Summary
The Toxoplasma gondii locus mitochondrial association factor 1 (MAF1) encodes multiple paralogs, some of which mediate host mitochondrial association (HMA). Previous work showed that HMA was a trait that arose in T. gondii through neofunctionalization of an ancestral MAF1 ortholog. Structural analysis of HMA-competent and incompetent MAF1 paralogs (MAF1b and MAF1a, respectively) revealed that both paralogs harbor an ADP ribose binding macro domain, with comparatively low (micromolar) affinity for ADP ribose. Replacing the 16 C-terminal residues of MAF1b with those of MAF1a abrogated HMA, and we also show that only three residues in the C-terminal helix are required for MAF1-mediated HMA. Importantly these same three residues are also required for the in vivo growth advantage conferred by MAF1b, providing a definitive link between in vivo proliferation and manipulation of host mitochondria. Co-immunoprecipitation assays reveal that the ability to interact with the mitochondrial MICOS complex is shared by HMA-competent and incompetent MAF1 paralogs and mutants. The weak ADPr coordination and ability to interact with the MICOS complex shared between divergent paralogs may represent modular ancestral functions for this tandemly expanded and diversified T. gondii locus.
Keywords: T. gondii, mitochondrial association factor 1, host mitochondrial association, MICOS, neofunctionalization, MAF1
Introduction
Tight associations between pathogen-containing vacuoles and host organelles such as mitochondria have been described in a variety of intracellular pathogens, including Chlamydia psittaci [1], Legionella pneumophila [2], Hammondia hammondi [3] and Toxoplasma gondii [4–6]. While these phenotypes have been known for decades, the underlying molecular mechanisms are poorly understood. In most cases the pathogen molecules required for organellar association have not been identified, nor have their cognate binding partners in the host. This has hindered our ability to understand the relevance of this intimate association between the pathogen-containing vacuole and the host mitochondrion to infection outcome.
In T. gondii, we and others have identified the parasite locus that is required for HMA, Mitochondrial Association Factor 1B (MAF1b), and shown that expression of MAF1b increases cytokine signaling [4] during the acute phase of in vivo infections. In infected cell lysates and cells expressing MAF1b ectopically, MAF1b protein interacts with host mitochondrial outer membrane proteins belonging to the MICOS complex, which may be involved in the ability of MAF1b to mediate HMA [7]. During the acute phase of mouse infections, MAF1b-expressing parasites outcompete their MAF1b-null counterparts [3], implying that MAF1b plays an important role in determining infection outcome.
The T. gondii MAF1 locus encodes multiple tandemly duplicated paralogs that vary both in sequence and copy number across T. gondii strains [3, 8]. TgMAF1 paralogs fall into two broad groups, which we have defined as ‘A’ and ‘B’ based on residue percent identity [3]. All nonpseudogenized MAF1 genes sequenced to date contain a signal peptide, a transmembrane (TM) domain and a large C-terminal region that lacks identifiable sequence homology to any known proteins ([3, 4] and Fig 1A). MAF1 paralogs harbor a repetitive, proline-rich region between the putative TM and C-terminal region that broadly distinguishes the ‘A’ and ‘B’ paralog groups. In genetic complementation experiments, only MAF1b paralogs are capable of complementing HMA(−) Type II strains, while complementation with MAF1a has no effect on HMA [3]. Numerous polymorphisms further distinguish TgMAF1RHa1 and TgMAF1RHb1, including in the C-terminus, which is more divergent across paralogs compared to the N-terminus [3]. Since MAF1b paralogs are present only in those species that are capable of mediating HMA and all other strains only harbor MAF1 paralogs most similar to the non-functional “A” copies, we hypothesized that the ability of MAF1b to intimately interact with host mitochondria evolved by neofunctionalization of an ancestral version of MAF1. While the ancestral role of MAF1a has yet to be determined, these non-functional (with respect to HMA) ancestral paralogs represent a robust comparative tool to further probe the mechanism, and ultimately the function, of HMA in T. gondii.
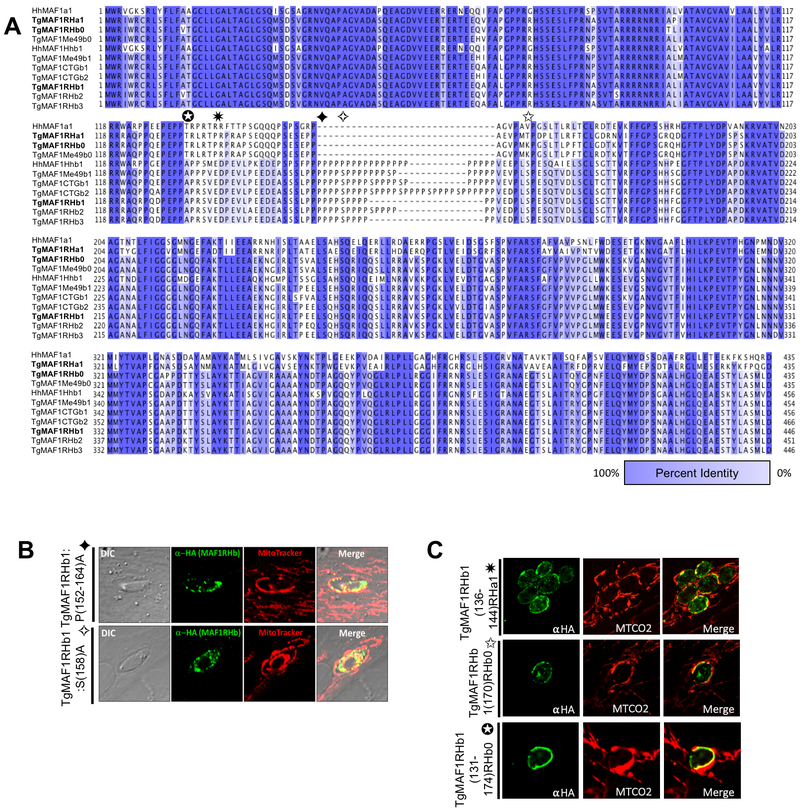
Figure 1. Proline-rich region of TgMAF1RHb1 is not required for HMA.
(A) Alignment of MAF1 isoforms from T. gondii and H. hammondi based on percent identity. Dark blue and white indicate 100% and 0% percent identity, respectively. Alignments were visualized in JalView after alignment using ClustalOmega. Bold sequences refer to isoforms utilized for mutational analysis. Colored boxes correspond to the boundaries of the indicated mutation. Domain architecture is indicated above alignment and corresponds to figure 2B. (B) HFFs were labeled with MitoTracker and infected with TgMe49 parasites expressing an HA epitope tagged TgMAF1RHb1 isoform with a disrupted proline-rich domain through replacement of prolines (P) or serines (S) with alanines (A). Cells were fixed at 18 hpi and visualized by confocal microscopy. (C) HFFs were infected with TgMe49 parasites expressing an HA tagged mutated TgMAF1RHb1 isoform. Site-directed mutations were made to the HMA(+), TgMAF1RHb1 isoform to the aligned sequence in the HMA(−) paralog TgMAF1RHa1 and TgMAF1RHb0. Cells were fixed at 18 hpi and visualized with confocal microscopy. Immunofluorescence staining was performed with antibodies against both the HA epitope tag and the mitochondrial protein, cytochrome c oxidase II (MTCO2). All mutations to the proline-rich and flanking region were unable to disrupt TgMAF1RHb1-driven HMA.
In the present study we first determined the overall structures of representative members of the A and B paralog groups using X-ray crystallography. These data allowed us to map the detailed structural differences between the A and B paralogs and identify specific residues within the C-terminal helix of MAF1a and MAF1b, that through mutagenesis studies, we showed are required for MAF1b to mediate HMA. We further leveraged these mutants to provide the first direct evidence that the C-terminus of a MAF1b paralog is essential, not only to HMA itself, but to the in vivo selective advantage conferred by HMA(+) T. gondii. We have also uncovered two putative ancestral functions for the MAF1 gene family that are shared by MAF1a and MAF1b paralogs, providing further support for the theory that mitochondrial association arose in T. gondii via neofunctionalization of an ancestral version of MAF1.
Results
The TgMAF1RHb1-specific proline-rich domain is dispensable for MAF1 function in HMA.
Type II T. gondii strains (including TgME49 and PRU) are HMA-negative and express undetectable levels of TgMAF1RHb1 [3]. Complementation of TgME49 and the near relative Neospora caninum, with TgMAF1RHb1 but not TgMAF1RHa1, confers the HMA phenotype [3]. To identify regions of TgMAF1RHb1 that are required for HMA we pursued a hypothesis-driven approach by comparing all sequenced MAF1 paralogs [3]. Sequences were aligned using Clustal-Omega and visualized in JalView (Fig 1A). Consistent with previous observations, the sequences cluster into two main groups [3], distinguished by the absence (‘A’ group) or presence (‘B’ group) of a proline-rich stretch between the putative transmembrane and the C-terminal region. To investigate the significance of the prolines in this region of TgMAF1RHb1, prolines 152–157 and 159–164 were mutated to alanine residues. However, these mutant paralogs were still capable of complementing HMA in TgMe49 parasites (Fig 1B). In an effort to disrupt the proline-rich region using an alignment-guided approach, separate mutations were made to the region flanking either side of the proline-rich region. Additionally, a third construct was made where the entirety of the proline-rich region, residues 131–174, were replaced with the aligned sequence in TgMAF1RHb0 (HMA(−)) which is nearly identical to TgMAF1RHa1 in this region (Fig 1A). Consistent with our point mutation analyses, this particular chimeric construct was still capable of mediating HMA in T. gondii strain TgME49 (Fig 1C). Additionally, work investigating the phosphoproteome following T. gondii infection identified significant MAF1 phosphorylation after secretion into the host cell [9]. To test the role of possible phosphorylation of the serines found within the MAF1b-specific proline-rich region, we mutated TgMAF1RHb1 serine 158 to alanine. This mutation did not disrupt the ability of TgMAF1RHb1 to confer HMA in TgMe49 parasites (Fig 1B). Overall, these data demonstrate that the proline-rich region is dispensable for HMA, despite its presence in all functional MAF1b paralogs.
The C-terminal regions of both TgMAF1Rha1 and b1 adopt an α/β globular structure with homology to ADP-ribose binding macro-domains.
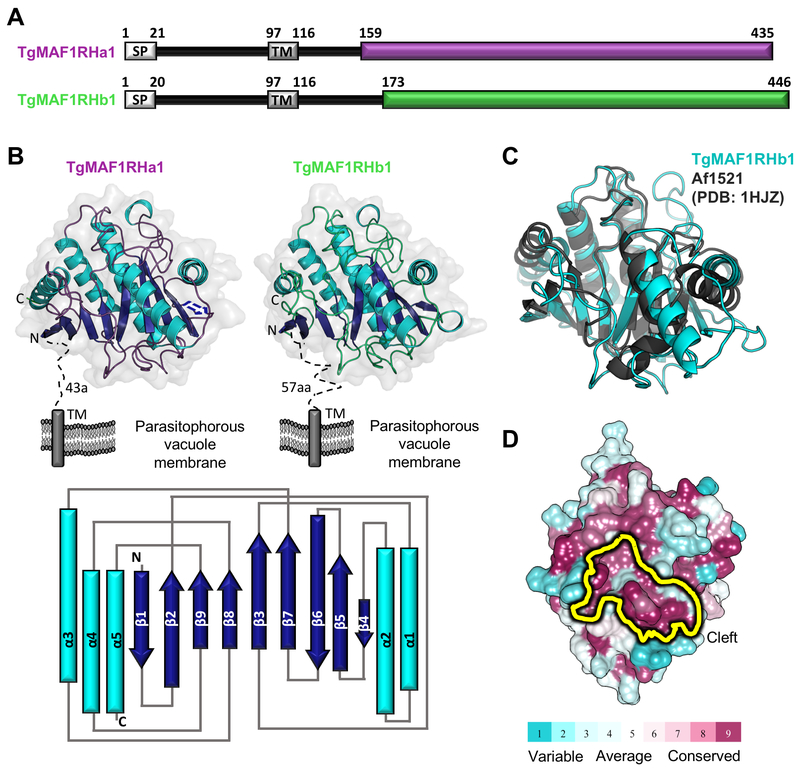
Aside from the proline-rich region characteristic of TgMAF1RHb1 paralogs, there are no other N-terminal regions in the TgMAF1 ectodomain with significant levels of polymorphism. We therefore turned our focus to the C-terminal region of the TgMAF1 ectodomain, which harbors numerous amino acid polymorphisms between the ‘A’ and ‘B’ paralog groups (e.g. TgMAF1RHb1 and TgMAF1RHa1 are 57% identical in the C-terminal region) (Fig 1A). To best guide our approach of correlating sequence differences between TgMAF1RHa1 and TgMAF1RHb1 to their functional differences in genetic complementation, we determined the X-ray crystal structures of the C-terminal domains of TgMAF1RHa1 and TgMAF1RHb1 [3]. Constructs encoding the predicted C-terminal domain of TgMAF1RHa1 (residues 159–435) and TgMAF1RHb1 (residues 173–443: note, the C-terminal Met444, Leu445 and Asp446 residues are not included in the crystallization construct due to protein stability problems) (Genbank Accession numbers SCA78655 and AMN92247, respectively) were recombinantly produced in E. coli, purified to homogeneity and crystallized for X-ray diffraction experiments (Fig 2A). The structure of TgMAF1RHb1 was phased by bromide single wavelength anomalous dispersion and refined to a resolution of 1.60 Å. Overall, the structure is well ordered with clear electron density extending from the Ser173 through Ser443 and including two C-terminal alanine residues derived from the expression vector. The 2.10 Å resolution structure of TgMAF1RHa1 was solved by molecular replacement using TgMAF1RHb1 as the search model with all three molecules in the asymmetric unit well-ordered. Despite possessing only 57% sequence identity within the C-terminal domain, structural overlays clearly showed that TgMAF1RHa1 and TgMAF1RHb1 adopt an overall similar architecture, with an rmsd of 0.6 Å over 240 Cα atoms (Fig 2B). Notably, these are the first structures of a MAF1 protein from any apicomplexan parasite.
Figure 2. The C-terminal region of TgMAF1 proteins adopts a conserved, well-ordered globular domain.
(A) Predicted domain architecture of TgMAF1RHa1 and TgMAF1RHb1. SP, signal peptide; TM, transmembrane. Colored boxes (TgMAF1RHa1- deep purple; TgMAF1RHb1-limegreen) indicate C-terminal region with strongly predicted secondary structure elements. Numbers correspond to amino acid positions. (B) Top: Tertiary structure of TgMAF1RHa1 and TgMAF1RHb1 colored based on secondary structure elements, with helices in cyan, strands in dark blue and loops colored as in (A). Dotted lines indicate unmolded regions and predicted features. The orientation of the proteins with respect to the parasitophorous vacuole membrane is shown. Bottom: Topology diagram of TgMAF1RHa1 and TgMAF1RHb1 colored as in Top. (C) Overlay of TgMAF1RHb1 (cyan) with Af1521 (dark grey; PDB ID 1HJZ) showing conservation of the core macro-domain architecture. (D) Mapping of conserved (burgundy) and variable (cyan) residues of TgMAF1RHa1 and TgMAF1RHb1 homologs onto the TgMAF1RHb1 surface using ConSurf [55]. The conserved cleft region is indicated in yellow.
Structural analysis of TgMAF1RHa1 and TgMAF1RHb1 revealed a compact, single domain with mixed α/β structure with a central, slightly curved 8-stranded β-sheet of mixed parallel and anti-parallel strands bound on one side by a 3 helical bundle and on the other side by a pair of helices (Fig 2B). Intriguingly, a DALI [10] structural homology search revealed significant similarity to macroH2A non-histone domains (also known as macro-domains) with the macro-domain from Archaeoglobulus fulgidus 1521 (Af1521) [11] identified as the most closely related structure (Z-score of 16; rmsd of 2.5 Å over 180 aligned Cα positions) (Fig 2C). A central feature of macro-domains is their ability to coordinate ADP-ribose (ADPr) and its derivatives through a surface cleft [12–14] that is conserved in both TgMAF1RHa1 and TgMAF1RHb1. In fact, sequences of all apicomplexan MAF1 homologs mapped onto the TgMAF1RHb1 core structure revealed the cleft as one of the most evolutionary conserved regions, supporting that this cleft is performing an important function (Fig 2D).
ADP-ribose forms a low affinity complex with TgMAF1RHa1 and TgMAF1RHb1.
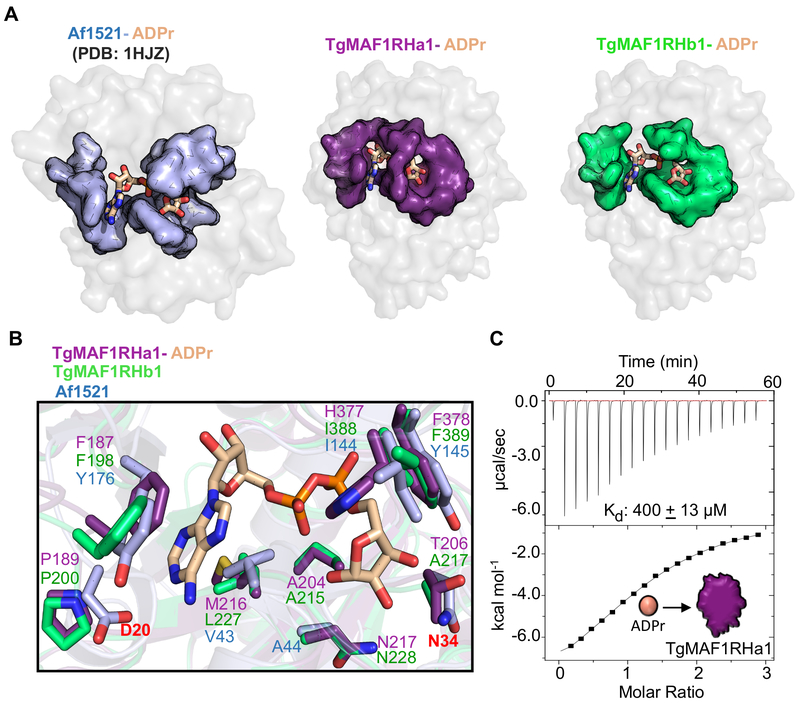
The striking resemblance to a canonical macro-domain and the presence of a well conserved surface cleft led us to hypothesize that TgMAF1s may be able to accommodate ADPr or a similar small molecule. Further support for this hypothesis was based on the observation that the structural homolog Af1521 was able to form a stable complex with ADPr (Figs 3A and B).
Figure 3. Structural characterization of TgMAF1RHa1/b1 reveals that ADP-ribose coordination by the macro-domains may be vestigial.
(A) Co-structures of TgMAF1RHa1, TgMAF1RHb1 (grey surface) and ADP-ribose (ADPr) (wheat ball-and-stick coloured by element), and Af1521 (PDB id- 1HJZ), highlighting the surface cleft of TgMAF1RHa1 (Left- coloured deep purple), TgMAF1RHb1 (Right- coloured limegreen) and Af1521 (Left- coloured light blue). (B) The residues coordinating ADPr in Af1521, TgMAF1RHa1 and TgMAF1RHb1 is shown based on the overlay of TgMAF1RHa1 (deep purple), TgMAF1RHb1 (limegreen), and Af1521 (lightblue) with ADPr (shown as wheat stick coloured by element). (C) Representative ITC binding isotherm of ADPr titrating into TgMAF1RHa1. The ITC clearly shows a low affinity binding with a Kd of ~400μM.
To investigate the ability of the TgMAF1 macro-domain to bind adenine nucleotide derivatives, we first determined co-structures of ADPr bound to TgMAF1RHa1 and TgMAF1RHb1 to 2.7 Å and 1.65 Å resolution, respectively. Stabilizing ADPr in the surface cleft is a Phe198 (TgMAF1RHb1) or Phe187 (TgMAF1RHa1) that stacks onto one side of the ADPr adenine ring and the side chains of Leu227 (TgMAF1RHb1) or Met216 (TgMAF1RHa1) that pack against the opposite side (Fig 3B). In addition, the second phosphate fits into a specific pocket formed by Ile388 and Phe389 (TgMAF1RHb1) or His377 and Phe378 (TgMAF1RHa1) (Fig 3B). Structural overlays revealed that residues in Af1521 known to be crucial for ADPr binding are not conserved in the TgMAF1 proteins; specifically, the strongly conserved Asp (labeled red in Fig 3B) for selectivity of adenine-based nucleotides [12, 15], and the Asn (labeled red in Fig 3B) shown to be critical for phosphatase activity on ADP-ribose-1”-phosphate [15, 16] are not conserved (Fig 3B). Thus, it appears that both TgMAF1RHa1 and TgMAF1RHb1 have lost key residues involved in coordinating ADPr suggesting, at minimum, a weaker binding affinity, which we next measured by isothermal titration calorimetry (ITC). Binding data was only obtained for TgMAF1RHa1, since the high salt required for TgMAF1RHb1 stability obscured complex formation. A Kd of approximately 400 μM was measured for TgMAF1RHa1 (Fig 3C), which is far weaker than the 130 nM Kd measured between ADPr and Af1521 [13]. It is worth noting that poly(ADPr) or poly-A binding does not necessarily require ADPr to bind with high affinity to form a functional complex [15, 17]. Thus, we cannot rule out the possibility that TgMAF1RHa1 and/or TgMAF1RHb1 are capable of binding these anionic polymers in a biologically relevant setting. Nor can we rule out that TgMAF1RHb1 may be capable of binding other nucleotides or even oligonucleotides such as poly(A) or poly(ADP-ribose), as has been shown for other Macro-domain containing proteins [13, 17]. However, the low affinity complexes observed here in both MAF1a and MAF1b paralog classes are consistent with the hypothesis that ADPr binding is not relevant to HMA.
Unique residues in the C-terminal helix of TgMAF1RHb1 are required for HMA.
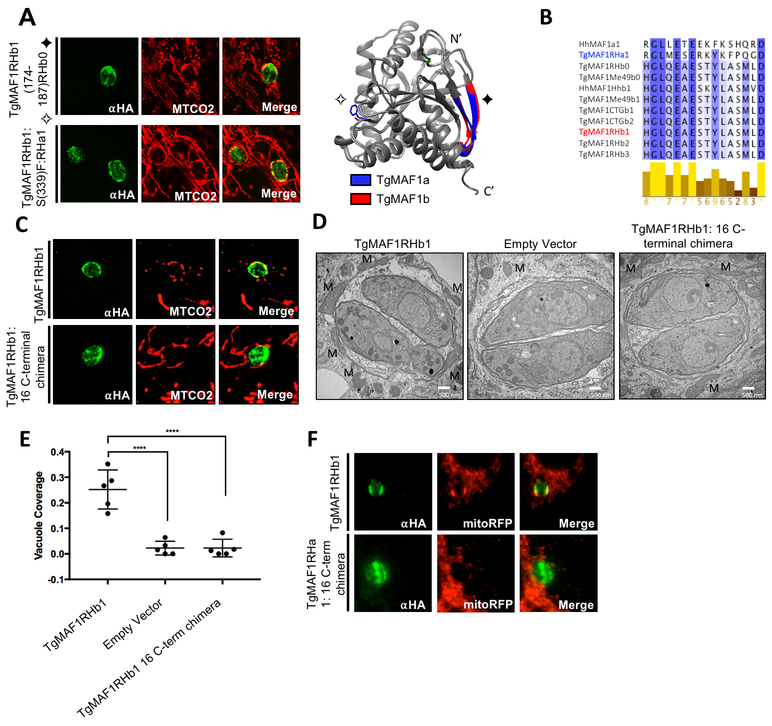
The structures of TgMAF1RHa1/b1 allowed us to identify additional residues for mutational studies by interrogating differences between the C- termini of both paralogs. We chose candidate residues by identifying residues that were 1) outward facing based on structural analysis and 2) consistently polymorphic among all members of the ‘A’ and ‘B’ groups. We first mutated peripheral TgMAF1RHb1-specific β-sheet (residues 174–187) to their aligned sequence in TgMAF1RHb0. Additionally, we mutated the outward-facing Ser339 to the aligned TgMAF1RHa1 phenylalanine residue. Both mutant constructs sufficiently conferred HMA when expressed in TgMe49 parasites (Fig 4A), indicating that these residues were not required for HMA.
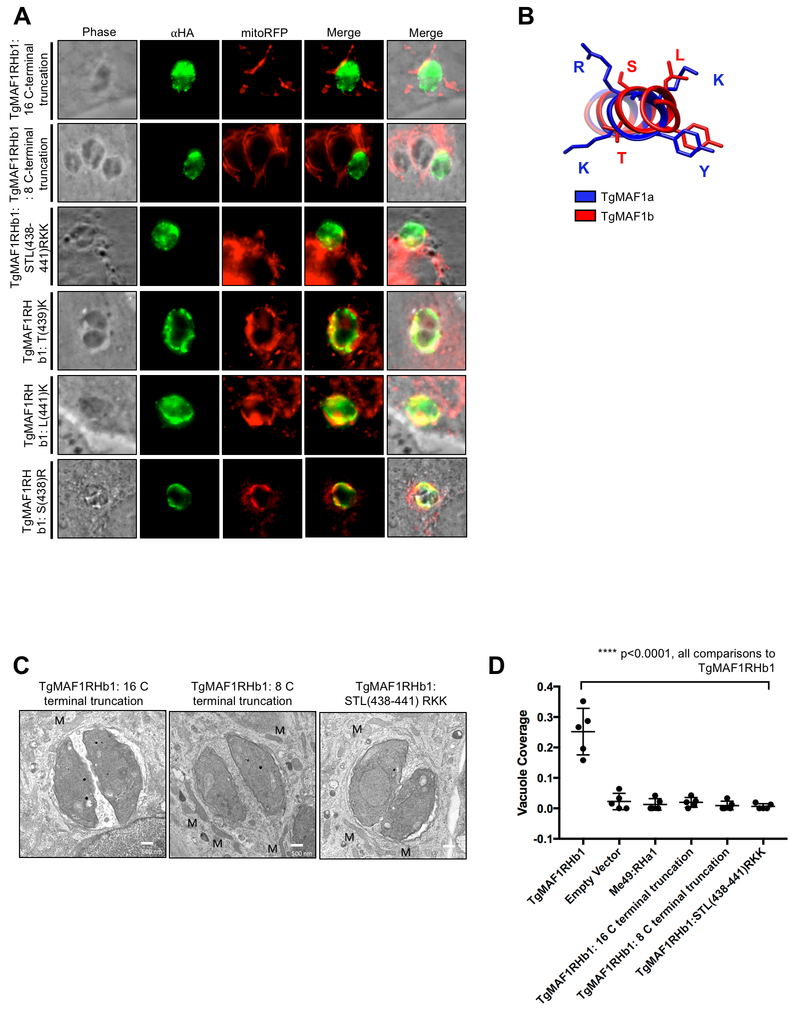
Figure 4. TgMAF1RHb1 16 C-terminal residues are required for HMA.
(A) HFFs were infected with TgMe49 parasites expressing an HA tagged mutated TgMAF1RHb1 isoform. Mutations were introduced using site-directed mutagenesis in an effort to disrupt key outward facing amino acids. Cells were fixed at 18 hpi and visualized using confocal microscopy. Immunofluorescence staining was performed with antibodies against the HA epitope tag and the mitochondrial protein, cytochrome c oxidase II (MTCO2). Symbols on mutation titles correspond to mutations in the structural overlay of both MAF1a and MAF1b generated in UCSF Chimera. (B) Alignment of the 16 C-terminal residues of MAF1 isoforms. Dark blue and white indicate 100% and 0% percent identity, respectively. Alignments were visualized in JalView after alignment using Clustal-Omega. Yellow bars below alignment show residue conservation across MAF1 isoforms. (C) HFFs were infected with TgMe49 parasites expressing either a WT HA-TgMAF1RHb1 isoform or a TgMAF1THb1 16 C-terminal chimeric form. The C-terminal chimera contained TgMAF1RHb1 residues 1–430 and TgMAF1RHa1 residues 420–435. Cells were fixed and treated with the same conditions outlined in 4A. (D) HFFs were infected with isolated TgMe49 clones transfected with either HA-TgMAF1RHb1, TgMAF1RHb1-C-term chimera or empty vector. Cells were fixed 18 hpi and processed for transmission electron microscopy. Mitochondria (M) are labeled. (E) Quantification of percent vacuole coverage, determined by electron microscopy. ****p<0.0001, one way ANOVA. (F) Normal rat kidney epithelial cells expressing RFP in the mitochondrial matrix (NRK mitoRFPs) were infected with TgMe49 parasites expressing either a WT HA-TgMAF1RHb1 isoform or a TgMAF1RHa1 16 C-terminal chimeric form. The C-terminal chimera is TgMAF1RHa1 residues 1–419 and TgMAF1RHb1 residues 431–446. Cells were fixed 18 hpi and visualized using epi-fluorescent microscopy. Immunofluorescence staining was performed with antibodies against the HA epitope tag.
We then focused on the 16 C-terminal residues of cloned MAF1b paralogs that are 94% similar to one another, but harbor multiple polymorphisms that distinguish the A and B lineages (Fig 4B). To investigate the significance of the C-terminus in MAF1-mediated HMA, we mutated the 16 C-terminal residues of TgMAF1RHb1 to those found in TgMAF1RHa1 to create a chimeric construct. Excitingly, the TgMAF1RHb1 16 C-terminal chimera was incapable of conferring HMA in TgME49, indicating a central role for the C-terminus in HMA (Fig 4C). When we examined TgME49 expressing the TgMAF1Rhb1 16 C-terminal chimera, we found that, as for the WT TgMe49, there was no significant HMA based on quantification of % vacuole coverage using EM (Fig 4D,E). The TgMAF1RHb1 16 C-terminal chimera shows similar vacuole coverage to an TgMe49 empty vector control further confirming the requirement of this region for HMA (Fig 4E). We then investigated the sufficiency of the C-terminus of TgMAF1RHb1 in driving HMA. Using site-directed mutagenesis we created a TgMAF1RHa1 chimeric construct with the 16 C-terminal residues of TgMAF1RHb1. The TgMAF1RHa1 16 C-terminal chimera did not drive HMA upon expression in TgMe49 parasites (Fig 4F), showing that while the 16 C-terminal residues of TgMAF1RHb1 are necessary for HMA, when expressed in a Type II genetic background they are not sufficient.
Three C-terminal residues in TgMAF1RHb1 are required for HMA.
As described above and shown in Figure 1A, the 16 C-terminal residues of TgMAF1RHb1 and TgMAF1RHa1 form the α5 helix that is unique to each TgMAF1RHb1 and TgMAF1RHa1 paralog. To further interrogate this region, truncation mutations were made to the C-terminus of TgMAF1RHb1 in an effort to abrogate HMA without disrupting proper MAF1 localization. Truncation of both the 16 and 8 C-terminal residues resulted in TgMAF1RHb1 products that did not confer HMA in TgMe49 parasites, confirming the requirement of these residues (Fig 5A and 5C). The structural differences of both the TgMAF1RHb1 and TgMAF1RHa1 helices and their opposing HMA phenotypes suggests residue-specific function within this region. Ten of the 16 C-terminal residues of this helix are polymorphic between the A and B paralogs, making it difficult to determine which residues to investigate based on primary sequence alone. However, when we examined the structure of the α5 helix, we identified a trio of basic residues unique to the C-terminus of TgMAF1RHa1, specifically Arg427, Lys428 and Lys430 (Fig 5B). These are strikingly different than their uncharged counterparts in TgMAF1RHb1 (Ser438, Thr439, and Leu441) (Fig 5B). To investigate the importance of these residues in HMA we mutated the STL residues in TgMAF1RHb1 to RKK, and this mutant was completely unable to mediate HMA when expressed in Type II T. gondii (Figs 5A and 5C). Using electron microscopy, we found that the percentage of the vacuole with interacting mitochondria in parasites expressing the RKK mutant is nearly indistinguishable from wild type Type II T. gondii (Fig 5D). When we mutated each residue individually (Ser→Arg, Thr→Lys and Leu→Lys), all 3 constructs could still mediate HMA when expressed in Type II T. gondii (Fig 5A). Additionally, mutation of both the Ser438 and Leu441 to Arg427 and Lys430 which lie on the same face of the C-terminal alpha helix also mediated HMA when expressed in Type II parasites (Fig 5A), suggesting the collective requirement of all three uncharged MAF1b residues (Ser438, Thr439 and Leu441) for HMA to occur.
Figure 5. Three residues in the C-terminus of TgMAF1RHb1 are required for HMA.
(A) NRK mitoRFP cells were infected with TgMe49 parasites expressing HA-TgMAF1RHb1 16 C-terminal mutant variants. Cells were fixed 18 hpi and visualized using epi-fluorescent microscopy. Immunofluorescence staining was performed with antibodies against the HA epitope tag. (B) TgMAF1RHa1 and TgMAF1RHb1 are highlighted in blue and red respectively which correspond to the color in the ribbon rendering of the C-terminal structures. Structure was visualized in UCSF chimera. (C) HFFs were infected with isolated TgMe49 clones transfected with each of the HA-TgMAF1RHb1 16 C-terminal mutants. Cells were fixed 18 hpi and processed for transmission electron microscopy. Mitochondria are labeled. (D) Quantification of percent vacuole coverage, determined by electron microscopy. ****p<0.0001, one way ANOVA (“all comparisons” means each comparison of TgMAF1RHb1 C-terminal mutant and TgMAF1RHa1 to TgMAF1RHb1 is significant)
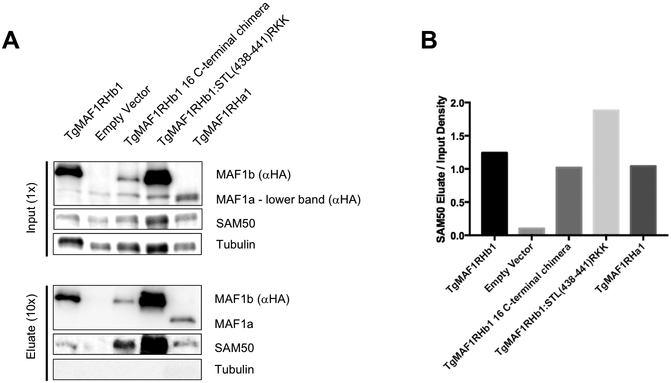
TgMAF1RHb1 mutants retain binding to SAM50.
TgMAF1RHb1 has recently been shown to interact with the mitochondrial intermembrane space bridging (MIB) complex which consists of proteins spanning both the outer and inner mitochondrial membranes. One MIB component on the cytosolic face of the outer mitochondrial membrane interacting, directly or indirectly, with TgMAF1RHb1 is SAM50 [18]. These data suggest SAM50 might function as the protein TgMAF1RHb1 uses to anchor itself to the mitochondria. If interactions with SAM50 are the sole basis for mediating HMA, neither TgMAF1RHa1 nor our TgMAF1RHb1 with the chimeric C-terminus should interact with SAM50. To investigate the ability of each of the C-terminal mutants and TgMAF1RHa1 to interact with SAM50, we conducted co-immunoprecipitations using HA conjugated beads. Consistent with published data we found that HA pulldowns from TgME49 clones expressing N-terminally HA-tagged TgMAF1RHb1 co-precipitated SAM50, while cytosolic host protein (aTubulin) showed no evidence for association with TgMAF1RHb1 (Fig 6A). Interestingly, however, we found that immunoprecipitations of TgMAF1RHa1, the TgMAF1RHb1 C-terminal truncation mutant, and the TgMAF1RHb1:STL→RKK mutant also specifically pulled down SAM50 (Fig 6A). Together, these data demonstrate that the specificity of the interaction between MAF1 and SAM50 is not dependent on the 16 C-terminal residues, and that MAF1-SAM50 interactions are not sufficient to mediate HMA.
Figure 6. TgMAF1RHb1 mutants bind to SAM50.
(A) Immunoprecipitation of lysed HFFs infected with cloned TgMe49: TgMAF1RHb1 16 C-terminal mutants, TgMe49:TgMAF1RHb1 and TgMe49:EV. Lysates were incubated with HA-conjugated beads and eluted with LDS sample buffer. Western blotting analysis was performed with the listed primary antibodies and HRP-conjugated secondary antibodies. Each 16 C-terminal mutant is able to bind to SAM50 (B) Densitometric input/eluate quantification of SAM50 western blot in panel A using ImageJ (NIH). Pulldowns of TgMAF1RHb1 C-terminal mutant infections have been repeated three times. Pulldowns of TgMAF1RHa1 infection was only performed once.
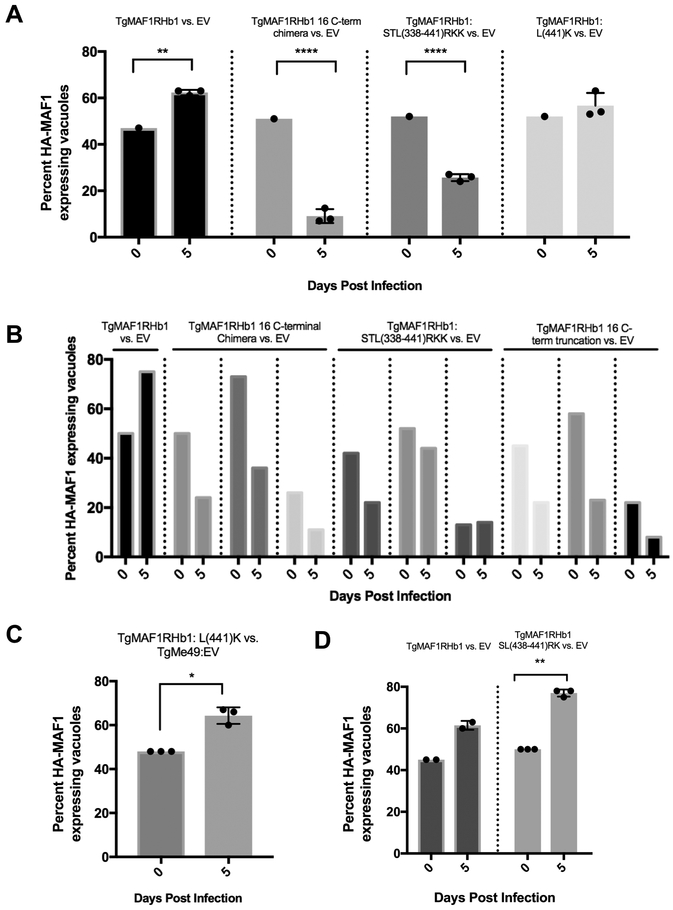
HMA-functional paralogs confer in vivo competitive advantage but TgMAF1RHb1 C-terminal mutants do not.
Previous work found that expression of TgMAF1RHb1 and not TgMAF1RHa1 in a Type II T. gondii strain provides a competitive advantage in an in vivo infection [3]. This growth advantage is not present during growth in vitro [3]. To test the in vivo selective advantage of expressing the C-terminal mutants, we performed in vivo competition assays by infecting mice with clonal lines expressing different HA-tagged WT and mutant TgMAF1RHa1 and TgMAF1RHb1 constructs with the same standard reference strain expressing only the empty vector (TgME49:EV). Mice were infected with total of 105 tachyzoites and the initial percent of parasites expressing HA-tagged MAF1 was quantified by immunofluorescence assay (IFA). Consistent with previous results [3], TgMAF1RHb1-expressing parasites competed effectively with the reference strain (Fig 7A, B), while parasites expressing the TgMAF1RHb1 16 amino acid truncation (Fig 7B), 16 amino acid C-terminal chimera (Figs 7A and B) or the 3 amino acid C-terminal chimera (STL→RKK; Fig 7A, B), competed less effectively with the reference strain. In contrast, one of the single amino acid mutants (Leu441Lys), that is HMA(+) (see Fig 5A) had a competitive index similar to parasites expressing WT TgMAF1RHb1 (Fig 7A, far right). It should be noted that in vivo competition assays are sensitive to parasite passage history, and we used a generic reference strain that was not passage matched to the strains expressing mutant MAF1b genes (although all of the HA-tagged lines were carefully passage matched). To further validate the competitive advantage of this mutant, a mixed, non-clonal population of Type II parasites expressing TgMAF1RHb1:Leu441Lys (HMA(+)) was injected into three Balb/C mice and the Leu441Lys expressing parasites were found to have a significant in vivo growth rate as compared to their WT TgMe49 counterparts (Fig 7C). Additionally, TgMe49 parasites expressing TgMAF1RHb1:SL(438/441)RK also outcompeted their passage-matched TgMe49:EV clone (Fig 7D). These data show that the in vivo competitive advantage conferred by expressing TgMAF1RHb1 in Type II T. gondii is almost certainly due to its impact on HMA, rather than the introduction of an additional copy of MAF1b.
Figure 7. HMA-functional paralog, TgMAF1RHb1 confers in vivo competitive advantage but TgMAF1RHb1 C-terminal mutants do not.
(A) Three mice per control or mutant were infected intraperitoneally (IP) with 50/50 mix of TgMe49 parasites expressing either WT or C-terminal TgMAF1RHb1 mutants and TgMe49:EV parasites (i.e. 50% TgMe49:TgMAF1 16 C-terminal chimera vs. 50% TgMe49 empty vector). HFFs were also infected with each input mixed population, fixed at 18 hpi and visualized utilizing epi-fluorescent microscopy. Immunofluorescence staining was performed with antibodies against the HA epitope tag. Following a five day in vivo infection, extracted peritoneal content was used to infect a monolayer of HFFs. Cells were fixed at 18 hpi and visualized utilizing epi-fluorescent microscopy. Immunofluorescence staining was performed with antibodies against the HA epitope tag. Both input (0 dpi) and output (5 dpi) populations were quantified by measuring the percent of MAF1-HA expressing parasites. **p=0.0038, ****p<0.0001 two-way ANOVA (Sidak test). (B) Similar procedure as A, however input parasites populations were mixes of 25/75, 50/50, and 75/25 and only one mouse was infected for each of the mixes. (C) Similar procedure to A and B. Three mice were IP infected with a natural mixed population of TgMe49:TgMAF1RHb1:L(441)K parasites. HFFs were infected with both input populations and peritoneal population after a five day infection. Cells were fixed and probed for parasites expressing HA-epitope tag by IFA (Paired t-test, *p=0.0174). (D) Similar procedure to A and B. Three mice were IP infected with a 50/50 mixture of TgMe49:EV and TgMe49:TgMAF1RHb1:SL(438/441)RK clones. HFFs were infected with both input populations and peritoneal population after a five day infection. Cells were fixed and probed for parasites expressing HA-epitope tag by IFA (Paired t-test, **p=0.0014).
Discussion
The ability to closely associate with host cell mitochondria has evolved independently in multiple intracellular pathogens including Legionella pneumophila, Chlamydia psittaci, and T. gondii [1, 2, 4–6]. However, in most organisms the importance of this phenotype is unknown. In the case of T. gondii, the pathogen gene product required for this phenomenon has been identified as MAF1b, a gene present in multiple copies within a tandemly expanded locus bearing extensive intra-strain and species sequence variation [3, 8]. In previous work we have shown that this variation has phenotypic consequences both with respect to HMA and infectivity in vivo [3]. In the present study we have exploited MAF1 locus diversity by using comparative structural biology and genetic complementation to identify regions of MAF1b required to mediate HMA.
Both MAF1 paralogs harbor vestigial ADP-ribose binding domains in their C-termini.
Paramount to our interrogation of the MAF1 locus was firstly characterizing two distinct MAF1 paralogs with opposing HMA phenotypes through crystallographic and in vitro mutational studies: TgMAF1RHa1 (TgMAF1RHa1; “HMA(−)”) and TgMAF1RHb1 (TgMAF1RHb1; “HMA(+)”) [3]. Given the fact that this domain binds ADPr with comparatively low affinity [13], it is likely that this domain is a pseudo ADPr binding domain. ADP-ribosylation is an essential post translational modification (PTM) that regulates a wide range of cellular processes including host immune pathways [13]. Pathogens such as the hepatitis E virus (HEV) and SARS evolved an antagonistic ADPr binding macrodomain which reverses ADP-ribosylation events in order to subvert the host immune response [19]. Many poly-ADP-ribose polymerases (PARPs) members which catalyze this PTM have been shown to undergo recurrent positive diversifying selection in mammals which suggests a host-pathogen molecular arms race. These systems explain a possible ancestral role for the ADPr binding domain of MAF1 in T. gondii [19, 20].
In addition to an expanded family of Macro-domain containing proteins, the T. gondii genome also harbors a large number of rhoptry pseudokinases that have lost catalytic activity. Multiple rhoptry pseudokinases are found within tandemly expanded gene clusters (e.g., ROP2/3/4 locus [21, 22] and ROP5 [8, 23–25]). MAF1 may represent a new locus, which has been modified from an existing protein domain for the purpose of manipulating the host cell. Interestingly the repurposing of this locus likely occurred in the most recent common ancestor of T. gondii, H.hammondi and N. caninum since all three parasite species harbor at least one MAF1 paralog, and based on sequence comparisons they all appear to have retained the pseudo-ADPr binding domain. To confirm whether the sequence comparison translated into structural conservation, we generated high confidence 3D models of HhMAF1a/b and NcMAF1b (Fig S1). Despite a similar charge distribution and high sequence conservation of the surface cleft forming residues, MAF1 paralogs appear to have lost the ability for high affinity coordination of ADPr and its derivatives, consistent with our hypothesis that the cleft region may be a vestigial domain. The polymorphic 16 residue C-terminal region, however, exhibits a remarkably divergent surface charge, suggesting that the evolutionary pressure on the C- terminal region is comparatively more than the cleft region, consistent with the importance of this C-terminal helix in gaining a central role in mediating HMA.
Structure-function analyses suggest that the C-terminus of TgMAF1RHa1 has three residues that prevent HMA.
We exploited sequence diversity across MAF1 paralog classes to ultimately determine that three residues in the C-terminus of MAF1b are necessary, but not sufficient, to mediate HMA. These data are consistent with previous studies of MAF1b, which found that C-terminally tagged MAF1b paralogs did not confer HMA in TgMe49 parasites [4]. Interestingly, the 16 C-terminal residues of the A and B paralogs that comprise the α5 helix differ at 10 of 16 amino acid positions within a given T. gondii strain, and these differences are highly conserved across divergent T. gondii strains and between T. gondii and H. hammondi. Notably, the C-terminal α5 helices in TgMAF1RHa1 and TgMAF1RHb1 adopt slightly different positions with respect to the body of the macro-domain, which may be due to sequence polymorphisms. With respect to HMA, only three of the 10 polymorphic residues of TgMAF1RHb1 are required. We hypothesize that Ser438, Thr439 and Leu441 in MAF1b, which differ markedly from the structurally analogous Arg427, Lys428 and Lys430 residues in MAF1a, present an optimal surface enabling coordination of a yet unidentified protein partner that gives rise to the HMA phenotype. The basic patch in MAF1a may also support non-specific molecular interactions with anionic biomolecules that effectively disrupt recruitment of HMA proteins. In either scenario, the significant differences in size and charge between the STL versus RKK residues appear to have profound effects on the ability of a given MAF1 paralog to drive HMA.
TgMAF1RHb1 mutants that do not mediate HMA still associate with members of the MICOS complex.
TgMAF1RHb1 exists primarily within the parasitophorous vacuole membrane (PVM) and has been found to interact with members of the MICOS complex in cellular lysates [18]. The MICOS complex spans the mitochondrial inner membrane and is responsible for the structural integrity of the mitochondria [26, 27]. Additionally, TgMAF1RHb1 pulls down SAM50 on the outer mitochondrial membrane, which interacts with the MICOS complex, forming the membrane inner bridge complex (MIB) [18]. Upon RNAi knockdown of SAM50 and MIC60, the ability of TgMAF1RHb1 to drive HMA was diminished [18], suggesting that associations between TgMAF1RHb1 and the MICOS complex are required for HMA. In the present study we found that all tested MAF1 paralogs (e.g., A and B) and HMA-deficient MAF1b mutants were capable of pulling down SAM50, although we did not determine precise binding affinities between the a and b paralogs and therefore do not know if their binding to SAM50 is equally avid. An exciting potential outcome from these data is that the ability to interact with host SAM50 may represent an ancestral function of MAF1, but that other interactions are necessary for effective HMA. The chimeric MAF1b/a constructs are perfect tools to identify the precise interactions necessary for this intriguing host cell manipulation phenotype [28, 29].
MAF1b confers in vivo competitive advantage and replication rate.
Parasites expressing HMA-driving MAF1b paralogs do confer a selective advantage during mouse infections [3], but the direct link between HMA itself and this phenotype was lacking. In the present study our results provide the most convincing evidence to date that HMA itself (rather than other effects mediated by ectopic expression of MAF1b) confers increased parasite replication and/or survival in vivo. The competitive index of the Leu→Lys single mutant (which is HMA(+)) was clearly superior to that of the STL→RKK triple mutant (which is HMA(−); Figs 7A and B). Arguably, loss of HMA function provides the parasites with a measurable disadvantage in vivo when competed with a WT TgMe49 strain continuously cultured in the lab. These data directly link HMA itself to increased in vivo parasite survival and/or growth. The selective advantage conferred by HMA is consistent with the fact that MAF1b paralogs show signs of positive, diversifying selection (defined previously in [3]), while MAF1a paralogs are highly conserved between strains and across species (see Fig 1A). Somewhat paradoxically, TgMAF1RHb1 expression in Type II parasites is also associated with an increased pro-inflammatory response in vitro (mouse embryonic fibroblasts) and in vivo, including the differential regulation of key Type 1 IFNs and pro-inflammatory cytokines in mice [4]. It is possible that increased cytokine production may either recruit more cells that are hospitable to T. gondii and/or block recruitment of cells that are more lethal to T. gondii (such as GR1+ macrophages; [30, 31]). It is also possible that the differential cytokine response is a direct result of a gene dosage effect of Type II parasites retaining their endogenous MAF1 locus and exogenously expressing an additional MAF1b paralog. This response is independent of the HMA phenotype and represents another ancestral function that is shared in both MAF1a and MAF1b paralogs.
In summary, using structure-function analyses we have identified three residues in T. gondii MAF1b that are required for MAF1b-mediated HMA, and have definitively linked HMA to increased parasite proliferation in vivo. In the process we have identified at least two functions associated with all MAF1 paralogs (ADP ribose coordination and association with the MICOS complex), which may represent the ancestral function of MAF1 prior to its neofunctionalization. This study has also enabled us to generate new MAF1 mutants that will serve as valuable reagents to probe the importance of HMA for T. gondii mediated host cell manipulation.
Materials and Methods
TgMAF1RHa1 and TgMAF1RHb1 cloning, protein production and purification
Constructs encoding the predicted C-terminal region of TgMAF1RHa1c (Genbank accession no. KU761333) (re-annotated TgMAF1RHb1; Ser173 to Ser443) and TgMAF1RHb1 (Genbank accession no. KU761342) (re-annotated TgMAF1RHa1; Thr159 to Asp435) were cloned, produced and purified as previously described [32]. Each protein was in a final buffer of HBS (20 mM Hepes pH 7.5, 150–300 mM NaCl) with 1% glycerol and 1 mM dithiothreitol.
Crystallization and data collection
Crystals of TgMAF1RHb1 were initially identified in the PEG/Ion Screen (Hampton Research) using sitting drops at 295 K. The final, refined drops consisted of 1.2 μL TgMAF1RHb1 at 20 mg/mL with 1.2 μL of reservoir solution (0.2 M ammonium sulfate, 20% PEG3350) and were equilibrated against 120 μL of reservoir solution. For phase determination, TgMAF1RHb1 crystals were soaked in a final cryoprotectant of reservoir solution with 12.5% glycerol and 1M NaBr for 3 min before flash cooling directly in liquid nitrogen. For co-crystallization with ADP-ribose (ADPr) (Sigma), TgMAF1RHb1 was crystallized in the presence of 5 mM ADPr and the cryopreservation solution contained reservoir solution with 12.5% glycerol and 10 mM ADPr. Diffraction data were collected on beamline 08B1–1 at the Canadian Light Source (CLS) for bromide-derivatized crystals, and on beamline 12–2 at the Stanford Synchrotron Radiation Lightsource (SSRL) for ADPr bound TgMAF1RHb1.
Crystals of TgMAF1RHa1 were initially identified in the Index screen (Hampton Research) using sitting drops at 295 K. The final, refined drops consisted of 1.0 μL TgMAF1RHa1 at 6.2 mg/mL with 1.0 μL of reservoir solution (0.9 M ammonium sulfate, 0.1 M Hepes pH 7.0, 0.5% PEG8000, 3% 2-methyl-2,4-pentanediol) and were equilibrated against 120 μL of reservoir solution. Crystals were cryopreserved in 80% saturated lithium sulfate and flash cooled in liquid nitrogen. A subset of crystals was soaked with 10 mM ADPr prior to cryopreservation. Diffraction data for TgMAF1RHa1 were collected on beamline 08ID-1 at the CLS, and on beamline 11–1 at SSRL for ADPr bound TgMAF1RHa1.
Data processing, structure solution and refinement
Diffraction data for TgMAF1 crystals were collected and processed to 1.60 Å (TgMAF1RHb1-Br), 1.65 Å (TgMAF1RHb1-ADPr), 2.10 Å (TgMAF1RHa1), and 2.70 Å (TgMAF1RHa1-ADPr) resolution using Imosflm [33], Scala [34] and Aimless [35] in the CCP4 suite of programs [36]. The structure of TgMAF1RHb1 was phased by bromide single wavelength anomalous dispersion. A total of 18 Br sites were identified and refined using the ShelxC/D/E pipeline [37]. High quality phases were obtained following density modification in dm [38] and enabled building and registering of approximately 85% of the backbone using buccaneer [39]. The TgMAF1RHb1-ADPr structure was solved by molecular replacement using the final refined TgMAF1RHb1 structure as the search model in PHASER [40], while the TgMAF1RHa1 structures were solved using a chainsaw trimmed model of TgMAF1RHb1 as the search model [41]. For each structure, COOT [42] was used for manual model building and selection of solvent atoms, and the models were refined in Phenix.Refine [43]. Complete structural validation was performed in Molprobity [44]. For each dataset, 5% of the reflections were set aside for calculation of Rfree. Data collection and refinement statistics are presented in Table 1.
Table 1.
Data collection statistics
| TgMAF1RHb1-Br | TgMAF1RHb1-ADPr | TgMAF1RHa1 | TgMAF1Rha1-ADPr | |
|---|---|---|---|---|
| A. Data collection statistics | ||||
| Space group | P212121 | P212121 | P21 | P21 |
| a, b, c (Å) | 45.09, 62.11, 89.21 | 45.47, 62.36, 89.69 | 79.89, 49.67, 114.75 | 78.23, 49.52, 115.71 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 96.09, 90 | 90, 96.63, 90 |
| Wavelength (Å) | 0.9195 | 0.9795 | 0.9794 | 0.9795 |
| Resolution (Å) | 45.09–1.60 (1.69–1.60) | 51.20–1.65 (1.68–1.65) | 48.86–2.10 (2.16–2.10) | 27.40–2.70 (2.86–2.70) |
| Measured reflections | 280,473 (26,900) | 114,544 (5,631) | 197,078 (14,313) | 117,630 (16,227) |
| Unique reflections | 33,786 (4,786) | 29,065 (1,489) | 51,862 (3,990) | 23,134 (3,436) |
| Redundancy | 8.3 (5.6) | 3.9 (3.8) | 3.8 (3.6) | 5.1 (4.7) |
| Completeness (%) | 99.8 (98.9) | 93.4 (97.4) | 98.5 (92.4) | 94.4 (87.9) |
| I/σ(I) | 12.7 (3.5) | 7.6 (2.2) | 11.5 (3.0) | 11.6 (2.2) |
| Rmerge (%) | 10.3 (47.5) | 9.4 (55.3) | 8.0 (44.4) | 9.0 (61.1) |
| B.Refinement statistics | ||||
| Resolution (Å) | 44.60–1.60 | 40.56–1.65 | 48.86–2.10 | 27.40–2.70 |
| Rwork / Rfree (%) | 17.0/21.0 | 20.5/22.8 | 21.2/24.4 | 23.3/28.3 |
| No. of atoms | ||||
| Protein (A/B/C) | 2057 | 2012 | 2114/2081/2099 | 2047/2005/1949 |
| Solvent/Br/Na | 257/11/3 | 105 | 159 | 16 |
| Glycerol or Sulfate | 18 | N/A | 70 | N/A |
| ADPribose | N/A | 36 | N/A | 72 |
| Average B-values (Å2) | ||||
| Protein (A/B/C) | 14.2 | 22.3 | 30.5/36.8/33.4 | 55.1/63.9/63.6 |
| Solvent/Br/Na | 24.0/26.9/32.9 | 28.4 | 31.5 | 42.6 |
| Glycerol or Sulfate | 23.0 | N/A | 47.8 | N/A |
| ADPribose | N/A | 33.4 | N/A | 68.6 |
| r.m.s. deviation from ideality | ||||
| Bond lengths (Å) | 0.008 | 0.012 | 0.003 | 0.003 |
| Bond angles (°) | 1.17 | 1.02 | 0.67 | 0.67 |
| Ramachandran statistics (%) | ||||
| Most favoured | 99.3 | 98.9 | 98.5 | 96.5 |
| Allowed | 0.7 | 1.1 | 1.5 | 3.5 |
| Disallowed | 0.0 | 0.0 | 0.0 | 0.0 |
Values in parentheses are for the highest resolution shell
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the following codes: TgMAF1RHb1-Br – 6BXR; TgMAF1RHb1-ADPr – 6BXW; TgMAF1RHa1 – 6BXS; TgMAF1RHa1-ADPr – 6BXT.
In-silico homology modeling of HhMAF1a1/b1 and NcMAF1RHa1
The crystal structure of TgMAF1RHa1 was used as a modeling template for HhMAF1RHa1 and NcMAF1RHa1 paralogs while the structure of TgMAF1RHb1 was used as the template for HhMAF1RHb1. The sequence identity between target and template was 40% or greater for all the MAF1 paralogs. Using Modeller 9v18 [45], 10 models for each MAF1 paralog was generated and the best model chosen based on the low value of normalized discrete optimized protein energy (DOPE). The assessment of the final model was carried out with Ramachandran statistics [46], QMEAN [47], and ProSA[48] (Supplementary Table 1).
Cell maintenance and parasite infection
TgMe49 in these experiments were regularly passed in human foreskin fibroblasts (HFFs) and incubated at 37°C in 5% CO2. NRK mito-RFP cells were a kind gift from Jennifer Lippincott-Schwartz (NIH, Bethesda, MD) [49]. Both cell types were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 50 μg/ml of penicillin and streptomycin, 10% FBS, and 2mM glutamine (cDMEM).
Expression of MAF1 mutants and transgenic parasites
Parental plasmid used for cloning all TgMAF1RHb1 constructs contains the HXGPRT gene. TgMAF1RHb1 proline-alanine mutations were constructed using short overlap extension (SOE) PCR. TgMAF1RHb1 and TgMAF1RHa1 C-terminal mutants were generated using a site-directed mutagenesis kit (NEB Q5 site-directed mutagenesis). All constructs were confirmed by sanger sequencing methods using TgMAF1RHb1 specific primers (Genewiz). The constructs contain the endogenous TgMAF1RHb1 promoter followed by the start codon, signal peptide, N-terminal hemagglutinin (HA) epitope tag and end at the stop codon. Transgenic lines were generated using TgMe49ΔHXGPRT:Luciferase parasites that were transfected with 55 μg of DNA linearized with HindIII. A T25 flask of TgMe49ΔHXGPRT:Luciferase parasites was scraped and passed through a 25 and 27 gauge needle followed by centrifugation for 10 minutes at 800xg. 2×107 parasites were re-suspended in Cytomix (120 mM KCl; 0.15 mM CaCl2; 10 mM KPO4; 25 mM Hepes, 2mM EDTA, 5mM MgCl2; pH to 7.6), GSH and ATP. Parasites were electroporated with 1.6Kv and 25μF. Following 24 hours of growth in cDMEM, parasites were selected with mycophenolic acid (MPA)/xanthine. Selected populations were then cloned via limited serial dilution in a 96-well plate. Cloned parasites were confirmed through immunofluorescence assays (IFA) by probing for HA epitope tag.
Immunofluorescence assays and microscopy
HFFs and NRK mito-RFP cells were grown to 100% and 60% confluency, respectively, on 0.7 cm2 8-well glass chamber slide system (ThermoFisher Scientific) in cDMEM. Monolayers were infected at an MOI of 1 with transgenic parasites. Cell were fixed at 18 hpi with 4% paraformaldehyde for 15 min. and blocked/permeabilized with blocking buffer (5% BSA, 0.1% Triton X-100, PBS). HFFs and NRK mito-RFP cells were then probed with anti-HA rat monoclonal antibody (3F10 clone, Roche) diluted to 0.1 μg/mL in blocking buffer (see above) for 1 hour at room temperature while shaking. HFFs were also incubated in anti-MTCO2 mouse monoclonal antibody and cells were washed with PBS. HFFs were incubated in 488 goat anti-rat and 594 goat anti-mouse secondary antibody (Life Technologies Alexa Fluor H+L) for 1 hour at room temperature while shaking, followed by PBS washes. NRK mito-RFP cells were incubated in 488 goat anti-rat secondary antibody for 1 hour followed by PBS washes. HFFs and NRK mito-RFP were then mounted in Vectashield mounting media (Vector laboratories) and sealed with cover glass. Slides were visualized using both confocal and epifluorescence microscopy. Images were taken of the three channels: 488 (anti-HA), 594 (anti-MTCO2 and mito-RFP) and DIC/phase. Images were cropped and merged using ImageJ (NIH).
MAF1 paralog alignments and C-terminal structural views
All MAF1 paralog sequences were obtained from GenBank (NCBI accession numbers: SCA78755.1, ANN02899.1, AMN92255.1, AMN92254.1, AMN92252.1, AMN92247.1, AMN92246.1, AMN92253.1, AMN92250.1, AMN92249.1, AMN92248.1) and aligned by percent identity using Clustal Omega. Alignment was visualized using JalView [50]. TgMAF1RHb1 and TgMAF1RHa1 primary amino acid sequences were visualized using UCSF chimera software [51, 52].
TEM and quantification of vacuole coverage
HFFs were infected with TgMAF1RHb1 C-terminal mutants. At 18 hpi cells were fixed with 2.5% glutaraldehyde in PBS for 1 hour at room temperature, washed three times with PBS for 10 minutes, post-fixed for 1 hour at 4C in 1% OsO4 with 1% potassium ferricyanide, and washed three times with PBS. Samples were then dehydrated in a graded series of alcohol for ten minutes with three changes in 100% ethanol for 15 minutes and changed three times in epon for 1 hour each. Following the removal of epon, samples were covered with resin and polymerized at 37°C overnight and then 48 hours at 60°C (protocol: Center for Biological Imagining - CBI, University of Pittsburgh, Pittsburgh, PA, USA). Samples were cross sectioned and processed by the CBI. Five vacuoles containing two parasites were imaged for each of the infections. Vacuoles were traced in ImageJ and the percent of the total distance around the vacuole in direct contact with the host mitochondria was quantified for each of the mutants and controls [53].
Immunoprecipitation and immunoblotting
HFFs were infected with cloned TgMe49 parasites expressing either a TgMAF1RHb1 C-terminal mutant, TgMe49:TgMAF1RHb1, TgMe49:Empty Vector, or TgMe49:TgMAF1RHa1 at an MOI of 2. Cell were lysed in IP lysis buffer (50mM Tris - pH 8.0, 150 mM NaCl, 1% IGEPAL CA-630, 0.05% Tween 20) and treated with complete protease inhibitors (Roche) on ice. The insoluble fraction was pelleted at 700xg for 10 minutes at 4°C and the soluble fraction was incubated with Pierce anti-HA magnetic beads (Thermo scientific) for 2 hours at room temperature using a rotator. Beads were washed five times with IP lysis buffer and eluted by boiling in LDS sample buffer (Thermo scientific). Both input and eluate fractions were resolved on 10% SDS-PAGE gel and transferred to nitrocellulose membrane. Membranes were blocked in 5% BSA in PBST and probed with primary antibodies to HA, SAM50, and Tubulin followed by goat horse radish peroxidase (HRP) conjugated secondary antibodies. Bands were visualized with SuperSignal West Pico chemiluminescent substrates (Thermo Scientific). Antibodies used for these experiments: Anti-HA high affinity rat monoclonal antibody (clone 3F10) – Roche, Goat Anti-Rat IgG H&L HRP – Abcam, Anti-SAMM50 antibody – Abcam, Goat Anti-Rabbit IgG HRP – Southern Biotech and Goat Anti-mouse IgG HRP – Southern Biotech.
In vivo competition assay
Mice were 12-wk-old Balb/C female mice (Jackson Labs). Using the same mutant and control parasite clones previously listed, we created a 1:1 mix of TgMe49:EV and TgMe49:TgMAF1RHb1, TgMe49:EV and TgMAF1RHb1 16 C-term chimera, TgMe49:EV and TgMAF1RHb1 STL(438–441)RKK, and TgMe49:EV and TgMAF1RHb1 L(441)K. TgME49:EV served as a baseline control and was not passage matched to the experimental strain. Three mice were intraperitoneally (IP) infected with 105 tachyzoites for each of the mixes allowing us to have three biological replicates for each treatment group. Coverslips with confluent HFFs were infected with the same mixed population preparations and probed for HA tag to quantify the input percentage of each mix. Parasite burden was measured daily over the course of the five day infection using in vivo bioluminescence imaging [54]. On day 6, each mouse was sacrificed and a peritoneal lavage was performed with PBS. A fraction of the peritoneal content was used to infect a confluent monolayer of HFFs, fixed at 18 hpi, and probed for HA tag for output percentage quantification.
Ethics Statement
Animal experiments were conducted according to the guidelines of the American Veterinary Medical Association. Accordingly, all euthanasia of animals was carried out using controlled exposure to CO2. All animal protocols were approved by the local Institutional Animal Care Committee at the University of Pittsburgh under IACUC protocol #12010130.
Supplementary Material
Acknowledgments
The authors would like to thank the technical support staff at the Stanford Synchrotron Radiation Lightsource, Canadian Light Source and the University of Pittsburgh Center for Biological Imaging. This work was supported by Canadian Institutes of Health Research Grant 148596 to M.J.B., National Institutes of Health (NIH) grant AI114655 to J.P.B. and AI73756 to J.C.B., and National Science Foundation Graduate Student Research Fellowship to L.F.P. M.J.B. gratefully acknowledges the Canada Research Chair program for salary support. We would also like to thank Sarah Sokol, Rachel Coombs, Sheen Wong, and Elizabeth Rudzki for editing this manuscript.
References
- 1.Matsumoto A, Bessho H, Uehira K, Suda T. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J Electron Microsc (Tokyo). 1991;40(5):356–63. [PubMed] [Google Scholar]
- 2.Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158(4):1319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adomako-Ankomah Y, English ED, Danielson JJ, Pernas LF, Parker ML, Boulanger MJ, et al. Host Mitochondrial Association Evolved in the Human Parasite Toxoplasma gondii via Neofunctionalization of a Gene Duplicate. Genetics. 2016;203(1):283–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pernas L, Adomako-Ankomah Y, Shastri AJ, Ewald SE, Treeck M, Boyle JP, et al. Toxoplasma effector MAF1 mediates recruitment of host mitochondria and impacts the host response. PLoS Biol. 2014;12(4):e1001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinai AP, Webster P, Joiner KA. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J Cell Sci. 1997;110 (Pt 17):2117–28. [DOI] [PubMed] [Google Scholar]
- 6.Jones TC, Yeh S, Hirsch J. The Interaction Between Toxoplasma Gondii and Mammalian Cells. J Exp Med. 1971;136(5):1157–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly FD, Wei BM, Cygan AM, Parker MP, Boulanger MJ, Boothroyd JC. Toxoplasma gondii MAF1b Binds the Host Cell MIB Complex To Mediate Mitochondrial Association. mSphere. 2017;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adomako-Ankomah Y, Wier GM, Borges AL, Wand HE, Boyle JP. Differential locus expansion distinguishes Toxoplasmatinae species and closely related strains of Toxoplasma gondii. MBio. 2014;5(1):e01003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe. 2011;10(4):410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen MD, Buckle AM, Cordell SC, Lowe J, Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J Mol Biol. 2003;330(3):503–11. [DOI] [PubMed] [Google Scholar]
- 12.Han W, Li X, Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat Res. 2011;727(3):86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkauskaite E, Jankevicius G, Ahel I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol Cell. 2015;58(6):935–46. [DOI] [PubMed] [Google Scholar]
- 15.Malet H, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, et al. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol. 2009;83(13):6534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol. 2006;80(17):8493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol. 2009;385(1):212–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly FD, Wei BM, Cygan AM, Parker ML, Boulanger MJ, Boothroyd JC. Toxoplasma gondii MAF1b Binds the Host Cell MIB Complex To Mediate Mitochondrial Association. mSphere. 2017;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, et al. Viral Macro Domains Reverse Protein ADP-Ribosylation. J Virol. 2016;90(19):8478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daugherty MD, Young JM, Kerns JA, Malik HS. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts. PLoS Genet. 2014;10(5):e1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boothroyd JC, Dubremetz JF. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat Rev Microbiol. 2008;6(1):79–88. [DOI] [PubMed] [Google Scholar]
- 22.Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, et al. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot Cell. 2011;10(9):1193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reese ML, Boothroyd JC. A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J Biol Chem. 2011;286(33):29366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behnke MS, Fentress SJ, Mashayekhi M, Li LX, Taylor GA, Sibley LD. The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog. 2012;8(11):e1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A. 2011;108(23):9631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozjak-Pavlovic V The MICOS complex of human mitochondria. Cell Tissue Res. 2017;367(1):83–93. [DOI] [PubMed] [Google Scholar]
- 27.Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, et al. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30(21):4356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forne I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics. 2012;11(4):M111012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonkin ML, Beck JR, Bradley PJ, Boulanger MJ. The inner membrane complex sub-compartment proteins critical for replication of the apicomplexan parasite Toxoplasma gondii adopt a pleckstrin homology fold. J Biol Chem. 2014;289(20):13962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunay IR, Damatta RA, Fux B, Presti R, Greco S, Colonna M, et al. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29(2):306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gay G, Braun L, Brenier-Pinchart MP, Vollaire J, Josserand V, Bertini RL, et al. Toxoplasma gondii TgIST co-opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-gamma-mediated host defenses. J Exp Med. 2016;213(9):1779–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adomako-Ankomah Y, English ED, Danielson JJ, Pernas LF, Parker ML, Boulanger MJ, et al. Host mitochondrial association evolved in the human parasite Toxoplasma gondii via neofunctionalization of a gene duplicate. Genetics. 2016;203(1):283–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans PR. Scaling and assessment of data quality. Acta Cryst. 2005;D(62):72–82. [DOI] [PubMed] [Google Scholar]
- 35.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowtan K dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–8. [Google Scholar]
- 39.Cowtan K Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Cryst. 2004;D60:1229–36. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb B, Sali A. Protein structure modeling with MODELLER. Protein Structure Prediction. 2014:1–15. [DOI] [PubMed] [Google Scholar]
- 46.RAMPAGE Server. http://ravenbioccam.ac.uk/rampage.php.
- 47.Benkert P, Tosatto SC, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins: Structure, Function, and Bioinformatics. 2008;71(1):261–77. [DOI] [PubMed] [Google Scholar]
- 48.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research. 2007;35(suppl_2):W407–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitra K, Lippincott-Schwartz J. Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol. 2010;Chapter 4:Unit 4 25 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. [DOI] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walzer KA, Adomako-Ankomah Y, Dam RA, Herrmann DC, Schares G, Dubey JP, et al. Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc Natl Acad Sci U S A. 2013;110(18):7446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic acids research. 2010;38(suppl_2):W529–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.