Abstract
Many industrial activities produce H2S, which is toxic at high levels and odorous at even very low levels. Chemolithotrophic sulfur-oxidizing bacteria are often used in its remediation. Recently, we have reported that many heterotrophic bacteria can use sulfide:quinone oxidoreductase and persulfide dioxygenase to oxidize H2S to thiosulfate and sulfite. These bacteria may also potentially be used in H2S biotreatment. Here we report how various heterotrophic bacteria with these enzymes were cultured with organic compounds and the cells were able to rapidly oxidize H2S to zero-valence sulfur and thiosulfate, causing no apparent acidification. Some also converted the produced thiosulfate to tetrathionate. The rates of sulfide oxidation by some of the tested bacteria in suspension, ranging from 8 to 50 µmol min−1 g−1 of cell dry weight at pH 7.4, sufficient for H2S biotreatment. The immobilized bacteria removed H2S as efficiently as the bacteria in suspension, and the inclusion of Fe3O4 nanoparticles during immobilization resulted in increased efficiency for sulfide removal, in part due to chemical oxidation H2S by Fe3O4. Thus, heterotrophic bacteria may be used for H2S biotreatment under aerobic conditions.
Electronic supplementary material
The online version of this article (10.1007/s10532-018-9849-6) contains supplementary material, which is available to authorized users.
Keywords: Sulfide oxidation, Heterotrophic bacteria, Immobilized cells, Sulfide:quinone oxidoreductase
Introduction
H2S is produced by various industrial and natural activities, such as petroleum refining, methane-containing biogas production, wastewater treatment, and food processing (Eikum and Storhaug 1986; Hughes et al. 2009; Janssen et al. 2009). H2S is malodorous at low levels and toxic at high levels, inhibiting aerobic respiration to humans and microorganisms (Nicholls and Kim 1982). It can also be problematic in sewer systems, causing corrosion (Zhang et al. 2008). Under neutral pH, H2S and HS− are the major species of sulfide, as pKa1 is 6.9 and pKa2 is > 12 (Kabil and Banerjee 2010).
Biofiltration is often used in H2S removal from waste gas or waste water because of its effectiveness, low energy consumption and minimal by-product generation compared to chemical and physical treatments (Sorokin 1994). Researchers have tried different reactor designs, various packing materials and nutrients for biofiltration to achieve stable efficiency of H2S elimination (Ben Jaber et al. 2016a, b; Gerrity et al. 2016; Li et al. 2008). However, biofiltration is limited to the use of a few of chemolithotrophic sulfur-oxidizing bacteria, such as Thiobacillus spp., Acidithiobacillus spp. and green sulfur bacterium (Pokorna and Zabranska 2015). A problem of using these sulfur-oxidizing bacteria is the production of sulfuric acid, leading to acidification of the liquid phase; however, the problem can be prevented if oxygen supply is restricted (Dolejs et al. 2015; Janssen et al. 1998; Sorokin et al. 2008; Wang et al. 2016). When acidification occurs, alkaline materials are added to neutralize the acidified liquid, which increases the cost (Gerrity et al. 2016; Mora et al. 2015; Pokorna and Zabranska 2015). Although some sulfide oxidizing bacteria such as Acidithiobacillus spp. can tolerate acidic conditions (Ben Jaber et al. 2016b), the acid effluent is also a source of pollution. Further, acidification does not favor H2S absorption for microbial consumption (Hughes et al. 2009). Consequently, a large reactor volume is required to increase the retention time of waste gas for efficient removal of H2S.
Although there were sporadic reports on utilizing heterotrophic bacteria for H2S removal under aerobic conditions, the genes and enzymes involved in the process were unknown (Chung et al. 1996). Recently, heterotrophic bacteria have been discovered to oxidize sulfide with sulfide:quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO) (Luebke et al. 2014; Xin et al. 2016). Bacteria with sqr and pdo genes are able to oxidize sulfide produced from sulfur-containing organic compounds, such as cysteine. When the genes are deleted, the mutant cannot oxidize the self-produced sulfide and release H2S to the gas phase (Xia et al. 2017). Many common soil bacteria, such as Bacillus spp. and Pseudomonas spp. possess sqr and pdo genes, and their efficiency to oxidize sulfide at low levels has been demonstrated (Xia et al. 2017). However, it is unclear whether all heterotrophic bacteria with sqr and pdo genes can rapidly oxidize sulfide at high levels and what they oxidize sulfide to. Thus, here we want to make a thorough investigation on the possibility of these bacteria for applications in H2S biotreatment. Here, we investigated whether heterotrophic bacteria could effectively oxidize exogenous H2S.
Materials and methods
Bacterial strains and culture condition
The bacterial strains used in this study are listed in Table 1. Lysogeny broth (LB) medium was used for culturing most bacteria, and D-sorbitol medium (DM) was used for Gluconobacter oxydans 621H (Yang et al. 2008). Zunongwangia profunda SM-A87 was cultivated in a medium composed of 10 g L−1 peptone, 5 g L−1 yeast extract and artificial sea water (Qin et al. 2007). Most of the bacteria were incubated at 30 °C, and E. coli BL21 was incubated at 37 °C.
Table 1.
The rates of sulfide oxidation by tested and reported bacteria
| Strains | Sulfide oxidation ratea | Reaction condition |
|---|---|---|
| T. thiooxidans | 158b | Aerobic |
| T. denitrificans | 80c | Anaerobic and nitrate |
| A. thioparus TK-m | > 59d | Aerobic |
| G. oxydans 621H | 50.1 ± 6.7e | Aerobic |
| T. ferrooxidans | 48b | Aerobic |
| Chlorobium | 12f | Light |
| P. aeruginosa PAO1 | 9.5 ± 0.5e | Aerobic |
| P. putida S16 | 8.4 ± 0.5e | Aerobic |
| B. cepacia ATCC 25416 | 5.6 ± 1.1e | Aerobic |
| S. marcescens ATCC 13880 | 1.5 ± 0.2e | Aerobic |
| Z. profunda SM-A87 | 0.25 ± 0.04e | Aerobic |
| B. cereus ATCC 10876 | 0.19 ± 0.02e | Aerobic |
| S. aureus ATCC 6538P | 0.08 ± 0.01e | Aerobic |
aThe tested bacteria are heterotrophs with SQR and PDO, and the reported bacteria are chemolithotrophs or phototroph. The unit of sulfide oxidation rate is µmol min−1 g−1 of cell dry weight
bOprime et al. (2001)
cSublette and Sylvester (1987)
dKanagawa and Mikami (1989)
eOur data
fKim and Chang (1991)
Sulfide oxidation analysis
Bacteria were cultivated for about 24 h when the bacteria reached to stationary phase and OD600nm was greater than 4. The cells were harvested by centrifugation (5000×g, 10 min) and resuspended to a turbidity of 2 at 600 nm in 50 mM HEPES buffer, pH 7.4 containing 50 μM diethylenetriaminepentaacetic acid (DTPA) to minimize spontaneous sulfide oxidation (Hughes et al. 2009). 10 mL of the cell suspension was transferred to a 50-mL centrifuge tube. Freshly prepared NaHS (Sigma; CAS: 207683-19-0) solution was added to initiate the reaction. The tube was capped tightly and incubated at 30 °C with shaking of 180 rpm. Sulfide was analyzed at various time intervals by using a diamine reagent (Fogo and Popowsky 1949). After the reaction, the cells were harvested again, washed with deionized water and lyophilized for 12 h to obtain cell dry weights (Xia et al. 2012). Sulfide oxidation rate (q) was determined by the following equation:
where C0 and Ct were the sulfide concentrations (µM) at the beginning and the time of sampling; V was the volume of solution (mL), m was the dry weight (g) of cells, and T was the incubation time (min). Sulfide oxidation analysis with immobilized cells was similar in 50 mM HEPES buffer, pH 7.4.
The sulfide oxidation experiments with multiple sulfide additions were done in a mineral medium (MM). One liter of MM contained 0.3 g of (NH4)2SO4, 1.5 g of Na2HPO4, 0.5 g of KH2PO4, 0.095 g of MgCl2, 0.033 g of CaCl2, 5 g of glucose, 0.1 g of pantothenic acid (Gupta et al. 2001) and 1 mL of a trace mineral solution. The trace mineral solution contained 4.5 g L−1 FeSO4·7H2O, 1.44 g L−1 ZnSO4·7H2O, 0.86 g L−1 MnSO4·H2O, 0.16 g L−1 CuSO4, 0.28 g L−1 CoSO4·7H2O, 0.06 g L−1 H3BO3 and 10 mL L−1 H2SO4 (conc.). The suspended or immobilized cells were in 50-mL centrifuge tubes containing 10 mL of MM and incubated at 30 °C, 180 rpm. Sulfide was added to 2 mM to initiate the reaction in each cycle. After each reaction, the immobilized cells were washed with water and re-incubated in fresh MM at 30 °C, 180 rpm. Each reaction cycle was carried out in every 12 h.
End-products analysis
Bacteria were harvested when they were cultivated to stationary phase and resuspended in 50 mM HEPES buffer to a turbidity of 8 at 600 nm. Ten mL of the cell suspension was transferred to a 50-mL centrifuge tube. Freshly prepared sodium sulfide was added to initiate the reaction. The tube was capped tightly and incubated at 30 °C, 180 rpm. The products and sulfide were analyzed at various time intervals.
To test the changes in pH after sulfide oxidation by magnetically immobilized cells, the experiments were done in the 50-mL centrifuge tube containing 10 mL of unbuffered 0.9% NaCl (pH 7) at 30 °C, 180 rpm. Sulfide was added to 2 mM to initiate the reaction in each cycle, and the next cycle started when sulfide was completely oxidized. The pH was tested at the beginning and the end of each cycle. The products were detected at the end of each cycle.
Cellular zero-valence sulfur, including polysulfide, persulfide, and elemental sulfur, was detected by the cyanolysis method (Xin et al. 2016). Sulfate, thiosulfate and sulfite were detected by using ion chromatography (ICS-1100 system; Dionex) with a mobile phase of 20 mM KOH at a flow rate 1 mL per min. The retention times of sulfite, sulfate and thiosulfate were 7.4 min, 7.9 min and 25.7 min, respectively (Liu et al. 2014). For tetrathionate detection, ion chromatography (ICS-1100 system; Dionex) was used with a moblie phase of 15 mM KOH at a flow rate 0.9 mL per min.
Preparation of Fe3O4 nanoparticles
The Fe3O4 nanoparticles were prepared according to a reported method with some modifications (Wang et al. 2007). 58.75 g of FeCl3·6H2O and 21.5 g of FeCl2·4H2O were dissolved in 1.5 L of distilled water at 30 °C with a gentle stream of N2 bubbling to minimize the oxidation of Fe2+. NH3 (8 M) was then slowly injected into the mixture with vigorous stirring until the pH reached 10. After precipitation, the Fe3O4 particles were repeatedly washed with distilled water until the pH became constant and they were lyophilized for 24 h to remove water. The lyophilized product was pulverized in an agate mortar to form Fe3O4 powder. 1.5 g of Fe3O4 powder was added into 10 mL of distilled water and dispersed by ultrasonic disruption (20 kHz; 10 min; Q125; QSonica) to form a stable suspension.
Preparation of nonmagnetically immobilized and magnetically immobilized cells
The nonmagnetically and magnetically immobilized cells were prepared according to a reported method (Wang et al. 2007) with some modifications. Briefly, bacteria were harvested when they were cultivated to stationary phase and resuspended in a small volume of distilled water. The alginate gel (3% [wt/vol]) and cell suspension were mixed at a ratio of cell wet weight to dry alginate powder of 3 (wt/wt). Nonmagnetically immobilized cells were formed by extruding the mixture through a syringe into 0.2 M CaCl2 and incubated for 2 h to let the beads harden. For preparing magnetically immobilized cells, an 80 µL/mL Fe3O4 particle suspension was added to the above-mentioned mixture of alginate and cell suspension, and the procedure was the same as above. Nonmagnetically immobilized inactive cells and magnetically immobilized inactive cells were prepared in the same way by using heat-inactivated (boiling water 30 min) cells.
Scanning electron microscope (SEM)
The alginic gel beads were first fixed with 2.5% of glutaraldehyde in 0.2 M phosphate buffer (pH 7.0) for 12 h, washed three times in the 0.2 M phosphate buffer (pH 7.0). The treated beads was further fixed with 1% of OsO4 for 2 h and washed three times in the 0.2 M phosphate buffer. The specimen was dehydrated by using a series of ethanol (30%, 50%, 70%, 80%, 90%, 95% and 100%) for 15 to 20 min at each soaking. In the end, the specimen was dehydrated in Alpha 1–2 LD plus freeze dryer (CHRIST, Germany) for 75 min, coated with gold, and observed under SEM by using a QUANTA FEG 250 Scanning Microscope (FEI company, USA).
Analysis of TsdA in sequenced bacterial genomes and construction of SQRs and PDOs phylogenetic trees
A microbial genomic protein sequence set from NCBI updated until November 11, 2017 was downloaded for thiosulfate dehydrogenase (TsdA) search. The query sequences of TsdA were reported TsdA(Denkmann et al. 2012; Kurth et al. 2016) and were used to search the database by using Standalone BLASTP algorithm with conventional criteria (e-value ≤ 1 e−10, coveryage ≥ 40%, identity ≥ 30%) to obtain TsdA candidates from 8286 bacterial genomes. A conserved domain COG3258 of TsdAs was used as standard feature for further filtration of TsdA candidates. The selected candidates were then manually screened by reported conserved amino acid sequences (Denkmann et al. 2012). The candidates were combined with the query TsdAs for phylogenetic tree analysis by using ClustalW for alignment and MEGA version 7.0 program for neighbor-joining tree building with a pairwise deletion, p-distance distribution, and bootstrap analysis of 1000 repeats as parameters(Kumar et al. 2016). The candidates that were in the same clade with the query TsdAs were identified as putative TsdA. The phylogenetic trees of 11 SQRs and 9 PDO (Table S1) were constructed by using a neighbor-joining analysis with the MEGA version 7.0 program, running a pairwise deletion, p-distance distribution, and bootstrap analysis of 1000 repeats.
Results
Heterotrophic bacteria with sqr and pdo oxidize exogenous sulfide
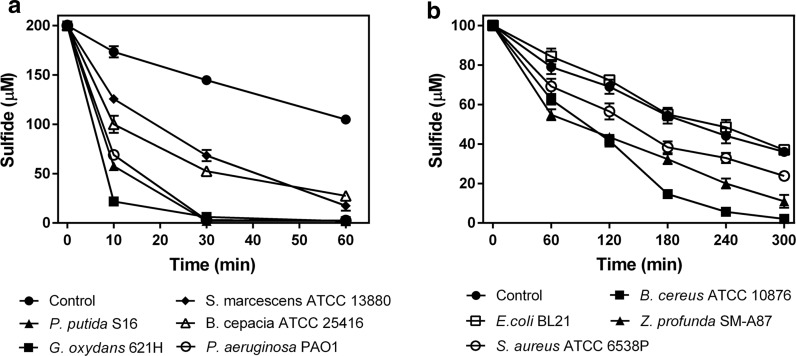
We selected eight heterotrophic bacteria containing sqr and pdo (Xia et al. 2017), grew them in a rich medium, harvested cells, and tested the resting cells for the oxidation of exogenous sulfide. They all oxidized sulfide, but the control E. coli BL21(DE3) without sqr and pdo did not (Fig. 1a, b). Most sulfide-oxidizers had higher activities of sulfide oxidation at the stationary phase of growth than at the log phase, but Gluconobacter oxydans 621H had similar rates at all phases of growth. Thus, all the cells were cultured to the stationary phase, harvested, and used for sulfide oxidation. The results showed all tested heterotrophic bacteria with sqr and pdo oxidized sulfide. Gluconobacter oxydans 621H, P. putida S16, P. aeruginosa PAO1 and B. cepacia ATCC 25416 were more efficient for sulfide oxidation (Fig. 1a) than others (Fig. 1b). The slow decrease of sulfide in the control was attributed to volatilization and abiotic oxidation of sulfide (Xin et al. 2016). The sulfide oxidation rates of these heterotrophic bacteria with sqr and pdo ranged from 0.1 to 50 µmol min−1 g−1 of cell dry weight, showing that some of the tested heterotrophic bacteria had rates of sulfide removal sufficient for H2S biotreatment, as the unit rates are not too far behind those reported rates of chemolithotrophic bacteria (Table 1).
Fig. 1.
Heterotrophic bacteria containing sqr and pdo oxidized exogenous sulfide. Cells were cultured in LB, harvested, wash and re-suspended at OD600nm of 2 in 50 mM HEPES buffer, pH 7.4, containing 50 µM DPTA. NaHS was added to 100 or 200 µM to initiate the reaction. a Six bacteria oxidized exogenous sulfide rapidly. b Three bacteria slowly oxidized exogenous sulfide; E. coli did not. Control contained no cells. Averages (n ≥ 3) with standard deviations (error bar) were shown
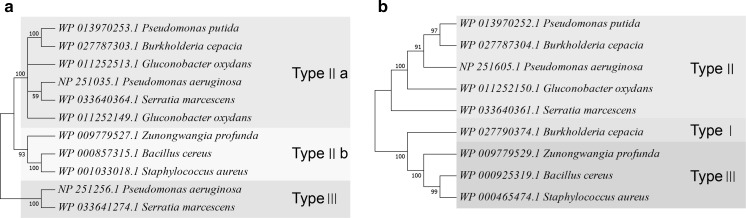
SQRs are characterized into six types, and the PDOs are classified into three (Gregersen et al. 2011; Xia et al. 2017). To evaluate whether the types of SQRs and PDOs may affect the degradation rate, we constructed phylogenetic trees with the 11 SQRs and nine PDOs from the eight tested bacteria. The SQRs were grouped into type II and type III, and the type II SQRs were further divided into types IIa and types IIb. The fast sulfide-oxidizing bacteria G. oxydans 621H, P. putida S16, P. aeruginosa PAO1, and B. cepacia ATCC 25416 all contained types IIa SQRs. The PDOs were mapped into all three types, and the fast sulfide-oxidizing bacteria all contained the type II PDOs (Fig. 2b). Interestingly, G. oxydans 621H had two sqr genes, encoding two type IIa SQRs, and one of them was adjacently linked to a pdo gene on genome. The sqr and pdo genes of P. putida S16 and B. cepacia ATCC 25416 were also adjacently linked on genome (Fig. 2a, b). However, the slow sulfide-oxidizing bacterium S. marcescens ATCC 13880 also harbored type IIa SQR and type II PDO (Fig. 2a, b). The results suggest that bacteria with type IIa SQRs and type II PDOs are likely fast sulfide oxidizers, but they need to be verified.
Fig. 2.
The phylogenetic trees of SQRs and PDOs from the eight heterotrophic bacteria. 11 SQRs (a) and 9 PDOs (b) were used for phylogenetic tree construction with reference sequences
The products of H2S oxidation
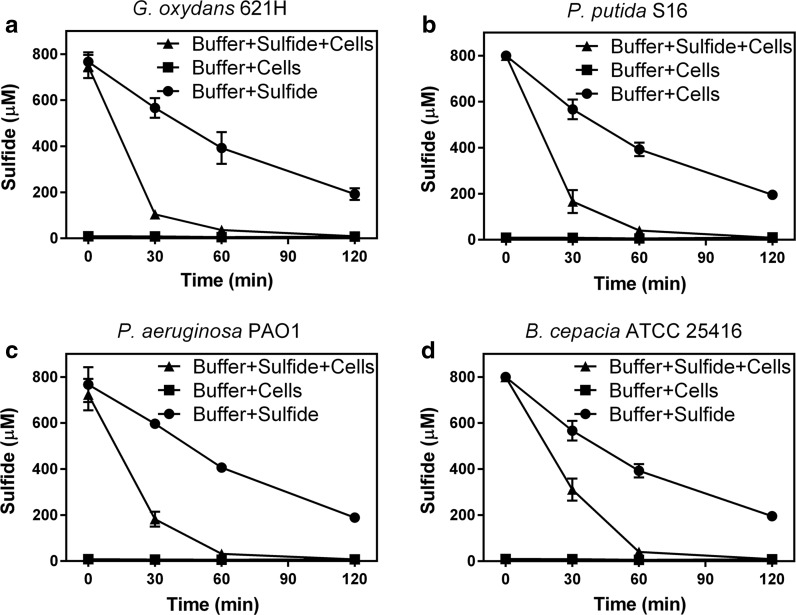
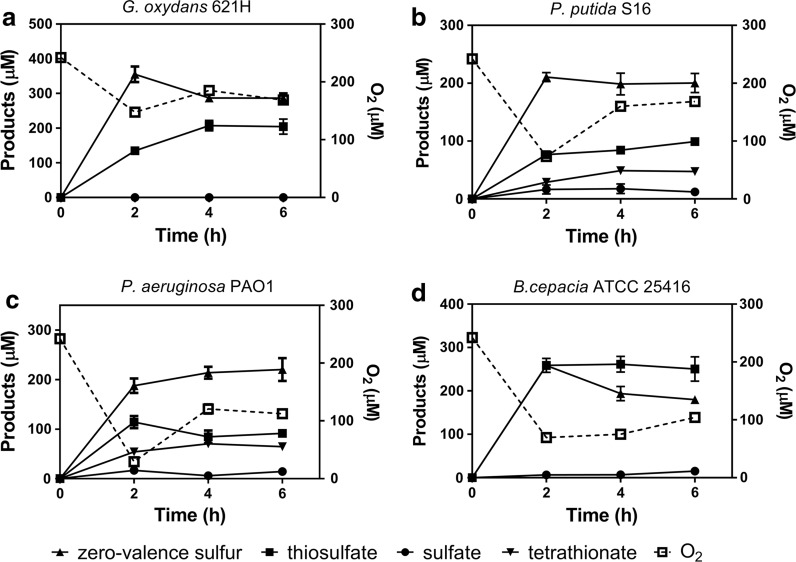
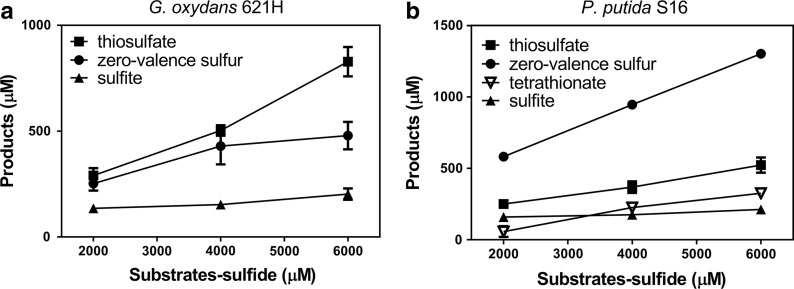
Four bacteria that rapidly oxidized sulfide (Fig. 1a) were used to analyze the end products from sulfide oxidation. To facilitate the detection of end products, sulfide was added to resuspended cells to a final concentration of 750 ± 50 µM, which was completely oxidized within 2 h (Fig. 3). The products were then detected. Sulfite was not detectable in all the samples. Thiosulfate and zero-valence sulfur were the main products of G. oxydans 621H and B. cepacia ATCC 25,416 (Fig. 4). Pseudomonas putida S16, P. aeruginosa PAO1 also produced tetrathionate (Fig. 4). Within the first 2 h of the reaction, sulfide was completely consumed by the tested bacteria, and the oxygen in the reaction mixture was decreased from about 240 μM to different levels, ranging from 25 μM with P. aeruginosa PAO1 to 150 μM with G. oxydans 621H (Fig. 4). After the first sampling at 2 h of incubation, further reduction in oxygen content was not observed. The increase in dissolved oxygen was likely due to sampling, during which the tube was opened. The product composition did not change much after the first 2-h incubation. Gluconobacter oxydans 621H that consumed the least amount of oxygen accumulated the most zero-valence sulfur (Fig. 4a). The two Pseudomonas spp. that consumed the most oxygen in the first two h also produced tetrathionate (Fig. 4b, c).
Fig. 3.
H2S oxidation by heterotrophic bacteria. Cells were harvested in LB, wash and re-suspended in 50 mM HEPES buffer (pH 7.4) at OD600nm of 8. NaHS was added to 750 ± 50 µM to initiate the reaction. a G. oxydans 621H; b P. putida S16; c P. aeruginosa PAO1; d B. cepacia ATCC 25416; sulfide only (closed square); cells only (closed square); cells and sulfide (closed triangle). All data are average of at least three samples with standard deviation (error bar)
Fig. 4.
The products of H2S oxidation by selected bacteria. The conditions were the same as described in Fig. 1 legend. Sulfide was completely oxidized within 2 h and products were analyzed. a G. oxydans 621H; b P. putida S16; c P. aeruginosa PAO1; d B. cepacia ATCC 25416. Averages (n ≥ 3) with standard deviations (error bar) were shown
Bacteria with TsdA can convert thiosulfate to tetrathionate
The production of tetrathionate is likely due to the presence of TsdA in P. putida S16 and P. aeruginosa PAO1. Pseudomonas spp. are known to use thiosulfate dehydrogenase (TsdA) to oxidize thiosulfate to tetrathionate, which may provide some supplemental energy (Denkmann et al. 2012; Sorokin et al. 1999). Thus, the distribution of TsdA were analyzed among 8286 microbial genomic protein sequences (NCBI updated until November 11, 2017) by using BLAST search, and then confirmed with the conserved domain and conserved amino acid sequence and phylogenetic tree analysis (Denkmann et al. 2012). 1275 identified TsdA distributed in 1112 bacterial genomes, including 553 Betaproteobacteria, 294 Gammaproteobacteria, 115 Alphaproteobacteria, 91 Epsilonproteobacteria, 11 Bacilli, 9 Synechococcales, 8 Flavobateria, 7 Sphingobacteria, 6 Deltaproteobacteria, and other classes with a few genomes containing TsdA. Of the Gammaproteobacteria, there are 178 Pseudomonas genomes contained TsdA species. Thus, when bacteria with TsdA are used for biotreatment, tetrathionate may also be produced.
Cell growth and sulfide oxidation are coupled
Pseudomonas putida S16 consumed 1 g of glucose and 0.29 g of (NH4)2SO4 to yield 0.24 g of biomass as our test in MM (Eq. 1). Then, the harvested P. putida S16 cells (0.24 g dry weight) oxidized 726 μmol of H2S to about 242 μmol of S0, 121 μmol of H2S2O3, 61 μmol of H2S4O3 in a buffer in 6 h (Fig. 4) (Eq. 2). Here the bacterial cells were for catalysis, and no carbon source was provided to support growth.
| 1 |
| 2 |
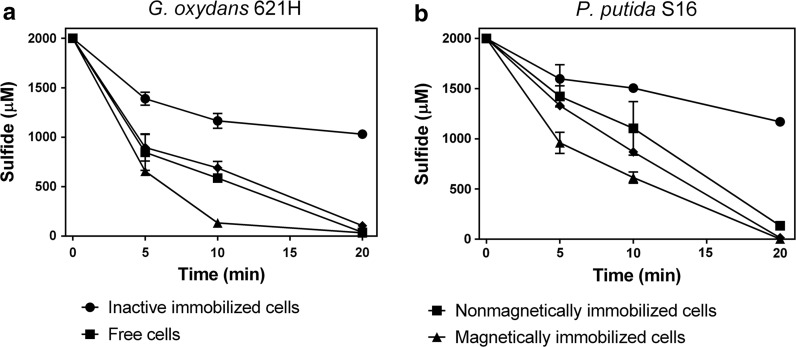
Sulfide oxidation by immobilized cells and free cells
We immobilized G. oxydans 621H and P. putida S16 into alginate gel beads as they showed the high rates of sulfide oxidation (Fig. 1, Table 1), and the immobilized cells had equivalent sulfide-oxidation rates to those of the suspended cells (Fig. 5). When cells were coimmobolized with the Fe3O4 nanoparticles into gel beads, the activity of sulfide oxidation increased by about 30% (Fig. 5). Further research found that Fe3O4 nanoparticles-containing beads without cells also catalyzed sulfide oxidation, consuming 400 μM more sulfide than the control with the beads containing no Fe3O4 in 40 min (Fig. S3), suggesting the ferric iron in Fe3O4 nanoparticles contributes to the increased sulfide oxidation rate.
Fig. 5.
Sulfide oxidation by free cells, immobilized cells, and magnetically immobilized cells. The experiments were carried out in the 50-mL centrifuge tube containing 10 mL of 50 mM HEPES buffer (pH 7.4) at 30 °C, 180 rpm. The biomass was equivalent between free cells (OD600 of 8) and immobilized cells. NaHS was added to 2 mM to initiate the reaction. a G. oxydans 621H immobilized cells and free cells; b P. putida S16 immobilized cells and free cells. Averages (n ≥ 3) with standard deviations (error bar) were shown
The sulfide oxidation products by immobilized cells of G. oxydans 621H and P. putida S16 containing Fe3O4 nanoparticles were also mainly zero-valence sulfur and thiosulfate (Fig. 6). The total amount of the detectable sulfur was less than half of the added sulfide, and the rest might be trapped inside the gel beads, which could not be detectable without releasing from the beads. Different from the products of free cells (Fig. 4), there was also a small amount of sulfite in the products of immobilized cells with Fe3O4 nanoparticles. Further research found that the same amount of Fe3O4 powder without cells reacted with sulfide under aerobic conditions to produce about 20% sulfite, 60% thiosulfate and 20% zero-valence sulfur, suggesting that Fe3O4 affected the product composition. However, the reaction rate with Fe3O4 was slower, at about 25% of that by immobilized G. oxydans 621H with Fe3O4 nanoparticles.
Fig. 6.
The products of magnetically immobilized cells. The experiments were carried out in the 50-mL centrifuge tube containing 10 mL of 0.9% NaCl at 30 °C on a reciprocal shaker at 200 rpm. NaHS was added to 2 mM to initiate the reaction in each cycle and the products were detected at the end of each cycle. a G. oxydans 621H magnetically immobilized cells; b P. putida S16 magnetically immobilized cells. Averages (n ≥ 3) with standard deviations (error bar) were shown
No acidification after sulfide oxidation
Acidification during sulfide biotreatment is a potential problem when sulfuric acid is produced by chemolithotrophs with sufficient supply of O2 (Pokorna and Zabranska 2015). When immobilized G. oxydans 621H and P. putida S16 with Fe3O4 nanoparticles were tested, the pH in the liquid phase (0.9% NaCl solution) was slightly increased after three cycles of sulfide oxidation, consuming total 6 mM NaHS (Table 2). The lack of acidification is likely due to the production of zero-valence sulfur, thiosulfate, and tetrathionate instead of sulfate (Fig. 6).
Table 2.
The change of pH after sulfide oxidation
| Strains | pH | |||
|---|---|---|---|---|
| Initiation | First cycle | Second cycle | Third cycle | |
| G. oxydans 621H | 7.38 ± 0.03 | 7.67 ± 0.10 | 7.97 ± 0.15 | 8.17 ± 0.15 |
| P. putida S16 | 7.13 ± 0.06 | 7.43 ± 0.12 | 7.67 ± 0.06 | 7.95 ± 0.05 |
The experiments were carried out in the 50-mL centrifuge tube containing 10 mL of 0.9% NaCl and magnetically immobilized cells at 30 °C, 180 rpm. NaHS was added to 2 mM to initiate the reaction in each cycle and the pH was tested at the beginning and the end of each cycle
The sulfide oxidation activities of immobilized cells are increased after repeated use and culturing
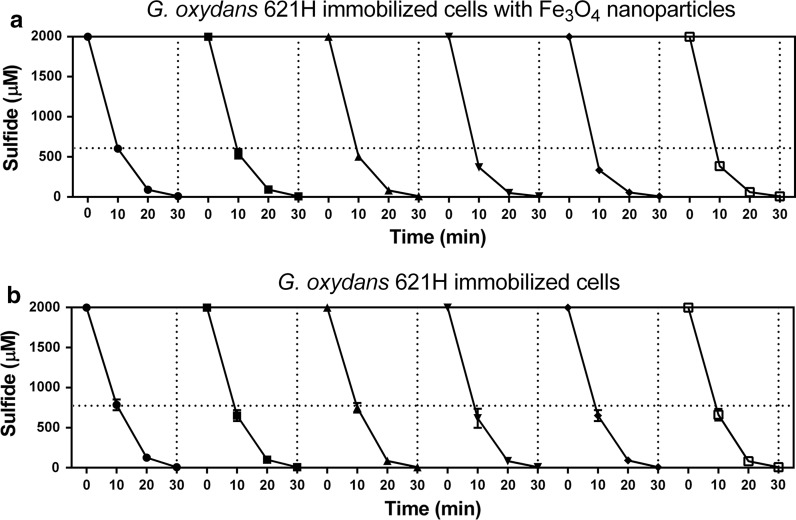
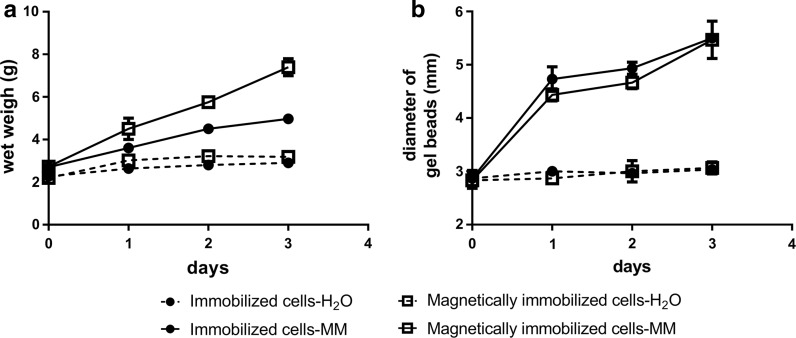
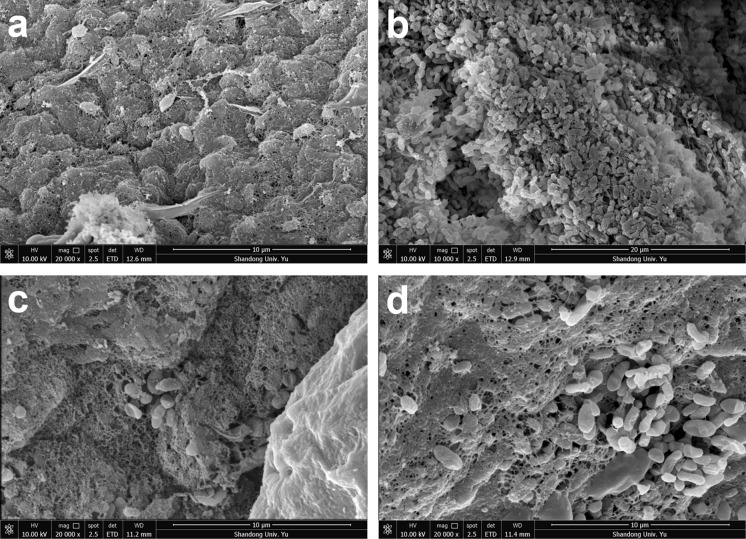
The activities of immobilized cells with or without Fe3O4 nanoparticles were repeatedly tested six times in 3 days, and the beads were culture in MM with 0.5% of glucose between testing. As shown in Fig. 7a, the sulfide oxidation activities of immobolized cells with Fe3O4 nanoparticles were obviously increased after 6 days’ use; they oxidized 2000 μM sulfide to about 600 μM in 10 min during the first cycle. After 3 days of sulfide-oxidation and culturing in MM, they oxidized 2000 μM sulfide to less than 400 μM in 10 min (Fig. 7a). Further, the sulfide oxidation activities of immobilized cells without Fe3O4 nanoparticles also increased to a less degree after three days (Fig. 7b). We then measured the wet weigh and diameter of gel beads over the 3 days. The diameters of the immobilized-cell beads with or without Fe3O4 nanoparticles increased to the same degree (Figs. 8b, S4), but the wet weigh of the beads with Fe3O4 nanoparticles increased more than the beads without (Fig. 8a). The increase was due to cell growth because the beads incubated in distilled water did not increase in both size or weight (Fig. 8). The results suggests that the gel beads with Fe3O4 nanoparticles has greater biomass carrying capacity. This possibility was further investigated by using scanning electron microscope (SEM) images of G. oxydans 621H immobilized in alginate gel beads (Fig. 9). After 3 days’ culturing, G. oxydans 621H had more biomass in gel beads with Fe3O4 nanoparticles than that in gel beads without Fe3O4 nanoparticles, both inside (Fig. 9a, b) and on the surface (Fig. 9c, d). More loose pores were observed in gel beads with Fe3O4 nanoparticles (Fig. 9), which may facilitate nutrient transfer and provide space for cells to grow.
Fig. 7.
The sulfide oxidation activity of immobilized cells was increased after repeated use and culturing. The experiments were carried out in 50-mL centrifuge tube containing 10 mL of MM at 30 °C, 180 rpm. NaHS was added to 2 mM to initiate the reaction in each cycle and was completely oxidized within 40 min. After each reaction, the immobilized cells were washed with water and re-incubated in fresh MM with glucose. The cycle of sulfide oxidation was performed every 12 h. a G. oxydans 621H immobilized cells with Fe3O4 nanoparticles; b G. oxydans 621H immobilized cells. Averages (n ≥ 3) with standard deviations (error bar) were shown
Fig. 8.
The change of wet weigh and diameter of the gel beads with immobilied G. oxydans 621H cells during culturing in distilled waster or MM. The cells were immobilized in alginate gel beads with or without Fe3O4 nanoparticles. The experiments were carried out in 50-mL centrifuge tubes containing 10 mL of MM or distilled water at 30 °C, 180 rpm. a The change of wet weigh of immobilized-cell beads. b The change of diameters of immobilized-cell beads. Averages (n ≥ 3) with standard deviations (error bar) were shown
Fig. 9.
SEM images of immobilized G. oxydans 621H cells. The cells were immobilized in alginate gel beads or alginate gel beads with Fe3O4 nanoparticles. After 3 days of sulfide oxidation and incubation (Fig. 7 legend), the beads were analzyed by SEM. The G. oxydans 621H cells were embedded in the gel beads. a Inside of the gel beads; b inside of the gel beads with Fe3O4 nanoparticles; c surface of the gel beads; d surface of the gel beads with Fe3O4 nanoparticle
Discussion
The common presence of sulfide-oxidizing activities in heterotrophic bacteria has only recently been recognized (Luebke et al. 2014; Xia et al. 2017). In this report, some of them are shown to be effective for sulfide oxidation (Fig. 1), and the finding may promote their use in H2S biotreatment. Heterotrophs rely on organic compounds as the source of both carbon and energy source for growth, and they are likely oxidize H2S for detoxification (Luebke et al. 2014; Xia et al. 2017). Sulfur-oxidizing chemolithotrophs oxidize sulfide to gain energy for growth; and anaerobic photoautotrophic bacteria use H2S to provide the reducing power for photosynthesis (Syed et al. 2006); they are commonly applied in the biological treatment of H2S under aerobic and anaerobic conditions (Ferrera et al. 2004; Janssen et al. 1997; Krayzelova et al. 2015). The finding that some heterotrophic bacteria can rapidly oxidize sulfide offers additional choices for H2S biotreatment.
As previously reported, the photoautotrophic Chlorobium has a sulfide removal rate of 12 µmol min−1 g−1 of cell dry weight (Kim and Chang 1991). The maximum sulfide oxidation rate of Thiobacillus denitrificans is reported as high as 80 µmol min−1 g−1 of cell dry weight under anaerobic conditions, using nitrate as the electron acceptor (Sublette and Sylvester 1987). Acidithiobacillus spp. can tolerate acidic condition, and they have also been used for H2S biotreatment, although their preferred substrates are metal sulfides or elemental sulfur (Ben Jaber et al. 2016b). Under aerobic and acidic conditions, Acidithiobacillus thioparus TK-m oxidizes H2S at a rate higher than 59 µmol min−1 g−1 of cell dry weight, which is estimated based on the H2S load rate for complete removal (Kanagawa and Mikami 1989). Thiobacillus thiooxidans and T. ferrooxidans show sulfide removal rates of about 158 and 48 µmol min−1 g−1 of cell dry weight, respectively, under acidic conditions (Oprime et al. 2001). Gluconobacter oxydans 621H showed rate of sulfide removal closely behind those of chemolithotrophs, and Pseudomonas spp. also had adequate rates for sulfide oxidaiton (Table 1). Thus, heterotrophic bacteria have the potential in sulfide biotreatment, especially when they can grow fast on consuming organic compounds and co-oxidize H2S. They can also be cultured, harvested, and used an agent for H2S treatment in reactor or in the field.
Immobilized cells can effectively avoid the loss of biomass and possess better stability compared to the suspended cells in sulfide removal reactor (Kim et al. 2008). Fe3O4 nanoparticles can be economically produced, and they can be directly used for oxidizing sulfide in sewers (Lin et al. 2017). Further, Fe3O4 nanoparticles are widely used as additives for bacterial immobilization because the nanoparticles can reduce the mass transfer resistance inside the gel beads and facilitate bioremediation of organic pollutants and removal of heavy metals (Wang et al. 2007; Zhang 2003). We demonstrate that the addition of Fe3O4 nanoparticles into gel beads produces more space inside the beads, facilitating nutrient transfer, providing space for growth, and resulting in an increased sulfide oxidation activity in repeated use and culturing (Figs. 7, 8, 9). The Fe3O4 nanoparticles also catalyzed chemical oxidation of sulfide, possibly due to the presence of Fe3+ in Fe3O4 (Li et al. 2008). On the basis of 30% sulfide oxidation by Fe3+, theoretically only 7% of Fe3O4 was consumed after six oxidaton cycles of total 12 mM sulfide (Fig. 7).
Most significantly, we showed G. oxydans 621H and P. putida S16 immobilized cells oxidize sulfide without causing acidification, probably due to the production of thiosulfate and zero-valence sulfur instead of sulfuric acid. The oxidation of H2S to zero-valence sulfur consumes protons, which could be the reason for the slight increase of pH. In our test, O2 was not a limiting factor, especially for G. oxydans 621H that mainly produced zero-valence sulfur. Even when it was initially consumed before the first sampling time point, it was reintroduced during sampling. In comparison, a strict control of O2 levels is required to minimize sulfuric acid production by sulfur-oxidizing chemotrophic bacteria (Pokorna and Zabranska 2015).
The production of thiosulfate and zero-valence sulfur is in agreement with our previous report that recombinant E. coli with SQR and POD oxidize sulfide to sulfite and thiosulfate (Xin et al. 2016). Interestingly, these wild type bacteria with SQR and PDO did not accumulate sulfite. This is likely due the slow polysulfide oxidation in these bacteria, and the produced sulfite rapidly reacts with the accumulated polysulfide to produce thiosulfate (Xin et al. 2016). We also detected tetrathionate as a major product after sulfide oxidation by P. putida S16 and P. aeruginosa PAO1, which is likely due to the presence of TsdA in the bacteria. Thus, the production of zero-valence sulfur, thiosulfate, and tetrathionate may prevent acidification after immediate sulfide oxidation. However, thiosulfate and tetrathionate may be further oxidized by microorganisms (Lenk et al. 2012), and the oxidation may be minimized by eliminating or inhibiting bacteria with TsdA.
H2S emission is a problem in sewer systems mainly due to its corrosion of concrete pipes. Traditional chemical methods of remediation bring huge costs to urban governance (Zhang et al. 2008). Considering sulfide oxidation by heterotrophic bacteria does not cause apparent acidification and the magnetically immobilized heterotrophs with Fe3O4 nanoparticles are conveniently recyclable, our finding may provide a new way to control the H2S corrosion in sewer systems. Further, many heterotrophic bacteria have the ability to degrade organic pollutants, such as pesticides, and to immobilize heavy metals (Cycoń et al. 2017; Mulligan 2005). Therefore, heterotrophic bacteria may potentially degrade organic pollutants as well as removing sulfide.
In conclusion, we identified that heterotrophic bacteria with sqr and pdo had the ability to oxidize exogenous sulfides and some of them have comparable oxidation rates with those of chemolithotrophic bacteria. In addition, the fast growth and no-acidification offer some advantages for H2S removal. Since these heterotrophic bacteria with sqr and pdo are abundant and diverse in nature (Xia et al. 2017), many of them may potentially be used for sulfide biotreatment. Thus, this report may promote the use of heterotrophic bacteria in H2S biotreatment. At least, they can be used as alternative choices rather than chemolithotrophs for H2S bioremediation. Further, since they are common in nature (Xia et al. 2017), they may be simply used for in situ H2S oxidation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The work was financially supported by grants from the National Natural Science Foundation of China (31770126, 21477062), and the State Key Laboratory of Microbial Technology at Shandong University. The authors declare that they have no conflicts of interest with the contents of this article.
Contributor Information
Honglei Liu, Phone: + 86 159-6664-2788, Email: lhl@sdu.edu.cn.
Luying Xun, Phone: +1 509-335-2787, Email: luying_xun@vetmed.wsu.edu.
References
- Ben Jaber M, Couvert A, Amrane A, Rouxel F, Le Cloirec P, Dumont E. Biofiltration of H2S in air—Experimental comparisons of original packing materials and modeling. Biochem Eng J. 2016;112:153–160. doi: 10.1016/j.bej.2016.04.020. [DOI] [Google Scholar]
- Ben Jaber M, Couvert A, Amrane A, Rouxel F, Le Cloirec P, Dumont E. Biofiltration of high concentration of H2S in waste air under extreme acidic conditions. New Biotechnol. 2016;33:136–143. doi: 10.1016/j.nbt.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Chung Y-C, Huang C, Tseng C-P. Biodegradation of hydrogen sulfide by a laboratory-scale immobilized Pseudomonas putida CH11 biofilter. Biotechnol Prog. 1996;12:773–778. doi: 10.1021/bp960058a. [DOI] [PubMed] [Google Scholar]
- Cycoń M, Mrozik A, Piotrowska-Seget Z. Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: a review. Chemosphere. 2017;172:52–71. doi: 10.1016/j.chemosphere.2016.12.129. [DOI] [PubMed] [Google Scholar]
- Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IAC, Dahl C. Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ Microbiol. 2012;14:2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x. [DOI] [PubMed] [Google Scholar]
- Dolejs P, Paclík L, Maca J, Pokorna D, Zabranska J, Bartacek J. Effect of S/N ratio on sulfide removal by autotrophic denitrification. Appl Microbiol Biotechnol. 2015;99:2383–2392. doi: 10.1007/s00253-014-6140-6. [DOI] [PubMed] [Google Scholar]
- Eikum A, Storhaug R. Odour problems related to waste water and sludge treatment. In: Nielsen VC, Voorburg JH, L’Hermite P, editors. Odour prevention and control of organic sludge and livestock farming. London: Taylor & Francis Group; 1986. [Google Scholar]
- Ferrera I, Sánchez O, Mas J. A new non-aerated illuminated packed-column reactor for the development of sulfide-oxidizing biofilms. Appl Microbiol Biotechnol. 2004;64:659–664. doi: 10.1007/s00253-004-1581-y. [DOI] [PubMed] [Google Scholar]
- Fogo JK, Popowsky M. Spectrophotometric determination of hydrogen sulfide. Anal Chem. 1949;21:732–734. doi: 10.1021/ac60030a028. [DOI] [Google Scholar]
- Gerrity S, Kennelly C, Clifford E, Collins G. Hydrogen sulfide oxidation in novel horizontal-flow biofilm reactors dominated by an Acidithiobacillus and a Thiobacillus species. Environ Technol. 2016;37:2252–2264. doi: 10.1080/09593330.2016.1147609. [DOI] [PubMed] [Google Scholar]
- Gregersen L, Bryant D, Frigaard N-U. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol. 2011 doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Singh VK, Qazi GN, Kumar A. Gluconobacter oxydans: its biotechnological applications. J Mol Microbiol Biotechnol. 2001;3:445–456. [PubMed] [Google Scholar]
- Hughes MN, Centelles MN, Moore KP. Making and working with hydrogen sulfide: the chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: a review. Free Radic Biol Med. 2009;47:1346–1353. doi: 10.1016/j.freeradbiomed.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Janssen AJH, Ma SC, Lens P, Lettinga G. Performance of a sulfide-oxidizing expanded-bed reactor supplied with dissolved oxygen. Biotechnol Bioeng. 1997;53:32–40. doi: 10.1002/(SICI)1097-0290(19970105)53:1<32::AID-BIT6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Janssen A, Meijer S, Bontsema J, Lettinga G. Application of the redox potential for controling a sulfide oxidizing bioreactor. Biotechnol Bioeng. 1998;60:147–155. doi: 10.1002/(SICI)1097-0290(19981020)60:2<147::AID-BIT2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Janssen AJH, Lens PNL, Stams AJM, et al. Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci Total Environ. 2009;407:1333–1343. doi: 10.1016/j.scitotenv.2008.09.054. [DOI] [PubMed] [Google Scholar]
- Kabil O, Banerjee R. Redox biochemistry of hydrogen sulfide. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa T, Mikami E. Removal of methanethiol, dimethyl sulfide, dimethyl disulfide, and hydrogen sulfide from contaminated air by Thiobacillus thioparus TK-m. Appl Environ Microbiol. 1989;55:555–558. doi: 10.1128/aem.55.3.555-558.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BW, Chang HN. Removal of hydrogen sulfide by Chlorobium thiosulfatophilum in immobilized-cell and sulfur-settling free-cell recycle reactors. Biotechnol Prog. 1991;7:495–500. doi: 10.1021/bp00012a003. [DOI] [PubMed] [Google Scholar]
- Kim JH, Rene ER, Park HS. Biological oxidation of hydrogen sulfide under steady and transient state conditions in an immobilized cell biofilter. Bioresour Technol. 2008;99:583–588. doi: 10.1016/j.biortech.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Krayzelova L, Bartacek J, Díaz I, Jeison D, Volcke EIP, Jenicek P. Microaeration for hydrogen sulfide removal during anaerobic treatment: a review. Rev Environ Sci Biotechnol. 2015;14:703–725. doi: 10.1007/s11157-015-9386-2. [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth JM, Brito JA, Reuter J, et al. Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. J Biol Chem. 2016;291:24804–24818. doi: 10.1074/jbc.M116.753863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk S, Moraru C, Hahnke S, et al. Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J. 2012;6:2178–2187. doi: 10.1038/ismej.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sun T, Zhu N, Cao X, Jia J. Comparative study of using different materials as bacterial carriers to treat hydrogen sulfide. Appl Microbiol Biotechnol. 2008;81:579–588. doi: 10.1007/s00253-008-1745-2. [DOI] [PubMed] [Google Scholar]
- Lin H-W, Couvreur K, Donose BC, Rabaey K, Yuan Z, Pikaar I. Electrochemical production of magnetite nanoparticles for sulfide control in sewers. Environ Sci Technol. 2017;51:12229–12234. doi: 10.1021/acs.est.7b01748. [DOI] [PubMed] [Google Scholar]
- Liu H, Xin Y, Xun L. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Environ Microbiol. 2014;80:1799–1806. doi: 10.1128/aem.03281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol. 2014;94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Fernández M, Gómez JM, Cantero D, Lafuente J, Gamisans X, Gabriel D. Kinetic and stoichiometric characterization of anoxic sulfide oxidation by SO-NR mixed cultures from anoxic biotrickling filters. Appl Microbiol Biotechnol. 2015;99:77–87. doi: 10.1007/s00253-014-5688-5. [DOI] [PubMed] [Google Scholar]
- Mulligan CN. Environmental applications for biosurfactants. Environ Pollut. 2005;133:183–198. doi: 10.1016/j.envpol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Nicholls P, Kim J-K. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can J Biochem. 1982;60:613–623. doi: 10.1139/o82-076. [DOI] [PubMed] [Google Scholar]
- Oprime MEAG, Garcia O, Jr, Cardoso AA. Oxidation of H2S in acid solution by Thiobacillus ferrooxidans and Thiobacillus thiooxidans. Process Biochem. 2001;37:111–114. doi: 10.1016/S0032-9592(01)00179-0. [DOI] [Google Scholar]
- Pokorna D, Zabranska J. Sulfur-oxidizing bacteria in environmental technology. Biotechnol Adv. 2015;33:1246–1259. doi: 10.1016/j.biotechadv.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Qin Q-L, Zhao D-L, Wang J, Chen X-L, Dang H-Y, Li T-G, Zhang Y-Z, Gao P-J. Wangia profunda gen. nov., sp. nov., a novel marine bacterium of the family Flavobacteriaceae isolated from southern Okinawa Trough deep-sea sediment. FEMS Microbiol Lett. 2007;271:53–58. doi: 10.1111/j.1574-6968.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- Sorokin DY. Use of microorganisms in protection of environments from pollution by sulfur compounds. Microbiology. 1994;63:533–547. [Google Scholar]
- Sorokin DY, Teske A, Robertson LA, Kuenen JG. Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria, belonging to the Pseudomonas stutzeri group. FEMS Microbiol Ecol. 1999;30:113–123. doi: 10.1111/j.1574-6941.1999.tb00640.x. [DOI] [PubMed] [Google Scholar]
- Sorokin D, van den Bosch PL, Abbas B, Janssen AJ, Muyzer G. Microbiological analysis of the population of extremely haloalkaliphilic sulfur-oxidizing bacteria dominating in lab-scale sulfide-removing bioreactors. Appl Microbiol Biotechnol. 2008;80:965–975. doi: 10.1007/s00253-008-1598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sublette KL, Sylvester ND. Oxidation of hydrogen sulfide by continuous cultures of Thiobacillus denitrificans. Biotechnol Bioeng. 1987;29:753–758. doi: 10.1002/bit.260290613. [DOI] [PubMed] [Google Scholar]
- Syed M, Soreanu G, Falletta P, Béland M. Removal of hydrogen sulfide from gas streams using biological processes—a review. Can Biosyst Eng. 2006;48:2.1–2.14. [Google Scholar]
- Wang X, Gai Z, Yu B, Feng J, Xu C, Yuan Y, Lin Z, Xu P. Degradation of carbazole by microbial cells immobilized in magnetic gellan gum gel beads. Appl Environ Microbiol. 2007;73:6421–6428. doi: 10.1128/AEM.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang Y, Zhang T, Zhou J. Effect of dissolved oxygen on elemental sulfur generation in sulfide and nitrate removal process: characterization, pathway, and microbial community analysis. Appl Microbiol Biotechnol. 2016;100:2895–2905. doi: 10.1007/s00253-015-7146-4. [DOI] [PubMed] [Google Scholar]
- Xia Y, Wübbeler JH, Qi Q, Steinbüchel A. Employing a recombinant strain of Advenella mimigardefordensis for biotechnical production of homopolythioesters from 3,3′-dithiodipropionic acid. Appl Environ Microbiol. 2012;78:3286–3297. doi: 10.1128/aem.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Lu C, Hou N, Xin Y, Liu J, Liu H, Xun L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017 doi: 10.1038/ismej.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Liu H, Cui F, Liu H, Xun L. Recombinant Escherichia coli with sulfide: quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol. 2016;18:5123–5136. doi: 10.1111/1462-2920.13511. [DOI] [PubMed] [Google Scholar]
- Yang X-P, Wei L-J, Lin J-P, Yin B, Wei D-Z. Membrane-bound pyrroloquinoline quinone-dependent dehydrogenase in Gluconobacter oxydans M5, responsible for production of 6-(2-hydroxyethyl) amino-6-deoxy-l-sorbose. Appl Environ Microbiol. 2008;74:5250–5253. doi: 10.1128/aem.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-X. Nanoscale iron particles for environmental remediation: an overview. J Nanopart Res. 2003;5:323–332. doi: 10.1023/a:1025520116015. [DOI] [Google Scholar]
- Zhang L, De Schryver P, De Gusseme B, De Muynck W, Boon N, Verstraete W. Chemical and biological technologies for hydrogen sulfide emission control in sewer systems: a review. Water Res. 2008;42:1–12. doi: 10.1016/j.watres.2007.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.