Abstract
Whole exome sequencing (WES) is utilized in diagnostic odyssey cases to identify the underlying genetic cause associated with complex phenotypes. Recent publications suggest that WES reveals the genetic cause in ~25% of these cases and is most successful when applied to children with neurological disease. The residual 75% of cases remain genetically elusive until more information becomes available in the literature or functional studies are pursued. WES performed on three families with presumed ciliopathy diagnoses, including orofaciodigital (OFD) syndrome, fetal encephalocele, or Joubert-related disorder, identified compound heterozygous variants in C2CD3. Biallelic variants in C2CD3 have previously been associated with ciliopathies, including OFD syndrome type 14 (OFD14; MIM: 615948). As three of the six identified variants were predicted to affect splicing, exon-skipping analysis using either RNA sequencing or PCR-based methods were completed to determine the pathogenicity of these variants, and showed that each of the splicing variants led to a frameshifted protein product. Using these studies in combination with the 2015 ACMG guidelines, each of the six identified variants were classified as either pathogenic or likely pathogenic, and are therefore likely responsible for our patients’ phenotypes. Each of the families had a distinct clinical phenotype and severity of disease, extending from lethal to viable. These findings highlight that there is a broad phenotypic spectrum associated with C2CD3-mediated disease and not all patients present with the typical features of OFD14.
Introduction
Ciliopathies are disorders of primary cilia that lead to a variety of phenotypes, including retinal degeneration, renal/liver disease, neuronal anomalies, skeletal abnormalities, and laterality defects, reflecting the multiple roles that cilia play during development. These diseases have a spectrum of severities, from viable multiorgan anomalies to early onset lethality [1, 2]. Ciliopathy-associated proteins are mainly associated with the formation and function of cilia with variants in >70 genes attributing to the phenotypic constellation [1]. Ciliopathies can be further divided into different subtypes: Joubert syndrome with neurological, renal, and retinal phenotypes; orofaciodigital (OFD) syndrome with neuronal, oral, facial, and digital anomalies; Leber’s congenital amaurosis with retinal anomalies; Bardet–Biedl syndrome with retinal, renal, digital, neurological phenotypes, and obesity; Alström syndrome with retinal, cardiac, neurological, auditory phenotypes, obesity, and insulin resistance; Jeune asphyxiating thoracic dystrophy with chondrodysplasia and respiratory insufficiency; Ellis van Creveld syndrome (EVC) with chondroectodermal dysplasia, digital, and facial anomalies; and Sensenbrenner syndrome, similar to EVC but with renal and facial anomalies [1, 3]. Despite the distinct disease groups, these syndromes have overlapping phenotypic features and genetic etiologies.
C2CD3 encodes for the C2 calcium-dependent domain containing three proteins involved in centriole elongation and cilia formation [4]. Recently, homozygous and compound heterozygous truncating and non-truncating variants in C2CD3 have been associated with ciliopathy-related disorders. Initially, three cases were reported: a sibling pair with homozygous c.184C>T, p.(Arg62*) variants, and an unrelated case with compound heterozygous variants, p.[(Cys1029Gly)];[Ala1305Valfs*3] [5]. These cases presented with canonical OFD along with microcephaly and brain malformations. Based on these findings, a new subclass of orofaciodigital, type 14 (OFD14; MIM: 615948) was created, which expanded the traditional phenotypic spectrum of OFD, with malformations of the face, oral cavity, and digits, to include microcephaly and cerebral malformations [5]. Subsequently, two reports described three families diagnosed with Joubert syndrome to be caused by biallelic C2CD3 variants. These studies included one patient homozygous for c.184C>T, p.(Arg62*) and a second patient compound heterozygous for c.[4951+1G>T];[5267G>A] (p.[?];[(Gly1756Glu)]), both of which also had oral frenulae and/or cleft palate, suggestive of an OFD-like phenotype [6], as well as two affected siblings with c.[5227G>T];[5929C>T] (p.[(Gly1743Cys)];[(Arg1977*)]) [7]. Finally, a recent report identified two affected fetuses with skeletal dysplasia due to c.[195G>C];[1492del] (p.[(Trp65Cys)];[(Ile477*)]) [8]. Taken together, these findings indicate that biallelic C2CD3 variants, irrespective of genotype, lead to a relatively broad spectrum of ciliopathy phenotypes.
Here, we describe three probands who presented with ciliopathy spectrum phenotypes, with clinical diagnoses of OFD, fetal encephalocele, or Joubert-related disorder. Whole exome sequencing (WES) was pursued in all three cases to elucidate the disease etiology. In all three cases, biallelic variants were identified in C2CD3. These cases illustrate how a comprehensive screening strategy can be valuable to determining the primary defect in phenotypically complex diseases, like ciliopathies, and broaden the phenotypic spectrum associated with C2CD3 variants.
Materials/subjects and methods
Study subjects
Family 1 was seen at Mayo Clinic and referred to the Center for Individualized Medicine by Clinical Genomics for further variant interpretation and functional validation studies. Family 2 was identified in Rhode Island and samples were forwarded to Mayo Clinic for ciliopathy gene screening. Family 3 was seen at the Royal University Hospital in Saskatoon, Canada. The families were consented to studies approved by the Mayo Clinic Institutional Review Board (IRB-12-009346 and IRB-285-00) and permission was obtained to publish photographs (where applicable) and the pertinent clinical information.
Whole exome sequencing
Clinical WES for the proband in Family 1 was performed at Baylor Genetics (custom capture), for the proband in Family 2 research WES was performed at the Mayo Clinic Medical Genome Facility (Agilent SureSelect v5 WES kit for capture), and for the proband in Family 3 clinical WES was performed at GeneDx (Agilent Clinical Research Exome kit for capture). WES for each case was completed on an Illumia HiSeq platform with 100 basepair paired-end reads. Quality control metrics of the sequencing data were generally achieved: >70% of reads aligned to target, 95% of target bases covered at >20×, 85% of target bases covered at >40×, mean coverage of the target bases was >100× for Baylor Genetics clinical exome, or variants with a quality score >20 and a read depth >10× for the Mayo Clinic research exome, or the mean depth of coverage was 64× and 94.6% of the exome was covered by at least 10× coverage for the GeneDx clinical exome. Bi-directional sequence was generated, aligned to reference gene sequences based on the Human Genome build GRCh37/UCSC hg19 and analyzed for sequence variants using either Baylor Genetics developed tools (Baylor Genetics clinical exome) or an in-house developed workflow following GATK best practice standards [9], and subsequently mined using GoldenHelix SNP and Variation Suite (Mayo Clinic research exome) where all genes were screened and evaluated for classic recessive, de novo + parental allele, and compound heterozygous inheritance at ≤0.5% minor allele frequency in the publically available databases (de novo ≤1%), or using a custom-developed analysis tool (Xome Analyzer; GeneDx clinical exome). Variants related to patient phenotypes were confirmed by capillary sequencing for patient and parental samples. C2CD3 variants were annotated using hg19, NM_001286577.1, NG_041791.1. The variants identified in C2CD3 have been submitted to the gene variant database at www.LOVD.nl/C2CD3 (individual IDs: 00163842, 00163924, and 00163925).
Skin biopsy, fibroblast culture, RNA isolation, and RNA sequencing for the proband in Family 1
A fibroblast sample was harvested from surgical waste (removed digit) from the proband in Family 1. The biopsy was minced, transferred to the bottom of a T-25 culture flask, and cultured in minimal essential media supplemented with 10% fetal bovine serum, 1% non-essential amino acids, and 1% antibiotics/antimycotics. Cultures were maintained at 37 °C in a 5% CO2 and 5% O2 incubator and media was replaced after 1 week. Once the fibroblasts had grown to ~50% confluency, cells were trypsinized and replated into a T-75 flask. Once the T-75 reached 80–90% confluency, the cells were harvested for RNA. For translation inhibition, 6 h before harvesting, the cultured fibroblasts in T-75 flask were incubated with puromycin readymade solution (Sigma Aldrich, St. Louis, MO), at a final concentration of 100 µg/mL. Cells from flasks untreated and treated with puromycin were scraped, counted, and resuspended in 0.75 mL of Qiazol solution (Qiagen, Germantown, MD) for 0.25 mL of sample, and frozen at −80 °C. RNA was isolated using the miRNeasy Mini Kit (Qiagen, Germantown, MD) following the standard protocol.
RNA libraries were prepared according to the manufacturer’s instructions for the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA). Unique indexes, included in the standard TruSeq Kits (12-Set A and 12-Set B) were incorporated at the adapter ligation step for multiplex sample loading on the flow cells. The concentration and size distribution of the libraries were determined on an Agilent Bioanalyzer DNA 1000 chip (Santa Clara, CA). A final quantification, using Qubit fluorometry (Invitrogen, Carlsbad, CA), was done to confirm sample concentration.
Two RNA-seq libraries per lane were loaded onto flow cells at concentrations of 8–10 pM to generate cluster densities of 700,000–800,000/mm2 following Illumina’s standard protocol using either the Illumina cBot or HiSeq 2500 and TruSeq Rapid Paired-End cluster kit version 1. The flow cells were sequenced as 100 basepair paired-end reads on an Illumina HiSeq 2500 using TruSeq Rapid SBS sequencing kit version 1 and HCS version 2.0.12.0 data collection software. Base-calling was performed using Illumina’s RTA version 1.17.21.3.
RNA-sequencing analysis was performed using MAP-RSeq [10]. Reads were aligned to the human genome (hg19) and transcriptome using Tophat2 [11] running Bowtie (v1) [12]. Gene and exon level read counts were generated using HiSeq [13] and BedTools [14], respectively.
PCR method for cDNA analysis for the proband in Family 2
RNA was isolated from the proband’s kidney (Family 2) using a standard TRIzol protocol. Kidney tissue from a PKD1 patient was used as a control. cDNA was synthesized using the Invitrogen Superscript III protocol. The splice alteration was assessed using a standard PCR protocol with primers spanning C2CD3 exons 16–20 and resolved on a 2% agarose gel.
Statistical methods
R [15] and lme4 [16] were utilized to apply linear mixed-effects models for comparing reads per kilobase per million mapped reads (RPKM) values, with group (patient vs. control) and variants as fixed effects, and subject and replication as random effects. Values in bar graphs were compared using a Student’s T test.
Clinical descriptions
Family 1
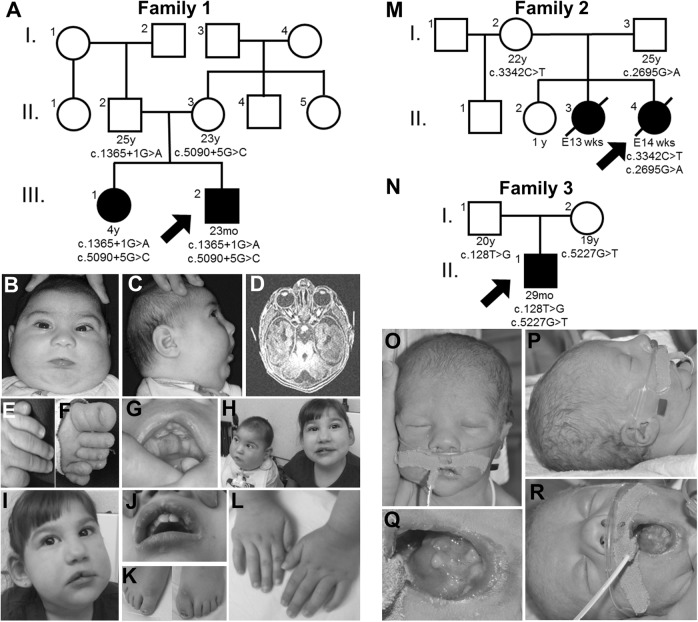
The proband in Family 1 had anomalies noted prenatally including unilateral upper extremity (left) and bilateral lower extremity postaxial polydactyly, suspected cleft palate, micrognathia, and microcephaly. The pregnancy was complicated by intrauterine growth restriction (IUGR) and polyhydramnios. This child was the product of the second pregnancy born at 38-6/7 weeks via spontaneous vaginal delivery (Fig. 1a, III.2). The delivery was complicated by non-reassuring fetal signs and respiratory distress with Apgar scores of 4 and 8 at 1 and 5 min, respectively. His birth length, weight, and head circumference were 47.5 cm (11th centile), 2.43 kg (<2nd centile), and 30 cm (<2nd centile), respectively. Clinical examination after birth revealed submucous cleft palate, incomplete cleft of the upper lip, maxillary and mandibular alveolar clefting, natal tooth on mandibular alveolus, lingual masses (lipomas and fibroma with ossification), microretrognathia, bilateral cryptorchidism, hypotonia, microcephaly, bilateral retinal colobomas, and postaxial polydactyly of the feet and left hand (Fig. 1b–h).
Fig. 1.
Pedigrees and representative images for Families 1, 2, and 3. a Pedigree for Family 1 highlighting that the proband (III.2) and his sister (III.1) are similarly affected (filled squares/circles). For all pedigrees, ages are in years (y), months (mo), and embryonic weeks (E; wks) and listed when available. The DNA variants identified in C2CD3 are shown in tested individuals to show segregation and inheritance. b-g Representative images of the proband in Family 1 at 6 months of age highlighting microcephaly and facial dysmorphisms (b, c), molar tooth sign (d), polydactyly (e, f), and tongue hamartoma and incomplete cleft lip (g). h Affected siblings together at 15 months (proband) and 4 years of age (sister). i–l Representative images of the proband’s sister at 4 years of age highlighting facial dysmorphisms and disproportionately large ears (i), incomplete cleft lip (j), and surgically corrected polydactyly of feet and hands (k, l). (m) Pedigree for Family 2 highlighting the proband (II.4) and sister (II.3) are affected. (n) Pedigree for Family 3 highlighting the proband (II.1) is affected. o–r Representative images of the proband, Family 3, highlighting anomalies of the face, ears (o, p), and tongue (q, r)
Due to a suspected ciliopathy, a brain MRI, renal ultrasound, and echocardiogram were completed. MRI demonstrated a Dandy–Walker malformation (DWM), “molar tooth” sign (MTS) (Fig. 1d), bilateral symmetric hyposulcation of the brain with rudimentary/absent sylvian fissures, insula and operculum, subependymal gray matter heterotopia, and rudimentary appearing hippocampal formations. The renal ultrasound showed bilateral pelviectasis and echocardiogram identified a small patent ductus arteriosus (PDA). At 23 months of age, the patient was tracheostomy and gastrostomy dependent, had global developmental delays, microcephaly, and sleep-disordered breathing.
The proband’s sister in Family 1 (Fig. 1a, III.1) had a history of similar congenital abnormalities including microcephaly, ankyloglossia, bifid tongue, tongue hamartomas, incomplete cleft of the upper lip, polydactyly of all four extremities, and bilateral optic nerve colobomas (Fig. 1h–l). She also had global developmental delay and microcephaly (<3rd centile). On MRI, she had a simplified gyral pattern, limbic system hypoplasia, mild cerebellar hypoplasia, and gray matter heterotopia. She carried a clinical diagnosis of OFD and had normal SNP array and karyotype, and negative molecular testing of OFD1.
Family 2
The mother was a 22-year-old with her fourth pregnancy (II.4; the third with the proband’s father; Fig. 1m). A Level II ultrasound at 14 weeks identified a posterior encephalocele and fetal echocardiogram detected a ventricular septal defect (VSD). The fetus was terminated and the postmortem examination noted multiple abnormalities, including occipital encephalocele, sloping forehead, microretrognathia, a VSD, short webbed neck, and bifid tongue. The fetus had a normal karyotype and although polydactyly was suspected by ultrasound examination, this was not confirmed on fetal examination. Microscopic analysis, including of the kidney and liver, was normal.
A pregnancy 6 months earlier had similar abnormalities (II.3). A Level II ultrasound at 13 weeks identified a large and apparently open occipital encephalocele with protruding brain tissue, leading to the decision to terminate the pregnancy. From the perinatal examination, a 1 cm vertebral defect in the lumbosacral area was noted. The karyotype was normal, and microscopic analysis of the heart, kidneys, eye, and adrenal gland did not show any abnormalities. No material was collected for DNA analysis. The other two children (II.1 and II.2) were normal.
Family 3
The mother was a 17-year-old with her first pregnancy, and used tobacco throughout the pregnancy and marijuana very early in the pregnancy. Her first ultrasound at 13 weeks had concerns for increased nuchal translucency (3.6 mm). Subsequent ultrasounds were done regularly due to fetal anomalies including IUGR, shortening of the long bones suggestive of skeletal dysplasia, bilateral ventriculomegaly, polyhydramnios, prefrontal edema, prominent upper lip, micrognathia, and a small stomach with no bubble. MRI revealed enlarged lateral ventricles associated with mild 3rd and 4th ventriculomegaly and suspected hypoplasia of cerebellar vermis raising suspicion of a DWM, as well as a small stomach, large pharynx and hypopharynx, and normal thorax with no cardiac or renal anomalies. At 26 weeks, amniocentisis was performed and the karyotype was that of a normal male.
The mother went into spontaneous labor at 38-3/7 weeks and the proband was delivered vaginally (Fig. 1m, II.1). Apgar scores were 6 and 8 at five and ten minutes, respectively. The birthweight, length, and head circumference were in the 10th (2.85 kg), 10th, and 50th centiles, respectively. The initial exam revealed significant micrognathia, malformed nodular tongue with absence of the anterior 3rd of the tongue, tight frenulum, bilaterally low-set posteriorly rotated ears, bulbous nose with long vertical crease (Fig. 1n–q), and very mild epispadias. Skeletal survey was normal. Mild respiratory distress and tonic posturing with myoclonic jerks were noted around 1 day of age with initial abnormal EEG; however, follow-up EEG was normal and seizures were ruled out. Brain MRI showed MTS, dilation of 3rd and 4th ventricles, midline cerebellar cleft, thinning of the corpus callosum with atrophy of the posterior aspect of the body and absence of the splenium; consistent with a diagnosis of Joubert syndrome. Renal ultrasound showed normal development of kidney and liver.
At 13 months of age, he continued to have significant paroxysmal events, requiring external chest compressions every 10 days, often triggered by vagal stimulation. At last follow-up, he was fed via G-tube to prevent aspiration, and had bilateral retina colobomas. He had reduced tone throughout; however, upper extremities were stiff. He continued to have developmental delays. Microarray and Joubert syndrome panel (sequencing and deletion duplication analysis of 13 genes) were normal.
Results
Whole exome sequencing
WES was completed on three probands with an underlying ciliopathy in which the genetic cause had not yet been identified. WES revealed six C2CD3 variants, with three predicted splice variants: c.1365+1G>A, c.5090+5G>C, and c.3342C>T, and three missense variants: c.2695G>A (p.(Val899Met)), c.5227G>T (p.(Gly1743Cys)), and c.128T>G (p.(Leu43Arg)), in the three probands (Table 1). Each proband inherited two C2CD3 variants and segregation analysis indicated that they were inherited in trans (Fig. 1). The six variants were either not present in gnomAD, or at an extremely low frequency (<0.001%), and all six variants were predicted by ≥3 in silico tools to either affect protein function, or disrupt splicing (Table 1). The proband in Family 1 had a similarly affected sister, who also harbored the same two C2CD3 variants. Samples were unavailable for Family 2’s similarly affected sibling. While five of the six variants were novel, one of the identified missense variants, c.5227G>T, p.(Gly1743Cys), had been previously reported in trans with a nonsense variant (c.5929C>T, p.Arg1977*), in two siblings affected with Joubert syndrome with severe global developmental delay and swallowing difficulties, oromotor apraxia, and gastrostomy (Table 2; [7, 17–25]). Because the pathogenicity of the three splice variants, c.1365+1G>A, c.5090+5G>C, and c.3342C>T, was not known, functional studies were pursued to determine the impact of these variants on C2CD3 transcripts.
Table 1.
Variants identified in C2CD3 in Families 1, 2, and 3
| Chr.11 position | cDNA variant | RNA variant | Protein variant | Location | Genotype | Inheritance | gnomAD MAF | dbSNP ID | In silico predictions & conservation | ACMG classification |
|---|---|---|---|---|---|---|---|---|---|---|
| Family 1, proband | ||||||||||
| 73834032 | c.1365+1G>A | r.1217_1365del | p.Ser406Argfs*23 | Intron 8 | Het | Paternal | 0.000004 | NR | Predicted loss of donor: SpliceSiteFinder-like, MaxEntScan, NNSPLICE, human splicing finder | Pathogenic |
| 73768446 | c.5090+5G>C | r.4951_5090del | p.Ser1652Tyrfs*26 | Intron 25 | Het | Maternal | NR | NR | Predicted loss of donor: SpliceSiteFinder-like, MaxEntScan, NNSPLICE | Pathogenic |
| Family 2, proband | ||||||||||
| 73811607 | c.2695G>A | NA | p.(Val899Met) | Exon 15 | Het | Paternal | NR | NR | GV/GD Score: 65.28/20.52 SIFT: deleterious Polyphen2: probably damaging Mutation taster: disease causing Highly conserved amino acid |
Likely pathogenic |
| 73804863 | c.3342C>T | r.3160_3344del | p.Gly1054Aspfs*7 | Exon 18 | Het | Maternal | 0.000004 | rs147520824 | Predicted loss of donor: SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer | Pathogenic |
| Family 3, proband | ||||||||||
| 73879586 | c.128T>G | NA | p.(Leu43Arg) | Exon 2 | Het | Paternal | NR | NR | GV/GD Score: 353.86/0.00 SIFT: deleterious Polyphen2: probably damaging Mutation taster: disease causing Highly conserved amino acid across mammals |
Likely pathogenic |
| 73760516 | c.5227G>T | NA | p.(Gly1743Cys) | Exon 27 | Het | Maternal | 0.000004 | NR | GV/GD Score: 0.00/158.23 SIFT: deleterious Polyphen2: Prob. damaging Mutation taster: disease causing Highly conserved amino acid |
Pathogenic |
Chr chromosome, Het heterozygous, MAF minor allele frequency, NA not available, NR not reported
Data are annotated using hg19, NM_001286577.1, NG_041791.1
Table 2.
Genotypic and phenotypic description of novel and previously described cases with C2CD3-mediated ciliopathy
| C2CD3 variants and inheritance | Sex | Age at Dx | Clinical diagnosis | Neuronal | Oral | Facial | Digital | Retinal | Renal & hepatic | Skeletal | Other | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 proband | c.[1365+1G>A];[5090+5G>C] r.[1217_1365del];[1491_5090del] p.[Ser406Argfs*23]; [Ser1652Tyrfs*26] |
M | Prenatal | OFD | DWM; MTS; hyposulcation; subependymal gray matter heterotopia; rudimentary appearing hippocampal formations; DD; hypotonia; microcephaly | Submucous palate, upper lip, maxillary and mandibular alveolar clefting; natal tooth; tongue hamartoma | Microretrognathia; facial dysmorphism | Unilateral hand and bilateral foot postaxial polydactyly | Retinal colobomas | Pelviectasis | None | IUGR, bilateral cryptorchidism PDA; sleep apnea |
| Family 1 sister | c.[1365+1G>A];[5090+5G>C] r.[1217_1365del];[1491_5090del] p.[Ser406Argfs*23]; [Ser1652Tyrfs*26] |
F | UK | OFD | Simplified gyral pattern; limbic system and cerebella hypoplasia; gray matter heterotopia; DD; microcephaly | Ankyloglosssia; bifid tongue with hamartoma; incomplete cleft of upper lip; high narrow palate | Epicanthal folds; mild synophrys; disproportionately large ears | Postaxial polydactyly of hands; pre- and post-axial polydactyly of feet | Optic nerve colobomas; cortical vision impairment; myopia; nystagmus | UK | UK | sleep apnea |
| Family 2 proband | c.[2695G>A];[3342C>T] r.[?];[3160_3344del] p.[Val899Met];[Gly1054Aspfs*7] | F | Prenatal | Fetal encephalocele | Occipital encephalaocele | Bifid tongue | Sloping forehead; microretrognathia; short webbed neck | None | None | None | None | VSD; hypoplastic thyroid |
| Family 2 sister | UK | F | Prenatal | Fetal encephalocele | Encephalocele with brain tissue protrusion | UK | UK | None | None | None | Lumbosacral vertebral defect | NR |
| Family 3 proband | c.[128T>G]; [5227G>T] p.[Leu43Arg];[Gly1743Cys] | M | Prenatal | Joubert-related disorder | MTS; DWM dilation of 3rd and 4th ventricle; midline cerebellar cleft; thin corpus callosum; DD; hypotonia | Malformed lobulated tongue; tight frenulum | Micrognathia; posteriorly rotated low-set ears; bulbous nose with vertical crease | None | Retinal colobomas | None | Shortening of long bones in utero | IUGR; mild epispadias; hypoapnea |
| Thauvin-Robinet (case 1) | c.[184C>T];[184C>T] p.[Arg62*];[Arg62*] | M | UK | OFD14 | MTS; incomplete corpus callosum agenesis; subarachnoid cysts; myelinization defect; ID; microcephaly | Lobulated tongue with hamartoma; cleft tongue; cleft palate; buccal frenulae; absent epiglottis; supernumary teeth | Facial dysmorphism | Bilateral hand postaxial polydactyly; bilateral broad hallux | Retinopathy | None | NR | micropenis |
| Thauvin-Robinet (case 2) | c.[184C>T];[184C>T] p.[Arg62*];[Arg62*] | F | Prenatal | OFD14 | Similar to Thauvin-Robinet case 1 | Similar to Thauvin-Robinet case 1 | Similar to Thauvin-Robinet case 1 | Similar to Thauvin-Robinet case 1 | Similar to Thauvin-Robinet case 1 | Similar to Thauvin-Robinet case 1 | Similar to Thauvin-Robinet case 1 | Cardiac malformation; neonatal death |
| Thauvin-Robinet (case 3) | c.[3085T>G];[3911-2A>T] r.[?];[3911_3914del] p.[Cys1029Gly];[Ala1304Valfs*3] | M | Prenatal | OFD14 | Vermian hypoplasia; mild agenesis of corpus callosum; subarachnoid cyst; microcephaly | Buccal frenulae | Facial dysmorphism | Bilateral hand post axial polydactyly; bilateral broad hallux | NR | Right renal hypoplasia | NR | micropenis |
| Bachmann-Gagescu (case 4) | c.[184C>T];[184C>T] p.[Arg62*];[Arg62*] | UK | UK | Joubert syndrome/OFD-like | UK | Oral frenulae and/or cleft palate | UK | UK | UK | UK | UK | UK |
| Bachmann-Gagescu (case 5) | c.[4951+1G>T];[5267G>A] p.[?];[Gly1756Glu] | UK | UK | Joubert syndrome/OFD-like | UK | Oral frenulae and/or cleft palate | UK | UK | UK | UK | UK | UK |
| Srour (case 6) | c.[5227G>T];[5929C>T] p.[Gly1743Cys];[Arg1977*] | M | UK | Joubert syndrome | MTS; severe global DD; hypotonia; ataxia | NR | NR | NR | NR | None | None | NR |
| Srour (case 7) | c.[5227G>T];[5929C>T] p.[Gly1743Cys];[Arg1977*] | M | UK | Joubert syndrome | MTS; severe global DD; hypotonia; ataxia | NR | NR | NR | Oculomotor apraxia | None | None | Respiratory abnormalities |
| Cortes (case 8) | c.[195G>C];[1429del] p.[Trp65Cys];[Ile477*] | F | Prenatal | Skeletal dysplasia | DWM | High arched palate | Hypertelorism, broad nasal bridge; dysplastic ears; micrognathia | Preaxial polydactyly of feet presenting as duplicated bent hallux; brachydactyly | Micropthalmia | Renal cysts | Short tubular bones; short ribs; trident acetabula | Anterior anus; atypical lung lobulation; mobile cecum |
| Cortes (case 9) | c.[195G>C];[1429del] p.[Trp65Cys];[Ile477*] | M | Prenatal | Skeletal dysplasia | Holoprosencephaly; DWM; dilated cisterna magnum and vermis aplasia; cerebellar hypoplasia; polymicrogyria; hydrocephalus | NR | Facial dysmorphism | Preaxial polydactyly of feet, pre- and post-axial polydactyly of hands | NR | Renal cysts | Short, bent tubular bones; hypoplastic tibia; short ribs; trident acetabula | ASD; atypical lung lobulation; mobile cecum; micropenis/ ambiguous genitalia |
ASD atrial septal defect, DD developmental delay, DWM Dandy–Walker malformation, Dx diagnosis, ID intellectual disability, MTS molar tooth sign, NA not available, NR not reported, PDA patent ductus arteriosis, UK unknown, VSD ventricular septal defect
Data are annotated using hg19, NM_001286577.1, NG_041791.1
RNA sequencing for the proband in Family 1 revealed reduced C2CD3 transcript expression and exon skipping for c.1365+1G>A and c.5090+5G>A
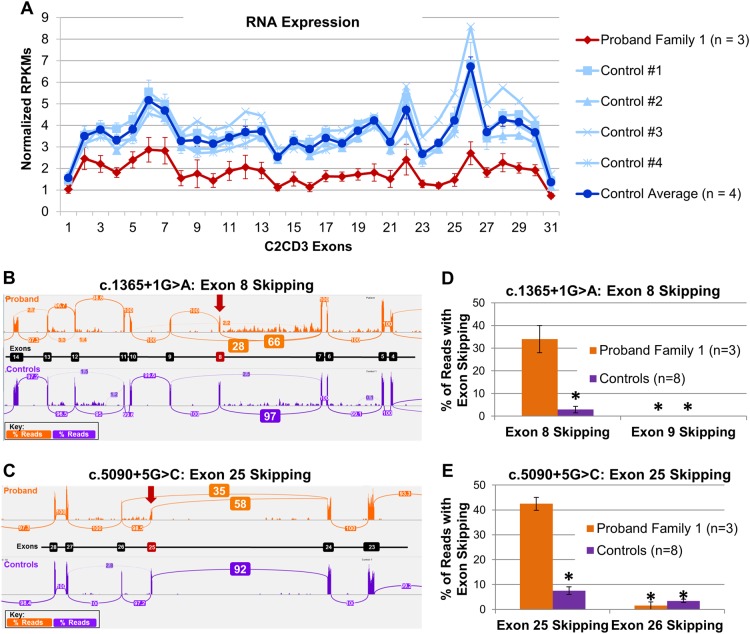
For the proband in Family 1, RNA sequencing was performed to examine gene expression, intron retention, and exon skipping for both of the putative splice variants, c.1365+1G>A and c.5090+5G>A. To examine overall RNA expression for C2CD3, RPKMs for each exon of the gene were plotted and showed reduced expression relative to tissue-matched controls throughout the entire C2CD3 gene (Fig. 2a). Using a linear mixed-effect model, we found that the proband (Family 1) had significantly lower RPKM values, 1.7328 ± 0.2297 (± standard error; χ2 (1) = 8.9825; p = 0.002726; Fig. 2a). As the proband (Family 1) had statistically reduced expression of C2CD3, we hypothesized that both alleles were likely affected by nonsense-mediated decay.
Fig. 2.
RNA expression and exon-skipping analysis for c.1365+1G>A and c.5090+5G>C variants in the proband in Family 1. a RNA-sequencing results showing the normalized Reads Per Kilobase Per Million Mapped Reads (RPKMs) for each exon in C2CD3, with the proband (Family 1) displayed in red, individual controls in light blue, and control average in blue. These results show a significant decrease in expression of C2CD3. b For c.1365+1G>A, sashimi plots show skipping of exon 8 in our proband (orange) when compared to a representative control (purple). Skipping of exon 8 (r.1217_1365del) leads to an out-of-frame protein product, p.Ser406Argfs*23. Values shown in boxes represent the splicing percentage. c For c.5090+5G>C, sashimi plots show skipping of exon 25 in our proband (orange) when compared to a representative control (purple). Skipping of exon 25 (r.1491_5090del) leads to an out-of-frame protein product, p.Ser1652Tyrfs*26. Values shown in boxes represent the splicing percentage. Bar graph highlighting the difference in exon-skipping events in the proband (orange) vs. controls (purple) for exon 8 (d) and for exon 25 (e) are statistically significant p < 0.0001
In an attempt to elucidate the cause of reduced RNA expression, we examined the rate of intron retention. At the exon 8–intron 8 boundary where the c.1365+1G>A variant resides, there were no read-through events into the intron (data not shown). Interestingly, at the exon 25–intron 25 boundary where the c.5090+5G>C variant resides, there were a handful of read-through events into the intron, which were not present in control samples (Supplementary Fig. 1). However, these findings were not statistically significant and therefore not responsible for the reduction in transcript expression as seen in Fig. 2a.
Next, we utilized the RNA-sequencing data to examine exon-skipping events for the proband (Family 1). Sashimi plots showed that the c.1365+1G>A variant led to normal splicing 66% of the time; however 28% of the reads spliced directly from exon 7 to exon 9, skipping exon 8, which was not observed in our controls (Fig. 2b). Skipping of exon 8 (r.1217_1365del) led to an out-of-frame protein product with a premature stop codon, p.Ser406Argfs*23. Sashimi plots also showed that the c.5090+5G>C variant led to normal splicing 58% of the time, however 35% of the reads resulted in splicing from exon 24 to exon 26, skipping exon 25. Skipping of exon 25 (r.1491_5090del) also led to an out-of-frame protein product with a premature stop codon, p.Ser1652Tyrfs*26 (Fig. 2c). Quantification of skipped reads for each variant are shown in bar graphs and are statistically distinct from controls (Fig. 2d, e).
Puromycin blocks translation, which in turn blocks nonsense-mediated decay; therefore, application of puromycin can “rescue” transcripts undergoing nonsense-mediated decay (NMD). Fibroblasts cultured from the proband (Family 1) and two control samples that were exposed to puromycin resulted in RPKM values that were no longer statistically distinct from our controls (Supplementary Fig. 2), suggesting “rescue” of the mRNA transcripts undergoing nonsense-mediated decay. This “rescue” effect was seen for all exons except 8 and 25 (Supplementary Fig. 3), which is consistent with the exon-skipping events observed in the Sashimi plots from our untreated RNA samples. This data provides evidence that the exon-skipping events, resulting in out-of-frame transcripts, lead to NMD, which in turn led to the reduced transcript expression of C2CD3 in the proband (Family 1; Fig. 2a).
PCR-based cDNA examination of the c.3342C>T C2CD3 variant from the proband in Family 2 revealed an impact on splicing
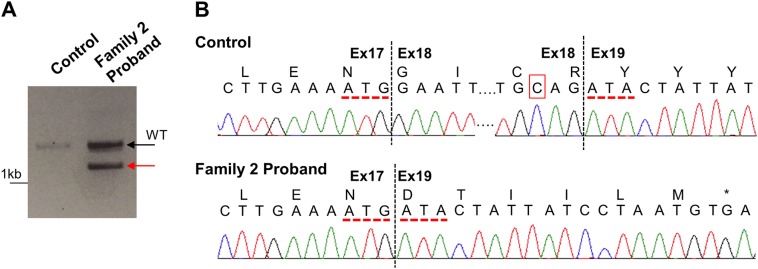
The c.3342C>T variant is located three nucleotides before the end of exon 18 and predicted to have an impact on splicing by several in silico algorithms (Table 1); therefore, functional characterization was pursued. cDNA analysis using RNA isolated from a control and a kidney sample from the proband (Family 2) was completed to examine exons 16–20. As observed in Fig. 3a, the control sample amplified a single WT product (black arrow), while the proband (Family 2) also had an alternative splice product (red arrow). Sanger sequencing completed on the cDNA PCR products showed normal splicing from exon 17 to exon 18 and exon 18 to exon 19 for the WT product (Fig. 3b). The alternative product (red arrow) from the proband (Family 2) showed skipping of exon 18 (r.3160_3344del), which resulted in a frameshift with a premature stop codon, p.Gly1054Aspfs*7.
Fig. 3.
cDNA analysis of c.3342C>T in the proband in Family 2. a cDNA analysis was completed using RNA isolated from control and the proband (Family 2) kidney samples with primers spanning exons 16–20. While the control sample amplified a single WT band (black arrow), the proband (Family 2) had an alternative splice product (red arrow) in addition to the WT band. b Sanger sequencing analysis of the cDNA PCR products revealed that the WT band of the control and of the proband (Family 2) showed correct splicing across the intron/exon 19 junction resulting in a cDNA that included exons 17–19 (top). The alternative splice band of the proband (Family 2) showed skipping of exon 18 (r.3160_3344del), resulting in a frameshift and a premature stop codon six amino acids later (p.Gly1054Aspfs*7). Red dotted lines indicate exon 17/19 junctions; red box in control sample indicates the position of the c.3342C>T variant
Discussion
Our examination of three families (five patients) with varied ciliopathy phenotypes showed that their phenotypes were due to biallelic variants in C2CD3. The variants were splicing or missense changes and scattered across the protein (Fig. 4). Five of the variants were novel, but one, c.5227G>T, p.(Gly1743Cys), was previously identified in trans with a nonsense variant, c.5929C>T, p.Arg1977* in two siblings with Joubert syndrome (Tables 1 and 2) [7]. The cDNA analysis convincingly showed that the splicing changes (even the atypical ones) resulted in the generation of frameshifted proteins with high levels of penetrance. Taken altogether, using the RNA/cDNA analyses, segregation analysis, in silico predictions, population evaluation, and the ciliopathy phenotype, we were able to establish that all six identified variants were either pathogenic or likely pathogenic using the 2015 ACMG guidelines (Table 1). These classifications allowed for a genetic diagnosis of a C2CD3-mediated ciliopathy to be made for each of the three probands, one affected sibling (Fig. 1a; Family 1: III.1), and inferred for the other affected sibling where no sample was available (Fig. 1m; Family 2: II.3).
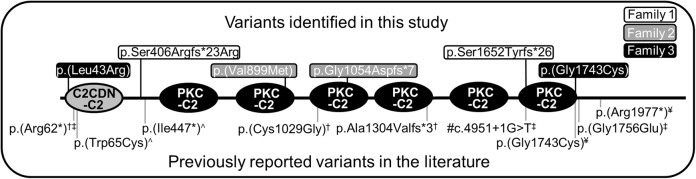
Fig. 4.
Protein domain structure of C2CD3 highlighting variants identified in this study and in the literature. The C2CDN-C2 (gray circle) and PKC-C2 domains (black circles) are shown on the protein topology for NM_001286577.1. The location of each of the identified variants identified in this study is highlighted on the top of the topology (Family 1 is in a white box, Family 2 is in a gray box, Family 3 is in a black box). Previously reported variants in the literature are shown on the bottom of the topology. #The precise protein product resulting from the c.4951+1G>T has not been established, so the location of this variant in the protein structure is putative. †[5], ‡[6], ¥[7], ˄[8]
While examining the clinical characteristics of our three probands and the two similarly affected siblings, we found significant overlap; however, not all of the phenotypes were the same and in some instances different organ systems were involved (Table 2). In addition, viability was strikingly different between the families. All of our patients had neuronal (five of five), most had oral (four of five) and facial anomalies (four of five), three had retinal phenotypes, two had digital phenotypes, congenital heart defects, and skeletal findings, and the affected males had genital anomalies. Interestingly, renal disease, nephronophthisis or polycystic kidney disease, common in many ciliopathies, were not found in our cases with the exception of pelviectasis in the proband in Family 1. Most of the observed phenotypes have been described in the context of ciliopathies, although the severity varied, especially for the neuronal phenotypes. Interestingly, phenotypes were unique between families and the variants themselves could not predict the phenotypic presentation. Despite differences observed between probands, not surprisingly affected siblings within our study had the most phenotypic overlap. The WES did not identify other clear ciliopathy alleles that may be modulating the phenotype.
Similar trends of phenotypic diversity have been observed in the five other described families (nine patients; Tables 2 and 3). Following the initial publication of three cases, OFD14 was defined as an autosomal recessive disorder caused by loss-of-function variants in C2CD3 in patients with OFD, malformations of the face, oral cavity, and digits, with microcephaly and cerebral malformations [5]. Subsequently, six additional ciliopathy cases described as Joubert syndrome or skeletal dysplasias have been identified with biallelic variants in C2CD3 (Table 2; Fig. 4) [6–8]. Interestingly, like the cases described herein, these cases did not all clearly fit within the clinical description of OFD14; rather, these cases fell along a ciliopathy spectrum, ranging from Joubert (cases 6–7), to OFD14 (cases 1–3), to skeletal dysplasias (cases 8–9; Table 2, Fig. 4) [6–8]. Similar to the cases in our study, previously published sibling pairs had similar clinical presentations. While all cases published and herein presented with features consistent with ciliopathies, the only truly consistent finding across all was brain malformations (Table 3). Interestingly, the first description of C2CD3-mediated OFD14 included cerebral malformations along with microcephaly as key features of the disorder; however, only 5 of 12 cases described have shown to be microcephalic. While these numbers could be slightly underestimated, as it is not clear if all features were assessed for in each case, not all cases of C2CD3-mediated ciliopathy present with microcephaly. Most cases presented with either MTS, DWM, or both, which is supportive of Joubert syndrome and related disorders. Oral, facial, digital, and retinal findings were common however not uniform across cases, whereas renal and skeletal anomalies were more rare. While Table 3 can start to inform features that may be present in cases with C2CD3-mediated ciliopathy, additional cases are necessary to really understand the phenotypic spectrum.
Table 3.
Phenotypic summary of newly described and previously described cases (n = 12)a
| Clinical feature | Number | Percentage |
|---|---|---|
| Neuronal | 12 of 12 | 100 |
| MTS | 6 of 12 | 50 |
| DWM | 4 of 12 | 33 |
| Brain malformation | 12 of 12 | 100 |
| Microcephalyb | 5 of 12 | 42 |
| Encephalocele | 2 of 12 | 17 |
| Hypotonia | 4 of 12 | 33 |
| ID/DD | 7 of 12 | 58 |
| Oral | 8 of 12 | 67 |
| Abnormality or clefting of lips or palate | 5 of 12 | 42 |
| Abnormality of the tongue | 6 of 12 | 50 |
| Facial | 9 of 12 | 75 |
| Micrognathia | 4 of 12 | 33 |
| Ear anomalies | 3 of 12 | 25 |
| Digital | 7 of 12 | 58 |
| Retinal | 7 of 12 | 58 |
| Renal & hepatic | 4 of 12 | 33 |
| Skeletal | 4 of 12 | 33 |
| Other | 10 of 12 | 83 |
| Respiratory | 6 of 12 | 50 |
| Genital anomalies | 5 of 12 | 42 |
| Cardiac anomalies | 3 of 12 | 25 |
aFor Bachmann-Gagescu et al. [6] a detailed description of the patients was not available; therefore, those two cases were excluded from these analyses
bMicrocephaly was previously described as a key feature for OFD14. While we have listed 5 of 12, the siblings in Family 2 were terminated in utero and it is not clear whether or not head circumference was measured at the time of autopsy.
On a genotype basis, most cases have at least one missense allele (and therefore may not be completely inactivating), but three were homozygous for a nonsense variant. The variants are also spread throughout the protein (Fig. 4). With so few cases and such varied phenotypes (Table 2, Fig. 4), it is difficult to identify specific genotype–phenotype correlations and determine whether the types of variants or the location within the protein has an impact on the patient's phenotype and disease manifestation. However, because siblings presented similarly, it is interesting to hypothesize that the C2CD3 genotype and the underlying genetic background play a role in the patient’s phenotype.
C2CD3 encodes a C2 calcium-dependent domain containing three proteins. A previously described C2cd3 knockout mouse model named Hearty showed strikingly similar phenotypes to patients with biallelic C2CD3 variants [4, 26]. A chicken with an OFD phenotype, talpid2, due to variants in C2cd3 has also been examined to understand the OFD phenotype [27, 28]. To further understand the mechanism of disease, Thauvin-Robinet and colleagues performed functional studies and identified that C2CD3 is involved in centriole elongation and cilia formation [5]. Interestingly, OFD1, which has been previously associated with OFD syndrome, acts as an antagonist to C2CD3. Taken together, Thauvin-Robinet and colleagues suggested that regulation of centriole length is a pathogenic mechanism related to ciliopathy phenotypes.
Overall, the phenotypic findings within this study and those previously described present a phenomenon that is commonplace in ciliopathies and occurs frequently in the next-generation sequencing era. Before genotypic information was known for many genetic diseases, these diseases were defined phenotypically, which was appropriate based on the available information. Based on phenotypic descriptions, different disease entities were created, like Joubert syndrome, skeletal dysplasias, OFD, and so on, defined using the organ systems involved and the phenotypic presentation of the affected individuals. In the ciliopathies, we now find that multiple genes cause the “same” disease and that the same gene causes multiple different “diseases” [1]. Now diseases can be described by the genetic cause, although as seen here, the resulting phenotypes can be quite varied, likely due to genetic and non-genetic inputs beyond the disease gene. With our expanding knowledge of genotypic information and a better understanding of human disease mechanisms, decisions will have to be made as to whether diseases should be primarily defined by their phenotypic sequelae, their underlying genotype, or if a combined naming system makes most sense.
Based on the findings of this study along with previously published cases, we confidently conclude that biallelic variants in C2CD3 are capable of causing a spectrum of ciliopathy disorders. The identification of the etiology in these syndromic diseases has allowed for more patients to receive a firm diagnosis for their underlying ciliopathy, which in some cases can be used for family planning strategies and a better understanding of disease progression and severity. As more C2CD3 cases are identified, the scientific community will better understand the role of C2CD3 in ciliopathies and whether genotype–phenotype correlations exist.
Electronic supplementary material
Acknowledgements
Mayo Clinic Center for Individualized Medicine Functional Validation Program, DK059597 (PCH).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0222-3) contains supplementary material, which is available to authorized users.
References
- 1.Braun DA, Hildebrandt F: Ciliopathies. Cold Spring Harb Perspect Biol. 2017;9:a028191. [DOI] [PMC free article] [PubMed]
- 2.Bruel AL, Franco B, Duffourd Y, et al. Fifteen years of research on oral-facial-digital syndromes: from 1 to 16 causal genes. J Med Genet. 2017;54:371–80. doi: 10.1136/jmedgenet-2016-104436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters AM, Beales PL. Ciliopathies: an expanding disease spectrum. Pediatr Nephrol. 2011;26:1039–56. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoover AN, Wynkoop A, Zeng H, Jia J, Niswander LA, Liu A. C2cd3 is required for cilia formation and Hedgehog signaling in mouse. Development. 2008;135:4049–58. doi: 10.1242/dev.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thauvin-Robinet C, Lee JS, Lopez E, et al. The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat Genet. 2014;46:905–11. doi: 10.1038/ng.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann-Gagescu R, Dempsey JC, Phelps IG, et al. Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J Med Genet. 2015;52:514–22. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srour M, Hamdan FF, McKnight D, et al. Joubert syndrome in French Canadians and identification of mutations in CEP104. Am J Hum Genet. 2015;97:744–53. doi: 10.1016/j.ajhg.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortés CR, McInerney-Leo AM, Vogel I, et al. Mutations in human C2CD3 cause skeletal dysplasia and provide new insights into phenotypic and cellular consequences of altered C2CD3 function. Sci Rep. 2016;6:24083. doi: 10.1038/srep24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asmann YW, Middha S, Hossain A, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics. 2012;28:277–8. doi: 10.1093/bioinformatics/btr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalari KR, Nair AA, Bhavsar JD, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinforma. 2014;27:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14: R36. [DOI] [PMC free article] [PubMed]
- 12.Langmead B. Aligning short sequencing reads with bowtie. Cur Protoc Bioinforma. 2010;32:11.17.11–11.17.14. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders S, Pyl PT, Huber W. Htseq - a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan AR. Bedtools: the swiss-army tool for genome feature analysis. Curr Protoc Bioinforma. 2014;47:1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R C Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/; 2015.
- 16.Bates D, Maechler M, Bolker B, Walker S. Fitting linear mixed-effects models using. lme4 J Stat Softw. 2015;67:1–48. [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–90. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 23.Tavtigian SV, Deffenbaugh AM, Yin L, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 25.Zhang MQ. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–32. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 26.Zohn IE, Anderson KV, Niswander LA. Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res A Clin Mol Teratol. 2005;73:583–90. doi: 10.1002/bdra.20164. [DOI] [PubMed] [Google Scholar]
- 27.Chang CF, Schock EN, O’Hare EA, et al. The cellular and molecular etiology of the craniofacial defects in the avian ciliopathic mutant talpid2. Development. 2014;141:3003–12. doi: 10.1242/dev.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schock EN, Chang CF, Struve JN, et al. Using the avian mutant talpid2 as a disease model for understanding the oral-facial phenotypes of oral-facial-digital syndrome. Dis Model Mech. 2015;8:855–66. doi: 10.1242/dmm.020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.