Abstract
Purpose
A growing body of evidence shows an association between DNA repair protein genotypes and susceptibility to various cancers. However, few studies have assessed the contribution of the genotype of XRCC3, a homologous repair gene, to the occurrence or prognosis of childhood acute lymphoblastic leukemia (ALL). In this study, we investigated the contribution of seven XRCC3 polymorphisms to childhood ALL.
Patients and methods
We recruited 266 patients with childhood ALL and 266 healthy controls. Genomic DNA was isolated from peripheral blood samples. The XRCC3 rs1799794, rs45603942, rs1799796, rs861530, rs28903081, rs861539, and rs3212057 polymorphic genotypes of each subject were determined through conventional polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis.
Results
Genotypes with the rs861539 polymorphism were significantly associated with the risk of childhood ALL. The allelic distribution analyses suggested a significant association between the T allele at rs861539 with an increased risk of childhood ALL in the Taiwanese population. Polymorphic variants of XRCC3 at rs3212057 or rs28903081 did not exist in the study population. XRCC3 rs1799794, rs45603942, rs1799796, and rs861530 were not significantly associated with the risk of childhood ALL in the Taiwanese population.
Conclusion
Our findings suggest that XRCC3 genotypes with polymorphisms at rs861539 may play a role in determining individual susceptibility to childhood ALL in this Taiwanese population. The polymorphism may be a potential detector and predictor of childhood ALL.
Keywords: acute lymphoblastic leukemia, ALL, childhood, genotype XRCC3, polymorphism, Taiwan
Introduction
Leukemia is the most common cancer among children worldwide, and the majority of leukemia cases develop in the absence of any known predisposing factor. However, several germline genetic mutations, which can either be inherited or occur de novo in the patient’s germline, are associated with the development of leukemia.1 The possibility that genetic factors play a crucial role in the development of childhood acute lymphoblastic leukemia (ALL) is supported by the following evidence. First, Down’s syndrome and Fanconi anemia, which are both inherited genetic human diseases, are associated with an increased risk of ALL.2,3 Second, genetic mutations in several cancer-related genes, such as p53, N-ras, and PHF6, are prevalent in patients with ALL.4 Third, only a small fraction of children who have been exposed to relevant environmental factors develop ALL. These findings indicate that genomic predisposition likely has a greater contribution to the development of childhood ALL than environmental factors.5 Discovering useful biomarkers for early detection and prediction of childhood ALL may help improve surveillance guidelines for affected children and their families. In addition, these markers may help deepen our understanding of the genetic polymorphisms related to the etiology of ALL.
XRCC3 encodes the DNA repair protein XRCC3, which is a member of the homologous recombination DNA repair subpathway that removes double-stranded breaks from the human genome.6 Subtle polymorphic variations among individuals determine the differences in the human phenotypes. Among all known single-nucleotide polymorphisms (SNPs), the famous rs861539 C/T polymorphism (also called Thr241Met, T241M, C18067T, and C722T) of the XRCC3 gene is the most commonly studied because of its association with several human diseases, such as cancers.7–10 The T variants of XRCC3 rs861539 are associated with an increased risk of nasopharyngeal carcinoma11 and lung cancer,12 but are not associated with oral cancer.13 The findings for breast cancer are equivocal, as several studies have reported a positive association14–16 while others have not.17,18 However, the contribution of XRCC3 to childhood ALL has not been well investigated,19–21 and current studies have found no significant association. In 2014, Smolkova et al19 investigated the contribution of XRCC3 genotypes together with those of NBN and PPP1R13B to the susceptibility of childhood ALL. They found that no genotype of rs1799794, rs861530, and rs861539 on XRCC3 individually, or any haplotype or diplotype among them, was associated with altered risk of childhood ALL in a representative population of 460 pediatric ALL cases and 552 healthy controls. They successfully developed a haplotype-based methodology to examine the contribution of a candidate gene to a specific human disease, such as XRCC3 in childhood ALL in this case. In the current study, we not only evaluated the three SNPs (rs1799794, rs861530, and rs861539) they investigated, but also explored four additional SNPs (rs45603942, rs3212057, rs1799796, and rs28903081). The role of XRCC3 rs1799794, rs861530, and rs861539 should be validated in many populations with representative samples, and whether other SNPs also contribute to the susceptibility of childhood ALL should also be investigated. Thus, we performed this study to determine whether XRCC3 genotypes serve as novel biomarkers of childhood ALL in a representative Taiwan population. Specifically, we examined the association between the rs1799794, rs45603942, rs861530, rs3212057, rs1799796, rs861539, and rs28903081 polymorphisms of XRCC3 and the risk of childhood ALL.
Materials and methods
Patients and controls
This study protocol was approved by the Institutional Review Board of China Medical University Hospital (DMR103-IRB-153). Written informed consent was obtained from one or both parents of all participants. Briefly, children with ALL <18 years were identified and entered into the study by pediatric clinicians with pathological confirmation, regardless of the age or stage they were diagnosed. Thus, 266 children who had been diagnosed with ALL were recruited from the Department of Pediatrics of China Medical University Hospital (Taichung City) in Taiwan during 2005–2010. All children completed a detailed history taking and provided peripheral blood samples. In addition, an equal number of age- and gender-matched healthy volunteers were recruited as the control group in accordance with the method for initial random sampling established by the Health Examination Cohort during 2005–2010 as published previously.22–24 Finally, a total of 457 Taiwanese volunteers aged <18 years were recruited into the study.
XRCC3 genotyping
Genomic DNA from the peripheral blood samples of each participant was extracted, aliquoted, and stored as described previously.25−27 Seven polymorphic sites were analyzed. Briefly, the seven polymorphic sites were genotyped through conventional polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) methodology. PCR was performed on the BioRad Mycycler (Bio-Rad Laboratories Inc., Hercules, CA, USA) following the manufacturer’s instructions. The PCR conditions for the genotyping experiments consisted of a 5 minutes initial cycle at 94°C, 40 cycles at 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds; and a final 10 minutes extension at 72°C. SNP-containing DNA amplicons were digested overnight with specific restriction endonucleases following the manufacturer’s instructions as our team published previously.28,29 Following digestion, each sample was immediately resolved through 3% agarose gel electrophoresis at 100 V for 20 minutes. All genotypic processing was repeated and performed blindly by at least two independent researchers, and the results were 100% concordant with each other. About 3%–5% of the SNP samples were subject to direct sequencing, and the consistency between PCR-RFLP recognition and direct sequencing was 100%. The success rate of PCR-RFLP was 100%.
Statistical analysis
We evaluated the sample size for the study group; the estimated sample size was 199 in each group with alpha =5%, power =80%, the proportion of homozygous mutants in the case group =10%, and difference =10%. This assessment indicates that the sample size for the present study was adequate and sufficient. Pearson’s chi-squared test without the Yates’ correction or Fisher’s exact test was used to compare the distribution of XRCC3 genotypes between the case and control groups. The association between the XRCC3 polymorphisms and childhood ALL risk was estimated by computing ORs and 95% CIs through unconditional logistic regression analysis after adjusting for possible confounding factors if needed.
Results
Comparison of the basic demographic characteristics of the childhood ALL cases and controls
The frequency distributions of the demographic characteristics of the 266 childhood ALL cases and 266 controls are summarized and compared in Table 1. Age and gender did not differ between the case and control groups because the case and control groups had been closely matched during subject recruitment (P>0.05) (Table 1).
Table 1.
Distribution of selected demographics of the 266 childhood ALL patients and the 266 matched controls
| Characteristics | Controls (n=266) | Patients (n=266) | P-value | ||||
|---|---|---|---|---|---|---|---|
| n | % | Mean (SD) | n | % | Mean (SD) | ||
| Age at onset (years) | 8.3 (4.8) | 7.0 (4.4) | 0.6483a | ||||
| Gender | 1.0000b | ||||||
| Male | 148 | 55.6 | 148 | 55.6 | |||
| Female | 118 | 44.4 | 118 | 44.4 | |||
Notes:
Based on Student’s t-test;
Based on chi-squared test.
Abbreviation: ALL, acute lymphoblastic leukemia.
Analysis of the association between the seven XRCC3 SNPs and the risk of childhood ALL
With the PCR-RFLP methods introduced in Table 2, the distributions of the frequencies of the XRCC3 polymorphic genotypes in the childhood ALL cases and controls are summarized and compared in Table 3. First, the XRCC3 rs3212057 and rs28903081 polymorphisms were not found in the cases or the controls. All subjects were of the same wild-type genotype at these polymorphic sites (Table 3). Second, the crude ORs for individuals with variant CT and TT genotypes at XRCC3 rs861539 were 2.31 (95% CI =1.30–4.12, P=0.0037) and 2.44 (95% CI =0.91–6.53, P=0.0675) compared with those for individuals carrying the CC wild-type genotype (Table 3). The P-value for the trend analysis was also significant (P=0.0039) (Table 3). The distributions of XRCC3 rs1799794, rs45603942, rs861530, and rs1799796 in the case and control groups did not differ (Table 3). A representative RFLP electrophoresis analysis of the PCR product for XRCC3 rs861539 is shown in Figure 1.
Table 2.
Summary of the rs numbers, designed primers, amplicon length before and after enzyme digestion, and the specific restriction enzymes according to their allelic genotypes for all the seven XRCC3 SNPs
| rs number | Primer sequencea | Restriction enzyme | Amplicon length (bp) | Allelic genotypes and enzymatic fragment sizes |
|---|---|---|---|---|
|
| ||||
| rs1799794 | F: 5′-CACACTGCGGTCTTGCAGTG-3′ | BtsCI | 505 | G: 505 bp |
| R: 5′-CAGGCTGGGTCTGGATACAA-3′ | A: 289+216 bp | |||
| rs45603942 | F: 5′-GGGATGCAGGTTCAACTGAC-3′ | AluI | 352 | C: 352 bp |
| R: 5′-AACTTGGACTGTGTCAAGCA-3′ | T: 187+165 bp | |||
| rs861530 | F: 5′-CCGAGGAACGTGCTGAACTT-3′ | FatI | 497 | G: 497 bp |
| R: 5′-CTCCCTAACAGCCTCCATGT-3′ | A: 293+204 bp | |||
| rs3212057 | F: 5′-CCATGACCGCAGGCACTTGT-3′ | HpyCH4III | 455 | G: 455 bp |
| R: 5′-AGAACGCGACAAGGATGGTA-3′ | A: 235+220 bp | |||
| rs1799796 | F:5′-GGAACCAGTTGTGTGAGCCT-3′ | AluI | 430 | G: 430 bp |
| R: 5′-CCTGGTTGATGCACAGCACA-3′ | A: 226+204 bp | |||
| rs861539 | F: 5′-GACACCTTGTTGGAGTGTGT-3′ | FatI | 358 | C: 358 bp |
| R: 5′-GTCTTCTCGATGGTTAGGCA-3′ | T: 200+158 bp | |||
| rs28903081 | F: 5′-CTGCTTCCTGTTTCTCAGGT-3′ | BstUI | 198 | A: 198 bp |
| R: 5′-GCACTGATCGTGTAGGAACA-3′ | G: 102+96 bp | |||
Notes:
F and R indicate forward and reverse primers, respectively.
Abbreviation: SNPs, single-nucleotide polymorphisms.
Table 3.
Distribution of XRCC3 genotypes among the childhood ALL and healthy controls
| Genotype | Controls (n=266) | % | Patients (n=266) | % | P-valuea | OR (95% CI) |
|---|---|---|---|---|---|---|
|
| ||||||
| rs1799794 | ||||||
| GG | 63 | 23.7 | 67 | 25.2 | 1.00 (ref) | |
| AG | 150 | 56.4 | 144 | 54.1 | 0.6270 | 0.90 (0.60–1.36) |
| AA | 53 | 19.9 | 55 | 20.7 | 0.9250 | 0.98 (0.59–1.63) |
| Ptrend | 0.8682 | |||||
| rs45603942 | ||||||
| CC | 242 | 91.0 | 239 | 89.9 | 1.00 (ref) | |
| CT | 16 | 6.0 | 16 | 6.0 | 0.9727 | 1.01 (0.50–2.07) |
| TT | 8 | 3.0 | 11 | 4.1 | 0.4829 | 1.39 (0.55–3.52) |
| Ptrend | 0.7818 | |||||
| rs861530 | ||||||
| AA | 77 | 29.0 | 80 | 30.1 | 1.00 (ref) | |
| AG | 144 | 54.1 | 147 | 55.2 | 0.9292 | 0.98 (0.67–1.45) |
| GG | 45 | 16.9 | 39 | 14.7 | 0.5030 | 0.83 (0.49–1.42) |
| Ptrend | 0.7723 | |||||
| rs3212057 | ||||||
| GG | 266 | 100.0 | 266 | 100.0 | 1.00 (ref) | |
| AG | 0 | 0.0 | 0 | 0.0 | ||
| AA | 0 | 0.0 | 0 | 0.0 | ||
| Ptrend | 1.0000 | |||||
| rs1799796 | ||||||
| AA | 121 | 45.5 | 116 | 43.6 | 1.00 (ref) | |
| AG | 129 | 48.5 | 130 | 48.9 | 0.7813 | 1.05 (0.74–1.50) |
| GG | 16 | 6.0 | 20 | 7.5 | 0.4598 | 1.30 (0.64–2.64) |
| Ptrend | 0.7581 | |||||
| rs861539 | ||||||
| CC | 241 | 90.6 | 214 | 80.4 | 1.00 (ref) | |
| CT | 19 | 7.1 | 39 | 14.7 | 0.0037* | 2.31 (1.30–4.12) |
| TT | 6 | 2.3 | 13 | 4.9 | 0.0675 | 2.44 (0.91–6.53) |
| Ptrend | 0.0039* | |||||
| rs28903081 | ||||||
| GG | 266 | 100.0 | 266 | 100.0 | 1.00 (ref) | |
| AG | 0 | 0.0 | 0 | 0.0 | ||
| AA | 0 | 0.0 | 0 | 0.0 | ||
| Ptrend | 1.0000 | |||||
Notes:
P-value based on chi-squared test.
Statistically significant.
Abbreviation: ALL, acute lymphoblastic leukemia.
Figure 1.
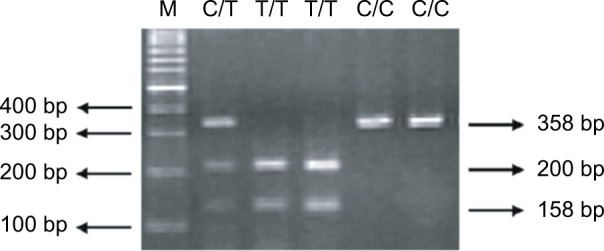
Restriction fragment length polymorphism electrophoresis analysis of the polymerase chain reaction product of XRCC3 rs861539.
Notes: The CC wild-type genotype is shown in lanes 5 and 6. Lanes 3 and 4 show the TT homovariant genotype, whereas lane 2 shows the C/T heterovariant genotype, respectively. Lane 1 (marked as M) is the DNA 100 bp molecular weight marker.
Analysis of the association between the XRCC3 allelic types at the seven SNP sites and the risk of childhood ALL
Supporting the findings in Table 3, the distribution of the XRCC3 rs861539 allelic frequencies differed significantly between the childhood ALL cases and the control group (Table 4). The crude OR of the subjects carrying the T allele at XRCC3 rs861539 was 2.25 (95% CI =1.44–3.51, P=0.0003, Table 4). XRCC3 rs1799794, rs45603942, rs861530, and rs1799796 were not associated with an increased risk of childhood ALL (Table 4).
Table 4.
Distribution of XRCC3 alleles among the childhood ALL patients and healthy controls
| Allele | Controls | % | Patients | % | P-valuea | OR (95% CI) |
|---|---|---|---|---|---|---|
|
| ||||||
| rs1799794 | ||||||
| Allele G | 276 | 51.9 | 278 | 52.3 | 1.00 (ref) | |
| Allele A | 256 | 48.1 | 254 | 47.7 | 0.9023 | 0.99 (0.77–1.25) |
| rs45603942 | ||||||
| Allele C | 500 | 94.0 | 494 | 92.9 | 1.00 (ref) | |
| Allele T | 32 | 6.0 | 38 | 7.1 | 0.4581 | 1.20 (0.74–1.95) |
| rs861530 | ||||||
| Allele A | 298 | 56.0 | 307 | 57.7 | 1.00 (ref) | |
| Allele G | 234 | 44.0 | 225 | 42.3 | 0.5775 | 0.93 (0.73–1.19) |
| rs1799796 | ||||||
| Allele A | 371 | 69.7 | 362 | 68.0 | 1.00 (ref) | |
| Allele G | 161 | 30.3 | 170 | 32.0 | 0.5512 | 1.08 (0.83–1.40) |
| rs861539 | ||||||
| Allele C | 501 | 94.2 | 467 | 87.8 | 1.00 (ref) | |
| Allele T | 31 | 5.8 | 65 | 12.2 | 0.0003* | 2.25 (1.44–3.51) |
Note:
P-value based on chi-squared test.
Statistically significant.
Abbreviation: ALL, acute lymphoblastic leukemia.
Stratification analysis for XRCC3 rs861539 according to age and gender
We further evaluated the effects of XRCC3 rs861539 on susceptibility to childhood ALL by stratifying their age and gender, and the results are presented in Tables 5 and 6, respectively. The association between the XRCC3 rs861539 genotype and childhood ALL was significant for children aged <3.5 years, but not for those ≥3.5 years (Table 5). The association between the XRCC3 rs861539 genotype and childhood ALL was slightly significant for boys and girls (Table 6).
Table 5.
ORs for XRCC3 rs861539 genotype and childhood ALL after stratification by age
| Genotype | <3.5 years old, n
|
OR (95% CI)a
|
aOR (95% CI)b
|
P-valuec | ≥3.5 years old, n
|
OR (95% CI)a
|
aOR (95% CI)b
|
P-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | |||||||
|
| ||||||||||
| AA | 121 | 99 | 1.00 (ref) | 1.00 (ref) | 120 | 115 | 1.00 (ref) | 1.00 (ref) | ||
| AG | 9 | 24 | 3.26 | 3.14 | 0.0030* | 10 | 15 | 1.57 | 1.49 | 0.2929 |
| (1.45–7.33) | (1.59–6.78) | (0.68–3.63) | (0.63–3.27) | |||||||
| GG | 3 | 10 | 4.07 | 3.98 | 0.0025* | 3 | 3 | 1.04 | 1.16 | 0.9589 |
| (1.09–15.21) | (1.14–11.94) | (0.21–5.28) | (0.33–6.48) | |||||||
| Ptrend | 0.0017* | 0.5751 | ||||||||
Notes:
Multivariate logistic regression analysis;
Multivariate logistic regression analysis after adjusted of gender;
P-value based on chi-squared test.
Statistically significant.
Abbreviation: ALL, acute lymphoblastic leukemia.
Table 6.
ORs for XRCC3 rs861539 genotype and childhood ALL after stratification by gender
| Genotype | Males, n | OR (95% CI)a | aOR (95% CI)b | P-valuec | Females, n | OR (95% CI)a | aOR (95% CI)b | P-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Controls | Cases | Controls | Cases | |||||||
|
| ||||||||||
| AA | 135 | 114 | 1.00 (ref) | 1.00 (ref) | 106 | 100 | 1.00 (ref) | 1.00 (ref) | ||
| AG | 9 | 25 | 3.29 | 3.07 | 0.0024* | 10 | 14 | 1.48 | 1.54 | 0.3640 |
| (1.48–7.33) | (1.63–6.59) | (0.63–3.49) | (0.61–3.67) | |||||||
| GG | 4 | 9 | 2.66 | 2.25 | 0.0987 | 2 | 4 | 2.12 | 2.23 | 0.3814 |
| (0.80–8.88) | (0.58–9.47) | (0.38–11.83) | (0.51–13.38) | |||||||
| Ptrend | 0.1492 | 0.0012* | ||||||||
Notes:
Multivariate logistic regression analysis;
Multivariate logistic regression analysis after adjusted of age;
P-value based on chi-squared test.
Statistically significant.
Abbreviation: ALL, acute lymphoblastic leukemia.
Discussion
Adducts or strand breaks induced by DNA-damaging agents are systematically and efficiently removed immediately after they are detected. Removal of damaged DNA sites is the main protective mechanism of living cells against potentially harmful genomic modifications.30,31 Hundreds of thousands of DNA adducts and strand breaks are repaired promptly and rapidly or genomic instability intensifies and becomes irreversible, resulting in cell death or carcino genesis. Several studies since 200032 have investigated the contribution of DNA repair gene polymorphisms to childhood ALL and identified useful biomarkers, such as XRCC1 codon 194,33,34 XRCC1 codon 399,33,35 human 8-oxoG DNA glycosylase 1 codon 326,36 and NBN codon 185,21 for early detection and prediction of childhood ALL susceptibility. We have also investigated the association among several DNA repair genes in Taiwan37–39 and first revealed that the XRCC4 G-1394T (rs6869366) and intron 3 DIP (rs28360071) genotypes were associated with an increased risk for childhood ALL.39 Several combined predictors, such as XRCC1 codon 399 plus OGG1 codon 326,40 and OGG1 codon 326 plus MUTYH codon 165, have also been proposed.40 These findings are inconclusive; thus, further multi-population and multi-institutional investigations should be conducted. The contribution of other DNA repair genes, such as XRCC3, has yet to be investigated.
XRCC3 is involved in the homologous recombination repair of double-stranded DNA and chromosomal fragmentation, translocations, and deletions.41 In 2003, Hinz et al reported that the hamster cell line irs1SF cannot produce normal cell-cycle checkpoint responses to double-strand break-induced programmed cell death because it is deficient in the homologous recombination repair gene XRCC3.42 However, the contribution of XRCC3 genotypes to cancer susceptibility remains largely unknown, and few researchers have investigated their role in childhood leukemia.20,21 In 2012, Erculj et al reported that carriers of the G allele at XRCC3 rs1799794 are at a lower risk for secondary neoplastic development than patients with the AA wild-type genotype.20 In 2014, Smolkova et al examined the genotype, haplotype, and diplotype of XRCC3 rs1799794, rs861530, and rs861539 but found no positive association.19 In 2015, Goricar et al reported that XRCC3 rs1799794 genotypes are not associated with the risk of childhood leukemia.21 We also did not find an association between SNP XRCC3 rs1799794 and susceptibility to childhood ALL (Tables 3 and 4). Previous studies have failed to detect a positive association between XRCC3 rs861539 and childhood ALL. Interestingly, we determined that XRCC3 rs861539 was significantly associated with the risk of childhood leukemia (Tables 3 and 4). The inconsistencies among the findings may lie in several factors. First, different populations were investigated, and the previous studies were investigating Caucasians, whereas we studied a Chinese population. Second, different sampling methods were used, so sampling bias may exist. We calculated sample size based on sampling and analyzing power >80%, which indicated that our samples are representative. In addition, the cases and controls were all Taiwanese, who have a very conservative genetic and geographic background.
Among the XRCC3 SNPs we investigated, rs1799794 and rs45603942 are promoter SNPs, rs861530 and rs1799796 are intronic SNPs, whereas rs3212057, rs861539, and rs28903081 are exonic SNPs. One of the highlights of the current study is that the XRCC3 rs861539 genotype was significantly associated with the risk of childhood ALL. XRCC3 rs861539 occurred in response to the polymorphism at amino acid 241, which was changed from threonine to methionine (or the nucleotide from C to T), resulting in impaired DNA repair function via a defect in the function of XRCC3 by removing the phosphorylation site.43 The XRCC3 homovariant Met/Met genotype has been identified with higher DNA double-strand break adducts after the same level of DNA damage challenges in the lymphocytes of investigated healthy subjects,7 while modifying the XRCC3 levels of the Thr241Met variant cells alters their DNA repair capacity.44 In addition, the XRCC3 heterovariant Thr241Met carriers who were exposed to ionizing radiation are associated with an increased number of micronuclei in their lymphocytes.45 In sum, DNA repair defects caused by XRCC3 rs861539 may affect DNA repair capacity and genomic stability, leading to variant susceptibilities to all types of cancers, including childhood ALL. All of the children with ALL investigated here may be at higher risk of recurrence and other types of cancers during their life, particularly those with the XRCC3 rs861539 variant genotypes.
Some limitations of this study should be discussed. First, phenotypic data, including the XRCC3 mRNA or protein expression levels are not currently available for further analysis. Second, DNA repair capacity measured with the patients’ blood sample according to their XRCC3 rs861539 different genotypes was also not available. In the future, the complete correlation among patient status, cell type, genetic type, and phenotype will help provide further insight into the role of XRCC3 in the initiation and development of childhood ALL.
In conclusion, this study investigated the contribution of XRCC3 genotypes with up to seven different SNPs to child hood ALL. In contrast to previous reports, our study showed that the XRCC3 rs861539 genotypes were significantly associated with increased susceptibility to childhood ALL in a Taiwanese population. Additional studies that elucidate the contribution of the genotypes of other DNA repair enzymes to the development of childhood ALL may promote early detection of childhood ALL. In addition, the XRCC3 genotype–phenotype correlation will further our understanding of the etiology of childhood ALL.
Acknowledgments
The authors are grateful to Hsin-Ting Li, Yu-Shih Wang, and Huai-Mei Hsu for their excellent technical assistance. All the participants in this study are appreciated. This study was supported mainly by Taoyuan General Hospital, Ministry of Health and Welfare, Taiwan, ROC to Dr Hsu and Dr Pei (grant numbers: PTH10740 and PTH10721) and partially by research grant from Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW107-TDU-B-212-123004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Stieglitz E, Loh ML. Genetic predispositions to childhood leukemia. Ther Adv Hematol. 2013;4(4):270–290. doi: 10.1177/2040620713498161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwaan CM, Reinhardt D, Hitzler J, Vyas P. Acute leukemias in children with Down syndrome. Hematol Oncol Clin North Am. 2010;24(1):19–34. doi: 10.1016/j.hoc.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25(43):5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 4.Szczepański T, Harrison CJ, van Dongen JJ. Genetic aberrations in paediatric acute leukaemias and implications for management of patients. Lancet Oncol. 2010;11(9):880–889. doi: 10.1016/S1470-2045(09)70369-9. [DOI] [PubMed] [Google Scholar]
- 5.Kaatsch P. Epidemiology of childhood cancer. Cancer Treat Rev. 2010;36(4):277–285. doi: 10.1016/j.ctrv.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Brenneman MA, Weiss AE, Nickoloff JA, Chen DJ. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat Res. 2000;459(2):89–97. doi: 10.1016/s0921-8777(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 7.Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22(9):1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 8.Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73(2):138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Ke HG, Li J, Shen Y, et al. Prognostic significance of GSTP1, XRCC1 and XRCC3 polymorphisms in non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2012;13(9):4413–4416. doi: 10.7314/apjcp.2012.13.9.4413. [DOI] [PubMed] [Google Scholar]
- 10.Guo S, Li X, Gao M, Li Y, Song B, Niu W. The relationship between XRCC1 and XRCC3 gene polymorphisms and lung cancer risk in northeastern Chinese. PLoS One. 2013;8(2):e56213. doi: 10.1371/journal.pone.0056213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JC, Tsai CW, Hsu CM, et al. Contribution of double strand break repair gene XRCC3 genotypes to nasopharyngeal carcinoma risk in Taiwan. Chin J Physiol. 2015;58(1):64–71. doi: 10.4077/CJP.2015.BAD279. [DOI] [PubMed] [Google Scholar]
- 12.Chen HJ, Chang WS, Hsia TC, et al. Contribution of genotype of DNA double-strand break repair gene XRCC3, gender, and smoking behavior to lung cancer risk in Taiwan. Anticancer Res. 2015;35(7):3893–3899. [PubMed] [Google Scholar]
- 13.Tsai CW, Chang WS, Liu JC, Tsai MH, Lin CC, Bau DT. Contribution of DNA double-strand break repair gene XRCC3 genotypes to oral cancer susceptibility in Taiwan. Anticancer Res. 2014;34(6):2951–2956. [PubMed] [Google Scholar]
- 14.Qureshi Z, Mahjabeen I, Baig R, Kayani M. Correlation between selected XRCC2, XRCC3 and RAD51 gene polymorphisms and primary breast cancer in women in Pakistan. Asian Pac J Cancer Prev. 2014;15(23):10225–10229. doi: 10.7314/apjcp.2014.15.23.10225. [DOI] [PubMed] [Google Scholar]
- 15.Smolarz B, Makowska M, Samulak D, et al. Association between single nucleotide polymorphisms (SNPs) of XRCC2 and XRCC3 homologous recombination repair genes and triple-negative breast cancer in Polish women. Clin Exp Med. 2015;15(2):151–157. doi: 10.1007/s10238-014-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su CH, Chang WS, Hu PS, et al. Contribution of DNA double-strand break repair gene XRCC3 genotypes to triple-negative breast cancer risk. Cancer Genomics Proteomics. 2015;12(6):359–367. [PubMed] [Google Scholar]
- 17.Jacobsen NR, Nexø BA, Olsen A, et al. No association between the DNA repair gene XRCC3 T241M polymorphism and risk of skin cancer and breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12(6):584–585. [PubMed] [Google Scholar]
- 18.Loizidou MA, Michael T, Neuhausen SL, et al. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res Treat. 2008;112(3):575–579. doi: 10.1007/s10549-007-9881-4. [DOI] [PubMed] [Google Scholar]
- 19.Smolkova B, Dusinska M, Hemminki K. NBN and XRCC3 genetic variants in childhood acute lymphoblastic leukaemia. Cancer Epidemiol. 2014;38(5):563–568. doi: 10.1016/j.canep.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Erčulj N, Faganel Kotnik B, Debeljak M, Jazbec J, Dolžan V. DNA repair polymorphisms influence the risk of second neoplasm after treatment of childhood acute lymphoblastic leukemia. J Cancer Res Clin Oncol. 2012;138(11):1919–1930. doi: 10.1007/s00432-012-1265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goričar K, Erčulj N, Faganel Kotnik B, et al. The association of folate pathway and DNA repair polymorphisms with susceptibility to childhood acute lymphoblastic leukemia. Gene. 2015;562(2):203–209. doi: 10.1016/j.gene.2015.02.077. [DOI] [PubMed] [Google Scholar]
- 22.Pei JS, Chang WS, Hsu PC, et al. The association of flap endonuclease 1 genotypes with the risk of childhood leukemia. Cancer Genomics Proteomics. 2016;13(1):69–74. [PubMed] [Google Scholar]
- 23.Pei JS, Chou AK, Hsu PC, et al. Contribution of matrix metalloproteinase-7 genotypes to the risk of non-solid tumor, childhood leukemia. Anticancer Res. 2017;37(12):6679–6684. doi: 10.21873/anticanres.12126. [DOI] [PubMed] [Google Scholar]
- 24.Pei JS, Chang WS, Hsu PC, et al. The contribution of MMP-8 promoter genotypes to childhood leukemia. In Vivo. 2017;31(6):1059–1064. doi: 10.21873/invivo.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai YL, Gong CL, Fu CK, et al. The contribution of matrix metalloproteinase-1 genotypes to hepatocellular carcinoma susceptibility in Taiwan. Cancer Genomics Proteomics. 2017;14(2):119–125. doi: 10.21873/cgp.20024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang CL, Wang CH, Hsu CH, et al. Contribution of double-strand break repair gene nijmegen breakage syndrome 1 genotypes, gender difference and smoking status to Taiwanese lung cancer. Anticancer Res. 2017;37(5):2417–2423. doi: 10.21873/anticanres.11581. [DOI] [PubMed] [Google Scholar]
- 27.Yueh TC, Wu CN, Hung YW, et al. The Contribution of MMP-7 Genotypes to Colorectal Cancer Susceptibility in Taiwan. Cancer Genomics Proteomics. 2018;15(3):207–212. doi: 10.21873/cgp.20079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung YW, Tsai CW, Wu CN, et al. The contribution of matrix metalloproteinase-8 promoter polymorphism to oral cancer susceptibility. In Vivo. 2017;31(4):585–590. doi: 10.21873/invivo.11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen TC, Hsia TC, Chao CY, et al. The Contribution of MMP-8 Promoter Polymorphisms in Lung Cancer. Anticancer Res. 2017;37(7):3563–3567. doi: 10.21873/anticanres.11726. [DOI] [PubMed] [Google Scholar]
- 30.Moraes MC, Neto JB, Menck CF. DNA repair mechanisms protect our genome from carcinogenesis. Front Biosci. 2012;17:1362–1388. doi: 10.2741/3992. [DOI] [PubMed] [Google Scholar]
- 31.Nissar S, Sameer AS, Rasool R, Rashid F. DNA Repair Gene - XRCC1 in Relation to Genome Instability and Role in Colorectal Carcinogenesis. Oncol Res Treat. 2014;37(7-8):4–422. doi: 10.1159/000364898. [DOI] [PubMed] [Google Scholar]
- 32.Infante-Rivard C, Mathonnet G, Sinnett D. Risk of childhood leukemia associated with diagnostic irradiation and polymorphisms in DNA repair genes. Environ Health Perspect. 2000;108(6):495–498. doi: 10.1289/ehp.00108495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pakakasama S, Sirirat T, Kanchanachumpol S, et al. Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2007;48(1):16–20. doi: 10.1002/pbc.20742. [DOI] [PubMed] [Google Scholar]
- 34.Batar B, Güven M, Baris¸ S, Celkan T, Yildiz I. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33(6):759–763. doi: 10.1016/j.leukres.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Joseph T, Kusumakumary P, Chacko P, Abraham A, Pillai MR. DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett. 2005;217(1):17–24. doi: 10.1016/j.canlet.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Huang L, Rong L, et al. hOGG1 Ser326Cys polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Cancer Sci. 2011;102(6):1123–1127. doi: 10.1111/j.1349-7006.2011.01928.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang CH, Wu KH, Yang YL, et al. Association between Ataxia Telangiectasia Mutated gene polymorphisms and childhood leukemia in Taiwan. Chin J Physiol. 2011;54(6):413–418. doi: 10.4077/CJP.2011.AMM106. [DOI] [PubMed] [Google Scholar]
- 38.Pei JS, Lee YM, Lo HH, Hsu YN, Lin SS, Bau DT. Association of X-ray repair cross-complementing-6 genotypes with childhood leukemia. Anticancer Res. 2013;33(12):5395–5399. [PubMed] [Google Scholar]
- 39.Wu KH, Wang CH, Yang YL, et al. Significant association of XRCC4 single nucleotide polymorphisms with childhood leukemia in Taiwan. Anticancer Res. 2010;30(2):529–533. [PubMed] [Google Scholar]
- 40.Stanczyk M, Sliwinski T, Cuchra M, et al. The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep. 2011;38(1):445–451. doi: 10.1007/s11033-010-0127-x. [DOI] [PubMed] [Google Scholar]
- 41.Kurumizaka H, Ikawa S, Nakada M, et al. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc Natl Acad Sci USA. 2001;98(10):5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinz JM, Helleday T, Meuth M. Reduced apoptotic response to camptothecin in CHO cells deficient in XRCC3. Carcinogenesis. 2003;24(2):249–253. doi: 10.1093/carcin/24.2.249. [DOI] [PubMed] [Google Scholar]
- 43.Talar-Wojnarowska R, Ga˛siorowska A, Olakowski M, et al. Analysis of XRCC2 and XRCC3 gene polymorphisms in pancreatic cancer. Biomed Rep. 2016;4(2):236–240. doi: 10.3892/br.2015.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Araujo FD, Pierce AJ, Stark JM, Jasin M. Variant XRCC3 implicated in cancer is functional in homology-directed repair of double-strand breaks. Oncogene. 2002;21(26):4176–4180. doi: 10.1038/sj.onc.1205539. [DOI] [PubMed] [Google Scholar]
- 45.Angelini S, Kumar R, Carbone F, et al. Micronuclei in humans induced by exposure to low level of ionizing radiation: influence of polymorphisms in DNA repair genes. Mutat Res. 2005;570(1):105–117. doi: 10.1016/j.mrfmmm.2004.10.007. [DOI] [PubMed] [Google Scholar]


