Abstract
Background: Metaplastic breast cancer is one of the most therapeutically challenging forms of breast cancer because of its highly heterogeneous and chemoresistant nature. We have previously demonstrated that ribosomal protein L39 (RPL39) and its gain-of-function mutation A14V have oncogenic activity in triple-negative breast cancer and this activity may be mediated through inducible nitric oxide synthase (iNOS). The function of RPL39 and A14V in other breast cancer subtypes is currently unknown. The objective of this study was to determine the role and mechanism of action of RPL39 in metaplastic breast cancer.
Methods: Both competitive allele-specific and droplet digital polymerase chain reaction were used to determine the RPL39 A14V mutation rate in metaplastic breast cancer patient samples. The impact of RPL39 and iNOS expression on patient overall survival was estimated using the Kaplan-Meier method. Co-immunoprecipitation and immunoblot analyses were used for mechanistic evaluation of RPL39.
Results: The RPL39 A14V mutation rate was 97.5% (39/40 tumor samples). High RPL39 (hazard ratio = 0.71, 95% confidence interval = 0.55 to 0.91, P = .006) and iNOS expression (P = .003) were associated with reduced patient overall survival. iNOS inhibition with the pan-NOS inhibitor NG-methyl-L-arginine acetate decreased in vitro proliferation and migration, in vivo tumor growth in both BCM-4664 and BCM-3807 patient-derived xenograft models (P = .04 and P = .02, respectively), and in vitro and in vivo chemoresistance. Mechanistically, RPL39 mediated its cancer-promoting actions through iNOS signaling, which was driven by the RNA editing enzyme adenosine deaminase acting on RNA 1.
Conclusion: NOS inhibitors and RNA editing modulators may offer novel treatment options for metaplastic breast cancer.
Metaplastic breast carcinoma is a rare form of breast cancer, accounting for 0.2% to 5% of all breast cancers (1). However, it is one of the most chemotherapy-refractory breast cancer subtypes and is associated with a higher rate of recurrence and poorer overall survival (OS) (2). Metaplastic breast cancers exhibit diversity in their carcinomatous and sarcomatous features, heterologous elements, and histologic and stromal appearance (2–15). This diversity makes it a difficult disease to treat.
Metaplastic breast cancers remain a therapeutic dilemma primarily due to our limited understanding of its pathogenesis. Treatment guidelines for metaplastic breast cancer are yet to be established, and currently treatment is similar to basal-like triple-negative breast cancers (TNBCs) (16). Metaplastic breast cancers express several markers associated with basal-like cancers (eg, epidermal growth factor receptor and cytokeratin 5/6); however, unlike basal-like carcinomas, they are typically chemoresistant (17–19). Three-year survival rates are approximately 40% for node-positive patients compared with approximately 70% for basal-like TNBCs (15). Node-positive patients have poorer clinical outcome, with recurrence patterns tending to favor pulmonary metastasis (2). Using integrated genomics and proteomics approaches, metaplastic breast cancers were found to be closely related to the claudin-low breast cancer subtype (20). Claudin-low breast cancer is characterized by the loss of cell-cell adhesion genes (5). Compared with other breast cancer subtypes, metaplastic breast cancers and claudin-low breast cancers are enriched for stem cell–like and epithelial-to-mesenchymal transition (EMT) characteristics (20,21), which partially contribute to their chemoresistance. A better understanding of the molecular mechanisms underlying metaplastic breast cancer pathogenesis is essential to the discovery of novel therapeutic targets for treatment.
We recently identified a novel cancer gene, ribosomal protein L39 (RPL39), responsible for stem cell self-renewal, treatment resistance, and lung metastasis in TNBC (22). Mechanistically, RPL39 increased inducible nitric oxide synthase (iNOS)-mediated nitric oxide (NO) production (22). Furthermore, the RPL39 A14V mutation was found to be a predictor of early distant metastatic relapse to the lung and worse OS. Although RPL39 plays a critical role in TNBC, the function of this protein in other breast cancer subtypes is currently unknown. As metaplastic breast cancers display stem-like and EMT features, we investigated the role and mechanism of action of RPL39 in this breast cancer subtype using patient samples and in vitro and in vivo models.
Methods
Mutation Analysis
Previously identified RPL39 gene mutations were analyzed in tissues samples from 40 patients with pathologically confirmed metaplastic breast cancer. Genomic DNA isolated from patient tissue samples was analyzed for the presence of wild-type or mutant allele using a custom-designed competitive allele-specific TaqMan (CAST) polymerase chain reaction (PCR) assay (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. The results of the CAST PCR analysis were confirmed by droplet digital PCR (ddPCR). ddPCR was performed using a standard protocol with custom RPL39 (A14V) ddPCR probes and primers (Bio-Rad Laboratories, Hercules, CA) as detailed in the Supplementary Methods (available online). For all experiments, positive, negative, and no template controls were used in the pre-amplification and ddPCR steps. Wild-type RPL39 plasmid DNA was used as negative control to determine the cutoff for RPL39 A14V mutation.
Bioinformatics Analysis
The Kaplan-Meier plotter is capable of assessing the effect of 54 675 genes on survival using 10 188 cancer samples. These include 4142 breast, 1648 ovarian, 2437 lung, and 1065 gastric cancer patients with a mean follow-up of 69, 40, 49, and 33 months, respectively (24). Written informed consent was obtained from all study patients. The primary purpose of the tool is a meta-analysis-based biomarker assessment, and we used this for determining the impact of RPL39 on TNBC patient survival. Additionally, we used the Susan G. Komen foundation sequence database for normal breast tissue from the Susan G. Komen for the Cure Tissue Bank at Indiana University. The BAM files of twenty normal breast tissue samples were analyzed using Rsamtools pileup (25) within R (R Development Core Team) using the PileupParam of min base quality = 20 (Supplementary Table 1, available online) (26).
Immunoblot Analysis
Immunoblot analyses are detailed in the Supplementary Methods (available online).
Animal Studies
All animal studies were performed in accordance with our institutional animal use committee guidelines and are detailed in the Supplementary Methods (available online).
Statistical Analysis
Patient and tumor characteristics were summarized using descriptive statistics.
Association of patient OS with RPL39 A14V mutation status and iNOS expression level was determined using log-rank tests. OS was estimated using the Kaplan-Meier method, and differences were compared using the log-rank test. Fisher’s exact test was used for bivariate analysis of categorical factors. Pairwise differences in fractional abundance were analyzed with the Wilcoxon rank-sum test. The correlations between biomarkers and their statistical significance were assessed using Pearson’s correlation coefficient and Student’s t distribution, respectively.
For the animal studies, sample means and 95% confidence intervals (CIs) calculated as the mean plus or minus 1.96 times the standard error of the mean, which is equivalent to the 95% confidence interval, were plotted to visualize changes in tumor volume over time. Differences in tumor volume were compared using mixed-effects linear models that included a random subject effect with a heterogeneous autoregressive variance-covariance structure to account for intra-animal correlation across time points. The models contained fixed effects for treatment, time point, and the interaction of these main effects. Satterthwaite’s method was used to calculate the denominator degrees of freedom for the F-tests of the fixed and simple effects in order to account for the variance heterogeneity. For proliferation, migration, and apoptosis assays, the Student’s t test or one-way analysis of variance (ANOVA) was used to compare group differences. For all analyses, a two-tailed P value of less than .05 was considered statistically significant.
In Vitro Cellular Analyses
Cell proliferation, cellular nitrate, and nitrite, migration, and immunoprecipitation assays were performed as described in the Supplementary Methods (available online).
Immunohistochemistry
Immunohistochemical analyses are detailed in the Supplementary Methods (available online).
Results
Prevalence of RPL39 A14V Mutation in Metaplastic Breast Cancer
Utilizing a published database of 457 TNBC patients (24), we found that high RPL39 expression was directly correlated with reduced OS (hazard ratio = 0.71, 95% confidence interval [CI] = 0.55 to 0.91, P = .006) (Figure 1A). CAST PCR and ddPCR were used to determine RPL39 A14V mutation rate and fractional abundance in tumor samples from patients with histologically confirmed metaplastic breast cancer (n = 40) and basal-like TNBC (n = 40) and available clinical outcome data. RPL39 A14V mutation rate (97.5% [39/40] vs 0% [0/40]; Fisher’s exact test, P < .001) (Figure 1B) and fractional abundance (Wilcoxon rank-sum test, P = .03) (Figure 1C) were statistically significantly higher in metaplastic breast cancer compared with basal-like TNBC. To ensure that the metaplastic breast cancer patient cohort was not skewing the data, the frequency of common mutations identified in TNBC was evaluated. Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha H1047R and E545K mutations were present in 10% (4/40) of primary TNBCs and 17.5% (7/40) of metaplastic breast cancers, which is consistent with previously published rates (20). Together, these findings demonstrate the high prevalence of RPL39 A14V mutation in metaplastic breast cancer.
Figure 1.
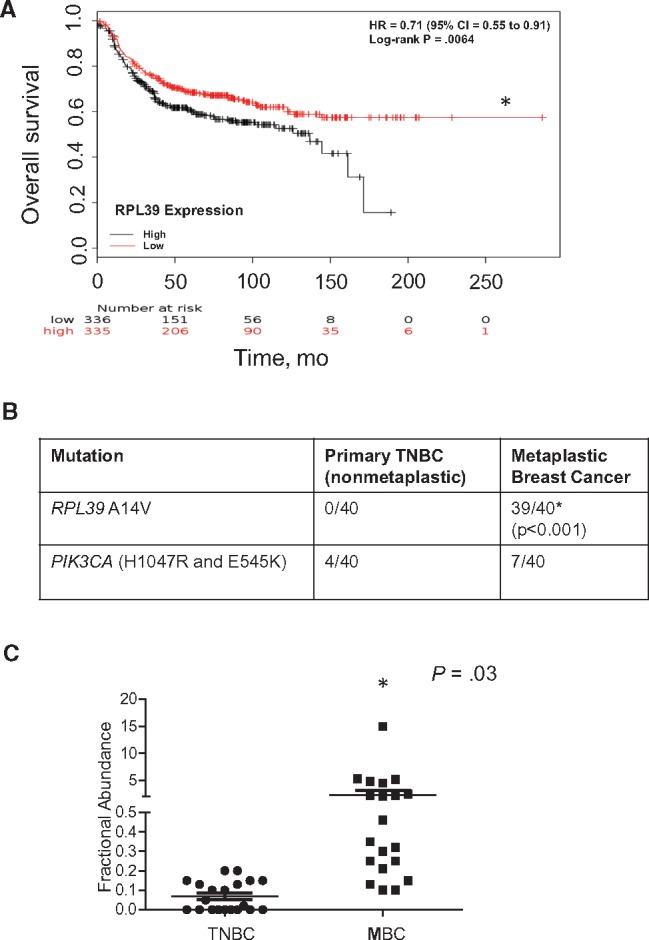
RPL39 (A14V) mutation status analyzed in metaplastic breast cancer. A) The relationship between ribosomal protein L39 (RPL39) expression and overall survival was assessed among 457 triple-negative breast cancer (TNBC) patients. The P value was calculated using a log-rank test. B)RPL39 A14V mutation rate was determined by competitive allele-specific TaqMan (CAST) polymerase chain reaction (PCR) analysis of tumor samples from patients with nonmetaplastic and metaplastic breast cancer (each n = 40). Fisher’s exact test was used to compare the mutation rate in metaplastic breast cancer vs nonmetaplastic breast cancer. PIK3CA mutations (H1047R and E545K) rates are also shown. C) The results of the CAST PCR analysis were confirmed by droplet digital PCR. Wilcoxon rank-sum test was used to determine statistically significant differences in the fractional abundance of the RPL39 A14V mutation between metaplastic breast cancer and TNBC. We used a threshold of 0.1% fractional abundance to define low RPL39 expression level. All statistical tests were two-sided. CI = confidence interval; HR = hazard ratio; MBC = metaplastic breast cancer; TNBC = triple-negative breast cancer.
Next, to confirm that the RPL39 A14V mutation was not a single-nucleotide polymorphism (SNP), RNA deep sequencing analysis was performed on 20 normal breast tissues. Results, presented as total counts for specified nucleotides within each sample (Supplementary Table 1, available online), indicated that RPL39 A14V is unlikely to be an SNP. We also analyzed the correlation between RPL39 A14V mutation and iNOS level. The correlation did not reach statistical significance because of the small sample size (Supplementary Figure 1 and Supplementary Table 2, available online). Given the rarity of metaplastic breast cancer, sufficient patient samples to accurately assess the correlation between RPL39 A14V mutation and iNOS level would be difficult to obtain.
Correlation of iNOS level With OS in Metaplastic Breast Cancer
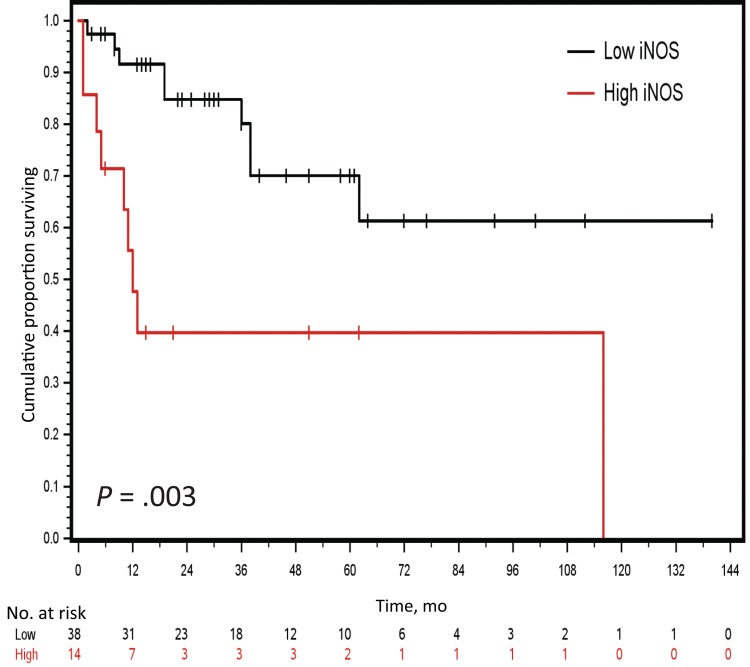
We previously reported that RPL39 regulates iNOS expression in TNBC (22) and downregulation of iNOS signaling reduces tumor growth (27). We and others have also reported that high iNOS expression level is associated with worse survival in TNBC patients (27,28). iNOS levels were immunohistochemically evaluated in the same cohort of metaplastic breast cancer patient tumor samples used for RPL39 mutation analysis. OS was statistically significantly shorter for metaplastic breast cancer patients in the high iNOS expression group compared with those in the low iNOS expression group (log-rank test, P = .003) (Figure 2). These results indicate that increased iNOS expression is a poor prognostic indicator in metaplastic breast cancer.
Figure 2.
Correlation between inducible nitric oxide synthase (iNOS) expression and patient overall survival in metaplastic breast cancer. iNOS expression levels were evaluated by immunohistochemical analysis in a cohort of metaplastic breast cancer patient tumor samples (n = 40). The graph presents Kaplan-Meier estimated survival curves for the high and low iNOS expression groups. A two-sided log-rank test was used to determine the statistical significance of the observed difference in survival. iNOS = inducible nitric oxide synthase.
Effect of iNOS Inhibition on Metaplastic Breast Cancer Cell Proliferation and Migration
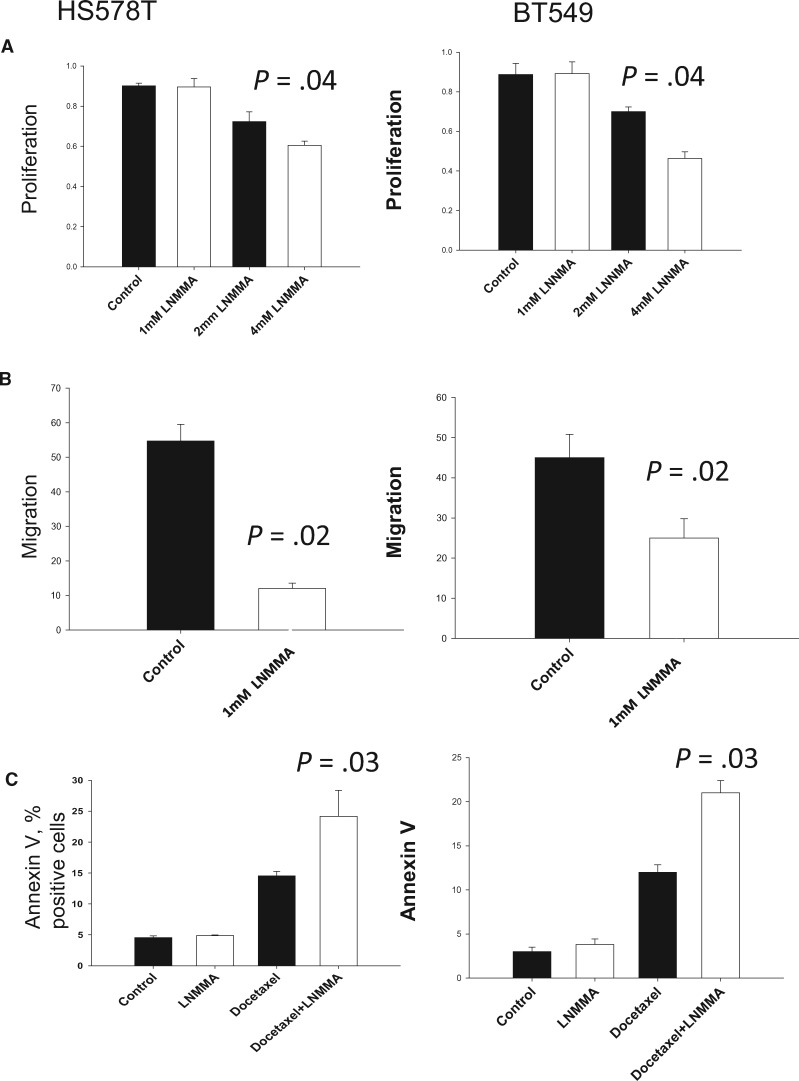
To determine the functional relevance of iNOS, the metaplastic-like breast cancer cell lines HS578T and BT549 were treated with the pan-NOS inhibitor NG-methyl-L-arginine acetate (L-NMMA) (27). The HS578T and BT549 cell lines have been reported to possess metaplastic genotypic characteristics (23) and do not carry the RPL39 A14V mutation. L-NMMA-mediated iNOS inhibition statistically significantly reduced cell proliferation and migration in a dose-dependent manner in both cells lines (one-way ANOVA, P = .04 and P = .02, respectively) (Figure 3, A and B). As metaplastic breast cancers are highly chemoresistant, we investigated whether L-NMMA increases the sensitivity of HS578T and BT549 cells to docetaxel. Compared with vehicle control cells, L-NMMA alone did not statistically significantly increase the number of apoptotic cells (Figure 3C). However, L-NMMA statistically significantly enhanced docetaxel-mediated apoptosis, as evidenced by the greater number of Annexin V–positive cells (one-way ANOVA, P = .03) (Figure 3C). Together, our findings indicate that iNOS inhibition demonstrates potent anticancer effects and decreases chemoresistance in metaplastic breast cancer cells. Additionally, we analyzed the effect of iNOS inhibition on RPL39 expression and found that RPL39 expression was reduced by both L-NMMA and iNOS siRNA (Supplementary Figure 2, available online).
Figure 3.
Role of inducible nitric oxide synthase (iNOS) inhibition in metaplastic breast cancer cell proliferation and migration in vitro. A) Metaplastic breast cancer cell lines HS578T and BT549 were treated with vehicle control or the indicated concentrations of L-NMMA for 72 hours. Proliferation was assessed by WST1 assay (HS578T: mean = 0.76, SD = 0.03, P = .04; BT549: mean = 0.74, SD = 0.01, P = .04). B) Migration by scratch assay was determined in HS578T and BT549 cells treated with vehicle control or 1 mM L-NMMA for 72 hours. (HS578T: mean = 12, SD = 1.5, P = .02; BT549: mean = 24, SD = 3.4, P = .02). C) HS578T and BT549 cells were treated with vehicle control, L-NMMA alone (1 mM), docetaxel alone (1 nM), or combined docetaxel and L-NMMA for 72 hours and the number of apoptotic cells determined by Annexin V staining (docetaxel vs combination therapy, HS578T, mean = 23.8, SD = 4.1, P = .03; BT549 mean = 22.1, SD = 1.8, P = .03). Data presented are based on three independent experiments, and error bars represent the standard deviation. P values were calculated using one-way analysis of variance, followed by the Tukey test for pairwise comparisons. All statistical tests were two-sided.
Effect of RPL39 on the In Vivo Response to L-NMMA in Metaplastic Breast Cancer
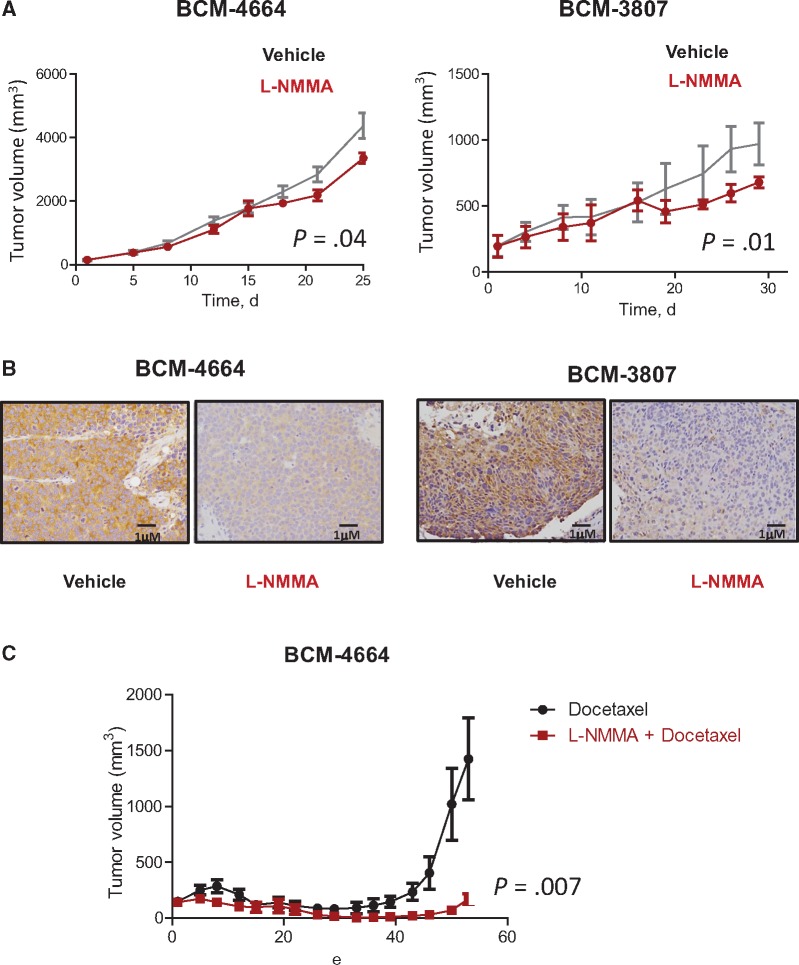
To investigate the effect of RPL39 on the in vivo response to L-NMMA, we used RPL39 A14V mutation–positive (BCM-4664) and –negative (BCM-3807) metaplastic breast cancer patient-derived xenografts (PDXs). Despite the difference in RPL39 mutation status, both BCM-4664 and BCM-3807 tumors showed high iNOS expression levels. L-NMMA statistically significantly reduced tumor volume in both the BCM-4664 and BCM-3807 models compared with the vehicle control (F-tests, P = .04 and P = .02, respectively) (Figure 4A). Compared with vehicle, L-NMMA substantially reduced iNOS expression in both the BCM-4664 and BCM-3807 models, verifying target engagement (Figure 4B). Furthermore, L-NMMA in combination with docetaxel statistically significantly reduced tumor volume in the BCM-4664 model compared with docetaxel alone (F-test, P = .007) (Figure 4C). As BCM-3807 is highly chemosensitive, combination effects were not observed (Supplementary Figure 3, available online). These findings suggest that iNOS-expressing metaplastic breast cancers are responsive to iNOS inhibition with L-NMMA regardless of RPL39 mutation status and mechanisms other than RPL39 A14V mutation may regulate iNOS expression.
Figure 4.
Testing the impact of inducible nitric oxide synthase (iNOS) inhibition on in vivo tumor growth and chemoresistance in metaplastic breast cancer. A) The effect of iNOS inhibition on in vivo tumor growth was determined using two metaplastic breast cancer patient-derived xenograft models, BCM-4664 and BCM-3807. Mice (n = 10 per group) were treated with vehicle (saline, intraperitoneal [i.p.], daily) or L-NMMA (200 mg/kg, i.p., daily) for 28 days. Tumor volume was measured twice weekly. P values were calculated using Student’s t test. Data are presented as the mean, with the error bars representing 95% confidence intervals (1.96SEM). B) Target engagement was verified by immunohistochemical analysis of iNOS expression in L-NMMA-treated BCM-4664 and BCM-3807 tumors. C) BCM-4664 mice were treated with docetaxel alone (20 mg/kg, i.p., once every 14 days) or in combination with L-NMMA for 42 days. Tumor volume was measured twice weekly. Data were analyzed using Student’s t test and presented as the mean ± 95% confidence interval (1.96 SEM). All statistical tests were two-sided.
Determination of RPL39 Signal Transduction Pathway in Metaplastic Breast Cancer
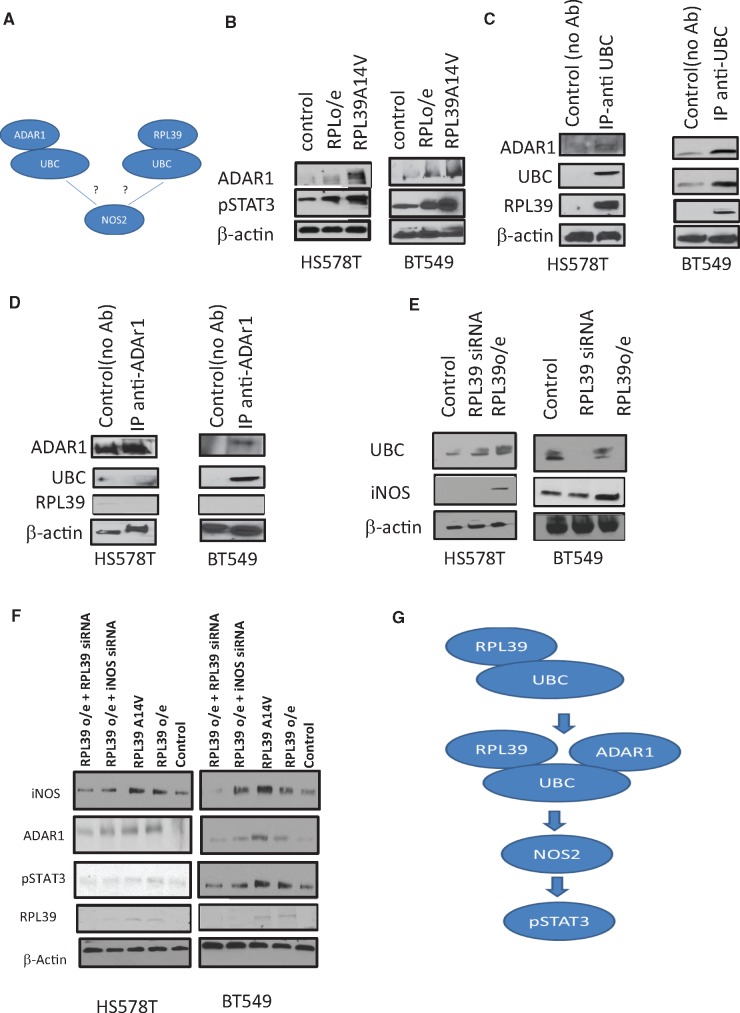
As our data indicate a role of RPL39 and iNOS in metaplastic breast cancer, we investigated the linking pathways using in silico data analysis (ingenuity pathway analysis) (Supplementary Figure 4, available online). Ingenuity pathway analysis identified a signaling pathway linking iNOS to ubiquitin C (UBC) and adenosine deaminase acting on RNA 1 (ADAR1). UBC is a ubiquitin-encoding gene that has been implicated in DNA repair and NOS signaling. The RNA-editing protein ADAR1 has recently been reported to play a role in cancer stem cell (CSC) self-renewal and to be associated with signal transducer and activator of transcription 3 (STAT3) pathway activation. Based on the results of the pathway analysis, we hypothesized that UBC may bind to both ADAR1 and RPL39 to activate iNOS signaling (Figure 5A). To investigate the in vivo interactions among these proteins, immunoblot analysis and co-immunoprecipitation assays were performed. Both RPL39 overexpression and A14V mutation increased ADAR1 and phosphorylated STAT3 expression (Figure 5B). Co-immunoprecipitation with anti-UBC revealed that UBC directly interacts with ADAR1 and RPL39 (Figure 5C), whereas co-immunoprecipitation with anti-ADAR1 showed that ADAR1 interacted with UBC but not RPL39 (Figure 5D). These data suggest an intermediary role for UBC in the RPL39 and ADAR1 interaction.
Figure 5.
Signal transduction pathway of RPL39 in metaplastic breast cancer. A) Ingenuity pathway analysis was done to identify potential links between RPL39 and iNOS. B) Immunoblot analysis of ADAR1 and phosphorylated STAT3 was performed in HS578T and BT549 cells transfected with a plasmid to overexpress RPL39 or RPL39 A14V. C) Ubiquitin C (UBC) was immunoprecipitated from HS578T and BT549 cell extracts. Immunoprecipitates were immunoblotted to detect the presence of the indicated proteins. D) ADAR1 was immunoprecipitated from HS578T and BT549 cell extracts. Immunoprecipitates were immunoblotted to detect the presence of the indicated proteins. E) Immunoblot analysis was performed to determine the expression of UBC, iNOS, and RPL39 in RPL39-overexpressing and siRNA-treated HS578T and BT549 cells. F) Immunoblot analysis was performed to determine the expression of iNOS, ADAR1, and phosphorylated STAT3 in iNOS and RPL39 in RPL39-overexpressing HS578T and BT549 cells treated with RPL39 or iNOS siRNA. G) Schematic of the proposed RPL39/UBC/ADAR1/iNOS/STAT3 signaling pathway is shown. β-actin served as a loading control in (B–F). ADAR1 = adenosine deaminase acting on RNA 1; IB = immunoblot; iNOS = inducible nitric oxide synthase; IP = immunoprecipitate; p = phosphorylated; RPL39 = ribosomal protein L39; STAT3 = signal transducer and activator of transcription 3; UBC = ubiquitin C.
Next, we deciphered the sequence of signaling events activated by RPL39. To further investigate the association between RPL39 and UBC, immunoblot analysis of RPL39-overexpressing and RPL39 siRNA-treated HS578T and BT549 cells was performed. RPL39 overexpression increased UBC expression in both BT549 and HS578T cells (Figure 5E). siRNA-mediated RPL39 downregulation decreased UBC expression in the BT549, but not the HS578T, cell line (Figure 5E). Changes in UBC expression mirrored those in iNOS expression (Figure 5E). We also tested the effect of RPL39 on ADAR1 expression. RPL39 overexpression and A14V mutation increased the expression of ADAR1 and phosphorylated STAT3 in HS578T and BT549 cells (Figure 5B). Furthermore, siRNA-mediated RPL39, but not iNOS, knockdown reduced ADAR1 expression in RPL39-overexpressing HS578T and BT549 cells, suggesting that ADAR1 is downstream of RPL39 and upstream of iNOS (Figure 5F). Furthermore, siRNA-mediated RPL39 and iNOS knockdown reduced phosphorylated STAT3 expression, suggesting that STAT3 is downstream of ADAR1 and iNOS (Figure 5F). A schematic of the proposed RPL39 signaling pathway is shown in Figure 5G. These data indicate that the RNA-editing enzyme ADAR1 drives iNOS signaling and a STAT3-dependent signaling pathway in metaplastic breast cancer.
Role of RPL39 in Metaplastic Breast Cancer Cell Proliferation and Migration
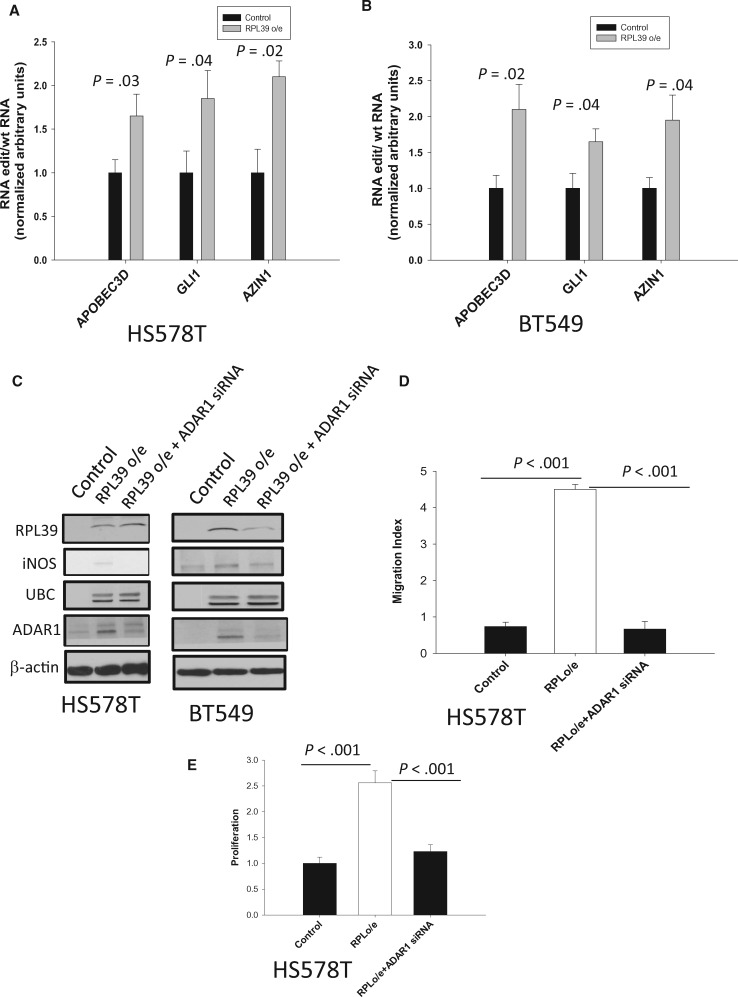
Whole-exome sequencing has previously shown the greatest fold-changes in RNA editing frequency in specific loci of CSCs (29). These sites were located within transcripts of the cytidine deaminase apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D (APOBEC3D); GLI family zinc finger 1(GLI1); antizyme inhibitor 1 (AZIN1); and ubiquitin ligase human homolog of mouse double minute 2 (MDM2). Analysis of editing rates from the RNA-seq data set showed increased RNA editing of APOBEC3D, and GLI1, AZIN1, and MDM2 in CSCs (29). RNA editing site-specific quantitative PCR also revealed increased RNA editing of APOBEC3D, GLI1, AZIN1, and MDM2 transcripts in lentiviral ADAR1-expressing cells (29).
As RPL39 increases ADAR1 expression, we investigated the role of RPL39 in the RNA editing of these genes. Overexpression of RPL39 in HS578T and BT549 cell lines statistically significantly increased the edited forms of APOBC3D, GLI1, and AZN1 RNA (Figure 6, A and B), but not MDM2 (data not shown), further confirming a role for RNA editing in the RPL39-mediated signaling pathway in metaplastic breast cancer.
Figure 6.
Role of RPL39 and RNA editing in metaplastic breast cancer cell proliferation and migration. A and B) RNA editing analysis is shown for ABOBEC3D, GLI1, and AZIN1 in RPL39-overexpressing HS578T and BT549 cells (HS578T cells: ABOBEC3D, mean = 16.1, SD = 2.8, P = .03; GLI1, mean = 17.2, SD = 3.1, P = .04; AZIN1, mean = 21.4, SD = 1.8, P = .02) (BT549 cells: ABOBEC3D, mean = 2.1, SD = 0.3, P = .02; GLI1, mean = 1.6, SD = 0.2, P = .04; AZIN1, mean = 1.9, SD = 0.2, P = .04). C) Immunoblot analysis with the indicated antibodies was performed using cell lysates from RPL39-overexpressing HS578T and BT549 cells treated with ADAR1-specific siRNA. D) Migration (P < .001) and (E) proliferation indices were measured in HS578T control cells, RPL39-overexpressing cells, and RPL39-overexpressing cells treated with ADAR1-specific siRNA (P < .001). Error bars represent standard deviation from the mean. Two-sided Student’s t test was used to calculate the P values. ADAR1 = adenosine deaminase acting on RNA 1; APOBEC3D = catalytic polypeptide-like 3D; AZIN1 = antizyme inhibitor 1; GLI1 = GLI family zinc finger 1; iNOS = inducible nitric oxide synthase; MDM2 = mouse double minute 2; RPL39 = ribosomal protein L39; UBC = ubiquitin C.
To determine whether RPL39-mediated iNOS signaling is modulated by ADAR1, iNOS expression was determined in RPL39-overexpressing HS578T and BT549 cells treated with ADAR1-specific siRNA. ADAR1 knockdown inhibited iNOS, but not RPL39, expression (Figure 6C), indicating that ADAR1 is downstream of RPL39 and upstream of iNOS. UBC expression was unaffected by ADAR1 knockdown, suggesting that UBC acts as bridge between RPL39 and ADAR1 rather than having a direct functional role. Additionally, ADAR1 knockdown affected cell migration (Figure 6D) and proliferation (Figure 6E) in RPL39-overexpressing cells. Together, these results indicate that RPL39 signals through an ADAR1/iNOS/STAT3 pathway in metaplastic breast cancer.
Discussion
Metaplastic breast cancer is an exceedingly rare, highly lethal, and chemotherapy-resistant TNBC (30). Targeted therapies for metaplastic breast cancer are currently not available; therefore, understanding the molecular mechanisms underlying its tumorigenesis is key to developing effective therapies. Hennesey et al. (21) demonstrated that metaplastic breast cancer tumors were phenotypically similar to claudin-low subtype, with an increased CSC component (20). Thus, we hypothesized that mutations previously described in CSCs may play a pivotal role in metaplastic breast cancer. In the present study, we found that the prevalence of the RPL39 A14V mutation is nearly ubiquitous in metaplastic breast cancer. High cytoplasmic iNOS level was associated with statistically significantly worse OS in patients with metaplastic breast cancer. These data provide evidence that iNOS is a valid therapeutic target for metaplastic breast cancer.
Previously, we identified the RPL39 A14V mutation in TNBC cells and found that it conferred a growth advantage (22). Additionally, RPL39 knockdown reduced the number of CD44+/CD24-/low and tumor-initiating cells, mammosphere formation efficiency, and lung metastases (22). Mechanistically, RPL39 increased iNOS-mediated NO production (22). High iNOS expression was associated with worse survival in patients with basal-like TNBC (27). Importantly, we demonstrated that L-NMMA-mediated iNOS inhibition suppressed TNBC tumorigenicity by decreasing proliferation, CSC self-renewal, and migration (27). In the present study, we found that iNOS inhibition with L-NMMA statistically significantly decreased proliferation and migration in vitro and statistically significantly enhanced docetaxel-mediated apoptosis in vitro in the metaplastic-like breast cancer cell lines HS578T and BT549. Our in vitro findings were corroborated in vivo. L-NMMA statistically significantly reduced tumor volume in the BCM-4664 (RPL39 A14V mutation–positive) and BCM-3807 (RPL39 A14V mutation–negative) metaplastic PDXs. Together, our in vitro and in vivo findings indicate that L-NMMA may be an effective therapeutic option for metaplastic breast cancer. L-NMMA decreased tumor volume in metaplastic PDXs regardless of RPL39 mutational status, suggesting that high iNOS expression in metaplastic breast cancer may be dependent on RPL39-independent pathways.
Our study is not without limitations. The number of patient samples in the study was small primarily because of the rarity of the disease. The small sample size prevented accurate analysis of the correlation of RPL39 and iNOS expression with survival. Furthermore, the impact of RPL39 on the iNOS-mediated NO pathway was limited to the in vitro analysis of cellular nitrate and nitrite levels. In vivo biophysical analysis of NO metabolites such as peroxinitrate, reactive nitrogen species, and reactive oxygen species in the context of the stromal microenvironment may provide additional mechanistic insights into the role of NO in metaplastic breast cancer. We plan in future studies to investigate the role of NO and related species in metaplastic breast cancer through comprehensive analysis involving biophysical, immunohistochemical, and mass spectrometric techniques.
We investigated the mechanism of action of RPL39 and iNOS in metaplastic breast cancer using in silico data analysis (ingenuity pathway analysis) and identified ADAR1 and UBC as important components of the RPL39/iNOS-mediated signaling pathway in metaplastic breast cancer. ADAR1 has recently been implicated in STAT3-dependent signaling in CSCs (29). This pathway may be directly edited in the CSC self-renewal of other cancers (29). In the present study, RNA editing was found to play a critical role in the RPL39/iNOS-mediated signaling pathway. Our results demonstrating the role of iNOS and RNA editing in metaplastic breast cancer provide a meaningful advancement in the development of potential therapeutic avenues for this highly lethal disease. The ability of ribosomal proteins to affect cellular translational capacity through RNA editing is a critical avenue of research for metaplastic breast cancer. Mutations in genes involved in ribosome function may play a crucial role in cancer survival and may allow for the development of novel targeted therapies for therapy-resistant cancers.
Funding
This work was funded by AUP-1010-0025 Chan Soon Shiong Institute of Advanced Health (JCC) and BCRF (JCC). Foundation Grants were awarded to JCC for studying novel therapeutics of breast cancer.
Notes
The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
References
- 1. Rayson D, Adjei AA, Suman VJ, et al. Metaplastic breast cancer: Prognosis and response to systemic therapy. Ann Oncol. 1999;104:413–419. [DOI] [PubMed] [Google Scholar]
- 2. Jung SY, Kim HY, Nam BH, et al. Worse prognosis of metaplastic breast cancer patients than other patients with triple-negative breast cancer. Breast Cancer Res Treat. 2010;1203:627–637. [DOI] [PubMed] [Google Scholar]
- 3. Hennessy BT, Giordano S, Broglio K, et al. Biphasic metaplastic sarcomatoid carcinoma of the breast. Ann Oncol. 2006;174:605–613. [DOI] [PubMed] [Google Scholar]
- 4. Hennessy BT, Krishnamurthy S, Giordano S, et al. Squamous cell carcinoma of the breast. J Clin Oncol. 2005;2331:7827–7835. [DOI] [PubMed] [Google Scholar]
- 5. Reis-Filho JS, Milanezi F, Steele D, et al. Metaplastic breast carcinomas are basal-like tumours. Histopathology. 2006;491:10–21. [DOI] [PubMed] [Google Scholar]
- 6. Wargotz ES, Deos PH, Norris HJ.. Metaplastic carcinomas of the breast. II. Spindle cell carcinoma. Hum Pathol. 1989;208:732–740. [DOI] [PubMed] [Google Scholar]
- 7. Wargotz ES, Norris HJ.. Metaplastic carcinomas of the breast. III. Carcinosarcoma. Cancer. 1989;647:1490–1499. [DOI] [PubMed] [Google Scholar]
- 8. Wargotz ES, Norris HJ.. Metaplastic carcinomas of the breast. I. Matrix-producing carcinoma. Hum Pathol. 1989;207:628–635. [DOI] [PubMed] [Google Scholar]
- 9. Wargotz ES, Norris HJ.. Metaplastic carcinomas of the breast: V. Metaplastic carcinoma with osteoclastic giant cells. Hum Pathol. 1990;2111:1142–1150. [DOI] [PubMed] [Google Scholar]
- 10. Wargotz ES, Norris HJ.. Metaplastic carcinomas of the breast. IV. Squamous cell carcinoma of ductal origin. Cancer. 1990;652:272–276. [DOI] [PubMed] [Google Scholar]
- 11. Wargotz ES, Norris HJ.. Metaplastic carcinomas and sarcomas of the breast. Am J Clin Pathol. 1991;966:781. [DOI] [PubMed] [Google Scholar]
- 12. Foschini MP, Dina RE, Eusebi V.. Sarcomatoid neoplasms of the breast: Proposed definitions for biphasic and monophasic sarcomatoid mammary carcinomas. Semin Diagn Pathol. 1993;102:128–136. [PubMed] [Google Scholar]
- 13. Gutman H, Pollock RE, Janjan NA, et al. Biologic distinctions and therapeutic implications of sarcomatoid metaplasia of epithelial carcinoma of the breast. J Am Coll Surg. 1995;1802:193–199. [PubMed] [Google Scholar]
- 14. Okada N, Hasebe T, Iwasaki M, et al. Metaplastic carcinoma of the breast. Hum Pathol. 2010;417:960–970. [DOI] [PubMed] [Google Scholar]
- 15. Bae SY, Lee SK, Koo MY, et al. The prognoses of metaplastic breast cancer patients compared to those of triple-negative breast cancer patients. Breast Cancer Res Treat. 2011;1262:471–478. [DOI] [PubMed] [Google Scholar]
- 16. Shah DR, Tseng WH, Martinez SR.. Treatment options for metaplastic breast cancer. ISRN Oncol. 2012;2012:706162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaufman MW, Marti JR, Gallager HS, et al. Carcinoma of the breast with pseudosarcomatous metaplasia. Cancer. 1984;539:1908–1917. [DOI] [PubMed] [Google Scholar]
- 18. Beatty JD, Atwood M, Tickman R, et al. Metaplastic breast cancer: Clinical significance. Am J Surg. 2006;1915:657–664. [DOI] [PubMed] [Google Scholar]
- 19. Chao TC, Wang CS, Chen SC, et al. Metaplastic carcinomas of the breast. J Surg Oncol. 1999;714:220–225. [DOI] [PubMed] [Google Scholar]
- 20. Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;6910:4116–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hennessy BT, Gonzalez-Angulo AM, Carey MS, et al. A systems approach to analysis of molecular complexity in breast cancer. Clin Cancer Res. 2009;152:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dave B, Granados-Principal S, Zhu R, et al. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc Natl Acad Sci U S A. 2014;11124:8838–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taube JH, Herschkowitz JI, Komurov K, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;10735:15449–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;1233:725–731. [DOI] [PubMed] [Google Scholar]
- 25. Delhomme N, Padioleau I, Furlong EE, et al. easyRNASeq: A bioconductor package for processing RNA-Seq data. Bioinformatics. 2012;2819:2532–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Obenchain V, Lawrence M, Carey V, et al. VariantAnnotation: A Bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014;3014:2076–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Granados-Principal S, Liu Y, Guevara ML, et al. Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 2015;17:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Glynn SA, Boersma BJ, Dorsey TH, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;12011:3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crews LA, Jiang Q, Zipeto MA, et al. An RNA editing fingerprint of cancer stem cell reprogramming. J Transl Med. 2015;13:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weigelt B, Kreike B, Reis-Filho JS.. Metaplastic breast carcinomas are basal-like breast cancers: A genomic profiling analysis. Breast Cancer Res Treat. 2009;1172:273–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.