Abstract
Pontocerebellar hypoplasia (PCH) is a heterogeneous neurodegenerative disorder with a prenatal onset. Using whole-exome sequencing, we identified variants in the gene Coenzyme A (CoA) synthase (COASY) gene, an enzyme essential in CoA synthesis, in four individuals from two families with PCH, prenatal onset microcephaly, and arthrogryposis. In family 1, compound heterozygous variants were identified in COASY: c.[1549_1550delAG]; [1486-3 C>G]. In family 2, all three affected siblings were homozygous for the c.1486-3 C>G variant. In both families, the variants segregated with the phenotype. RNA analysis showed that the c.1486-3 C>G variant leads to skipping of exon 7 with partial retention of intron 7, disturbing the reading frame and resulting in a premature stop codon (p.(Ala496Ilefs*20)). No CoA synthase protein was detected in patient cells by immunoblot analysis and CoA synthase activity was virtually absent. Partial CoA synthase defects were previously described as a cause of COASY Protein-Associated Neurodegeneration (CoPAN), a type of Neurodegeneration and Brain Iron Accumulation (NBIA). Here we demonstrate that near complete loss of function variants in COASY are associated with lethal PCH and arthrogryposis.
Introduction
Pontocerebellar hypoplasia (PCH) is a heterogeneous group of rare neurodegenerative disorders, characterized by hypoplasia of cerebellum and pons. Onset is often prenatal, and atrophy of supratentorial structures is variably present [1]. Based on clinical features, neuroimaging and genetic findings, the current classification comprises eleven distinct PCH subtypes (PCH1-11), with 17 associated genes listed in the OMIM database. Clinical and neuroradiological features show great variation between and within the different subtypes. Central motor impairments, epilepsy, and severe intellectual disability are common neurological findings. Presentation in the prenatal period with polyhydramnios and multiple congenital contractures (arthrogryposis) occurs in some subtypes [1, 2].
Here, we show that loss of function variants in COASY, which encodes the bifunctional enzyme Coenzyme A (CoA) synthase, essential in the biosynthesis of CoA, can also lead to a PCH like phenotype. Partial defects in COASY have already been described as a cause of COASY Protein- Associated Neurodegeneration (CoPAN[MIM: # 615643]). CoPAN is a type of Neurodegeneration and Brain Iron Accumulation (NBIA [MIM: #615643]), and is a progressive pediatric onset disorder characterized by iron accumulation in specific brain areas, typically the basal ganglia resulting in an “eye of the tiger” sign on brain magnetic resonance imaging (MRI). [3, 4] Clinical features include dystonia, spasticity, and cognitive decline [5].
Materials and methods
Whole-exome sequencing (WES) in family 1
Whole-exome capture was performed using the SeqCap EZExomev3 (Roche NimbleGen) followed by sequencing on a HiSeq 2500 platform (Illumina) according to the manufacturer’s recommendations for paired-end 125 bp reads. Sequence reads were aligned to the human reference genome (h19) with BWAMEM (bio-bwa.sourceforge.net/). Variants were called using the GATK3.2 software package (www.broadinstitute.org/gatk/) and filtered using Cartagenia Bench Lab NGS (Agilent). Exclusion criteria were < 5 reads or a frequency higher than 1% in public (ESP, dbSNP, 1000 Genomes) and/or in-house databases. We assessed de novo, homozygous or compound heterozygous variants located in exons or within ± 6 nt in the intron.
SNP array and WES family 2
A SNP microarray (Genome-Wide Human SNP array 5.0, Affymetrix) was performed on affected individual II-4. Homozygosity mapper was used for detection of homozygous regions [6].
Whole-exome capture was performed using the Agilent Sureselect v5 exome capture kit and sequencing on a 5500xl (Life Technologies). Reads were aligned to the human reference genome (hg19) using Lifescope tools and de-duplicated and realigned using Samtools and GATK. Single-nucleotide variants were called using FreeBayes and diBayes, whereas insertions and deletions were called by Atlas 2. Variants were annotated using a using a custom in-house pipeline and rare variants obtained after excluding variants with > 1% frequency in normal populations (ESP, 1000 Genomes) or an in-house database, and those residing beyond 15 nucleotides upstream or six nucleotides downstream (− 15/ + 6) of intro–exon boundaries. The genetic and clinical details from the affected individuals from family 1 and 2 were submitted to Decipher, with the following IDs: 368461 and 368464, respectively.
Cell culture, mRNA isolation, and cDNA analysis
Amnion cells and fibroblasts were cultured on standard growth medium. RNA was isolated with the RNeasy mini kit (Qiagen, Cat. No. 74106) from cells at 75% confluence. complementary DNA (cDNA) was synthesized with SuperScript III Reverse Transcriptase, according to the manufacturers protocol (Thermofisher). cDNA was amplified by PCR (see table S3 for primer sequence). Sanger sequencing was done using standard methods.
Immunoblot analysis
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1,5% Triton X-100, 0,5% Sodiumdeoxycholate, 0.1% sodium dodecyl sulfate) for 45 min on ice. Lysates were centrifuged for 15 min at 4 °C at maximum speed. Protein concentration was measured with the BCA protein kit. Equal amounts of protein extracts were loaded and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. COASY antibody (rabbit, monoclonal, Abcam EPR8246) and actin antibody (mouse, monoclonal, chemicon, MAB1501) for loading control were used.
In vitro COASY activity assay in cell homogenates
Cells of affected individuals and controls were homogenized by sonication in phosphate buffered saline. Cell homogenates (10 µl with a protein concentration between 2 and 3 mg/ml) were added to a medium containing 5 mM 4-phosphopantetheine (4’PP, SIC-021167 Syncom) in the presence of 2 mM ATP, 5 mM MgCl2, and 100 mM Tris pH 8.0 in a final volume of 50 µl, for 1 h at 37 °C. After termination of the reaction with perchloric acid (0.42 M final concentration), the reaction mixture was centrifuged (5 min, 20,000 g, 4 °C) and the supernatant brought to pH 4.0 using 0.6 M citrate (pH 4.0) plus 2 M KOH. The products formed, i.e., dephospho-CoA and CoA were analyzed by ultra-high performance liquid chromatography on a reversed phase column (2.2 µm Acclaim RSLC PolarAdvantage van Thermo Scientific product no. 072624) using a linear gradient of acetonitrile in 100 mM potassium phosphate (pH 4.0) under continuous monitoring of the absorbance at 210 and 260 nm.
Results
Clinical and genetic features of family 1
Individual II-1 from family 1 (Fig. 1a) was the first child of a non-consanguineous couple. An ultrasound at 20 weeks of gestation showed microcephaly with normal abdominal circumference and femur length, cerebellar hypoplasia, micrognatia, multiple contractures, and polyhydramnios. Autopsy after termination at 21 weeks of gestation showed severe microcephaly (head circumference 15.4 cm; far below the 5th percentile), a hypoplastic cerebellum (transcerebellar diameter was 1.45 cm; far below the 5th percentile) without a recognizable dentate nucleus, and hypoplasia of the brainstem and spinal cord. Total brain volume was very small: the weight of the total brain after fixation was only 23.8 gram (normal at 21 weeks of gestation: 52.19 ± 7.23 gram), with the cerebellum and brainstem together weighting only 1.1 gram. Specific iron staining revealed no signs of iron accumulation in the brain, specifically not in the basal ganglia. Multiple contractures were present: elbows were flexed with clenched fists and overlapping fingers, hips were exorotated with extended legs with one clubfoot and one rocker bottom foot. Dysmorphic features included a sloping forehead and micrognathia. Array CGH analysis identified no significant chromosomal microdeletions- or duplications. Disease-causing variants in PCH-associated genes CASK, RELN, TUBA1A, and EXOSC3 were excluded with Sanger sequencing.
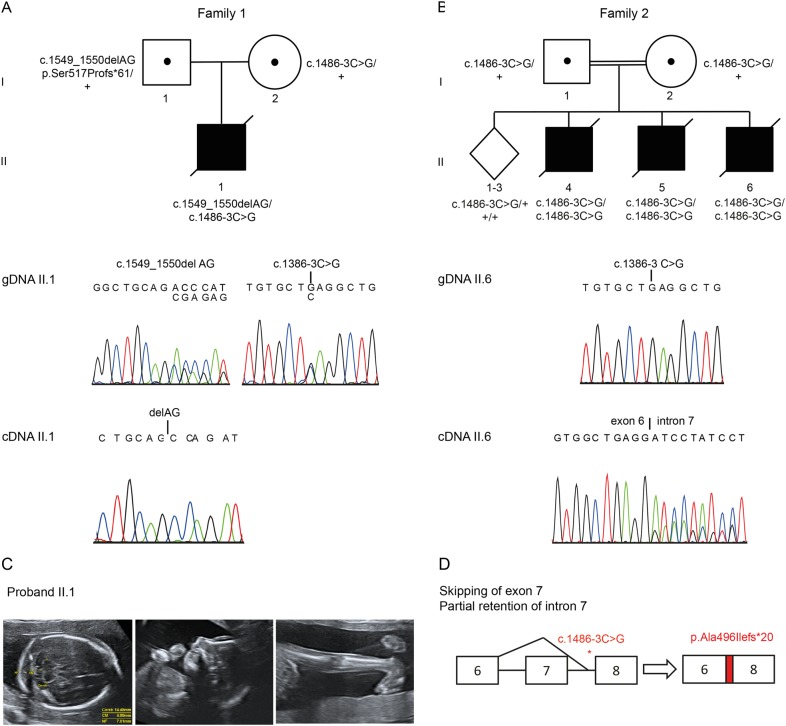
Fig. 1.
Identification of COASY variants. a Pedigrees of family 1 (left) and family 2 (right). Affected individuals are indicated by black filled symbols. Dotted symbols indicate carrier status. Wild type allele is indicated by + confirmation with Sanger sequence is shown below the pedigrees. b Left: sequence trace of COASY exon 8 of cDNA of patient II-1 from family 1, showing monoallelic expression of the paternally inherited frameshift mutation c.1549_1550delAG (p.Ser517Profs*61) in cDNA, indicating loss of expression of the maternal allele. Right: sequence trace of COASY cDNA from patient II-6 from family 2, homozygous for the c.1486-3 C > G variant in intron 7. It was shown that this variant leads to skipping of exon 7 and, by the activation of a cryptic acceptor site, partial retention of intron 7, disturbing the reading frame and resulting in a premature stop codon (c.1388_1485del98ins53; p.Ala496Ilefs*20). c Proband of family 1. Pregnancy was terminated at 22 weeks of gestation. Multiple contractures and severe microcephaly with a small cerebellum were observed. Cereb = Cerebellum, CM = Cisterna Magna, NF = Nuchal Fold. d Schematic representation of the splicing defect induced by the c.1486-3 C > G variant as described in c
Subsequently, DNA of the proband and parents was subjected to WES. We identified compound heterozygous variants in the COASY gene (GenBank: NM_025233.6) in the patient: c.[1549_1550delAG]; [1486-3 C>G]. The c.1549_1550del AG variant (paternally inherited) results in a frameshift (p.(Ser517Profs*61)) and is located in exon 8 (exons are numbered as in NG_034110.1). The c.1486-3 C>G variant in intron 7 is maternally inherited (Fig. 1b. Both variants are very rare: the frameshift variant has not been reported in the Genome Aggregation Database, whereas the c.1486-3 C>G variant has an allele frequency of 22 in 277,022 with no homozygous individuals reported. Sixteen of those 22 alleles are found in the South Asian population, suggesting a founder effect. Because the phenotype in our patient differed from the CoPAN phenotype that was previously associated with COASY variants, we used Genematcher to see whether similar cases could be encountered [7]. Indeed, a second family with an identical phenotype was identified.
Clinical and genetic features of family 2
Family 2 was a consanguineous couple of Pakistani descent with six children, three of whom were affected (Fig. 1a). In the first affected, microcephaly and a small cerebellum were detected by ultrasound at 28 weeks of gestation. Prenatal MRI confirmed these findings. After a normal at term delivery, a boy was born with a birth weight of 2200 g (below the 5th percentile) and severe microcephaly (head circumference 29.5 cm, far below the 5th percentile). He had multiple contractures and was jittery. Brain MRI showed hypoplasia of cerebellum, brainstem and spinal cord. Widening of cerebral sulci and dilatation of lateral ventricles suggested supratentorial atrophy. He died in the hospital at the age of 1 month of complications from severe PCH. In the following pregnancy, a small cerebellum was detected by ultrasound at 20 weeks of gestation. The cerebellar size was consistent with a gestational age of 17 weeks. An MRI at 22 weeks showed a small cerebellum, brainstem, spinal cord, and basal ganglia. Based upon these findings, the pregnancy was terminated. The subsequent pregnancy had a similar course.
Owing to the presence of parental consanguinity and a genetically heterogeneous recessive condition, a SNP microarray (Genome-Wide Human SNP array 5.0, Affymetrix) was performed on affected individual II-4. Six regions of homozygosity were identified (Supplementary Material, Table S1) that contained two known PCH genes: SEPSECS and RARS2. Sanger sequencing of the coding exons of these two genes did not identify any potentially disease-causing variants. Subsequently, affected individual II-5 underwent WES. Analysis of the homozygous variants residing in the regions of homozygosity identified in individual II-4 identified three variants (Supplementary Material, Table S2), including the c.1486-3 C>G variant in COASY. Segregation studies in the remaining five siblings demonstrated that all three affected siblings were homozygous for the COASY variant, whereas the three unaffected siblings were either heterozygous or homozygous for the wild-type allele (Fig. 1b).
Functional characterization of COASY mutations
To evaluate the effect of the identified variants on splicing, mRNA was extracted from amniocytes from individual II-1 from family 1 and fibroblasts from individual II-6 from family 2, reverse transcribed into cDNA and subjected to Sanger sequencing. Predominant expression from the paternal allele with the c.1549_1550delAG frameshift variant was observed in individual II-1 from family 1 (Fig. 1c). Despite the use of two different primer pairs, we only detected a minor signal representing the maternal allele harboring the c.1486-3 C>G variant. This suggests that the transcript from the allele containing the splice site variant likely undergoes nonsense mediated decay, whereas the transcript from the allele with the frameshift variant has a higher stability.
In individual II-6 (family 2), homozygous for the c.1486-3 C>G variant, analysis of cDNA fragments showed that this variant results in skipping of exon 7 and partial retention of intron 7 through the loss of the exon 7 donor splice site and the activation of a cryptic splice acceptor site 53 nucleotides upstream of exon 8, introducing a frameshift and premature stop codon (c.1388_1485delins(NG_034110.1:8532_8585)),p.Ala496Ilefs*20; Fig. 1c). In addition, a minor amount of correctly spliced transcript was detected. We assume that the different fate of the abnormal transcript of the allele with the splice site mutation in individual II-1 and II-6 is explained by a bias in PCR amplification of the transcripts, induced by the presence of an allele with higher stability in individual II-1.
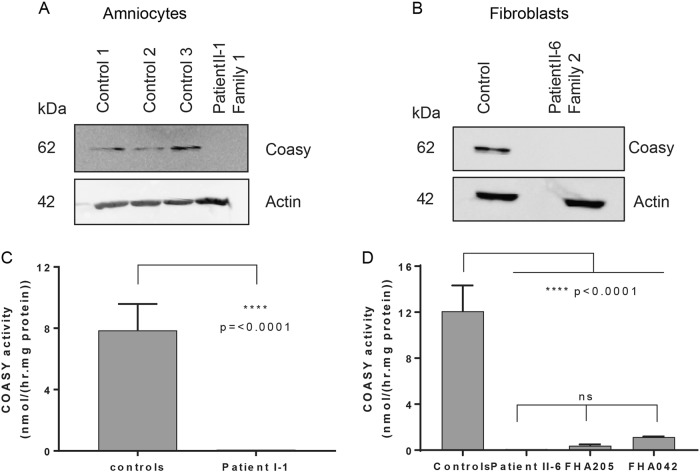
Analysis of COASY protein levels in amniocytes of individual II-1 from family 1 and fibroblasts from individual II-6 from family 2, revealed no detectable COASY protein on immunoblot (Fig. 2a, b).
Fig. 2.
Immunoblot and in vitro CoA synthesis in cells of affected individuals. a Immunoblot analysis of COASY in amniocytes of individual II-1 compared with controls, showing no COASY protein in the patient. b Immunoblot analysis of COASY in fibroblasts of individual II-6 compared with control, showing no COASY protein in the patient. c COASY activity in amniocytes of individual II-1 from family 1 compared with 4 controls. Mean ± SD of values from three experiments are shown. A t test was performed to determine the p value. d COASY activity in fibroblasts of individual II-6 from family 2 compared with four controls and two NBIA patients (FHA205 and FHA042). Mean ± SD of values from three experiments are shown. An one-way ANOVA was performed to detect statistically significant differences. Residual COASY activity was not significantly different in individual II-6 compared with the NBIA individuals
Subsequently, the enzymatic activity of COASY was assessed in cells from affected individual II-1 from family 1 and affected individual II-6 from family 2. In both patients, COASY activity was markedly reduced to ~ 1% of that in control amniocytes and fibroblasts, respectively (Fig. 2c, d). Fibroblasts of the individuals with NBIA and COASY pathogenic variants described by Dusi et al. [4] were included in the analysis for comparison. In fibroblasts of these patients, a small amount of residual activity was measured (3% and 9% compared with 1% in patient II-6; Fig. 2d).
Discussion
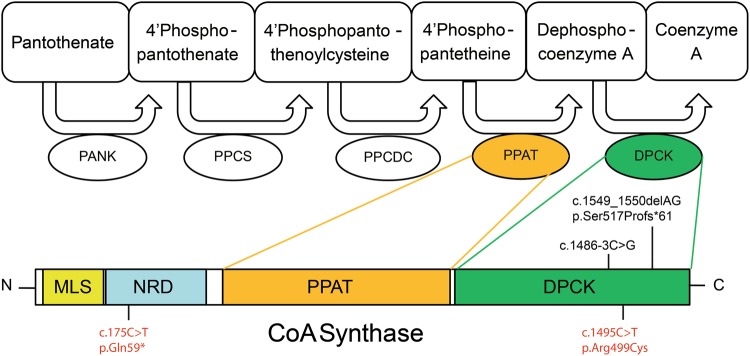
Taken together, we have identified biallelic variants in COASY in four patients from two families with PCH, severe prenatal onset microcephaly and arthrogryposis. We showed that no stable COASY protein was produced in patient cells, which is in agreement with the almost complete absence of COASY activity in vitro. The pathway of CoA synthesis is conserved in all organisms and consists of five enzymatic steps. First, pantothenic acid is phosphorylated to produce 4′PP by pantothenate kinase. Phosphopantothenoylcysteine synthase then conjugates 4’PP with cysteine [8]. The resulting 4′phosphopantothenoylcysteine is converted to 4′-phosphopantetheine [8]. The last two steps, adenylation and phosphorylation are catalyzed by COASY, which comprises both 4′PP adenyltransferase and dephospho-CoA kinase (DPCK) activity (Fig. 3). [9] As no other pathway is currently known, COASY is considered crucial for de novo CoA synthesis. CoA is an essential factor involved in numerous enzyme reactions including pyruvate dehydrogenase, 2-ketoglutarate dehydrogenase, acyl-CoA synthase, and thereby affect multiple fundamental metabolic processes such as the citric acid cycle, fatty acid oxidation, and amino-acid degradation. In addition, roles of CoA in protein acetylation, cell signaling, and DNA integrity have been implied recently [10–13]. Defects in both PANK2 and COASY have previously been associated with two types of NBIA: pantothenate kinase-associated neurodegeneration (PKAN) and CoPAN, respectively. Iron staining of the brain of affected individual II-1 from family 1 did not show iron accumulation. This might be explained by the early age of death (gestational age of 21 weeks. We show here that more severe COASY defects result in a lethal phenotype. The variants we report here are predicted to affect both the ubiquitously expressed alpha isoform as well as the brain specific beta isoform, as is the case with the disease-causing variants reported in individuals with CoPAN [4, 14, 15]. COASY gamma is a third isoform, predicted to contain the C-terminal region with the DPCK domain. We compared COASY enzyme activity in fibroblasts of individual II-6 of family 2 and the two individuals with CoPAN reported by Dusi et al. In these CoPAN individuals, a small amount of COASY protein was detected in fibroblasts, whereas in our patients' cells the protein was virtually absent. In line with this, the residual activity of COASY was somewhat higher in the CoPAN patients compared with the PCH patient (3% and 9% versus 1%). This result is based on three independent experiments, and although the difference is not statistically significant, we consider the lower residual activity of COASY as a trend that most likely contributes to the more severe phenotype in the patients we report in this paper.
Fig. 3.
Overview of CoA synthesis pathway and schematic representation of COASY gene. PPAT and DPCK activity comprised by COASY are colored orange and green, respectively. Variants identified in this study are depicted in black, variants found in other studies and associated with NBA are depicted in red
Regarding the fundamental function of CoA, it remains enigmatic how the affected children managed to survive until term. It is generally agreed that all intracellular CoA is either obtained via the five-step de novo synthesis described above, or from the uptake of 4′PP, which is subsequently converted to CoA by COASY [11, 16]. In the absence of functional COASY, both pathways fail. There are however several clues for the presence of a (yet unidentified) CoA transporter in the cellular membrane. In zebrafish embryos, the phenotype resulting from a morpholino knockdown of COASY expression could be rescued by both the addition of CoA to water and by injection of CoA in the brain ventricle [17]. Also the phenotype in both zebrafish- and hiPSC PKAN models could be rescued by the addition of CoA to the water and culture medium, respectively [18, 19]. In line with this, it seems reasonable to assume that the fetal CoA depletion is partially rescued by supplementation with maternal CoA. This would explain the in utero survival of our patients.
Electronic supplementary material
Acknowledgements
We thank both families for their participation in this study. We acknowledge Els Voorhoeve from the VU medical center in Amsterdam for providing the control fibroblasts and amniocytes for this study. We thank Eleonora Aronica for performing the iron staining on brain tissue of the affected individual from family 1. We thank Valeria Tiranti from the Istituto Neurologico Carla Besta for providing the fibroblasts of the individuals with NBIA. This work was supported by the Joshua Deeth Foundation.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0233-0) contains supplementary material, which is available to authorized users.
References
- 1.Namavar Y, Barth PG, Kasher PR, et al. Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain. 2011;134:143–56. doi: 10.1093/brain/awq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth PG Pontocerebellar hypoplasias. An overview of a group of inherited neurodegenerative disorders with fetal onset. Brain Dev. 1993;15:411–22. [DOI] [PubMed]
- 3.Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat Genet. 2001;28:345–9. doi: 10.1038/ng572. [DOI] [PubMed] [Google Scholar]
- 4.Dusi S, Valletta L, Haack TB, et al. Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation. Am J Hum Genet. 2014;94:11–22. doi: 10.1016/j.ajhg.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venco P, Dusi S, Valletta L, Tiranti V. Alteration of the coenzyme A biosynthetic pathway in neurodegeneration with brain iron accumulation syndromes: Table 1. Biochem Soc Trans. 2014;42:1069–74. doi: 10.1042/BST20140106. [DOI] [PubMed] [Google Scholar]
- 6.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper–An interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:593–9. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, De Crécy-Lagard V. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem. 2002;277:21431–9. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- 9.Aghajanian S, Worrall D. Identification and characterization of the gene encoding the human phosphopantetheine adenylyltransferase and dephospho-CoA kinase bifunctional enzyme (CoA) synthase. Biochem J. 2002;365:13–18. doi: 10.1042/bj20020569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosveld F, Rana A, Van Der Wouden PE, et al. De novo CoA biosynthesis is required to maintain DNA integrity during development of the Drosophila nervous system. Hum Mol Genet. 2008;17:2058–69. doi: 10.1093/hmg/ddn105. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan B, Sibon OCM. Coenzyme A, more than ‘just’ a metabolic cofactor. Biochem Soc Trans. 2014;42:1075–9. doi: 10.1042/BST20140125. [DOI] [PubMed] [Google Scholar]
- 12.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–50. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 13.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–21. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Evers C, Seitz A, Assmann B, et al. Diagnosis of CoPAN by whole exome sequencing: waking up a sleeping tiger’s eye. Am J Med Genet Part A. 2017;173:1878–86. doi: 10.1002/ajmg.a.38252. [DOI] [PubMed] [Google Scholar]
- 15.Annesi G, Gagliardi M, Iannello G, Quattrone A. Mutational analysis of COASY in an Italian patient with NBIA. Park Relat Disord. 2016;28:150–1. doi: 10.1016/j.parkreldis.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Sibon OCM, Strauss E. Coenzyme A: to make it or uptake it? Nat Rev Mol Cell Biol. 2016;17:605–6. doi: 10.1038/nrm.2016.110. [DOI] [PubMed] [Google Scholar]
- 17.Khatri D, Zizioli D, Tiso N, et al. Down-regulation of coasy, the gene associated with NBIA-VI, reduces Bmp signaling, perturbs dorso-ventral patterning and alters neuronal development in zebrafish. Sci Rep. 2016;6:1–15. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zizioli D, Tiso N, Guglielmi A, et al. Knock-down of pantothenate kinase 2 severely affects the development of the nervous and vascular system in zebrafish, providing new insights into PKAN disease. Neurobiol Dis. 2016;85:35–48. doi: 10.1016/j.nbd.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orellana DI, Santambrogio P, Rubio A, et al. Coenzyme A corrects pathological defects in human neurons of PANK2‐associated neurodegeneration. EMBO Mol Med. 2016;8:1197–211. doi: 10.15252/emmm.201606391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.