Mol. Phyl. Evol. 2017;107:564–575. doi: 10.1016/j.ympev.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Bocak L, Bocakova M, Hunt T, Vogler AP. Multiple ancient origins of neoteny in Lycidae (Coleoptera): consequences for ecology and macroevolution. Proc. R. Soc. B. 2008;275:2015–2023. doi: 10.1098/rspb.2008.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon DP, Hayward A. Why grow up? A perspective on insect strategies to avoid metamorphosis. Ecol. Entomol. 2016;41:505–515. doi: 10.1111/een.12313. [DOI] [Google Scholar]
- 21.Gould, S. J. Ontogeny and Phylogeny. Cambridge: Harvard University Press (1977).
- 22.Pollock DA, Normark BB. The life cycle of Micromalthus debilis LeConte (1878) (Coleoptera: Archostemata: Micromalthidae): historical review and evolutionary perspective. J. Zool. Syst. Evol. Res. 2002;40:105–112. doi: 10.1046/j.1439-0469.2002.00183.x. [DOI] [Google Scholar]
- 23.Jordal BH, Beaver RA, Normark BB, Farrell BD. Extraordinary sex ratios and the evolution of male neoteny in sib-mating Ozopemon beetles. Biol. J. Linn. Soc. 2002;75:353–360. doi: 10.1111/j.1095-8312.2002.tb02076.x. [DOI] [Google Scholar]
- 24.Kiselyova T, McHugh JV. A phylogenetic study of Dermestidae (Coleoptera) based on larval morphology. Syst. Entomol. 2006;31:469–507. doi: 10.1111/j.1365-3113.2006.00335.x. [DOI] [Google Scholar]
- 25.Bocak L, Grebennikov VV, Masek M. A new species of Dexoris (Coleoptera: Lycidae) and parallel evolution of brachyptery in the soft-bodied elateroid beetles. Zootaxa. 2013;3721:495–500. doi: 10.11646/zootaxa.3721.5.5. [DOI] [PubMed] [Google Scholar]
- 26.Naoki T, Bocak L, Ghani IA. Discovery of a new species of the brachelytrous net-winged beetle genus Alyculus (Coleoptera: Lycidae) from Peninsular Malaysia. Zootaxa. 2015;4144:145–150. doi: 10.11646/zootaxa.4144.1.11. [DOI] [PubMed] [Google Scholar]
- 27.Wong ATC. A new species of neotenous beetle, Duliticola hoiseni (Insecta: Coleoptera: Cantharoidea: Lycidae) from Peninsular Malaysia and Singapore. Raffl. Bull. Zool. 1996;44:173–187. [Google Scholar]
- 28.Cicero JM. Ontophylogenetics of cantharoid larviforms (Coleoptera: Cantharoidea) Coleopt. Bull. 1988;42:105–151. [Google Scholar]
- 29.Miller, R. S. A revision of the Leptolycini (Coleoptera: Lycidae) with a discussion of paedomorphosis. PhD Thesis. Columbus: The Ohio State University (1991).
- 30.Jeng, M. L. Comprehensive phylogenetics, systematics, and evolution of neoteny of Lampyridae (Insecta: Coleoptera). PhD thesis, Lawrence: University of Kansas (2008).
- 31.Bocak L, Brlik M. Revision of the family Omalisidae (Coleoptera, Elateroidea) Ins. Syst. Evol. 2008;39:189–212. doi: 10.1163/187631208788784101. [DOI] [Google Scholar]
- 32.Masek M, Ivie MA, Palata V, Bocak L. Molecular phylogeny and classification of Lyropaeini (Coleoptera: Lycidae) with description of larvae and new species of Lyropaeus. Raffl. Bull. Zool. 2014;62:136–145. [Google Scholar]
- 33.Masek M, Palata V, Bray TC, Bocak L. Molecular phylogeny reveals high diversity, geographic structure and limited ranges in neotenic net-winged beetles Platerodrilus (Coleoptera: Lycidae) PLoS One. 2015;10:e0123855. doi: 10.1371/journal.pone.0123855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bocek M, Fancello L, Motyka M, Bocakova M, Bocak L. The molecular phylogeny of Omalisidae (Coleoptera) defines the family limits and demonstrates low dispersal propensity and the ancient vicariance patterns. Syst. Entomol. 2018;43:250–261. doi: 10.1111/syen.12271. [DOI] [Google Scholar]
- 35.Bourgeois J. Monographie des Lycides de l’ancien-monde. L’Abeille. 1882;20:1–120. [Google Scholar]
- 36.Bocak L, Kundrata R, Andújar-Fernández C, Vogler AP. The discovery of Iberobaeniidae (Coleoptera: Elateroidea): a new family of beetles from Spain, with immatures detected by environmental DNA sequencing. Proc. R. Soc. B. 2016;283:20152350. doi: 10.1098/rspb.2015.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bocak L, et al. Building the Coleoptera tree- of-life for >8000 species: composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 2014;39:97–110. doi: 10.1111/syen.12037. [DOI] [Google Scholar]
- 38.Reddy S, et al. Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling. Syst. Biol. 2017;66:857–879. doi: 10.1093/sysbio/syx041. [DOI] [PubMed] [Google Scholar]
- 39.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prum RO, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526:569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 41.Bray TC, Bocak L. Slowly dispersing neotenic beetles can speciate on a penny coin and generate space-limited diversity in the tropical mountains. Sci. Rep. 2016;6:33579. doi: 10.1038/srep33579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kundrata R, Bocak L. Taxonomic review of Drilini (Elateridae: Agrypninae) in Cameroon reveals high morphological diversity, including the discovery of five new genera. Ins. Syst. Evol. 2017;48:441–492. [Google Scholar]
- 43.Ponomarenko, A. G. The geological history of beetles. Pp. 155–171. In Biology, Phylogeny, and Classification of Coleoptera: Papers Celebrating the 80th Birthday of Roy A. Crowson (eds by Pakaluk, J. and Slipinski, S. A.). Warszawa: Muzeum i Instytut Zoologii PAN (1995).
- 44.Doludenko, M. P., Ponomarenko, A. G. & Sakulina, G. V. La géologie du gisement unique de la faune et de la flore du jurassique supérieur d’Aulié (Karatau, Kazakhstan du Sud). Moscow: Academie des Sciences de l’URSS, Inst. Géologique (1990).
- 45.Moore BP, Brown WV. Identification of warning odour components, bitter principles and antifeedants in an aposematic beetle–Metriorrhynchus rhipidium (Coleoptera: Lycidae) Ins. Biochem. 1981;15:493–499. doi: 10.1016/0020-1790(81)90016-0. [DOI] [Google Scholar]
- 46.Eisner T, et al. Defensive chemistry of lycid beetles and of mimetic cerambycid beetles that feed on them. Chemoecology. 2008;18:109–119. doi: 10.1007/s00049-007-0398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motyka M, Kampova L, Bocak L. Phylogeny and evolution of Müllerian mimicry in aposematic Dilophotes: evidence for advergence and size-constraints in evolution of mimetic sexual dimorphism. Sci. Rep. 2018;8:3744. doi: 10.1038/s41598-018-22155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riddiford LM. How does juvenile hormone control insect metamorphosis and reproduction? Gen. Comp. Endocrin. 2012;179:477–484. doi: 10.1016/j.ygcen.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Jindra M, Palli SR, Riddiford LM. The Juvenile Hormone Signaling Pathway in InsectDevelopment. Ann. Rev. Entomol. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- 50.Gould, S. J. Wonderful Life: The Burgess Shale and the Nature of History. New York: Norton (1989).
- 51.Beatty J. Replaying Life’s Tape. J. Philos. 2006;103:336–362. doi: 10.5840/jphil2006103716. [DOI] [Google Scholar]
- 52.Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor, https://www.biorxiv.org/content/biorxiv/early/2018/04/09/274100.full.pdf (accessed on April 30th, 2018) (2018). [DOI] [PMC free article] [PubMed]
- 53.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vurture GW, et al. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li D, Liu C, Luo R, Sadakane K, Lam T. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 56.Li D, et al. MEGAHITv1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Bankevich A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl. 2):215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 59.Waterhouse RM, et al. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petersen M, et al. Orthograph: a versatile tool for mapping coding nucleotide sequences to clusters of orthologous genes. BMC Bioinformatics. 2017;18:111. doi: 10.1186/s12859-017-1529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye B, Zhang Y, Shu J, Wu H, Wang H. RNA-sequencing analysis of fungi-induced transcripts from the bamboo wireworm Melanotus cribricollis (Coleoptera: Elateridae) larvae. PLoS One. 2018;13:e019118. doi: 10.1371/journal.pone.0191187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K, Hong W, Jiao H, Zhao H. Transcriptome sequencing and phylogenetic analysis of four species of luminescent beetles. Sci. Rep. 2017;7:1814. doi: 10.1038/s41598-017-01835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amaral DT, Mitani Y, Ohmiya Y, Viviani VR. Organization and comparative analysis of the mitochondrial genomes of bioluminescent Elateroidea (Coleoptera: Polyphaga) Gene. 2016;586:254–262. doi: 10.1016/j.gene.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Fallon, T. R. et al. 2017 Firefly genomes illuminate parallel origins of bioluminescence in beetles, https://www.biorxiv.org/content/biorxiv/early/2018/02/25/237586.full.pdf. Accessed on June 22nd, 2018. [DOI] [PMC free article] [PubMed]
- 65.Fallon TR, Li F, Vicent MA, Weng J. Sulfoluciferin is Biosynthesized by a Specialized Luciferin Sulfotransferase in Fireflies. Biochemistry. 2016;55:3341–3344. doi: 10.1021/acs.biochem.6b00402. [DOI] [PubMed] [Google Scholar]
- 66.Zdobnov EM, et al. OrthoDBv9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 2016;45:D744–D749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poelchau M, et al. The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2015;43(Database issue):D714–719. doi: 10.1093/nar/gku983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKenna DD, et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2017;17:227. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keeling CI, et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14:R27. doi: 10.1186/gb-2013-14-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richards S, et al. Tribolium genome sequencing consortium). The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 71.Shelton JM, et al. Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics. 2015;16:734. doi: 10.1186/s12864-015-1911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 74.Peters RS, et al. Evolutionary History of the Hymenoptera. Curr. Biol. 2017;27:1013–1018. doi: 10.1016/j.cub.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 75.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Misof B, Misof KA. Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Syst. Biol. 2009;58:21–34. doi: 10.1093/sysbio/syp006. [DOI] [PubMed] [Google Scholar]
- 77.Kück P, et al. Parametric and non-parametric masking of randomness in sequence alignments can be improved and leads to better resolved trees. Front. Zool. 2010;7:10. doi: 10.1186/1742-9994-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kück P, Longo GC. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014;11:81. doi: 10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 81.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grunewald Stefan, Spillner Andreas, Bastkowski Sarah, Bogershausen Anja, Moulton Vincent. SuperQ: Computing Supernetworks from Quartets. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2013;10(1):151–160. doi: 10.1109/TCBB.2013.8. [DOI] [PubMed] [Google Scholar]
- 86.Bastkowski S, et al. SPECTRE. A suite of PhylogEnetiC tools for reticulate evolution. Bioinformatics. 2017;34:1056–1057. doi: 10.1093/bioinformatics/btx740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2005;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 88.Sayyari E, Mirarab S. Fast Coalescent-Based Computation of Local Branch Support from Quartet Frequencies. Mol. Biol. Evol. 2016;33:1654–1668. doi: 10.1093/molbev/msw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 90.Simon S, Blanke A, Meusemann K. Reanalyzing the Palaeoptera problem - The origin of insect flight remains obscure. Arthropod Struct. Dev. 2018;47:328–338. doi: 10.1016/j.asd.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Shen XX, Hittinger CT, Rokas A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nature Ecol. Evol. 2017;1:126. doi: 10.1038/s41559-017-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequences reported in this article can be accessed in GenBank under accessions AB123456-789.